ABSTRACT
Peroxisomes are ubiquitous organelles in eukaryotic cells that fulfill multiple important metabolisms. Pex13 and Pex14 are key components of the peroxisomal docking complex in yeasts and mammals. In the present work, we functionally characterized the homologues of Pex13 and Pex14 (Mopex13 and Mopex14) in the rice blast fungus Magnaporthe oryzae. Mopex13 and Mopex14 were peroxisomal membrane distributed and were both essential for the maintenance of Mopex14/17 on the peroxisomal membrane. Mopex13 and Mopex14 interacted with each other, and with Mopex14/17 and peroxisomal matrix protein receptors. Disruption of Mopex13 and Mopex14 resulted in a cytoplasmic distribution of peroxisomal matrix proteins and the Woronin body protein Hex1. In the ultrastructure of Δmopex13 and Δmopex14 cells, peroxisomes were detected on fewer occasions, and the Woronin bodies and related structures were dramatically affected. The Δmopex13 and Δmopex14 mutants were reduced in vegetative growth, conidial generation and mycelial melanization, in addition, Δmopex13 showed reduced conidial germination and appressorial formation and abnomal appressorial morphology. Both Δmopex13 and Δmopex14 were deficient in appressorial turgor and nonpathogenic to their hosts. The infection failures in Δmopex13 and Δmopex14 were also due to their reduced ability to degrade fatty acids and to endure reactive oxygen species and cell wall-disrupting compounds. Additionally, Mopex13 and Mopex14 were required for the sexual reproduction of the fungus. These data indicate that Mopex13 and Mopex14, as key components of the peroxisomal docking complex, are indispensable for peroxisomal biogenesis, fungal development and pathogenicity in the rice blast fungus.
Introduction
Rice blast, one of the most destructive diseases in rice, causes a 10–30% loss of rice yield each year, which is sufficient to feed more than 60 million people [Citation1]. The causal agent of the disease is Magnaporthe oryzae, an ascomycete fungus. The M. oryzae infection is initiated by a conidium landing on a rice leaf. The conidium attaches to the leaf surface by an adhesive released from its tip [Citation2,Citation3]. Then, a polarized germ tube arises from the conidium and differentiates into a swollen dome-shaped appressorium. The appressorium generates enormous turgor by accumulating high concentrations of glycerol [Citation4]. The appressorium has a strong cell wall enriched in chitin, while being equipped with a melanin layer on the inner surface that avoids the efflux of glycerol and results in appressorial turgor up to 8.0 MPa [Citation4,Citation5]. The cellular turgor generates a huge mechanical force. Then a narrow penetrating peg emerges from the base of the appressorium. The penetrating peg, with the aid of the mechanical force, ruptures the leaf cuticle and epidermal cell wall and differentiates subsequently into invasive hyphae that ramify within the host cell and spread from cell to cell, resulting in visible lesions and rice blast symptoms [Citation2]. The rice blast fungus infects not only rice but also a wide range of economically important crops such as barley, oats, rye grass and millet [Citation2]. A M. oryzae strain that jumped from forage grass to wheat has caused serious problems in South America [Citation6], and recently, wheat blast has arisen in Bangladesh, placing wheat production in Asia at risk [Citation7]. Therefore, a better understanding of the mechanism of M. oryzae infection is still urgently required.
Peroxisomes, a type of single membrane-bound organelles, were shown to be crucial for infection by M. oryzae, as well other fungal plant pathogens [Citation8–Citation16]. Peroxisomes are present in almost all eukaryotes, which accommodate various crucial metabolic processes, such as fatty acid β-oxidation and the disposal of reactive oxygen species (ROSs) [Citation17]. Defects in peroxisomes in humans lead to severe developmental diseases; for instance, Zellweger syndrome causes early death in childhood [Citation18,Citation19]. In filamentous fungi, in addition to fatty acid and ROS metabolism, peroxisomes also play roles in some specific biochemical pathways, such as the biosynthesis of β-lactam antibiotics [Citation20]. Furthermore, the Woronin body, a type of fungal-specific organelle which is required to plug the septal pores to prevent cytoplasmic leakage in response to hyphal damage, is also deemed to represent a special class of peroxisomes [Citation21].
The proteins constituting peroxisomes, including peroxisomal membrane proteins (PMPs) and matrix proteins, are synthesized in the cytosol and imported into the organelles posttranslationally. The proteins involved in the import machinery for these PMPs and matrix proteins are commonly termed peroxins. Peroxin-encoding genes are designated PEX genes [Citation17]. Thus far, more than thirty PEXs have been isolated in various types of organisms. Mutants with damage in PEX genes usually lack normal peroxisomes in conjunction with a misdistribution of peroxisomal proteins, which results in a series of metabolic disorders related to peroxisomes [Citation22]. In filamentous fungi, pex mutants show some phenotypes that are unique to other types of organisms. For example, the Penicillium chrysogenum pex5 mutant is deficient in asexual spore formation [Citation23], and the Podospora anserina pex2 mutant shows alterations in karyogamy, a process of sexual sporulation [Citation24]. Notably, in plant pathogenic fungi such as M. oryzae, Colletotrichum orbiculare (syn. C. lagenarium) and Fusarium graminearum, pex1, pex5, pex6, pex11, pex13 and pex19 mutants exhibit disorders in host infection and pathogenicity-related processes [Citation9,Citation11–Citation16,Citation25].
Peroxisomal matrix proteins usually possess one of two types of peroxisomal targeting signals (PTSs), PTS1 or PTS2 [Citation17,Citation26]. The PTS1-containing matrix proteins are recognized and combined with the PTS1 receptor Pex5, whereas the PTS2-containing proteins are recognized and loaded by Pex7 (in fungi, together with the coreceptor Pex20) [Citation27]. The matrix protein-loaded receptors dock at a peroxisomal docking complex on the peroxisomal membrane, and via interaction with the complex, the cargos are translocated and released into peroxisomes. The receptors are then exported back to the cytosol for further rounds of importation [Citation22,Citation26]. In some cases, the peroxisomal docking complex also plays roles in receptor recycling [Citation28]. The docking complex in plant and mammals is composed of two membrane-associated peroxins, Pex13 and Pex14, while in yeasts it also possesses a third component, Pex17 [Citation29,Citation30].
Filamentous fungi are equipped with the bulk of the known PEX genes [Citation27] and several of them have been characterized. The constitution, functions and mechanisms of peroxisomal docking complex have been only limitedly investigated in filamentous fungi. Genome data indicated that filamentous fungi conservedly have Pex13 and Pex14, the two main components of peroxisomal docking complex while lack a typical Pex17, instead, contain a specific peroxin, Pex14/17 (or Pex33), which exhibits similarities to both Pex14 and Pex17 [Citation31,Citation32].The deletion of PEX13 in C. orbiculare impaired both of the PTS1 and PTS2 import machinery and led to inability of the mutants to utilize fatty acids as a carbon source. Appressoria formed by the copex13 mutants were defective in melanization and lost the penetration ability on host plants [Citation16]. These findings suggested an important role of Pex13 in peroxisomal biogenesis and fungal pathogenicity. We have previously demonstrated the involvement of Pex14/17 in peroxisomal biogenesis, peroxisomal metabolism and infection in M. oryzae [Citation14]. Interestingly, however, the deficiency in Δmopex14/17 is mild in contrast to mutations of Pex5, Pex6 or Pex19 [Citation9,Citation11–Citation13]. This result may suggest two possibilities: one is that the peroxisomal docking complex plays an unimportant role in the import cycle of peroxisomal matrix proteins in M. oryzae, and the other is that Mopex14/17, as an accessary component in the peroxisomal docking complex, plays minor roles and thus is unable to fully reflect the significance of the complex. To further illuminate the roles of the peroxisomal docking complex in fungal development and pathogenicity and to more exhaustively understand peroxisomal biogenesis in filamentous fungi, in this study, we investigated Pex13 and Pex14 in the rice blast fungus.
Materials and methods
Strains, culture and transformation
The Magnaporthe oryzae strain Guy-11 [Citation33] was primarily used as the wild type in present work, and for sexual reproduction, the 2539 strain was used as a tester strain. The media, conditions and methods for fungal culture and Agrobacterium-mediated transformation were performed as described previously [Citation34–Citation36].
Sequence analysis
The DNA sequences of MoPEX13 and MoPEX14 were retrieved from the database of the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/), and the coding information for genes was confirmed by amplifying and sequencing the coding regions. The BLAST program was used to search for homologous genes, ClustalW was used for the sequence alignments, GeneDoc was used to draw the alignment maps and MEGA 5.0 was used to establish phylogenetic trees.
Nucleic acid manipulation
Isolation of genomic DNA and total RNA was performed as previously described [Citation35]. PCR, restriction digestion and gel electrophoresis were performed using standard procedures. Gene cloning was performed using ClonExpress II One Step Cloning Kit (Vazyme Biotech, Nanjing, China) according to the protocol. Quantitative RT-PCR was processed on a 7500 Fast Real-Time System (Applied Biosystems, Foster, CA, USA) using standard procedures with β-tubulin gene (MGG_00604) as the reference gene.
Gene deletion and mutant recovery
For gene deletion, at least 1.5 kb of the upstream and downstream fragments of each gene were amplified from genomic DNA and inserted into p1300-KO [Citation13] to generate the gene replacement vectors pKO-PEX13 and pKO-PEX14. The transformants that were resistant to hygromycin B were harvested and screened preliminarily by genomic PCR with primer sets 13cds2/13cds3 and 14cds2/14cds3. The potential deletants were further confirmed by multiple rounds of PCR amplification and transcriptional PCR. For double deletion of MoPEX14 and MoPEX14/17, pSKO-PEX14, a gene replacement vector for MoPEX14 bearing the chlorimuron-ethyl resistant gene (SUR), was constructed and introduced into Δmopex14/17–54, a previously obtained MoPEX14/17 mutant [Citation14], using similar procedures.
For mutant recovery, the genomic fragments containing the full-length ORFs, 1.5 kb upstream and 0.5 kb downstream of MoPEX13 and MoPEX14/17, were amplified and inserted into p1300BAR to generate the complementary vectors p1300BAR-13com and p1300BAR-14com, respectively. The resulting transformants were selected based on glufosinate–ammonium resistance and checked by genomic PCR. The candidate complement strains were further confirmed by reverse-transcription PCR. For each gene, two confirmed complement strains were used in the phenotypical analysis.
Construction of fluorescent fusions and fluorescence microscopy
To monitor the subcellular distribution of Mopex13 and Mopex14, the coding sequences of the genes were amplified from total RNA with the primer sets 13cds2/13cds3 and 14cds2/14cds3 and introduced into p1300BMGFP-C [Citation13] by SmaI/SacI and SmaI/EcoRI digestion, respectively, to generate p1300BMGFP-PEX13 and p1300BMGFP-PEX14. The plasmid p1300BMGFP-PEX5 containing GFP-Mopex5 was constructed in a similar manner and used to monitor the distribution of Mopex5. p1300NMCHA (containing mCherry-PTS1), p1300BMGFPB (containing PTS2-GFP), p1300BMGFP-PMP47 (containing GFP-Pmp47), p1300BMGFP-PEX17 (containing GFP-MoPEX14/17) and p1300NMCh-HEX1 (containing pNMCherry-HEX1) were used to monitor the distribution of PTS1-containing proteins, PTS2-proteins, Mopmp47, Mopex14/17 and Mohex1, respectively [Citation13–Citation15,Citation37].The vectors were transformed into the M. oryzae strains via ATMT and selected by glufosinate–ammonium or G418 resistance. The fluorescence of the transformants was detected under a Leica SP2 Confocal System (Mannheim, Germany) using previously described methods [Citation15].
Assays for conidiation, appressorial formation, turgor genesis, lipid utilization, cell well integrity, ROS resistance and sexual reproduction
The measurements of conidiation, appressorial formation, appressorial turgor genesis, lipid utilization, cell well integrity and ROS resistance were performed according to previously documented methods [Citation13,Citation14]. All the experiments were replicated at least three times.
For sexual reproduction, the wild type Guy-11 and Guy-11-derived mutants were cross-cultured with the tester strain 2539 and 2539-derived mutants at 22°C for 7 d and then grown in the light for four weeks. The perithecia present at the junctions between opposite mating-type strains were examined and counted.
Pathogenicity tests and infectious structure observation
The rice cultivar CO39 and barley cultivar ZJ-8 were used in the pathogenicity tests. Spray-inoculation of rice seedlings and barley leaves and droplet-inoculation of intact and wounded barley leaves were performed as described previously [Citation13,Citation14]. Heat treatment of barley leaves was processed by laying 20-μl droplets of 65°C water on the inoculation sites, which were naturally cooled for five min and removed before inoculation. The microscopic observation of infectious structures was performed using previously described procedures [Citation11,Citation15].
Transmission electron microscopy
For TEM analysis, the conidia were harvested from colonies cultured on minimal medium [Citation35] supplemented with 0.5% Tween 80 for 8–10 days, and the mycelia were collected by culturing the conidia in liquid minimal medium with 0.5% Tween 80 for 2 days. The conidial and mycelial samples respectively were pretreated as previously described [Citation38] and examined under a JEM-1230 electron microscope (JEOL, Tokyo, Japan).
Yeast two-hybrid assay
The yeast two-hybrid analysis was carried out with strain Y2HGold (Clontech, USA) using the methods in the user’s manual supplied with Yeastmaker™ Yeast Transformation System 2 (Cat. No. 630439, Clontech, USA).
Results
Pex13 and Pex14 orthologs in M. oryzae
The possible homologues of Pex13 (MGG_00157) and Pex14 (MGG_01028) in M. oryzae were retrieved by searching the databases (www.ncbi.nlm.nih.gov) using the homologues from yeasts and related fungal species, and assigned as Mopex13 and Mopex14, respectively. The real coding regions of the two genes were confirmed by amplifying the cDNA fragments and were found to be identical to the hypothetical versions in the genome database (). MoPEX13 has a 1465-bp open reading frame (ORF) with three exons and two introns, encoding a peptide of 441 amino acid residues that shows 43% identity to Pex13 from Saccharomyces cerevisiae (KZV09435) and 57% to Pex13 from Colletotrichum orbiculare (ENH77396). The Mopex13 protein contains two conserved transmembrane regions (TMD1 and TMD2) and a Src homology 3 (SH3) domain at the C-terminus ()), which were confirmed to be essential for Pex13 to dock the peroxisomal matrix protein receptor [Citation39]. The ORF of MoPEX14 is 1387-bp long with four exons and three introns, encoding a 361-amino-acid peptide with 24% identity to S. cerevisiae Pex14 (AJS05562) and 56% to C. orbiculare Pex14 (ENH79683). The putative transmembrane regions containing AXXXA and GXXXG motifs were found at residues 112–142 of Mopex14, which mediate the chromomeric oligomerization of Pex14 ()) [Citation40].
Figure 1. Sequences of Pex13 homologs and Pex14 homologs in fungal species. (a-b) The putative protein sequences of Pex13 and Pex14 homologs, Anpex13 (XP_659115) and Anpex14 (CBF76374.1) from Aspergillus nidulans, Copex13 (ENH77396.1) and Copex14 (ENH79683.1) from Colletotrichum orbiculare, Fgpex13 (XP_011316361) and Fgpex14 (XP_011328505.1) from Fusarium graminearum, Mopex13 (XP_003718970.1) and Mopex14 (XP_003717914.1) from Magnaporthe oryzae, Ncpex13 (XP_965021.3) and Ncpex14 (XP_957540.1) from Neurospora crassa, and Scpex13 (KZV09435.1) and Scpex14 (KZV11075.1) from Saccharomyces cerevisiae were retrieved and aligned using ClustalX. Identical amino acids are highlighted against a black background, conserved residues against a dark gray background, and similar residues against a light gray background. The putative conserved transmembrane domain 1 (TMD1) and 2 (TMD2) domain and SH3 domain in the Pex13 proteins are underlined in blue. The putative hydrophobic region and coiled-coil structure in the Pex14 proteins are underlined in red. The phylogenetic relationships of the Pex13 homologs (c) and the Pex14 homologs (d) respectively were calculated using the neighbor joining method in the MEGA 6.0 software.

Mopex13 and Mopex14 are distributed on the peroxisomal membrane and required for the peroxisomal distribution of Mopex14/17
To obtain experimental evidence for Mopex13 and Mopex14 as components of the peroxisomal docking complex in M. oryzae, we first tested whether they were distributed on the peroxisomal membrane. The GFP-tagged versions of Mopex13 and Mopex14 were respectively integrated into M. oryzae strain Guy-11. The transformants of GFP-Mopex13 and GFP-Mopex14 emitted fluorescence both in a punctate pattern, which generally overlapped with the red fluorescence of mCherry-PTS1 representing the peroxisomal matrix ()). In the enlarged images, GFP-Mopex13 and GFP-Mopex14 could be detected surrounding mCherry-PTS1. These results indicated that Mopex13 and Mopex14 were distributed on the peroxisomal membrane.
Figure 2. Cellular localization of Mopex13 and Mopex14. (a) Mopex13 and Mopex14 are distributed on the peroxisomal membrane. Fluorescent microscopic analysis of the cotransformants GFP-Mopex13/mCherry-PTS1 and GFP-Mopex14/mCherry-PTS1 in Magnaporthe oryzae wild type Guy-11. GFP-Mopex13 and GFP-Mopex14 were both distributed in a punctate pattern, overlaying well with the peroxisome marker mCherry-PTS1. (b) The deletion of Mopex13 and Mopex14 did not alter the peroxisomal distribution of each other but led to a cytoplasmic distribution of Mopex14/17. The lack of Mopex14/17 did not change the distribution of Mopex13 or Mopex14. Bars = 5 µm.
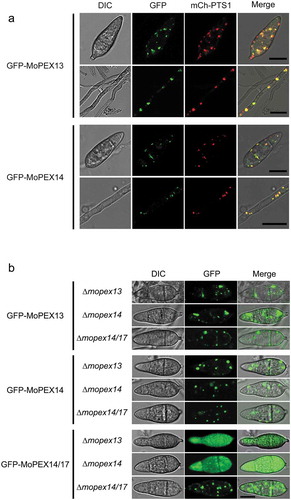
We then tested whether Mopex13, Mopex14 and Mopex14/17 as a complex could influence the subcellular distribution of each other. The gene deletion mutants ∆mopex13 and ∆mopex14 were constructed via homologous recombination strategies. Then, a GFP-tagged version of Mopex14/17, which was previously demonstrated to have a peroxisomal distribution in the wild type [Citation14], was integrated respectively into ∆mopex13 and ∆mopex14. In both mutants, the fluorescence of GFP-Mopex14/17 was altered to become completely cytoplasmic ()). This result indicated that the distribution of Mopex14/17 on the peroxisome membrane was dependent on Mopex13 and Mopex14. However, the peroxisomal distribution of GFP-Mopex13 and GFP-Mopex14 was unaffected by the deletion of MoPEX14/17, and Mopex13 and Mopex14 did not affect the distribution of each other.
Mopex13 and Mopex14 interact with each other and with Mopex14/17 and PTS receptors
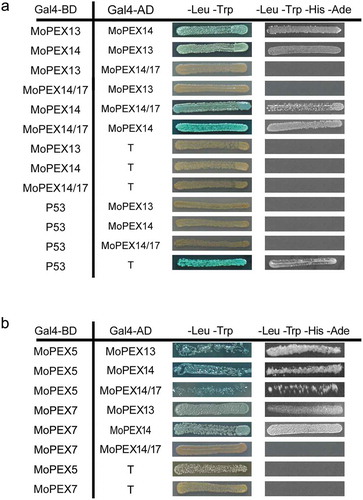
To further determine whether Mopex13, Mopex14 and Mopex14/17 could act as a complex and function in the docking of PTS receptors, we tested the interactions between Mopex13, Mopex14 and Mopex14/17 and with the PTS receptors using the yeast two-hybrid assay (). Mopex14 interacted with Mopex13 and Mopex14/17, suggesting that the proteins worked as a complex, although Mopex13 did not bind to Mopex14/17. Mopex13 and Mopex14 interacted with both the PTS1 receptor Mopex5 and the PTS2 receptor Mopex7, while Mopex14/17 interacted with Mopex7. The two-hybrid data supporting the conclusion that Mopex13, Mopex14 and Mopex14/17 were the components of peroxisomal docking complex in M. oryzae.
Figure 3. Interactions of Mopex13, Mopex14, Mopex14/17 and PTS receptors. The interactions between Mopex13, Mopex14 and Mopex14/17 (a), and the interactions of Mopex13, Mopex14 and Mopex14/17 with Mopex5 and Mopex7 (b) were analyzed by the yeast two-hybrid assay. The proteins were fused to the GAL4 DNA-binding domain (GAL4-BD) and the GAL4-activation domain (GAL4-AD), respectively. The indicated plasmid combinations were cotransformed into the Y2HGold yeast strain (Clontech). The transformants were tested for blue coloration on leucine/tryptophan double dropout plates presenting X-a-Gal and for viability on leucine/tryptophan/adenine/histidine quadruple dropout plates.
Mopex13 and Mopex14 play roles in import of peroxisomal matrix proteins and Woronin body protein HEX1
To reveal the roles played by Mopex13 and Mopex14 in peroxisome biogenesis, we tested the import of peroxisomal matrix proteins and PMPs in the mutants. The mCherry-PTS1 (mCherry fused to PTS1) and PTS2-GFP (GFP fused to PTS2) were integrated respectively into ∆mopex13, ∆mopex14 and the wild type. In the transformants derived from the wild type, both red and green fluorescence were detected predominantly as puncta, indicating the peroxisomal distribution of both PTS1- and PTS2-containing proteins (). In the transformants from ∆mopex13, mCherry-PTS1 and PTS2-GFP were both cytoplasmically distributed, demonstrating the inviability of the PTS1 and PTS2 peroxisomal import pathways. In ∆mopex14, however, although both mCherry-PTS1 and PTS2-GFP were largely present in the cytoplasm, fluorescent puncta were detected, representing the retained peroxisomal distribution. Consistent results were obtained when a PTS1 tagged GFP were used (Figure S1). These data indicated that Mopex13 was indispensable for the import of both PTS1 and PTS2 matrix proteins, while Mopex14 partially affected the PTS1 and PTS2 import pathways.
Figure 4. Mopex13 and Mopex14 are required for the distribution of peroxisomal matrix proteins and the Woronin body protein HEX1. The mCherry fused to PTS1 (mCh-PTS1), GFP fused to PTS2 (PTS2-GFP), Pmp47 labeled with GFP (GFP-Pmp47), HEX1 labeled with mCherry (mCh-HEX1), and Mopex5 labeled with GFP (GFP-MoPEX5) were respectively introduced into wild type Guy-11, the ∆mopex13 and ∆mopex14 mutants, as well as the ∆mopex14/17 mutants for GFP-MoPEX5. The peroxisomal distribution patterns of mCh-PTS1, PTS2-GFP, mCh-HEX1 were dramatically changed in the ∆mopex13 and ∆mopex14 mutants, while those of GFP-Pmp47 and GFP-MoPEX5 were unaltered compared to those in the wild type. The distribution of GFP-MoPEX5 was also unchanged in the ∆mopex14/17 mutant. Bars = 5 µm.
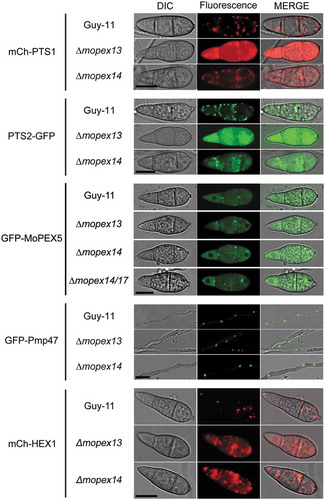
To further identify additional clues regarding the functional association of the Pex13-Pex14-Pex14/17 complex with matrix import, we compared the distribution profiles of PTS1 receptor Mopex5 in the wild type and in the ∆mopex13, ∆mopex14 and ∆mopex14/17 mutants. However, no difference was found. In either the wild type or the mutants, GFP-Mopex5 was detected bimodally in the cytosol and peroxisome-like structures, in accordance with its roles as a shuttle reporter.
To estimate whether Mopex13 and Mopex14 had roles in the import of PMPs, we detected the subcellular distribution of Mopmp47, a representative PMP in M. oryzae, in the mutants [Citation13]. In both the ∆mopex13 and ∆mopex14 mutants, the GFP-tagged Mopmp47 (GFP-Mopmp47) exhibited a punctate pattern without a notable difference from that in the wild type, supporting the normal peroxisomal distribution of Mopmp47 in the mutants, namely, Mopex13 and Mopex14 were dispensable for the import of PMPs.
To elucidate the effects of Mopex13 and Mopex14 on Woronin body formation, we examined the distribution of Hex1, the dominant protein in Woronin bodies [Citation41]. The mCherry-tagged Hex1 (mCh-HEX1) expressed in the wild type exhibited a punctate distribution and accumulated in the central region of the septa in conidial and hyphal cells. However, the distribution of mCherry-Hex1 was greatly altered in ∆mopex13 and ∆mopex14, in which the fluorescence was predominantly cytoplasmic, indicating that both Mopex13 and Mopex14 were required for proper distribution of Hex1 at Woronin bodies. Because Hex1 contains a typical PTS1 and its localization also rely on peroxisomal import pathway, the Woronin body is deemed as peroxisomal derived vesicle or specialized peroxisome [Citation41,Citation42]. Thus, the misdistribution of Hex1 in ∆mopex13 and ∆mopex14 is probably resulted from the defects in peroxisomal import of the mutants.
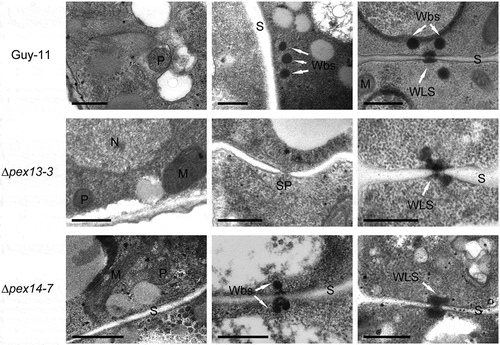
Structures of Woronin bodies were altered in ∆mopex13 and ∆mopex14 mutants
Considering the influence of Mopex13 and Mopex14 on the import of peroxisomal matrix proteins, we examined the structures of peroxisomes in the mutants by TEM analysis. In the wild type, electrondense round peroxisomes were detected in almost every electron microscopic section, mainly presenting at the periphery of the conidial cells. In ∆mopex13 and ∆mopex14, peroxisomes and peroxisome-like structures were still detectable but much less frequently than in the wild type cells. As peroxisome-derived organelles, Woronin bodies were also examined by TEM (). In the wild type, Woronin bodies were easily detectable. Typically, three to five independent Woronin bodies flank intact septa or injured ones that were sealed by Woronin body-related structures. In ∆mopex13 and ∆mopex14, Woronin body-like structures remained plugging the septal pores or associating at the edge of pores. However, separated Woronin bodies were not found beside either intact or injured septa. Additionally, in ∆mopex13, unsealed septal pores without any Woronin bodies could occasionally be observed. Taken together, the deletion of MoPEX13 or MoPEX14 negatively affected the presence of peroxisomes and the structure of Woronin bodies, but it did not completely eliminate the two types of organelles in M. oryzae cells.
Mopex13 and Mopex14 are required during vegetative development and conidiation in M. oryzae
When cultured on CM, ∆mopex13 and ∆mopex14 mutants developed at a significantly lower rate than the wild type and complemented strains (). The ∆mopex13 mutant formed flat and bald colonies. Cellular leakage and hyphal decay could also be observed more occasionally in the ∆mopex13 colonies. The ability of ∆mopex13 and ∆mopex14 to produce conidia was dramatically reduced. When cultured on CM for 6 days, the colonies of ∆mopex13 and ∆mopex14 produced only 2% and 10% the conidia of those of the wild type and complemented strains (). The reduction in vegetative growth of ∆mopex13 and ∆mopex14 was also confirmed by measuring the dry weight of mycelia cultured in the liquid CM ()). The melanin production of the mutants was also greatly reduced in the liquid cultures (,h)). These results indicated that Mopex13 and Mopex14 were involved in the vegetative growth of the fungus.
Figure 6. Vegetative growth, conidiation and melanization of the ∆mopex13 and ∆mopex14 mutants. The ∆mopex13 and ∆mopex14 mutants, wild type and complemented strains were cultured on 9-cm CM plates at 28°C for 10 days (a&b). The radical and aerial hyphal growth of the strains was compared (c). Conidiation of the strains was recoded and compared (d&e). Bar = 50 µm. The strains were cultured in CM liquids with 12-h light/12-h dark cycles at 28°C for 4 days, and then the mycelia were harvest and weighed (f). Melanization of the cultures was recorded and compared by measuring the absorbance value of the culture solutions at 415 nm (OD415) (g&h). Means and standard errors were calculated from three independent repeats. Single asterisks indicate significant differences at P < 0.05 compared with the wild-type, and double asterisks indicate significant differences at P < 0.01.
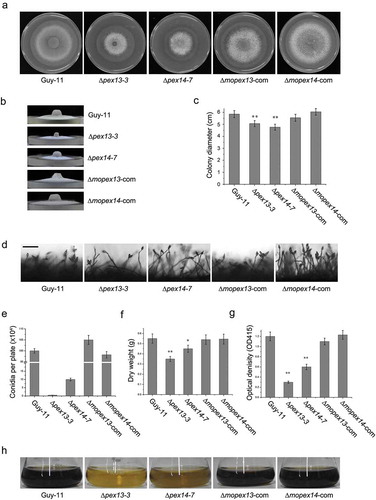
Mopex13 and Mopex14 are indispensable for host infection
To investigate the roles of Mopex13 and Mopex14 in plant disease, we performed a spray-inoculation of rice and barley seedlings. The leaves inoculated with the wild type and the complement strains generated large numbers of typical lesions (). In contrast, no lesions were found on ∆mopex13 and ∆mopex14-inoculated barley or rice leaves. Inoculation of detached barley leaves provided the same results. These results indicated that Mopex13 and Mopex14 were indispensable for the pathogenicity of M. oryzae.
Figure 7. Mopex13 and Mopex14 are indispensable for pathogenicity on rice and barley. (a) Two-week-old rice cultivar CO39 seedlings were spray-inoculated with the conidial suspension (1 × 105 conidia/ml). The symptoms were recorded at 7 days postinoculation (dpi). (b) Seven-day-old barley cultivar ZJ-8 seedlings were spray-inoculated with the conidial suspension (1 × 105 conidia/ml) and recorded at 3 dpi. (c) Detached barley leaves were inoculated with 20 μl of the conidial suspensions (upper, 1 × 105 conidia/ml; lower, 1 × 104 conidia/ml) and incubated for 4 days. (d) Droplet-inoculated barley leaves were sampled at 24, 36 and 48 hpi, discolored, and examined under a light microscope to detect the infectious structures. Bar = 20 µm.

The histological evidence of the pathogenic defects was gained by microscopic observation of the inflection structures on barely leaves (). Abundant bulbous invasive hyphae were visible at the inoculation sites of the wild type, which expanded quickly in the host tissue, and at 48 h post-inoculation (hpi), the infectious hyphae were fully established in the first infected cells and had ramified to neighboring cells. However, on the leaves inoculated with ∆mopex13 or ∆mopex14, despite the formation of appressoria similar to those of the wild type, no invasive hypha was found up to 48 hpi. These data indicated that the appressoria of ∆mopex13 and ∆mopex14 had lost the ability to penetrate the host cells.
Appressorial development is deficient in ∆mopex13 and ∆mopex14
The penetration failure suggested a deficiency in the appressoria of the mutants. Thus, we tested the appressorial development of the mutants on a plastic membrane under inducible conditions. The rates of conidial germination and appressorial formation were dramatically reduced in ∆mopex13 compared with wild type (). Furthermore, the appressoria of ∆mopex13 were smaller and less pigmented ()). Moreover, lipid droplets could be detected in the conidia of ∆mopex13 which formed appressoria and their germ tubes. ∆mopex14 showed unchanged rates of conidial germination and appressorial formation, as well as the microscopic appearance of appressoria, although the lipid droplets, as in ∆mopex13, were as well detected in its conidia and germ tubes. To determine whether the appressorial turgor pressure was altered in ∆mopex13 and ∆mopex14, we performed an incipient cytorrhysis assay ()). The collapse rates of the appressoria of ∆mopex14 treated with glycerol were increased compared with the wild type, suggesting a reduction of the turgor generation in ∆mopex14 appressoria. However, the appressoria of ∆mopex13 collapsed at a level lower than that of the wild type, and plasmolysis synchronized. This phenomenon might be due to an increased permeability to glycerol of the ∆mopex13 appressoria. Therefore, cytorrhysis assays were subsequently performed using polyethylene glycerol 8000 (PEG8000), which has a larger molecular size than glycerol, as the external inductive agent ()). As expected, more collapsed appressoria were observed in ∆mopex13 mutants treated with PEG8000 compared with the wild type. These results indicated that the turgor was reduced in ∆mopex13 and ∆mopex14 and that glycerol could cross the appressorial cell wall of ∆mopex13, likely resulting from the reduced pigmentation and deficiency of cell wall integrity. Corresponding with their involvement in appressorial morphogenesis, qRT-PCR indicated that MoPEX13 and MoPEX14 were increasingly transcribed during conidial germination and appressorial formation, with a peak at 6 ~ 8 h post-incubation, the key stage of appressorial differentiation ()). Taken together, these findings indicated that Mopex13 and Mopex14 were required to form normal morphologic and functional appressoria in M. oryzae. To further confirm whether this was one of the reasons for the penetration failure of the mutants and to determine whether the genes could also function at postpenetration stages, we compared the infections by the mutants on intact barley leaves and on wounded ones with cuticle removal by grinding (()). In contrast to the completely symptomless intact leaves, the wounded leaves inoculated with ∆mopex13 and ∆mopex14 displayed slight lesions, although they were less developed than those caused by the wild type. Correspondingly, infective hypha of ∆mopex13 and ∆mopex14 could be detected on the wounded leaves, although at a reduced level than those of the wild type. Taken together, we concluded that the requirement for Mopex13 and Mopex14 during host infection was related to both cuticle penetration and postpenetration developments within host cells.
Figure 8. Mopex13 and Mopex14 are required in appressorial development and host penetration. The conidia of ∆mopex13, ∆mopex14 and wild type harvested from 9-day-cultured CM plates and suspended at 1 × 105 conidia/ml were incubated to allow germination and appressorial formation. (a&b) The conidial germination rates and appressorial formation rates at sequential time points were calculated and statistically compared. (c) Microscopy of the appressorial development of the strains. (d&e) Incipient-cytorrhysis was performed to compare the appressorial turgor genesis in the 24-h appressoria of strains using glycerol or PEG8000 as the external solute. Means and standard errors were calculated from three independent replicates (at least 100 conidia each). (f) The relative transcription of MoPEX13 and MoPEX14 during appressorial development was examined by qRT-PCR using the tubulin gene as a reference. (g) Inoculation of cuticle-removed (wound) barley leaves with 1 × 105 conidia/ml conidial suspensions. The mutants partially regained the ability to cause lesions on wounded leaves. (h) Infectious structure on inoculated intact and wounded barley leaves.
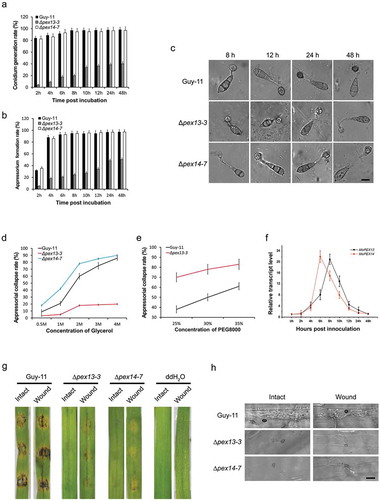
Defects in lipid degradation and migration in ∆mopex13 and ∆mopex14 mutants
Considering the importance of lipid metabolism in fungal infection and the indispensability of peroxisomes in lipid metabolism, we evaluated the ability of the ∆mopex13 and ∆mopex14 mutants to metabolize lipids. First, the relative transcription of the genes was tested using qRT-PCR, and the results indicated that MoPEX13 and MoPEX14 could be upregulated by lipids, such as triolein, olive oil, Tween 80 and sodium acetate ()). On media containing fatty acids (Tween 80 or olive oil) as the sole carbon source, the mutants lost the ability to survive, supporting the involvement of Mopex13 and Mopex14 in fatty acid degradation. Moreover, the mutants could utilize acetate, which represented the products of fatty acid oxidation and the initiator of the glyoxylate cycle. Thus, Mopex13 and Mopex14 were indispensable for the degradation of fatty acids, but not for subsequent biochemical processes, such as the glyoxylate cycle.
Figure 9. Mopex13 and Mopex14 are involved in lipid metabolism. (a) Lipid utilization by ∆mopex13 and ∆mopex14 and the wild-type strain Guy-11. The strains were cultured on minimal medium with glucose (1%), Tween 80 (0.5%), olive oil (0.5%) or sodium acetate (50 mM) as the sole carbon source for 5 days. (b) Relative transcription of MoPEX13 and MoPEX14 in the wild type cultured on minimal medium with glucose (1%) or with glucose (0.5%) together with Triolein (0.5%), Tween 80 (0.5%), olive oil (0.5%) or sodium acetate (50 mM). (c) Deletion of MoPEX13 or MoPEX14 delayed lipid mobilization and degradation during appressorium development. Conidia of the wild-type strain Guy-11 and mutants were incubated on a hydrophobic membrane and allowed to form appressoria. Samples at different time points were stained with Nile Red and examined microscopically. Bar = 5 μm. (d) Inoculation of barley leaves with conidial suspensions (1 × 105 conidia/ml) supplemented with glucose (2.5%). The presence of glucose partially improved the ability of the mutants to cause disease.
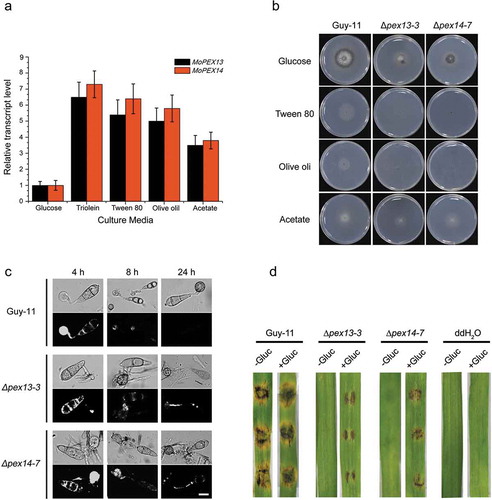
Next, we investigated whether lipid degradation was influenced in the mutants during appressorial development by Nile red staining ()). In the conidia and initially formed appressoria at 4 h post-incubation, lipids were visualized in both the mutants and wild type. After 8 h post-incubation, lipid droplets were scarcely observed in the conidia and appressoria in the wild type. However, in the Δmopex13 and Δmopex14 mutants, the numbers of lipid droplets remained detectable in the appressoria, germ tubes and conidia even at 24 h post-incubation. These data indicated that the lipid degradation and lipid mobilization from conidia to appressoria were impaired by the lack of Mopex13 or Mopex14.
The lipids in conidia provide an important source supporting appressorial morphogenesis and host infection ()). To further investigate whether the infection defects in Δmopex13 and Δmopex14 were caused by disordered lipid metabolism, we performed an inoculation with conidial suspensions supplemented with an additional carbon source. With the added glucose, both mutants showed a partial restoration of their pathogenicity on barley leaves. These results indicated that deficient lipid degradation played a role in the infection failure of ∆mopex13 and ∆mopex14.
Antioxidant ability and endurance to cell wall disturbances were reduced in ∆mopex13 and ∆mopex14 mutants
ROS elimination is another main metabolic feature of peroxisomes and is crucial for fungal infection. The ability of the mutants to degrade ROS was evaluated by comparing their growth on CM supplemented with oxidants. The tolerance of both ∆mopex13 and ∆mopex14 to H2O2, methyl viologen or Rose Bengal was significantly reduced, indicating the requirement for Mopex13 and Mopex14 in the antioxidant ability of M. oryzae (). Subsequently, we performed pathogenicity test on barley leaves which were briefly treated with heat before inoculation to inhibit their ROS generation ()). On these heat-treated leaves, ∆mopex13 and ∆mopex14 caused slight symptoms. Namely, the virulence of the mutants could be partially restored by inhibiting host cell ROS generation. These results suggest that the deletion of MoPEX13 or MoPEX14 reduces the ability of M. oryzae to remove ROS and that the excess ROS in host cells inhibit the development of mutants within the host tissue.
Figure 10. Deletion of MoPEX13 or MoPEX14 reduced the tolerance of the mutants to ROS. (a) The ∆mopex13, ∆mopex14 and wild type strains were cultured on complete medium in the presence of hydrogen peroxide (0.04%, v/v), methyl viologen (MV, 0.25 mg/ml) or Rose Bengal (RB, 100 μM). Olive oil (0.5%) or sodium acetate (50 mM) was used as the sole carbon source for 7 days. (b-d) The colony diameters of the tested strains were measured to calculate and compare significant differences in habitation rates. Means and standard errors were calculated from three independent replicates. Single asterisks indicate significant differences at P < 0.05 compared with the wild-type, and double asterisks indicate significant differences at P < 0.01. (e) Inoculation on barley leaves that were heat-treated to suppress ROS generation. The heat treatment improved the pathogenicity of the mutants on the barley leaves.
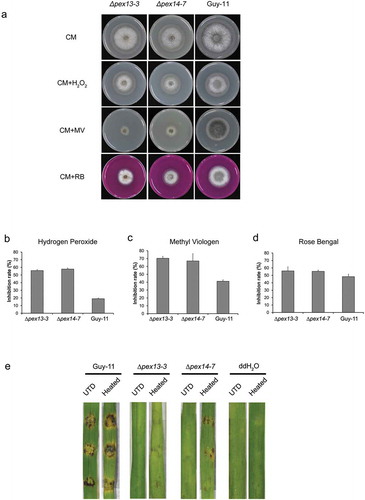
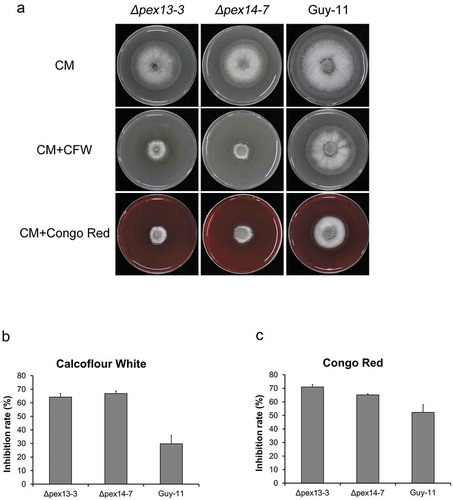
Cell wall biogenesis is another factor that is related to fungal pathogenicity and peroxisomal metabolism. To determine whether Mopex13 and Mopex14 were associated with the regulation of cell wall integrity in M. oryzae, we cultured the ∆mopex13 and ∆mopex14 mutants on CM containing Congo Red (CR) or Calcofluor white (CFW), two chemicals disturbing cell wall biogenesis (). Compared with the wild type Guy-11, the mutants exhibited an enhanced sensitivity to both agents, indicating that Mopex13 and Mopex14 played a role in cell wall integrity.
Figure 11. Deletion of MoPEX13 or MoPEX14 reduced the tolerance of the mutants to cell wall interference agents. (a) The ∆mopex13, ∆mopex14 and wild type strain were cultured on CM supplemented with Congo red (100 µg/ml) or Calcofluor white (50 µM) for 7 days. (b&c) Statistical comparison of the radial growth of the strains. Error bars represent the deviation from three replicates, and double asterisks indicate significant differences in comparison to the wild type at P < 0.01.
Mopex13 and Mopex14 are involved in the sexual reproduction of the fungus
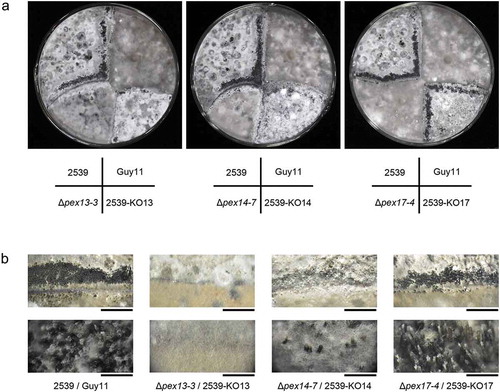
Next, we investigated the roles of Mopex13 and Mopex14 in sexual reproduction by mating the experimental strain with strain 2539, a tester strain with an opposite mating type to Guy-11. First, we generated the ∆mopex13 and ∆mopex14 mutants derived from 2539. Then, the wild type strains Guy-11 and 2539 and the mutants from the two strains were crossed cultured basing on their mating types. After four weeks, numerous perithecia were observed at the junctions of the wild type Guy-11 and 2539 (). In contrast, perithecia were hardly found in the crosses of ∆mopex13 from Guy-11 and that from 2539, and they were sparely formed in the cross of the wild type Guy-11 and ∆mopex13 from 2539. A similar situation was observed for the ∆mopex14 mutants. This result indicated that Mopex13 and Mopex14 were involved in the sexual reproduction of the fungus. In addition, Mopex14/17 was also tested but was found to be indispensable for sexual generation.
Figure 12. Mopex13 and Mopex14 are required in sexual reproduction of M. oryzae. The ∆mopex13, ∆mopex14 and ∆mopex14/17 mutants derived from Guy-11 strain and those from 2539 strain were cross-cultured on oatmeal agar plates at 22°C for 30 days. (a) The asci formed at the junction of the strains were recorded. (b) Magnified images of the junction areas of the strains. Bars in the upper panel = 5 mm; bars in the lower panel = 1000 μm.
Discussion
In the present work, we demonstrated that Mopex13 and Mopex14, as potential components of the peroxisomal docking complex, were crucial for the biogenesis of the peroxisome and Woronin bodies and were required for the development and pathogenicity of the rice blast fungus M. oryzae. These findings reinforce knowledge concerning peroxisomal biogenesis and the roles of peroxisomes in plant fungal pathogens.
Pex13, Pex14 and Pex17 are widely accepted as components of the peroxisomal docking complex [Citation26,Citation30,Citation43]; however, the situation remains complicated in different type of organisms. In yeasts, Pex13, Pex14 and Pex17 constitute the docking complex, play respective roles in the import of peroxisomal matrix proteins and cannot be substituted for one another. In mammals, in which Pex17 is absent, Pex13 and Pex14 form a docking complex and are sufficient to fulfil the import of matrix proteins [Citation26]. Filamentous fungi lack typical Pex17 but instead contain Pex14/17, a chimera resembling both Pex14 and Pex17 in sequence [Citation27,Citation31,Citation32]. Thus, the composition of the docking complex in filamentous fungi is mysterious, and it is possible that Pex14/17 function as a Pex17 orthologue or as a redundant protein to Pex14. To clarify these issues, we have previously characterized Pex14/17 in M. oryzae and confirmed that Mopex14/17 is distributed on the peroxisomal membrane and is required for the import of PTS1-containing proteins and peroxisomal metabolisms [Citation14]. In the present work, we demonstrated that Mopex13 and Mopex14 were also distributed on the peroxisomal membrane and further are required for the peroxisomal membrane distribution of Mopex14/17. Mopex13 and Mopex14 both interacted with Mopex14/17 and with each other. These data indicate that the three proteins constitute a complex on the peroxisomal membrane. In addition, Pex14/17 in P. chrysogenum can complement the defects in the S. cerevisiae Pex17 mutant, and the Pex17-like C-terminus of P. chrysogenum Pex14/17 other than its Pex14-like N-terminus is able to restore the defects in matrix protein import and sporulation in the P. chrysogenum Δpex14/17 mutant [Citation32]. These results indicate that Pex14/17 is analogous to Pex17 in function and support that Pex13, Pex14 and Pex14/17 in filamentous fungi compose the peroxisomal docking complex and may function in a manner resembling their counterparts in yeasts (Figure S2).
As the docking site for PTS1 and PTS2 receptors, the Pex13-Pex14-Pex17 complex physically exhibits interactions with the receptors. In S. cerevisiae and mammals, Pex13 binds to both import receptors, Pex5 and Pex7 [Citation39,Citation44]. Pex14 also binds to both receptors [Citation30,Citation43]. Two-hybrid data in Neurospora crassa also suggest that both Pex13 and Pex14 are able to bind the PTS1 and PTS2 import receptors [Citation45]. Our data agree with these investigations and showed that Mopex13 and Mopex14 interacted with Mopex5 and Mopex7. Moreover, Mopex14/17 also interacted directly or indirectly with the PTS1 receptor. These results indicate that the docking machinery centering on Pex13-Pex14 is shared by diverse eukaryotes. Interestingly, however, we did not observe an impact of the disruption of Mopex13, Mopex14 or Mopex/14/17 on the distribution of Mopex5 by the GFP fusion technique.
The disruption of MoPEX13 led to serious damage to the import of peroxisomal matrix proteins. GFP-PTS1 and PTS2-GFP were completely cytosolic in Δmopex13, and as a result, the mutant was disordered in peroxisome-related metabolism, such as fatty acid utilization and ROS degradation. These results are consistent with the situation in other species. For instance, the S. cerevisiae Pex13 mutant lost the ability to grow on methanol or oleate, and the peroxisomal matrix proteins in the mutants were mislocalized into the cytosol [Citation39,Citation44,Citation46]. The C. orbiculare Pex13 mutant is incapable of importing both PTS1- and PTS2-containing proteins [Citation16]. In the Podospora anserina Δpex13 mutant, GFP-PTS1 is completely cytoplasmically distributed [Citation47]. Additionally, the Aspergillus nidulans Pex13 mutant fails to import malate synthase and isocitrate lyase into peroxisomes via the PTS1 or PTS2 pathways [Citation48]. In higher organisms such as Arabidopsis thaliana and humans, Pex13 is also crucial for the import of matrix proteins [Citation39,Citation49]. Moreover, ∆mopex13 exhibited typical disorders, which have been observed in other M. oryzae pex mutants with severely impaired peroxisome biogenesis, such as reduced vegetative growth and sporulation, defective melanization and appressorial glycerol accumulation, as well as the inability to initiate host infection [Citation9,Citation11–Citation13,Citation25]. Taken together, these data suggest that Pex13 is conserved in yeasts, plants, mammals and fungi and is crucial in peroxisomal import for both PTS1- and PTS2-containing matrix proteins, fully reflecting that the peroxisomal docking complex plays key important roles in fungal development and pathogenicity.
Although Pex14 was deemed the central component of the peroxisomal docking complex [Citation43,Citation50], Mopex14 contributes less than Mopex13 to peroxisomal import in M. oryzae and the development of the fungus. For instance, in contrast to the completely cytosolic distribution of PTS1- and PTS2-containing proteins in ∆mopex13, trace amounts of peroxisomal distribution were detected in ∆mopex14. Deletion of MoPEX14 caused less severe defects than that of MoPEX13 in vegetative growth, conidiation, germination and appressorial formation. In previous investigations, the functions of Pex14 in matrix import and peroxisomal structures were also likely to be less conserved than Pex13 among different organisms. In S. cerevisiae and humans, Pex14 is an essential component of the import machinery of peroxisomal matrix proteins [Citation50,Citation51]. In Arabidopsis thaliana, Pex14 facilitates the import of matrix proteins but is not absolutely required [Citation52]. In the P. anserina pex14 mutant, both types of matrix proteins exhibit a remnant peroxisomal distribution in several developmental phases [Citation47]. In the pex14 mutant of N. crassa, the PTS1 proteins Hex1 and Mls1 are exclusively cytosolic, whereas Fox2 and Icl1, which lack a typical PTS, are detected in both the cytosol and peroxisomes [Citation45]. In addition, the C. orbiculare pex13 mutant lacks intact peroxisomes in conidia and appressoria [Citation16]. However, our TEM data indicated that peroxisomes were present in Δmopex13 and Δmopex14, although their amounts were likely reduced, and the punctate distribution of GFP-Mopmp47 further supported the presence of peroxisomes in the two mutants. These results may reflect the functional variation of the peroxisomal docking complex among different organisms.
The disruption of MoPEX13 or MoPEX14 did not alter the peroxisomal distribution of each other or of Mopmp47, suggesting that Pex13 and Pex14 were not required for peroxisomal targeting of PMPs. Consistently, in yeast ∆pex13 cells, Pex11 is immunodetected predominantly in fractions corresponding to peroxisomal membrane [Citation44]. In Pichia pastoris pex14 mutants, mPTS-EGFP is targeted to peroxisomal remnants [Citation53]. GFP-PEX2 is distributed on peroxisomal remnants in both ∆pex13 and ∆pex14 mutants of P. anserina [Citation47]. N. crassa Pex13 is distributed in a punctate pattern in the wild-type as well as in the pex14 mutant [Citation45]. Pex14 is detected mainly in the membrane fraction in the Arabidopsis Pex13 mutant, as in the wild type [Citation49]. These data support the independence of Pex13 and Pex14 in PMP targeting. However, interestingly in our analysis, the peroxisomal localization of Mopex14/17 was completely altered by the disruption of Mopex13 or Mopex14. Considering that the three proteins function as a complex, we deduced that ∆mopex13 and ∆mopex14 mutants lost the ability to maintain Mopex14/17 on the peroxisomal membrane, probably because Mopex13 and Mopex14 provide a docking site for Mopex14/17. Analogously, in some cases, Pex13 has also been found to be required for Pex14 localization in the peroxisomal membrane [Citation54].
Woronin bodies, a type of organelle that is specifically present in filamentous ascomycetes, generate by budding from peroxisomes [Citation42]. Hex1, the major protein in Woronin bodies, is first imported into peroxisomes and then translocated into Woronin bodies during the budding process [Citation55]. Compared with peroxisomal biogenesis, the mechanism responsible for the formation of Woronin bodies is much less well understood. We have previously demonstrated the involvement of Pex19 and Pex11 in the biogenesis of Woronin bodies in M. oryzae [Citation13,Citation15]. Deletion of MoPEX19 leads to the absence of peroxisomes and Woronin bodies and, hence, Hex1 dispersion in the cytosol of Δmopex19 cells [Citation13]. Mopex11A, as a main mediator of peroxisomal fission in M. oryzae, is required for the separation of Woronin bodies from peroxisomes [Citation15]. In the present work, we demonstrated that both Pex13 and Pex14 were indispensable for Woronin body formation in M. oryzae. In both the Δmopex13 and Δmopex14 mutants, Hex1 was predominantly cytosolic, in contrast to the punctate Woronin body distribution in the wild type. In the ultrastructural images, typical Woronin bodies were absent in the mutants, and the septum pores occasionally remained open, although some of them were plugged with Woronin body-related structures. Moreover, the cellular leakage in the mutants confirmed the functional disorders in Woronin bodies. In N. crassa, Pex14 and Pex14/17 were observed to affect the structure of Woronin bodies and the proper localization of Hex1 [Citation31,Citation45]. A. oryzae Pex11-1 was found to be required in the formation of Woronin bodies [Citation56]. These data suggested that quite a few proteins that function in peroxisomal biogenesis played roles in the generation of Woronin bodies and supported the association of the two organelles. Notably, filamentous ascomycetes have a specific peroxin, Pex22-like, which shows low homology to Pex22 in S. cerevisiae but contains a typical Pex4-binding domain conserved in Pex22 proteins [Citation27]. Fam1, the Pex22-like protein in C. orbiculare, has been shown to be able to complement the defect in peroxisomal matrix import of S. cerevisiae pex22 mutants [Citation57]. fam1 mutants are incapable of importing PTS-containing proteins and are defective in peroxisome function and pathogenicity. However, Fam1 is distributed not on the peroxisomal membrane but specifically on the membrane of Woronin bodies. These data may suggest that some PEX genes function in Woronin body formation in a different manner from their function in peroxisome biogenesis.
To date, seven peroxins, PEX1, PEX5, PEX6, PEX7, PEX11, PEX19 and PEX14/17, have been characterized in M. oryzae [Citation9,Citation11–Citation15,Citation25]. Together with these data, the investigation of MoPex13 and MoPex14 demonstrated the importance of peroxisomal biogenesis and peroxisome-enveloped metabolism in fungal pathogenicity. Similar to MoPEX1, MoPEX6 or MoPEX19, the disruption of MoPEX13 caused almost complete inability of the PTS1 and PTS2 pathways. Accordingly, Δmopex13, Δmopex1, Δmopex6 and Δmopex19 displayed severe developmental defects. They formed significantly smaller colonies with few aerial hyphae, produced much fewer conidia than the wild type, showed dramatically reduced conidial germination and appressorial formation, and completely lost their pathogenicity [Citation9,Citation12,Citation13,Citation25]. By contrast, the absence of Mopex14/17, which plays an accessory role in the import of PTS1 proteins, causes damage to development and pathogenicity at a lower level [Citation14]. The Δmopex14 defects in development were moderate, also probably because Δmopex14 retained a weak capacity to import matrix proteins. Therefore, the influence of the peroxins on fungal development and pathogenicity corresponded well with their contributions to peroxisomal biogenesis. The pathogenicity can thus be regarded as a barometer reflecting the importance of a gene in peroxisomal biogenesis in fungal pathogens. The absence of Mopex5, despite solely disabling the PTS1 pathway [Citation11], disrupted the development and pathogenicity of the fungus similarly to the disruption of MoPEX1, MoPEX6, MoPEX13 or MoPEX19. However, the defects of Δmopex7, in which the PTS2 pathway was specifically disabled, were less robust than those of Δmopex5 [Citation11]. These findings indicated that the impact of peroxisomal biogenesis on fungal development and pathogenicity was mainly associated with the PTS1 import pathway, in agreement with the viewpoint that the majority of peroxisomal matrix proteins are imported via this pathway.
The roles of the peroxisome in fungal pathogenicity are generally associated with lipid degradation, ROS elimination, and appressorial melanization and turgor genesis. Accordingly, the Δmopex13 and Δmopex14 mutants exhibited reduced growth on lipids as a sole carbon source, reduced endurance to oxidizing agents and formed lighter colored appressoria with decreased turgor. In the present work, we confirmed this association through the inoculation of conidia supplemented with carbohydrate nutrients, inoculation on wounded leaves, and, for the first time, inoculation on heat-treated leaves. Notably, however, the defects in lipid degradation, ROS elimination and appressorial functions still could not fully explain the infection failure of the mutants, because only very weak symptoms developed even following the inoculation of heat-treated wounded leaves with conidia supplemented with glucose. This finding may indicate an interesting possibility necessitating further confirmation that the mutant has defects in nutrient utilization within rice cells.
In the present work, we demonstrated a significantly reduced ability to produce the sexual generation in the Δmopex13 and the Δmopex14 mutants compared with the wild type strains. This is the first study to show that the peroxins in M. oryzae are required for sexual reproduction. We have previously examined Δmopex5 and Δmopex7 but found that neither showed altered ascus production [Citation11]. These results may suggest the possibility that the functional mechanism of peroxins is quite different in sexual reproduction than in pathogenicity. Evidence for this viewpoint has also been documented in other fungal species. In P. anserina, Pex3 and Pex19 functioning in peroxisome membrane biogenesis and RING–finger peroxins Pex2, Pex10 and Pex12 are all crucial for meiocyte formation [Citation24,Citation47,Citation58]. However, in docking peroxins of P. anserina, Pex13, but not Pex14 or Pex14/17, is required for meiocyte formation. Even the elimination of both Pex14/17 and Pex14 has no effect on meiocyte differentiation [Citation47]. In addition, although Pex5 and Pex7 are not required in meiocyte differentiation in P. anserina, the PTS2 coreceptor Pex20 is essential [Citation24,Citation47]. More interestingly, the deletion of PEX2 in A. nidulans does not affect meiotic development. Therefore, the role of peroxins in meiosis is not conserved in filamentous fungi, and the roles of peroxins and the peroxisome in the sexual reproduction of rice blast fungus requires a more thorough investigation.
In summary, we characterized Mopex13 and Mopex14, potential members of the peroxisome docking complex in the rice blast fungus M. oryzae. Both Mopex13 and Mopex14 are peroxisomal membrane proteins and interact with each other, with Mopex14/17 and with PTS1 and PTS2 receptors. Deletion of Mopex13 completely blocks the PTS1 and PTS2 matrix import pathways, and deletion of Mopex14 partially inactivates the two pathways. The MoPEX13 and MoPEX14 deletion mutants are deficient in peroxisome-related metabolism, exhibit multiple disorders in vegetative and infection related developments, and present a loss of pathogenicity.
Supplemental Material
Download TIFF Image (239.7 KB)Supplemental Material
Download TIFF Image (1.8 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Talbot NJ. On the trail of a cereal killer: exploring the biology of Magnaporthe grisea. Annu Rev Microbiol. 2003;57:177–202.
- Wilson RA, Talbot NJ. Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat Rev Microbiol. 2009;7(3):185–195.
- Hamer JE, Howard RJ, Chumley FG, et al. A mechanism for surface attachment in spores of a plant pathogenic fungus. Science. 1988;239(4837):288–290.
- De Jong JC, Mccormack BJ, Smirnoff N, et al. Glycerol generates turgor in rice blast. Nature. 1997;389(6648):244–245.
- Chumley FG, Valent B. Genetic analysis of melanin-deficient, nonpathogenic mutants of Magnaporthe grisea. Mol Plant Microbe Interact. 1990;3(3):135–143.
- Maciel JL, Ceresini PC, Castroagudin VL, et al. Population structure and pathotype diversity of the wheat blast pathogen Magnaporthe oryzae 25 years after its emergence in Brazil. Phytopathology. 2014;104(1):95–107.
- Islam MT, Croll D, Gladieux P, et al. Emergence of wheat blast in Bangladesh was caused by a South American lineage of Magnaporthe oryzae. BMC Biol. 2016;14(1):84.
- Kimura A, Takano Y, Furusawa I, et al. Peroxisomal metabolic function is required for appressorium-mediated plant infection by Colletotrichum lagenarium. Plant Cell. 2001;13(8):1945–1957.
- Ramos-Pamplona M, Naqvi NI. Host invasion during rice-blast disease requires carnitine-dependent transport of peroxisomal acetyl-CoA. Mol Microbiol. 2006;61(1):61–75.
- Min K, Son H, Lee J, et al. Peroxisome function is required for virulence and survival of Fusarium graminearum. Mol Plant Microbe Interact. 2012;25(12):1617–1627.
- Wang J, Zhang Z, Wang Y, et al. PTS1 peroxisomal import pathway plays shared and distinct roles to PTS2 pathway in development and pathogenicity of Magnaporthe oryzae. PLoS One. 2013;8(2):e55554.
- Wang ZY, Soanes DM, Kershaw MJ, et al. Functional analysis of lipid metabolism in Magnaporthe grisea reveals a requirement for peroxisomal fatty acid beta-oxidation during appressorium-mediated plant infection. Mol Plant Microbe Interact. 2007;20(5):475–491.
- Li L, Wang JY, Zhang Z, et al. MoPex19, which is essential for maintenance of peroxisomal structure and woronin bodies, is required for metabolism and development in the rice blast fungus. PLoS One. 2014;9(1):e85252.
- Li L, Wang JY, Chen HL, et al. Pex14/17, a filamentous fungus-specific peroxin, is required for the import of peroxisomal matrix proteins and full virulence of Magnaporthe oryzae. Mol Plant Pathol. 2016;18(9):1238.
- Wang JY, Li L, Zhang Z, et al. One of three Pex11 family members is required for peroxisomal proliferation and full virulence of the rice blast fungus Magnaporthe oryzae. PLoS One. 2015;10(7):e0134249.
- Fujihara N, Sakaguchi A, Tanaka S, et al. Peroxisome biogenesis factor PEX13 is required for appressorium-mediated plant infection by the anthracnose fungus Colletotrichum orbiculare. Mol Plant Microbe Interact. 2010;23(4):436–445.
- Lazarow P, Fujiki Y. Biogenesis of peroxisomes. Ann Rev Cell Biol. 1985;1(1):489–530.
- Faust PL, Banka D, Siriratsivawong R, et al. Peroxisome biogenesis disorders: the role of peroxisomes and metabolic dysfunction in developing brain. J Inherit Metab Dis. 2005;28(3):369–383.
- Wanders RJA, Waterham HR. Peroxisomal disorders: the single peroxisomal enzyme deficiencies. BBA-Mol Cell Res. 2006;1763(12):1707–1720.
- Meijer WH, Gidijala L, Fekken S, et al. Peroxisomes are required for efficient penicillin biosynthesis in Penicillium chrysogenum. Appl Environ Microbiol. 2010;76(17):5702–5709.
- Markham P, Collinge AJ. Woronin bodies of filamentous fungi. FEMS Microbiol Rev. 1987;46:1–11.
- Baker A, Sparkes IA. Peroxisome protein import: some answers, more questions. Curr Opin Plant Biol. 2005;8(6):640–647.
- Kiel J, Van Den Berg M, Bovenberg RAL, et al. Penicillium chrysogenum Pex5p mediates differential sorting of PTS1 proteins to microbodies of the methylotrophic yeast Hansenula polymorpha. Fungal Genet Biol. 2004;41(7):708–720.
- Bonnet C, Espagne E, Zickler D, et al. The peroxisomal import proteins PEX2, PEX5 and PEX7 are differently involved in Podospora anserina sexual cycle. Mol Microbiol. 2006;62(1):157–169.
- Deng SZ, Gu ZK, Yang N, et al. Identification and characterization of the peroxin 1 gene MoPEX1 required for infection-related morphogenesis and pathogenicity in Magnaporthe oryzae. Sci Rep-UK. 2016;6:36292.
- Subramani S. Protein translocation into peroxisomes. J Biol Chem. 1996;271(51):32483–32486.
- Kiel J, Veenhuis M, Van Der Klei IJ. PEX genes in fungal genomes: common, rare or redundant. Traffic. 2006;7(10):1291–1303.
- Rayapuram N, Subramani S. The importomer - A peroxisomal membrane complex involved in protein translocation into the peroxisome matrix. BBA-Mol Cell Res. 2006;1763(12):1613–1619.
- Urquhart AJ, Kennedy D, Gould SJ, et al. Interaction of Pex5p, the type 1 peroxisome targeting signal receptor, with the peroxisomal membrane proteins Pex14p and Pex13p. J Biol Chem. 2000;275(6):4127–4136.
- Huhse B, Rehling P, Albertini M, et al. Pex17p of Saccharomyces cerevisiae is a novel peroxin and component of the peroxisomal protein translocation machinery. J Cell Biol. 1998;140(1):49–60.
- Managadze D, Wurtz C, Wiese S, et al. Identification of PEX33, a novel component of the peroxisomal docking complex in the filamentous fungus Neurospora crassa. Eur J Cell Biol. 2010;89(12):955–964.
- Opalinski L, Kiel J, Homan TG, et al. Penicillium chrysogenum Pex14/17p-a novel component of the peroxisomal membrane that is important for penicillin production. FEBS J. 2010;277(15):3203–3218.
- Notteghem JL, Silue D. Distribution of the mating type alleles in Magnaporthe grisea populations pathogenic on rice. Phytopathology. 1992;82(4):421–424.
- Crawford MS, Chumley FG, Weaver CG, et al. Characterization of the heterokaryotic and vegetative diploid phases of Magnaporthe grisae. Genetics. 1986;114(4):1111–1129.
- Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell. 1993;5(11):1575–1590.
- Rho HS, Kang S, Lee YH. Agrobacterium tumefaciens-mediated transformation of the plant pathogenic fungus, Magnaporthe grisea. Mol Cells. 2001;12(3):407–411.
- Wang JY, Wu XY, Zhang Z, et al. Fluorescent co-localization of PTS1 and PTS2 and its application in analysis of the gene function and the peroxisomal dynamic in Magnaporthe oryzae. J Zhejiang Univ-Sc B. 2008;9(10):802–810.
- Liu XH, Lu JP, Zhang L, et al. Involvement of a Magnaporthe grisea serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis. Eukaryot Cell. 2007;6(6):997–1005.
- Gould SJ, Kalish JE, Morrell JC, et al. Pex13p is an SH3 protein of the peroxisome membrane and a docking factor for the predominantly cytoplasmic PTS1 receptor. J Cell Biol. 1996;135(1):85–95.
- Itoh R, Fujiki Y. Functional domains and dynamic assembly of the peroxin Pex14p, the entry site of matrix proteins. J Biol Chem. 2006;281(15):10196–10205.
- Soundararajan S, Jedd G, Li XL, et al. Woronin body function in Magnaporthe grisea is essential for efficient pathogenesis and for survival during nitrogen starvation stress. Plant Cell. 2004;16(6):1564–1574.
- Liu F, Ng SK, Lu Y, et al. Making two organelles from one: Woronin body biogenesis by peroxisomal protein sorting. J Cell Biol. 2008;180(2):325–339.
- Albertini M, Rehling P, Erdmann R, et al. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell. 1997;89(1):83–92.
- Erdmann R, Blobel G. Identification of Pex13p, a peroxisomal membrane receptor for the PTS1 recognition factor. J Cell Biol. 1996;135(1):111–121.
- Managadze D, Wurtz C, Sichting M, et al. The peroxin PEX14 of Neurospora crassa is essential for the biogenesis of both glyoxysomes and Woronin bodies. Traffic. 2007;8(6):687–701.
- Barnett P, Bottger G, Klein ATJ, et al. The peroxisomal membrane protein Pex13p shows a novel mode of SH3 interaction. Embo J. 2000;19(23):6382–6391.
- Peraza-Reyes L, Arnaise S, Zickler D, et al. The importomer peroxins are differentially required for peroxisome assembly and meiotic development in Podospora anserina: insights into a new peroxisome import pathway. Mol Microbiol. 2011;82(2):365–377.
- Hynes MJ, Murray SL, Khew GS, et al. Genetic analysis of the role of peroxisomes in the utilization of acetate and fatty acids in Aspergillus nidulans. Genetics. 2008;178(3):1355–1369.
- Mano S, Nakamori C, Nito K, et al. The Arabidopsis pex12 and pex13 mutants are defective in both PTS1- and PTS2-dependent protein transport to peroxisomes. Plant J. 2006;47(4):604–618.
- Williams C, Van Den Berg M, Distel B. Saccharomyces cerevisiae Pex14p contains two independent Pex5p binding sites, which are both essential for PTS1 protein import. FEBS Lett. 2005;579(16):3416–3420.
- Will GK, Soukupova M, Hong X, et al. Identification and characterization of the human orthologue of yeast pex14p. Mol Cell Biol. 1999;19(3):2265–2277.
- Monroe-Augustus M, Ramon NM, Ratzel SE, et al. Matrix proteins are inefficiently imported into Arabidopsis peroxisomes lacking the receptor-docking peroxin PEX14. Plant Mol Biol. 2011;77(1–2):1–15.
- Johnson MA, Snyder WB, Cereghino JL, et al. Pichia pastoris Pex14p, a phosphorylated peroxisomal membrane protein, is part of a PTS-receptor docking complex and interacts with many peroxins. Yeast. 2001;18(7):621–641.
- Girzalsky W, Rehling P, Stein K, et al. Involvement of Pex13p in Pex14p localization and peroxisomal targeting signal 2-dependent protein import into peroxisomes. J Cell Biol. 1999;144(6):1151–1162.
- Beck J, Ebel F. Characterization of the major Woronin body protein HexA of the human pathogenic mold Aspergillus fumigatus. Int J Med Microbiol. 2013;303(2):90–97.
- Escano CS, Juvvadi PR, Jin FJ, et al. Disruption of the Aopex11-1 gene involved in peroxisome proliferation leads to impaired Woronin body formation in Aspergillus oryzae. Eukaryot Cell. 2009;8(3):296–305.
- Kubo Y, Fujihara N, Harata K, et al. Colletotrichum orbiculare FAM1 encodes a novel woronin body-associated Pex22 peroxin required for appressorium-mediated plant infection. MBio. 2015;6(5):e01305–e01315.
- Peraza-Reyes L, Zickler D, Berteaux-Lecellier V. The peroxisome RING-finger complex is required for meiocyte formation in the fungus Podospora anserina. Traffic. 2008;9(11):1998–2009.