ABSTRACT
Streptococcus suis is an important zoonotic pathogen. The massive use of tylosin and other antibiotics in swine production has led to the emergence of resistant phenotypes of S. suis. However, there are no adequate measures available to address the problem of bacterial resistance. This study involved the use of 1/4 MIC (0.125 µg/mL) of tylosin to investigate resistance-related proteins by S. suis ATCC 700794. Our results showed that 171 proteins were differentially expressed in S. suis tested with 1/4 MIC (0.125 µg/mL) of tylosin using iTRAQ-based quantitative proteomic methods. TCS, heat shock protein and elongation factors were differentially expressed at 1/4 MIC (0.125 µg/mL) of tylosin compared to non treated, control cells. Using quantitative RT-PCR analysis, we verified the relationship between the differentially expressed proteins in S. suis with different MIC values. The data showed that expression profile for elongation factor G (fusA), elongation factor Ts (tsf), elongation factor Tu (tuf), putative histidine kinase of the competence regulon, ComD (comD), putative competence-damage inducible protein (cinA) and protein GrpE (grpE), observed in tylosin-resistant S. suis, correlated with that of S. suis ATCC 700794 at 1/4 MIC (0.125 µg/mL). The MIC of tylosin-resistant showed high-level resistance in terramycin, chlortetracycline, ofloxacin and enrofloxacin. Our findings demonstrated the importance of elongation factors, TCS and heat shock protein during development of tylosin resistance in S. suis. Thus, our study will provide insight into new drug targets and help reduce bacterial multidrug resistance through development of corresponding inhibitors.
Introduction
Tylosin is one of the most common macrolide antibiotics, which is effective against both Gram-positive and Gram-negative bacteria [Citation1]. It is widely used to prevent respiratory diseases caused by organisms, such as Mycoplasma, Staphylococcus aureus, Pseudomonas aeruginosa and Streptococcus suis [Citation2]. S. suis is an important zoonotic pathogen that causes a wide range of diseases in pigs, including meningitis, septicemia, pneumonia, endocarditis, and arthritis [Citation3]. With the massive use of tylosin, different parts of the world have reported widespread clinical resistance [Citation4,Citation5]. During 2010 to 2013, characterization of 227 S. suis strains isolated from pigs showed high levels of resistance to tylosin (93.8 %) in Korea [Citation6]. Moreover, 95.6% of the isolates were resistant to clindamycin, tilmicosin, tylosin, oxytetracycline and more classes of antimicrobials, indicating the high prevalence of multidrug resistant S. suis.
Generally, there are at least four classical antibiotic resistance mechanisms including inactivation of drugs via hydrolysis or modification, alteration or bypass of the drug target, permeability changes in the bacterial cell wall which restricts antimicrobial access to target sites, and active efflux of the antibiotic from the microbial cell. In addition to the above mechanisms, new gene acquisition, gene expression modifications, two-component regulatory systems (TCS), heat shock proteins, and bacterial elongation factor are involved in drug resistance [Citation7,Citation8].
TCS, the most widespread regulatory systems in bacteria [Citation9], are recognized by two-main protagonists: the histidine kinase and their cognate response regulator [Citation10]. The competence operon comCDE and putative competence-damage inducible protein (cinA) which are involved in the regulation of TCS, provided evidence for gene transfer events in Streptococcus pneumoniae [Citation11]. Meanwhile, previous studies have shown that TCS regulates antibiotic resistance in many pathogenic bacteria [Citation12–Citation15]. Therefore, TCS may play an important role in multidrug resistance. It is well known that elongation factors play important roles in forming bonds between the tRNA attached peptidyl-transferase and the next amino acid, ribosomal translation, causing premature dissociation of the peptidyl-tRNA from the ribosome and protein synthesis [Citation16–Citation19]. Previous studies have shown that elongation factors can not only inhibit bacterial protein synthesis using the antibiotics tetracycline, tylosin and fluoroquinolone [Citation20,Citation21], but also take part in imparting drug resistance in Escherichia coli. Moreover, elongation factors have been reported to be differentially expressed in tylosin-resistant Mycoplasma gallisepticum [Citation22–Citation24]. Thus, elongation factors are considered as potential targets for multi-drug resistant bacteria. In addition, heat shock proteins are involved in antibiotic resistance mechanism. Heat shock proteins are stress-responsive proteins which act as chaperonins to fold proteins. Many heat shock proteins (HSP) such as DnaJ, DnaK, GroEL, GroES, and GrpE have been differentially expressed in drug-resistant strains [Citation25–Citation28]. They play a role in the development of drug resistance by providing coping mechanism during the early stages of exposure to drugs in E. coli [Citation29].
Quantitative proteomics have considerably improved during the past decade and been employed in comparative analysis to study whole proteome-wide expression dynamics of cells grown under a variety of growth or stress conditions such as antibiotics [Citation30]. Researchers have shown that quantitative proteomic analysis can be used as an effective tool to find novel resistance mechanisms [Citation31]. However, changes in the proteomics profile of S. suis in response to tylosin have not been yet reported.
In our study, iTRAQ labeling-based quantitative proteomics was utilized to study the differentially expressed proteins in S. suis upon addition of tylosin in comparison to non-treated, control cells. Bioinformatics analysis revealed that proteins related to TCS, protein elongation factor and heat shock protein could be involved in tylosin resistance mechanism. In order to identify whether these proteins contributed to tylosin-resistance, S. suis was induced in the presence of sub-minimum inhibitory concentration (MIC; 0.125 µg/mL) of tylosin, in vitro. In the tylosin-resistant strain, candidate proteins were further verified for changes at mRNA level by qRT-PCR. The MICs of tetracycline and fluoroquinolone were determined to confirm resistance to these drugs in the strain and to establish the possibility of multidrug resistance in the presence of tylosin. Our findings demonstrated the importance of TCS, protein elongation factor and HSP in tylosin resistance of S. suis, thus providing insight into developing corresponding inhibitors to reduce bacterial resistance.
Materials and methods
Bacterial strain and cultivation
Streptococcus suis ATCC 700794 was purchased from American type culture collection. S.suis was grown in Todd-Hewitt broth (THB) (THB: Summus Ltd, Harbin, Heilongjiang, China) or Todd-Hewitt broth agar (THA) with 5% (v/v) fetal bovine serum (Sijiqing Ltd, Hangzhou, Zhejiang, China). The cultures were used for inducing the resistance ATCC 700794 (S-t-R 128) and the MIC assays.
Minimum inhibitory concentrations and growth rates
The MIC of S. suis to tylosin (98% purity, Solaibao Biotechnology Co., Ltd, Beijing, China) was determined by microtitre broth dilution method as recommended by the Clinical and Laboratory Standards Institute (CLSI). Cultures were diluted to 1 × 106 colony-forming units (CFU)/mL using THB. Finally, 100 μL of cell suspensions were inoculated into the wells of a 96-well plates (Corning Costar® 3599 Corning, NY, USA) containing serial dilutions of tylosin culture medium as previously reported [Citation32,Citation33]. At the same time, negative control was set up as outlined in a previous study [Citation33]. The inoculated microplates were incubated at 37°C for 24 h. The MICs of tylosin-resistant mutants to a variety of antibiotics, terramycin, chlortetracycline, ofloxacin, enrofloxacin, florfenicol and penicillin K (98% purity, Solaibao Biotechnology Co., Ltd, Beijing, China) were measured using the same procedure.
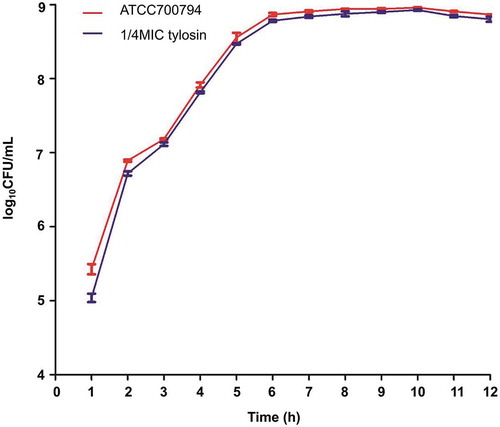
The growth rates of S. suis ATCC 700794 treated with and without 1/4 MIC (0.125 µg/mL) tylosin were analyzed [Citation33]. Briefly, S. suis ATCC 700794 treated without tylosin and with 1/4 MIC tylosin (0.125 µg/mL) were incubated at 37℃ for 12 h. Then, the samples were taken every hour and applied to the blood plate (THA with 5% (v/v) fetal bovine serum). All samples were diluted into 10−1, 10−2, 10−3, 10−4, 10−5, 10−6, 10−7 and 10−8 CFU/mL. 0.1 mL bacterial fluid from 10−5, 10−6 and 10−7 dilutions were taken and applied to the plate. The assays were repeated 3 times. These plates were cultured at 37℃ overnight. The number of colonies grown on the plate were counted on the 2nd day and there were 20 to 300 colonies per plate [Citation34] ().
iTRAQ analysis
Protein was extracted from S. suis cells under two different conditions at 37℃ for 24 h (cells treated with 1/4 MIC (0.125 µg/mL) of tylosin and non-treated cells) as described previously [Citation33]. The iTRAQ analysis was carried out at Shanghai Applied Protein Technology Co. Ltd (APT, Shanghai, China). Three biological replicates were evaluated to minimize experimental error. According to the FASP procedure and the reagent protocol [Citation33], iTRAQ-labeled peptides were fractionated by strong cation exchange (SCX) chromatography using the AKTA Purifier system. LC–MS/MS analysis was performed on a Q Exactive mass spectrometer coupled to EasynLC (Proxeon Biosystems, Thermo Fisher Scientific).
Bioinformatics analysis
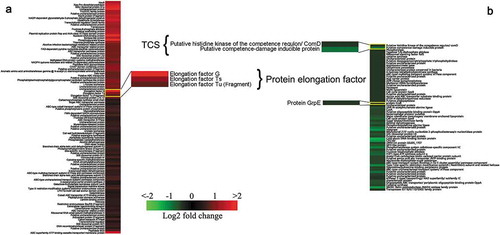
The sequence data of 171 differentially expressed proteins were retrieved from Uniprot KB. The differentially expressed patterns were illustrated using K-means clustering in conjunction with heat map () [Citation35]. The relationships between up-regulated and down-regulated proteins were analyzed using STRING database (http://string-db.org/). The creation of a PPI network based on experimental data was used to predict the interactions.
Selection of tylosin-resistant S. suis mutants
S. suis was used to select tylosin-resistant mutants [Citation32]. To select resistant mutants, we performed serial passaging in THB medium containing 1/4 MIC (0.125 µg/mL) of tylosin. MIC of tylosin was determined for S. suis and resistant strains were induced in vitro using the broth dilution method in 96-well plates. In these plates, each well contained diluted concentrations of tylosin with 0.125 μg/mL, 0.25 μg/mL, 0.5 μg/mL, 1 μg/mL, 2 μg/mL and 4 μg/mL. The inoculated microplates were incubated at 37℃ for 24 h before evaluated. In addition, the stability of tylosin resistance was tested by serial passage (10 times) on an antibiotic-free medium.
The quantitative RT-PCR (qRT-PCR)
S. suis was grown to mid-log phase and then the culture medium was supplemented with 1/4 MIC (0.125 µg/mL) of tylosin prior to further incubation at 37℃ for 24 h. Control cells were incubated in the absence of tylosin. Resistant strains at a drug resistance level of 16 μg/mL of tylosin (S-t-R 16) and 128 μg/mL of tylosin (S-t-R 128) were also incubated at 37℃ for 24 h. Total RNA extraction (Omega, Beijing, China) and cDNA synthesis (Takara, Dalian, China) were done according to the manufacturer’s instructions. To investigate the effect of tylosin on the expression of genes, six proteins were chosen (). The 16S rRNA of S. suis strain was selected as a reference gene for all the experiments and the primers of target genes are listed in [Citation36]. The reaction conditions were 94℃ for 10 min followed by 40 cycles of amplification at 94℃ for 15 s and 60℃ for 60 s [Citation36]. The assays were repeated 3 times.
Table 1. Identification of Differentially Expressed Proteins.
Table 2. Primers used for the quantitative qRT-PCR analysis.
Statistical analysis
Each set of values was paralleled with three sets of controls and compared with the control groups. Student’s t-test was used to calculate the differences between the two mean values, with p < 0.05, set as the statistical significance threshold.
Results
Bacterial growth under the influence of tylosin at sub-MIC
MIC of tylosin against S. suis ATCC 700794 was 0.5 µg/mL in the present study. Furthermore, after 10 h of incubation at 37℃, both non-treated and treated (1/4 MIC (0.125 µg/mL) of tylosin) S. suis ATCC 700794 reached stationary phase, indicating there was no effect of tylosin on the growth rates of S. suis strain ().
Analysis using iTRAQ
To investigate the effect of tylosin stress on S. suis, 1/4 MIC of tylosin was selected (0.125 µg/mL) for quantitative proteomic analysis. First, bacteria treated and untreated with tylosin were labeled with isobaric reagents. Following this, the samples were pooled, fractioned by SCX chromatography, separated by LC and analyzed by MS/MS. Our analysis detected 1501 proteins. A ratio of proteins with > 1.2 or < 0.8 (p-value<0.05) was categorized as differentially expressed proteins. A total number of 171 differentially expressed proteins were identified in 1/4 MIC (0.125 µg/mL) of tylosin treated cells when compared to non-treated cells. Due to tylosin stress, 105 proteins were up-regulated, and 66 proteins were down-regulated in S. suis treated with tylosin, in comparison to the non-treated sample. In hierarchical clustering analysis (), some proteins were found to have a significant change in their expression as visualized using the R studio software.
The resistance-related proteins like elongation factor G (G7SMG3), elongation factor Ts (G7SBK2), elongation factor Tu (Q9EZW2) were up-regulated and had fold-change values of 3.67, 2.27 and 1.74, respectively. Proteins like putative histidine kinase of the competence regulon, ComD (G5L3D2), putative competence-damage inducible protein (G7SLJ0), protein GrpE (A4VZB4) were down-regulated in tylosin-treated sample with fold-change values of 0.74, 0.56 and 0.72, respectively ().
Protein–Protein interaction analysis
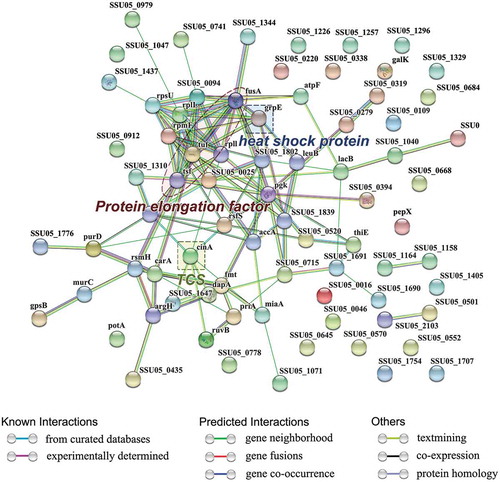
Protein-protein interaction and network analysis were performed on 171 differentially expressed proteins using the web-based tool STRING. Among these proteins, 77 proteins were included in the network and connected with each other. These proteins belonged to protein elongation factor, TCS and heat shock protein families (). The major networking proteins in the STRING analysis were EF-Tu, Ef-G, EF-Ts, putative competence-damage inducible protein and protein GrpE.
Figure 3. The network of significantly differentially expressed proteins (ratio>1.2 or <0.8 (p-value<0.05)) was analyzed by STRING. Small nodes represent proteins of unknown 3D structure; large nodes represent proteins with some 3D structure information or predicted. Coloured nodes represent query proteins and first shell of interactors; white nodes represent second shell of interactors. The blue lines represent database evidence; the purple lines represent experimental evidence; yellow lines represent text mining evidence; the black lines represent coexpression evidence; and green lines represent neighbourhood evidence.
MIC determination of resistant strain
The tylosin MIC against S. suis ATCC 700794 was 0.5 μg/mL. After 25 passages induction, the tested strain could tolerate 128 μg/mL, which MIC increased 256-fold in comparison with the parent strain. The resistance phenotype remained stable after 10 passages in antibiotic-free medium. In CLSI, there is no clear clinical breakpoint for tylosin against S. suis, but the strain which could amplify erm can represent the resistance to macrolide. In the previous studies, we know the gene of ermB can mediates high levels of resistance to tylosin and can be amplified in the resistant strain [Citation37–Citation41]. So, in our study, the ermB was used to verify whether the strain was resistant or sensitive to tylosin. The results showed that the strains S-t-R 16,S-t-R 128 could amplify ermB, but S. suis ATCC 700794 could not amplify ermB (Supplementary Material). So, the strains S-t-R 16,S-t-R 128 were tylosin-resistant, while S. suis ATCC 700794 was not resistant.
The susceptibility of S. suis to different kinds of antibiotics were determined using the in-vitro method recommended by the CLSI. The MICs of terramycin, chlortetracycline, ofloxacin and enrofloxacin indicated high-level resistance. However, the MICs of florfenicol and penicillin K were unchanged ().
Table 3. MICs of resistance strain Streptococcus suis ATCC 700794.
Investigation of changes in mRNA transcript levels in tylosin-resistant S. suis
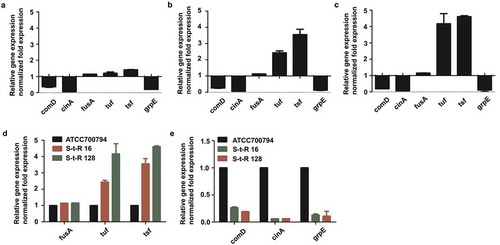
To explore whether the differential expression of proteins reflected a change at the transcription stage, mRNA levels of six proteins in treated with 1/4 MIC (0.125 µg/mL) of tylosin and non-treated S. suis were analyzed by qRT-PCR. Transcript levels of elongation factors (fusA, tuf, tsf), GrpE (grpE) and TCS (cinA, comD) were analyzed. Complementing the proteomics data, gene expressions of grpE, cinA and comD were down-regulated at mRNA level, while the gene expression of fusA, tuf and tsf were up-regulated. In order to verify whether the expressions of these proteins related to drug resistance, drug-resistant bacteria was induced and analyzed for changes at the transcription level. In the drug-resistant bacteria, the mRNA levels of fusA, tuf, tsf, comD and grpE were consistent with that of S. suis ATCC 700794 which was treated with 1/4 MIC (0.125 µg/mL) of tylosin ()). In order to confirm that the change was caused by tylosin, we detected the expression levels of fusA, tsf, tuf, cinA, comD and grpE in S. suis ATCC 700794, S-t-R 16 ()) and S-t-R 128 ()). With the increase in the level of resistance, the gene expression levels of fusA, tsf and tuf ()) increased, while that of cinA, comD and grpE decreased ()).
Figure 4. (a). Effect of 1/4 MIC of tylosin on mRNA expression of genes in Streptococcus suis ATCC 700794. Data are expressed as means ± standard deviations. The expression was normalized to 16S rRNA. Significantly different (*p < 0.05) compared to untreated control bacteria. (b). Effect on mRNA expression of genes in drug resistance level at 16 μg/mL of tylosin (S-t-R 16) Streptococcus suis ATCC 700794. Data are expressed as means ± standard deviations. The expression was normalized to 16S rRNA. Significantly different (*p < 0.05) compared to control bacteria. (c). Effect on mRNA expression of genes in drug resistance level at 128 μg/mL of tylosin (S-t-R 128) Streptococcus suis ATCC 700794. Significantly different (*p < 0.05) compared to untreated control bacteria. Data are expressed as means ± standard deviations. The expression was normalized to 16S rRNA. Significantly different (*p < 0.05) compared to control bacteria. (d). Effect on mRNA expression of fusA, tuf, tsf in Streptococcus suis ATCC 700794, drug resistance level at 16 μg/mL of tylosin (S-t-R 16) Streptococcus suis ATCC 700794 and drug resistance level at 128 μg/mL of tylosin (S-t-R 128) Streptococcus suis ATCC 700794. Data are expressed as means ± standard deviations. The expression was normalized to 16S rRNA. Significantly different (*p < 0.05) compared to control bacteria. (e). Effect on mRNA expression of comD, cinA, grpE in Streptococcus suis ATCC 700794, drug resistance level at 16 μg/mL of tylosin (S-t-R 16) Streptococcus suis ATCC 700794 and drug resistance level at 128 μg/mL of tylosin (S-t-R 128) Streptococcus suis ATCC 700794. Data are expressed as means ± standard deviations. The expression was normalized to 16S rRNA. Significantly different (*p < 0.05) compared to control bacteria.
Discussion
Generally, antibiotics are the first choice of treatment opted by veterinarians and farmers whenever there is an occurrence of bacterial disease. The long term use or abuse of antibiotics has often resulted in bacterial resistance, as in the cases of bacteria such as E. coli, Staphylococcus aureus and Coagulase-Negative Staphylococci [Citation1]. Thus, an understanding of the mechanisms involved in antibiotic resistance is necessary to extend the life of current antibiotics and enable the discovery of novel targets [Citation6]. The intricate molecular variations within bacterial pathogens associated with antibiotics, in response to treatment bacterial diseases, complicates the identification and quantification of functional proteins. So far, S. suis is a major public health issue and an emerging zoonotic agent in Southeast and East Asia [Citation42,Citation43]. In order to find appropriate inhibitors that can reduce bacterial resistance in tylosin-resistant S. suis, it is very important to focus on discovering specific target proteins which are related to tylosin resistance or multidrug resistance. In the last few years, proteomics methods have become a powerful tool for investigating antibiotic resistance mechanisms [Citation44]. In this study, we used an iTRAQ-based proteomic approach to gain a better understanding of proteins that are differentially expressed under tylosin stress.
In order to explore the differences in protein expression treated with 0.125 µg/mL tylosin and non-untreated S. suis, we observed proteins mainly belonging to protein elongation factor, two-component signal transduction system (TCS) and heat shock protein families based on our functional analysis using STRING. Previous studies suggest three types of elongation factors that required for prokaryotic cells translation are involved in drug resistance. Among them, the translation elongation Factor-G (EF-G) was reported to increase the resistance to kanamycin in E. coli via different point mutations in EF-G [Citation24,Citation45]. EF-Tu, another elongation factor, is the target of four families of antibiotics viz kirromycin, enacyloxin IIa, pulvomycin and GE2270A in E. coli [Citation46,Citation47]. Additionally, EF-G and EF-Tu have been identified as up-regulated proteins in tylosin-resistant M. gallisepticum [Citation27]. EF-TS has been displayed a significant change in the multidrug resistant E. coli strains [Citation23]. To find out whether these proteins were related to drug resistance in S. suis, strains S-t-R 16 and S-t-R 128 were induced in our experiment. The genes fusA (EF-G), tuf (EF-Tu) and tsf (EF-Ts) were up-regulated in strains S-t-R 16 and S-t-R 128, compared to the S. suis ATCC 700794. The expression of these elongation factor genes changed significantly with increase in the degree of resistance. This could be attributed to the function of tylosin. Previous research has reported that the MIC of ofloxacin is up-regulated in erythromycin-induced strain [Citation48] and the MIC of chlortetracycline increased 64 times in the tylosin-induced E. coli [Citation49]. In our experiment, the MICs of terramycin, chlortetracycline, ofloxacin and enrofloxacin were shown high-level resistance in the S-t-R 128 strain. Some studies have provided evidence that the “site” mutations of EF-G result in large changes in gene expression such as cyaA (adenylate cyclase) and topA (topoisomerase I) [Citation24]. The topA gene acts jointly with topoisomerase IV genes (parC and parE) and relaxes DNA supercoils in opposition to GyrAB activity, while the GyrAB protein is the most important site to form the resistance of fluoroquinolone [Citation21]. Previous studies have reported that the tetracycline (TC) antibiotics inhibit bacterial protein synthesis by preventing the attachment of aminoacyl tRNA to the ribosomal acceptor A site [Citation50]. The S-t-R 128 strain showed multidrug resistance to macrolides, quinolones and tetracycline that means the protein elongation factors may play important roles in conferring resistance to quinolones and tetracycline.
Natural transformation is a broadly conserved mechanism for horizontal gene transfer in bacterial species [Citation51]. In S. pneumoniae, several observations suggest that mosaic PBP genes was taken up at the physiological state. This sate is referred to as competence. Bacteria belonging to the Streptococcus genus, such as S. pneumoniae, Streptococcus mutans and S. suis are reported to be transformable species [Citation46,Citation52]. The TCS pathway, ComCDE, is involved in controlling competence. ComD and putative competence-damage inducible protein, which were significantly down-regulated in tylosin-treated sample, belong to the TCS of ComCDE.
In our study, comD and cinA gene transcripts were significantly down-regulated which was in agreement with to the proteomics data. Similar results were obtained in tylosin-resistant strain in which the expression levels of cinA and comD were significantly down-regulated. Additionally, down-regulation of these genes correlated with the level of drug resistance due to tylosin. Since, S. suis has been reported as the transformable species, ComCDE may exist in S. suis and play an important role in multidrug resistance.
A heat shock protein in Stenotrophomonas maltophilia acted as a stress-responsive protein by inducing quinolone resistance [Citation25]. Additionally, a study demonstrates role of the heat shock protein GrpE that has proved early exposure to these drugs in E. coli [Citation53]. In this study, GrpE was significantly down-regulated in tylosin resistant S. suis and this correlated with the level of drug resistance. Previous studies have also shown that expression levels of GrpE in tylosin resistant M. gallisepticum were significantly altered at various drug concentrations [Citation27]. Taken together, it is possible that GrpE may be involved in multidrug resistance.
We analyzed differentially expressed proteins of S. suis in response to tylosin stress by iTRAQ-based proteomic analysis. Several proteins showed significant changes in their level of expression upon tylosin stress in S. suis, including protein elongation factor, TCS and GrpE. Six drug resistance related proteins were determined in tylosin-resistant S. suis in response to induction with the drug and the corresponding changes in transcript levels were determined. The results showed that protein translation, elongation factor G, elongation factor Tu, elongation factor Ts, putative histidine kinase of the competence regulon, ComD, putative competence-damage inducible protein and protein GrpE were differentially expressed. The MICs of tetracycline and fluoroquinolone showed higher level of resistance compared to the sensitive bacteria. Based on these observations, we infer that these variations may be due to stress from the tylosin. Our data would help in identifying new target responsible for tylosin-resistant in S. suis and reducing antibiotic resistance by corresponding inhibitors.
Abbreviations
| S. suis | = | Streptococcus suis |
| MIC | = | Minimum inhibitory concentration |
| iTRAQ | = | isobaric tag for relative and absolute quantitation |
Author contributions
Y-HL designed the whole experiment. R-XC directed the completion of the experiment. X-XX, XL, Q-WQ, MC, FY, J-XM, X-RC, Y-HZ, B-OG, J-WL, Z-XL, Y-PX were supportive during the experiment.
Supplemental Material
Download MS Word (209.2 KB)Acknowledgments
The authors appreciate Shanghai Applied Protein Technology Co. Ltd for their support during the iTRAQ sequence data-base searching and data analysis.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplementary data for this article can be accessed here.
Additional information
Funding
References
- Sugawara A, Maruyama H, Shibusawa S, et al. 5-O-Mycaminosyltylonolide antibacterial derivatives: design, synthesis and bioactivity [Article]. J Antibiot. 2017 Aug;70(8):878–887. PubMed PMID: WOS:000406255800003; English.
- Qiu G, Rui YP, Zhang JL, et al. Macrolide-resistance selection in tibetan pigs with a high load of mycoplasma hyopneumoniae [Article; early access]. Microb Drug Resist 2018 Sep;24(7):1043–1049. PubMed PMID: WOS:000418582700001; English. DOI:10.1089/mdr.2017.0254
- Gottschalk M, Xu JG, Calzas C, et al. Streptococcus suis: a new emerging or an old neglected zoonotic pathogen? [Article]. Future Microbiol. 2010 Mar;5(3):371–391. PubMed PMID: WOS:000276191300011; English.
- Aarestrup FM, Carstensen B. Effect of tylosin used as a growth promoter on the occurrence of macrolide-resistant enterococci and staphylococci in pigs [; Research Support, Non-U.S. Gov’t]. Microbial Drug Resist (Larchmont, NY). 1998;4(4):307–312. PubMed PMID: MEDLINE:9988049; English.
- de Jong A, Simjee S, El Garch F, et al. Antimicrobial susceptibility of enterococci recovered from healthy cattle, pigs and chickens in nine EU countries (EASSA Study) to critically important antibiotics [Article]. Vet Microbiol. 2018 Mar;216:168–175. PubMed PMID: WOS:000428487800024; English.
- Gurung M, Tamang MD, Moon DC, et al. Molecular basis of resistance to selected antimicrobial agents in the emerging zoonotic pathogen streptococcus suis [Article]. J Clin Microbiol. 2015 Jul;53(7):2332–2336. PubMed PMID: WOS:000358287700045; English.
- Chen LF, Chiu C-T, Lo J-Y, et al. Clinical characteristics and antimicrobial susceptibility pattern of hospitalized patients with community acquired urinary tract infections at a regional hospital in Taiwan. Healthcare Infect. 2013 Dec;19(1):20–25. PubMed PMID: MEDLINE:25580164; English.
- Palmieri C, Varaldo PE, Facinelli B. Streptococcus suis, an emerging drug-resistant animal and human pathogen [Review]. Front Microbiol. 2011;2:6. PubMed PMID: WOS:000208863500243; English.
- Mitrophanov AY, Groisman EA. Signal integration in bacterial two-component regulatory systems [Review]. Genes Dev. 2008 Oct;22(19):2601–2611. PubMed PMID: WOS:000259700900003; English.
- Grebe TW, Stock JB. The histidine protein kinase superfamily [Comparative study;; Research support, Non-U.S. Gov’t; Research Support, U.S. Gov’t, P.H.S.; Review]. Adv Microb Physiol. 1999;41:139–227. PubMed PMID: MEDLINE:10500846; English.
- Hakenbeck R. Transformation in Streptococcus pneumoniae: mosaic genes and the regulation of competence [; Research Support, Non-U.S. Gov’t; Review]. Res Microbiol. 2000;151(6):453–456. PubMed PMID: MEDLINE:10961458; English.
- Sun JR, Chiang YS, Shang HS, et al. Phenotype microarray analysis of the AdeRS two-component system in Acinetobacter baumannii [Article]. Eur J Clin Microbiol Infect Dis. 2017 Dec;36(12):2343–2353. PubMed PMID: WOS:000415629700006; English.
- Zhang SM, Li XF, Wang X, et al. The two-component signal transduction system YvcPQ regulates the bacterial resistance to bacitracin in Bacillus thuringiensis [Article]. Arch Microbiol. 2016 Oct;198(8):773–784. PubMed PMID: WOS:000382029700007; English.
- Zhu LC, Lau GW. Inhibition of competence development, horizontal gene transfer and virulence in streptococcus pneumoniae by a modified competence stimulating peptide [Article]. PLoS Pathog. 2011 Sep;7(9):15. PubMed PMID: WOS:000295409000043; English.
- Poole K. Bacterial stress responses as determinants of antimicrobial resistance [Review]. J Antimicrob Chemother. 2012 Sep;67(9):2069–2089. PubMed PMID: WOS:000307832800003; English.
- Lysnyansky I, Ayling RD. Mycoplasma bovis: mechanisms of resistance and trends in antimicrobial susceptibility [Review]. Front Microbiol. 2016 Apr;7:7. PubMed PMID: WOS:000374836600001; English.
- Callens BF, Haesebrouck F, Maes D, et al. Clinical resistance and decreased susceptibility in streptococcus suis isolates from clinically healthy fattening pigs [Article]. Microb Drug Resist. 2013 Apr;19(2):146–151. PubMed PMID: WOS:000316736200011; English.
- Galante J, Ho ACY, Tingey S, et al. Quorum sensing and biofilms in the pathogen, streptococcus pneumoniae [Article]. Curr Pharm Design. 2015;21(1):25–30. PubMed PMID: WOS:000345434600005; English.
- Thirup SS, Van LB, Nielsen TK, et al. Structural outline of the detailed mechanism for elongation factor Ts-mediated guanine nucleotide exchange on elongation factor Tu [Article]. J Struct Biol. 2015 Jul;191(1):10–21. PubMed PMID: WOS:000357907700002; English.
- Aminov RI, Garrigues-Jeanjean N, Mackie RI. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins [Evaluation Studies;; Validation Studies]. Appl Environ Microbiol. 2001;67(1):22–32. PubMed PMID: MEDLINE:11133424; English.
- Kary SC, Yoneda JRK, Olshefsky SC, et al. The global regulatory cyclic AMP receptor protein (CRP) controls multifactorial fluoroquinolone susceptibility in salmonella enterica serovar typhimurium [Article]. Antimicrob Agents Chemother. 2017 Nov;61(11):14. PubMed PMID: WOS:000413558300026; English.
- Cetin R, Anborgh PH, Cool RH, et al. Functional role of the noncatalytic domains of elongation factor Tu in the interactions with ligands [; Research Support, Non-U.S. Gov’t]. Biochemistry. 1998 Jan;37(2):486–495. PubMed PMID: MEDLINE:9425069; English.
- Gautam A, Vinson HM, Gibbs PS, et al. Proteomic analysis of multidrug resistant Escherichia coli strains from scouring calves [Article]. Vet Microbiol. 2011 Aug;151(3–4):363–371. PubMed PMID: WOS:000293428400019; English.
- Mogre A, Veetil RT, Seshasayee ASN. Modulation of global transcriptional regulatory networks as a strategy for increasing kanamycin resistance of the translational elongation factor-G mutants in escherichia coli [Article]. G3-Genes Genomes Genet. 2017 Dec;7(12):3955–3966. PubMed PMID: WOS:000417073900012; English.
- Bemardini A, Corona F, Dias R, et al. The inactivation of RNase G reduces the Stenotrophomonas maltophilia susceptibility to quinolones by triggering the heat shock response [Article]. Front Microbiol. 2015 Oct;6:9. PubMed PMID: WOS:000363860400001; English.
- Kim JW, Kim HK, Kang GS, et al. The SAV1322 gene from Staphylococcus aureus: genomic and proteomic approaches to identification and characterization of gene function [Article]. BMC Microbiol. 2016 Sep;16:12. PubMed PMID: WOS:000383424600004; English.
- Xia X, Wu CM, Cui YW, et al. Proteomic analysis of tylosin-resistant Mycoplasma gallisepticum reveals enzymatic activities associated with resistance [Article]. Sci Rep. 2015 Nov;5:7. PubMed PMID: WOS:000365020600001; English.
- Sidjabat HE, Gien J, Kvaskoff D, et al. The use of SWATH to analyse the dynamic changes of bacterial proteome of carbapanemase-producing Escherichia coli under antibiotic pressure [Article]. Sci Rep. 2018 Mar;8:11. PubMed PMID: WOS:000426354200043; English.
- Goltermann L, Good L, Bentin T. Chaperonins fight aminoglycoside-induced protein misfolding and promote short-term tolerance in escherichia coli [Article]. J Biol Chem. 2013 Apr;288(15):10483–10489. PubMed PMID: WOS:000317565000028; English.
- Radhouani H, Pinto L, Poeta P, et al. After genomics, what proteomics tools could help us understand the antimicrobial resistance of Escherichia coli? [Review]. J Proteomics. 2012 Jun;75(10):2773–2789. PubMed PMID: WOS:000305266000003; English.
- Lee CR, Lee JH, Park KS, et al. Quantitative proteomic view associated with resistance to clinically important antibiotics in Gram-positive bacteria: a systematic review [Review]. Front Microbiol. 2015 Aug;6:29. PubMed PMID: WOS:000359819000001; English.
- Kim S, Lieberman TD, Kishony R. Alternating antibiotic treatments constrain evolutionary paths to multidrug resistance [Article]. Proc Natl Acad Sci U S A. 2014 Oct;111(40):14494–14499. PubMed PMID: WOS:000342633900052; English.
- Zhao YL, Zhou YH, Chen JQ, et al. Quantitative proteomic analysis of sub-MIC erythromycin inhibiting biofilm formation of S-suis in vitro [Article]. J Proteomics. 2015 Feb;116:1–14. PubMed PMID: WOS:000350922400001; English.
- Xiao M, Yang F, Wang X, et al. Establishment of concentration determination of staphylococcus aureus by spectrophotometry [Article]. Prog Vet Med. 2014;35(11):40–43. PubMed PMID: CSCD:5286640; Chinese.
- Shi ZH, Zhao C, Long W, et al. Microarray expression profile analysis of long non-coding RNAs in umbilical cord plasma reveals their potential role in gestational diabetes-induced macrosomia [Article]. Cell Physiol Biochem. 2015 May;36(2):542–554. PubMed PMID: WOS:000355944000012; English.
- Yang YB, Chen JQ, Zhao YL, et al. Sub-MICs of azithromycin decrease biofilm formation of streptococcus suis and increase capsular polysaccharide content of S. suis [Article]. Front Microbiol. 2016 Oct;7:8. PubMed PMID: WOS:000388535500002; English.
- Lonks JR. What is the clinical impact of macrolide resistance? Curr Infect Dis Rep. 2004;6(1):7–12. PubMed PMID: MEDLINE:14733843; English.
- Li MX, Cai C, Chen J, et al. Inducible expression of both ermb and ermt conferred high macrolide resistance in Streptococcus gallolyticus subsp pasteurianus isolates in China [Article]. Int J Mol Sci. 2016 Oct;17(10):11. PubMed PMID: WOS:000387768300180; English.
- Jiang CG, Li YH, Cai XH, et al. Erythromycin resistance of streptococcus suis isolates in vivo [Article]. Agric Sci China. 2009 Apr;8(4):502–507. PubMed PMID: WOS:000278075300017; English.
- Nishijima T, Saito Y, Aoki A, et al. Distribution of mefE and ermB genes in macrolide-resistant strains of Streptococcus pneumoniae and their variable susceptibility to various antibiotics. J Antimicrob Chemother. 1999;43(5):637–643. PubMed PMID: MEDLINE:10382884; English.
- Diawara I, Zerouali K, Katfy K, et al. Phenotypic and genotypic characterization of Streptococcus pneumoniae resistant to macrolide in Casablanca, Morocco [Article]. Infect Genet Evol. 2016 Jun;40:200–204. PubMed PMID: WOS:000374768000028; English.
- Gottschalk M, Segura M, Xu J. Streptococcus suis infections in humans: the Chinese experience and the situation in North America [; Research Support, Non-U.S. Gov’t; Review]. Anim Health Res Rev. 2007;8(1):29–45. PubMed PMID: MEDLINE:17692141; English.
- Sriskandan S, Slater JD. Invasive disease and toxic shock due to zoonotic Streptococcus suis: an emerging infection in the East? [Editorial Material]. PLoS Med. 2006 May;3(5):595–597. PubMed PMID: WOS:000238623900009; English.
- Yang XY, Xu JY, Wei QX, et al. Comparative proteomics of streptococcus pneumoniae response to vancomycin treatment [Article]. Omics. 2017 Sep;21(9):531–539. PubMed PMID: WOS:000411498000004; English.
- Borg A, Pavlov M, Ehrenberg M. Mechanism of fusidic acid inhibition of RRF- and EF-G-dependent splitting of the bacterial post-termination ribosome [Article]. Nucleic Acids Res. 2016 Apr;44(7):3264–3275. PubMed PMID: WOS:000375800200033; English.
- Kaur G, Rajesh S, Princy SA. Plausible drug targets in the streptococcus mutans quorum sensing pathways to combat dental biofilms and associated risks [Review]. Indian J Microbiol. 2015 Dec;55(4):349–356. PubMed PMID: WOS:000363949700001; English.
- Kobayashi T, Nonaka L, Maruyama F, et al. Molecular evidence for the ancient origin of the ribosomal protection protein that mediates tetracycline resistance in bacteria [Article]. J Mol Evol. 2007 Sep;65(3):228–235. PubMed PMID: WOS:000250065600003; English.
- Zanella A, Martino PA, Pratelli A, et al. Development of antibiotic resistance in Mycoplasma gallisepticum in vitro. Avian Pathol. 1998;27(6):591–596. PubMed PMID: MEDLINE:18484048; English.
- Cao LL, Zhou AG. Influence of kitasamycin, tylosin and chlortetracycline on E. coli resistance strain metabolism and drug resistance accumulation. Chin J Anim Sci. 2009;45(11):46–50.
- Aleksandrov A, Simonson T. Binding of tetracyclines to elongation factor Tu, the tet repressor, and the ribosome: a molecular dynamics simulation study [Article]. Biochemistry. 2008 Dec;47(51):13594–13603. PubMed PMID: WOS:000261768100016; English.
- Ellison CK, Dalia TN, Ceballos AV, et al. Retraction of DNA-bound type IV competence pili initiates DNA uptake during natural transformation in Vibrio cholerae [Article]. Nat Microbiol. 2018 Jul;3(7):773-+. PubMed PMID: WOS:000436530900006; English.
- Matsumoto-Nakano M. Role of Streptococcus mutans surface proteins for biofilm formation [; Review]. Jpn Dent Sci Rev. 2018 Feb;54(1):22–29. (Epub 2017 Sep). PubMed PMID: MEDLINE:29628998; English.
- Murakami J, Terao Y, Morisaki I, et al. Group A streptococcus adheres to pharyngeal epithelial cells with salivary proline-rich proteins via GrpE chaperone protein [Article]. J Biol Chem. 2012 Jun;287(26):22266–22275. PubMed PMID: WOS:000306418600064; English.