ABSTRACT
Serine protease autotransporters of Enterobacteriaceae (SPATEs) are secreted proteins that contribute to virulence and function as proteases, toxins, adhesins, and/or immunomodulators. An extra-intestinal pathogenic E. coli (ExPEC) O1:K1 strain, QT598, isolated from a turkey, was shown to contain vat, tsh, and three uncharacterized SPATE-encoding genes. Uncharacterized SPATEs: Sha (Serine-protease hemagglutinin autotransporter), TagB and TagC (tandem autotransporter genes B and C) were tested for activities including hemagglutination, autoaggregation, and cytotoxicity when expressed in E. coli K-12. Sha and TagB conferred autoaggregation and hemagglutination activities. TagB, TagC, and Sha all exhibited cytopathic effects on a bladder epithelial cell line. In QT598, tagB and tagC are tandemly encoded on a genomic island, and were present in 10% of UTI isolates and 4.7% of avian E. coli. Sha is encoded on a virulence plasmid and was present in 1% of UTI isolates and 20% of avian E. coli. To specifically examine the role of SPATEs for infection, the 5 SPATE genes were deleted from strain QT598 and tested for cytotoxicity. Loss of all five SPATEs abrogated the cytopathic effect on bladder epithelial cells, although derivatives producing any of the 5 SPATEs retained cytopathic activity. In mouse infections, sha gene-expression was up-regulated a mean of sixfold in the bladder compared to growth in vitro. Loss of either tagBC or sha did not reduce urinary tract colonization. Deletion of all 5 SPATEs, however, significantly reduced competitive colonization of the kidney supporting a cumulative role of SPATEs for QT598 in the mouse UTI model.
Introduction
Escherichia coli is a common commensal of the gastrointestinal tract of mammals and birds, and is also a versatile pathogen associated with a variety of intestinal and extra-intestinal infections. Pathogenic E. coli belong to two main groups: intestinal pathogenic E. coli, and extra-intestinal pathogenic E. coli (ExPEC) [Citation1,Citation2]. Among ExPEC, the strains have been classified into pathotypes based on the sites of infection or the animal species they have infected, although these different ExPEC subgroups often share certain traits [Citation3–Citation8]. Such pathotypes include neonatal meningitis E. coli (NMEC), uropathogenic E. coli (UPEC), and avian pathogenic E. coli (APEC) [Citation2,Citation9,Citation10]. Avian pathogenic E. coli (APEC) are a subset of ExPEC that cause respiratory infections and septicemia in poultry [Citation4,Citation10–Citation12]. The genomes of a number of APEC strains and their virulence plasmids have been sequenced and share similarities to some human ExPEC isolates and their plasmids [Citation13–Citation18]. The plasticity of the E. coli genome has led to the emergence of numerous combinations of genes that can be encoded on genomic islands or harbored on plasmids that can contribute to fitness, adaptability, and virulence of a variety of ExPEC strains [Citation19–Citation21].
APEC and human ExPEC strains share multiple virulence factors that promote survival and colonization of the host during extraintestinal infections. These include fimbriae, iron acquisition systems, autotransporter (AT) proteins, capsular polysaccharides, O-antigens, toxins and secretion systems [Citation1,Citation2,Citation9,Citation11,Citation12]. Most APEC strains also contain conjugative colicin V (ColV) or similar plasmids that encode multiple virulence genes that have been shown to contribute to virulence in poultry [Citation11,Citation22,Citation23], and also to urinary tract infection or systemic infection in rodent models [Citation6,Citation24,Citation25]. The shared battery of virulence genes and the close phylogenetic relatedness of some APEC and human ExPEC strains suggest that some APEC may be potential zoonotic pathogens for humans [Citation6,Citation7,Citation25–Citation28].
Among pathogenic E. coli virulence factors, AT proteins comprise a large family that falls into three main categories: SPATEs (Serine Protease Autotransporters of Enterobacteriaceae), trimeric AT proteins, and the self-associating autotransporters (SAATs), such as AIDA-1 and Antigen43 (Ag43) [Citation29–Citation31]. AT proteins are exported by the type V secretion system, which can be classified into 5 subgroups: Va for the monomeric autotransporters which includes SPATEs, Vb for the two-partner secretion system, Vc for the trimeric AT, Ve for the ATs that are homologous to both type Va and type Vb, and Vd for the intimins and invasins which have a reverse order of domains [Citation32]. The export process of the AT may also require additional proteins such as the BAM and TAM assembly systems [Citation33,Citation34]. SPATEs consist of three specific domains: (i) a signal peptide which translocates the protein from cytoplasm to periplasm by the Sec-dependant pathway (ii) a functional passenger domain which contains a conserved serine protease motif (GDSGS), and (iii) a β-barrel domain which is localized in the outer membrane acting as a pore-forming domain that translocates the passenger domain [Citation35]. SPATEs have been grouped into two main classes; class 1 SPATEs consist of cytotoxic proteins, whereas class 2 SPATEs represent immunomodulator proteins [Citation32]. Certain SPATEs including the secreted autotransporter toxin (Sat), vacuolating autotransporter protein (Vat), temperature-sensitive hemagglutinin (Tsh), which has also been called hemoglobin protease (Hbp) [Citation36], and protein involved in colonization (Pic) [Citation37] have been previously reported in APEC and human ExPEC.
The SPATEs comprise a diverse group of autotransporter proteins that contribute to the virulence of pathogenic E. coli and Shigella spp., and other Enterobacteria [Citation2,Citation22,Citation32,Citation37–Citation43]. Some SPATEs were shown to be important virulence factors in disseminated infection of ExPEC due to their proteolytic activity, which can promote the degradation of host cell substrates and elicit an inflammatory response [Citation32,Citation44]. In ExPEC, SPATE proteins have previously been characterized and have been shown to be associated with infections of both humans and other animals including poultry. SPATEs identified in uropathogenic E. coli include Sat [Citation44], Vat [Citation45,Citation46] and PicU [Citation41]. The sat gene encodes a vacuolating toxin and sat sequences were present in 55% of UPEC strains [Citation40] but were not identified in a collection of APEC isolates [Citation47]. PicU is homologous to the Pic protein identified in Shigella and enteroaggregative E. coli (EAEC) [Citation37]. PicU was found in 22% of UPEC isolates [Citation41] and 9% of APEC strains [Citation47]. The Vat autotransporter was first discovered in APEC [Citation45], was present in 60–70% of ExPEC from human infections [Citation46,Citation48] and 33% of APEC strains [Citation47]. The Vat toxin was shown to contribute to virulence, respiratory infection, and cellulitis in broiler chickens [Citation45]. Both pic and vat were shown to contribute to the fitness of UPEC in a mouse model of systemic infection [Citation43]. Tsh was the first SPATE identified in E. coli [Citation49] and was shown to contribute to the development of respiratory lesions in the air sacs of chickens [Citation22]. The tsh gene is located on ColV-type plasmids, was present in 50% of APEC strains [Citation47], is less commonly associated with human ExPEC, but can be associated with certain human ExPEC strains [Citation18,Citation50–Citation52].
In this report, analysis of the genome sequence of an APEC O1 strain, QT598, revealed that it contained 5 distinct SPATEs. Three of these, two chromosomally encoded SPATE genes (we name tagB and tagC) and a novel plasmid-encoded SPATE gene (sha) have not been previously characterized. The remaining two SPATEs were the previously characterized Vat and Tsh proteins. Herein, we have characterized the three novel SPATEs, determined their prevalence among avian and human urinary tract isolates, and investigated the role of these SPATEs for cytotoxicity and in the colonization of the murine urinary tract.
Results
Genomic analysis identifies five predicted SPATEs encoded by E. coli strain QT598
Strain QT598 was initially isolated from an infected turkey poult in France as MT156 [Citation53]. It is a phylogenetic group B2 strain belonging to serogroup O1, a common serogroup among ExPEC strains causing infections in both poultry and humans. This APEC strain was sequenced initially because it contains most of the known APEC-associated virulence genes and was previously found to be virulent in one-day-old chicks [Citation54]. QT598 belongs to sequence type (ST) 1385. Other strains belonging to ST1385 include other APEC O1 isolates, a canine urinary isolate, and environmental isolates (http://enterobase.warwick.ac.uk/). Interestingly, ST1385 strains are related to other STs including ST91, which contains some strains from human extra-intestinal infections and E. coli F54, an O18:K1 human fecal isolate sharing many virulence genes found in ExPEC from neonatal meningitis [Citation55].
The genome of QT598 contains five SPATE-encoding sequences (). Two of the SPATE genes, tsh and a novel SPATE which we have called sha (for serine-protease hemaglutinin autotransporter), are located on a ColV-type plasmid (pEC598). The vat gene was also identified on a genomic island. Finally, a genomic region was identified containing two distinct SPATE-encoding sequences in close proximity to each other, which we have named tagB and tagC (for tandem autotransporter genes B and C).
Figure 1. Regions containing the five SPATE-encoding genes in E. coli QT598.
The tsh and sha genes are located on a ColV-type plasmid (pEC598). The vat, tagB, and tagC genes are located on genomic islands. Arrows indicate open reading frames (ORFs). SPATE encoding ORFs and regulatory gene vatX are in red. Predicted full amino acid lengths and GC content of the SPATE ORFs are indicated below arrows. Blue ORFs are related to insertion elements, integrases, or mobile elements. Dark green ORFs are predicted fimbrial proteins. Light green ORFS are predicted EAL-domain proteins. Grey ORFS are hypothetical uncharacterized ORFs. Orange ORFs are hypothetical regulatory proteins. Purple ORFs are genes conserved in E. coli K-12 that border the SPATE-encoding genomic regions. Direct repeats (DR) are indicated for the region containing the tag AT genes.
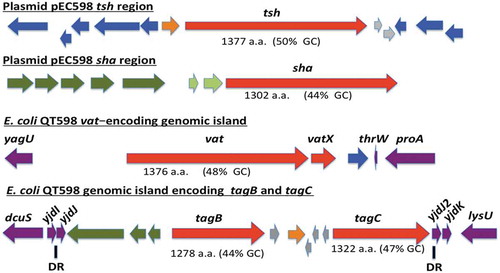
The tsh open reading frame on plasmid pEC598 shares highest identity to tsh from plasmid pACN001-B (accession number KC853435.1) [Citation56] and similar sequences in the NCBI database, differing in only 1 nucleotide, a Gly1177- Ser1177 substitution. Compared to the characterized Tsh (Hbp) proteins, Tsh from APEC strain χ7122 [Citation22] and hemoglobin protease (Hbp) from ExPEC strain EB1 [Citation36], TshQT598 contains 4 and 2 amino acid differences, respectively. In QT598, tsh is also flanked by sequences related to transposases and insertional sequences () that also flank tsh on other IncFII plasmids [Citation22,Citation36]. The sha gene is also located on pEC598 and has a 44% GC content. Sequence analysis of the sha gene revealed an open reading frame (ORF) of 3909 bp encoding a predicted precursor protein of 1302 amino acids with an N-terminal domain signal peptide (residues 1–51), a passenger domain (residues 52–1026) (predicted molecular mass of 105.7 kDa) containing a consensus serine protease motif 256GDSGS, and β-barrel domain (residues 1027–1302). Interestingly, the highly conserved SPATE cleavage site of two consecutive asparagines “EVNNLNK”, found between the passenger domain and the β-barrel of most SPATEs [Citation57], is absent in Sha. Sha is more closely related to Tsh and Vat proteins than to other SPATEs (). The global alignment of Sha with TshQT598 has 43% identity/56% similarity with 237 gaps, whereas the global alignment of Sha with Vat QT598 is 38% identity/52% similarity with 252 gaps.
Figure 2. Phylogenetic analysis of new SPATEs identified in the QT598 genome.
The evolutionary history of passenger domains of QT598 SPATEs (highlighted in yellow) as well as other characterized SPATEs was inferred using the Neighbor-Joining method [Citation98]. The optimal tree with the sum of branch length = 8.78918031 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the JTT matrix-based method [Citation99] and are in the units of the number of amino acid substitutions per site. The analysis involved 17 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 723 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 [Citation85]. Multiple sequence alignment was performed by Clustal W, and the tree was constructed using the Mega6 software with PhyML/bootstrapping. A cluster of cytotoxic SPATEs (class 1) comprise the green branches, while immunomodulator SPATEs (class 2) are in blue branches. DNA regions encoding SPATE protein sequences are available in NCBI database as follows: EspC, GenBank Accession No. AAC44731, TagB and TagC, MH899681; EspP, NP_052685; SigA, AF200692; Pet, SJK83553; Sat, AAG30168; EatA, CAI79539, SepA, Z48219; Pic, ALT57188; Boa, AAW66606; EaaA, AAF63237; Sha, MH899684; Vat, MH899682; TleA, KF494347; Tsh/Hbp, MH899683.
![Figure 2. Phylogenetic analysis of new SPATEs identified in the QT598 genome.The evolutionary history of passenger domains of QT598 SPATEs (highlighted in yellow) as well as other characterized SPATEs was inferred using the Neighbor-Joining method [Citation98]. The optimal tree with the sum of branch length = 8.78918031 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the JTT matrix-based method [Citation99] and are in the units of the number of amino acid substitutions per site. The analysis involved 17 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 723 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 [Citation85]. Multiple sequence alignment was performed by Clustal W, and the tree was constructed using the Mega6 software with PhyML/bootstrapping. A cluster of cytotoxic SPATEs (class 1) comprise the green branches, while immunomodulator SPATEs (class 2) are in blue branches. DNA regions encoding SPATE protein sequences are available in NCBI database as follows: EspC, GenBank Accession No. AAC44731, TagB and TagC, MH899681; EspP, NP_052685; SigA, AF200692; Pet, SJK83553; Sat, AAG30168; EatA, CAI79539, SepA, Z48219; Pic, ALT57188; Boa, AAW66606; EaaA, AAF63237; Sha, MH899684; Vat, MH899682; TleA, KF494347; Tsh/Hbp, MH899683.](/cms/asset/350e1e33-04f8-4e17-88c6-4445b6a62589/kvir_a_1624102_f0002_oc.jpg)
The vat gene from QT598 is present on a genomic island that includes the vatX regulatory gene, and is located between the E. coli conserved genes yagU and proA adjacent to the thrW-tRNA gene (). This is a conserved insertion site for vat-encoding genomic islands [Citation58]. VatQT598 is a predicted 1376 aa precursor with a single substitution (His534-Arg534) compared to Vat from UPEC strain CFT073 (accession number AAN78874.1). At least 41 predicted Vat protein sequences from different E. coli strains share an identical predicted VatQT598 sequence, indicating it is a common allelic variant of Vat (Supplemental Table 2). These entries include sequences from strains isolated from fecal samples and infections of poultry and two human UTIs that are labeled as “Hbp” or “SepA” proteins in the databank.
The two new chromosomal-encoded SPATE genes were named tagB and tagC, Tandem autotransporter genes (Tag) because of their tandem co-localization in the genome of QT598 as well as in various other E. coli strains such as: multidrug-resistant CTX-M-15-producing ST131 isolate E. coli JJ1886 (Accession number CP006784), porcine E. coli PCN033 (Accession number CP006632), and E. coli CI5 (Accession number CP011018). The tagB and tagC SPATE-encoding genes from QT598 are located on a genomic island between the E. coli conserved genes yjdI and yjdK (). This genomic island has a mean GC content of 41%, which is considerably lower than the 50% GC of E. coli, suggesting horizontal gene transfer. This genomic region is also bordered by direct repeat (DR) sequences that correspond to duplication of yjdJ sequences bordering each side of the genomic island (). Related genomic islands containing similar SPATE encoding genes at this insertion site are present in other E. coli genome sequences including antibiotic-resistant strains isolated from the urinary tract and other infections in humans (Supplemental Table 3). The predicted TagB and TagC proteins share the closest identity to the EaaA [Citation59] and EspC [Citation60], respectively (). TagB shares 46% identity/63% similarity to EaaA with 84 gaps over its full length. TagC shares 60% identity/74% similarity to EspC with 22 gaps. TagB comprises a predicted signal peptide (residues 1–58), a passenger domain from residues 59–1006 (predicted molecular mass of 101 kDa) containing a consensus serine protease motif 253GDSGS, and a β-barrel domain from residues 1007–1283. TagC comprises a predicted signal peptide (residue 1–53), a passenger domain from residues 55–1032 (predicted molecular mass of 105.14 kDa) with a consensus serine protease motif 250GDSGS, and β-barrel domain ranging between 1033 and 1309 residues. Both TagB and TagC contain the conserved twin asparagine (N-N) residues in the linker domain connecting passenger and β-barrel domains, EIN1006NLNDRM and EVN1032NLNKRM, respectively.
Prevalence of new SPATE genes in human uropathogenic and avian pathogenic E. coli
To determine the distribution of the SPATE sequences among E. coli clinical isolates, the presence of these three new SPATE sequences as well as vat, sat, and tsh were detected by PCR in a collection of UPEC isolates from Guadeloupe (697 isolates) [Citation61] and from avian pathogenic E. coli (299 isolates) [Citation22]. For the UPEC isolates, tagB sequences were present in 70 isolates (10%), whereas tagC sequences were present in 80 isolates (11.5%). Interestingly, 96.8% (69/70) of the tagB-positive isolates were also tagC-positive. Furthermore, 68 of the tagB isolates belonged to phylogenetic group B2, with one isolate from group B1 and one untypable isolate. The 11 isolates that contained tagC but not tagB sequences belonged to groups other than B2: B1 (3 isolates), D (3 isolates), F (4 isolates), or A (1 isolate). Sha was the least common sequence, and was present in only 6 UTI isolates (0.9%), all of which belonged to group B2 and were also vat-positive. Five of the sha-positive strains also contained tagB and tagC, whereas vat and sat sequences were more common and found in 333 isolates (47.8%) and 217 isolates (31.1%), respectively. The tsh gene was present in 41 UPEC isolates (5.9%). In summary, in UPEC, tagB and tagC were found together in a subset of strains belonging to phylogenetic group B2, although some strains belonging to other phylogenetic groups were only tagC positive.
With regards to the APEC strains, tagB sequences were present in 14 isolates (4.7%) and tagC sequences were present in 21 isolates (7%). All 14 tagB-positive APEC were also tagC-positive, and 13/14 of these belonged to phylogenetic group B2. Among these, 10 strains belonged to serogroup O1, 1 was serogroup O78, and 3 were of undetermined serogroup. Interestingly, among the 299 APEC strains that were screened, comprised of 109 from chickens, 175 from turkeys, and 15 from ducks, all of the tagB or tagC-positive isolates were exclusively from infections in turkeys. Overall, similar to UPEC, tagB, and tagC were specifically present in a subset of APEC strains, mainly belonging to group B2, although some strains belonging to other phylogenetic groups only contained tagC sequences.
The sha sequences were present in 61 APEC strains (20%). The majority, 42 strains, belonged to phylogenetic group A, 11 strains belonged to group B1, 5 strains belonged to group B2, and 3 strains belonged to group D. Among these sha-containing strains, 35 belonged to serogroup O78, 3 were from serogroup O1, 2 strains each belonged to serogroups O11, O54, O21, and O8, and one belonged to serogroup O55. The remaining 12 strains were from undetermined serogroups. The sha gene is, therefore, clearly more prevalent among APEC than UPEC in the subset of strains we analyzed.
Cloning and production of SPATEs in culture supernatants
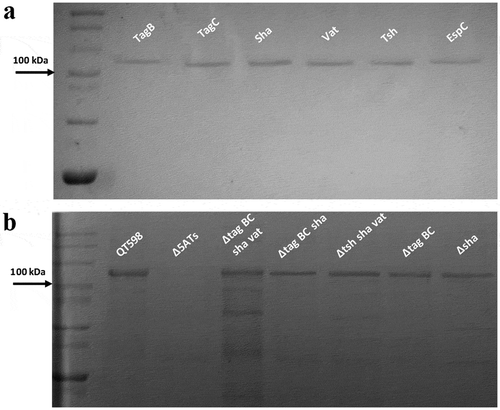
All five of the predicted SPATE-encoding genes and promoter regions were cloned to determine their expression and for use in a variety of phenotypic tests. Each of the five SPATE genes, when cloned into E. coli BL21, produced a high-molecular-weight protein (>100 kDa) in culture supernatants that corresponded to the expected product (). In addition, derivatives of strain QT598 wherein these SPATE-encoding genes were inactivated were generated. Analysis of supernatant fractions of QT598, by SDS-PAGE, revealed the presence of SPATE proteins expressed under laboratory conditions, as demonstrated by visualization of bands with a high molecular mass (>100 kDa) secreted in the external milieu. By contrast, no such bands were observed in the supernatant extracts of the SPATE-free, Δ5ATs, derivative of QT598 (). The purity of concentrated supernatant filtrate was also evaluated by silver staining (Supplemental Fig. S1). Protein bands of the newly identified SPATEs were extracted from gels and sampled by mass spectrometry for peptide analysis following trypsin digestion (Supplemental Fig. S2). For Sha, peptides corresponding to the mature secreted protein spanned from amino acids 52 to 1009. Despite not containing the twin asparagine (N-N) cleavage site present in most SPATEs, peptide profiles suggest the cleavage site from the β-barrel domain likely resides within the 1010–1020 region. This region contains two adjacent polar amino acids, Ser1015 and Asp1016, that may serve as the cleavage site. For TagB, peptide scans suggest that the mature secreted protein spans from amino acids 54 to 1006 based on the twin Asp1006-Asp1007 location. For TagC, peptide scans suggest that the mature secreted protein spans from at least amino acid 60 to 1026 with a predicted twin Asp1032-Asp1033 cleavage site. As expected, peptides corresponding to the predicted amino-terminal signal peptides and the carboxy-terminal predicted β-barrel domains of the Sha, TagB, and TagC SPATEs were not identified from peptide analysis of the secreted proteins (Supplemental Fig. S2).
Figure 3. Detection of SPATE proteins by SDS-PAGE A. SDS-PAGE analysis of cloned SPATE genes.
a. Clones expressing SPATE proteins were produced in the BL21 background with high-copy plasmid pBCsk+. Supernatants were filtered then concentrated through Amicon filters with 50 kDa cutoff. Samples containing 5 µg protein were migrated with protein marker (10–200 kDa) and stained with Coomassie blue (arrow represents 100 kDa size marker). b. Detection of SPATEs from supernatants of strain QT598 and various SPATE gene mutant derivatives. Supernatants from an overnight culture of the respective mutants were filtered, concentrated and run on SDS-polyacrylamide gels and stained with Coomassie blue to visualize proteins.
Cleavage of oligopeptides by SPATE proteins
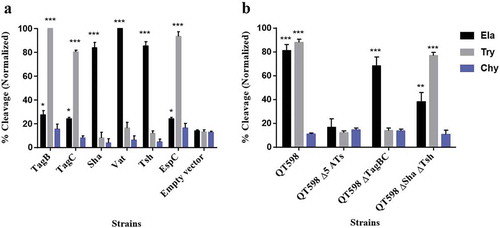
To determine the protease substrate cleavage specificity of the new SPATEs, we used synthetic polypeptides conjugated with pNA at the C-terminus. Purified proteins from supernatants of each SPATE were incubated with N-Succinyl-Ala-Ala-Ala-p-nitroanilide (elastase substrate), N-Benzoyl-L-arginine 4-nitroanilide (trypsin substrate) and N-succinyl-ala-ala-pro-phe-p-nitroanilide (chymotrypsin substrate) (Sigma-Aldrich, St. Louis, MO, USA). TagB and TagC demonstrated trypsin-like activity and efficiently cleaved N-Benzoyl-L-arginine 4-nitroanilide, similarly to the EspC protein (). By contrast, Sha demonstrated significant elastase-like activity toward N-Succinyl-Ala-Ala-Ala-p-nitroanilide, as did the Vat and Tsh proteins (). The cleavage activity of high-molecular-weight supernatant fractions from WT strain QT598 and SPATE mutant derivatives was also determined (). QT598 demonstrated both trypsin-like and elastase-like activity, whereas the Δ5ATs mutant had lost these activities. By contrast, a strain which had lost only tagBC demonstrated only elastase-like activity conferred by vat, tsh, and sha (). In addition, pre-incubation of these supernatants with PMSF eliminated or sharply inhibited oligo-peptide cleavage indicating the activity demonstrated was due to the SPATE proteins produced by the strains (Supplemental Fig. S3)
Figure 4. Oligopeptide cleavage profiles of SPATEs.
a. Enzymatic activity of cloned SPATEs. Five μg of each SPATE-containing supernatant was incubated at 37°C for 3 h with 1mM of synthetic oligopeptide specifically recognized by the following enzymatic activities: Elastase (Ela)-(N-Suc-Ala-Ala-Ala-pNA); Trypsin (Try)-(N-Ben-L-arginine-pNA); or Chymotrypsin (Chy)-(N-Suc-Ala-Ala-Pro-Phe-pNA). Absorbance at 410nm was normalized to the maximum absorbance value. b. Enzymatic activity of supernatants from strain QT598 and SPATE gene mutant derivatives. Samples were tested as described above. Data are the means of three independent experiments, and error bars represent the standard errors of the means (* p < 0.05, **p < 0.01, ***p < 0.001 one-way ANOVA with multiple comparisons vs pBCsk+ (A) or QT598Δ5ATs (B)).
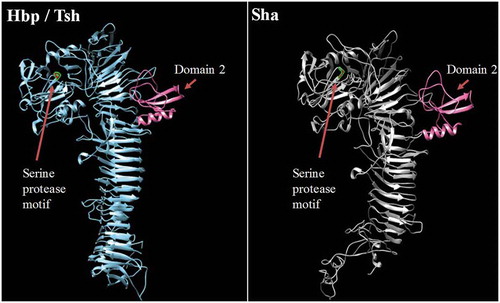
Multiple alignment of the new autotransporters with other SPATEs places TagC within the class 1, cytotoxic and enterotoxic SPATEs, along with the EspC SPATE from Enteropathogenic E. coli (EPEC) (). EspC was previously shown to cleave spectrin, Factor V, pepsin and hemoglobin [Citation42,Citation62] and as with TagC and TagB, similarly demonstrated trypsin-like protease activity (). The SPATE sharing closest identity to TagB is the class 2 SPATE EaaA, identified from commensal E. coli ECOR-9 [Citation59] (). Sha shares more identity to class 2 SPATEs Tsh/Hbp (66% identity) [Citation36,Citation63], TleA (60% identity) [Citation64] and Vat (56% identity) [Citation45]. As such, Sha is likely to share other properties more similar to Tsh (adhesin, hemagglutinin) and Vat (cytotoxin), and the elastase-like substrate specificity of Sha, also shown for Tsh and Vat, is in line with this.
To predict the 3D structure of the passenger domain of new SPATEs, we used the I-Tasser program to generate a 3D structure model and UCSF chimera to compare structures [Citation65,Citation66]. We found that the Sha protein is also predicted to contain the small domain 2 that was identified in Tsh/Hbp and was considered to be characteristic of class 2 SPATEs [Citation32] (). This domain was however absent in predicted models of TagB and TagC (Fig. S4).
Figure 5. Stereo ribbon diagram showing the predicted three-dimensional structure of the Sha SPATE passenger domain derived from the Hbp/Tsh crystal structure.
Crystal structure of the Heme-binding protein (Hbp) (PDB 1WXR), which is near identical to Tsh, was used to model a homologous structure based on alignment with the Sha protein sequence. The model was generated using the I-TASSER server with 100.0% confidence by the single highest scoring template. Sha is shown to harbor a conserved domain, domain 2 (shown in pink), which is characteristic of class 2 SPATEs.
Increased adherence to epithelial cells is mediated by SPATEs
Some AT proteins can promote adherence to host cells [Citation31,Citation67]. To investigate this with TagB, TagC, and Sha, the different SPATE encoding genes were cloned in the fim-negative E. coli K-12 strain ORN172 and tested for increased adherence to avian fibroblasts (CEC-32), human kidney (HEK-293) and bladder (5637) epithelial cells (). Clones expressing either TagB, TagC, Sha, or Vat adhered significantly to kidney cells, whereas Tsh and EspC did not significantly increase adherence (). For the bladder cells, all SPATEs tested except for Vat and EspC significantly increased adherence (). By contrast, for avian fibroblasts, only TagB and TagC increased adherence (). These cell culture-based results suggest that TagB, TagC, and Sha, as well as Vat and Tsh, may contribute to host cell interactions in the urinary tract, and that TagBC may also contribute to adherence to tissues in poultry. Although some individual SPATEs were shown to increase adherence to host cells, loss of all 5 SPATEs did not reduce adherence of strain QT598 (Fig. S5).
Figure 6. TagB, TagC, and Sha SPATEs promote adherence to the human kidney (HEK-293) and bladder (5637) epithelial, and avian fibroblast (CEC-32) cell lines.
Cell monolayers were infected with E. coli fim-negative ORN172 expressing SPATE proteins at a multiplicity of infection (MOI) of 10 and incubated at 37°C at 5% CO2 for 2 h. Adherent bacteria were enumerated by plating on LB agar. Empty vector (pBCsk+) was used as a negative-control and APEC MT78 [Citation80] as a positive control for adherence to cell lines. Data are the averages of three independent experiments. Error bars represent standard errors of the means. (*p < 0.05, **p < 0.01, ***p < 0.001 vs empty vector by one-way ANOVA).
![Figure 6. TagB, TagC, and Sha SPATEs promote adherence to the human kidney (HEK-293) and bladder (5637) epithelial, and avian fibroblast (CEC-32) cell lines.Cell monolayers were infected with E. coli fim-negative ORN172 expressing SPATE proteins at a multiplicity of infection (MOI) of 10 and incubated at 37°C at 5% CO2 for 2 h. Adherent bacteria were enumerated by plating on LB agar. Empty vector (pBCsk+) was used as a negative-control and APEC MT78 [Citation80] as a positive control for adherence to cell lines. Data are the averages of three independent experiments. Error bars represent standard errors of the means. (*p < 0.05, **p < 0.01, ***p < 0.001 vs empty vector by one-way ANOVA).](/cms/asset/f6b01afa-fcac-4a16-82c6-d55a740d5979/kvir_a_1624102_f0006_oc.jpg)
Sha, TagB, Vat, and Tsh are hemagglutinins
Tsh has been previously shown to hemagglutinate chicken and sheep erythrocytes [Citation22,Citation49,Citation68]. To assess whether other SPATEs also show hemagglutinin activity, we verified hemagglutination by the different SPATEs with erythrocytes from a variety of species. Interestingly, Sha, Tsh, and Vat all demonstrated hemagglutinin activities against sheep, bovine, pig, dog, chicken, turkey, rabbit, horse, and human blood (type O and A groups). In addition, TagB and TagC hemagglutinated sheep, bovine and pig erythrocytes, but not human, turkey, rabbit, dog, chicken, or horse erythrocytes (). However, the titer for TagC was very low for any erythrocytes tested and EspC demonstrated no hemagglutination.
Table 1. Hemagglutination activities of different SPATEs.
Table 2. Strains and plasmids used in this study.
TagB, TagC, and Sha mediate autoaggregation, but only Sha increases biofilm formation
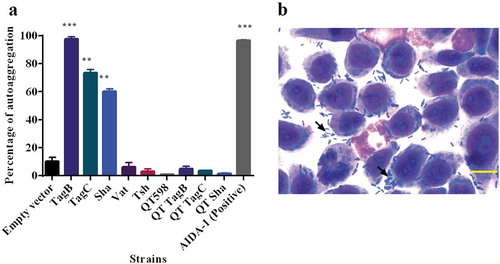
Some AT proteins such as AIDA-1 and Ag43 mediate cell–cell interactions and autoaggregation, which can contribute to virulence and facilitate host cell adherence [Citation69–Citation71]. We observed that the absorbance of clones expressing Sha, TagB, and TagC dropped rapidly similar to the positive control AIDA-1(). An aggregative adherence pattern was also observed on interaction with bladder epithelial cell culture (). As these SPATEs demonstrated autoaggregation, we were interested to know if these plasmids could also confer autoaggregation to ExPEC QT598. However, the introduction of these plasmids did not lead to an autoaggregation phenotype in QT598 ().
Figure 7. TagB, TagC, and Sha are autoaggregating proteins.
a. Clones of E. coli fim-negative strain ORN172 expressing SPATE proteins were grown 18 h and adjusted to OD600 of 1.5 and left to rest at 4°C. Samples were taken at 1 cm from the top surface of the cultures after 3 h to determine the change in OD600. Assays were performed in triplicate, and the rate of autoaggregation was determined by the mean decrease in OD after 3 h. The autoaggregation phenotype was absent when plasmids expressing tagB, tagC, or sha were introduced into APEC strain QT598 (QT TagB, QT TagC, QT Sha respectively). Empty vector (pBCsk+) was used as a negative control and the AIDA-1 AT as a positive control for autoaggregation. Error bars represent standard errors of the means (* p < 0.05, **p < 0.01, ***p < 0.001 compared to empty vector using one-way ANOVA). b. Giemsa stain of the 5637 bladder cell line infected with a tagB expressing clone after 2 h demonstrates an aggregative adherence pattern to bladder cells (arrow head). A similar pattern was found for tagC and sha expressing clones (not shown here). Bar represents 50 µm.
Proteins involved in autoaggregation can also increase biofilm formation [Citation69]. We, therefore, checked biofilm forming capacity of these new SPATE clones at different temperatures (25°C, 30°C, 37°C, and 42°C). TagB and TagC did not increase biofilm formation. However, Sha, Vat, and Tsh significantly increased biofilm production at lower temperatures (25°C and 30°C), but no significant differences in biofilm production were observed at higher temperatures of 37°C and 42°C ().
Figure 8. Sha, Vat, and Tsh promote biofilm formation.
Clones of E. coli fim-negative strain ORN172 expressing SPATE proteins were grown at different temperatures (25°C, 30°C, 37°C, and 42°C) in polystyrene plate wells for 48 h and then stained with crystal violet. Remaining crystal violet after washing with acetone was measured as absorbance at 595 nm. Data are the means of three independent experiments, and error bars represent standard errors of the means. Empty vector (pBCsk+) was used as a negative control, and a string biofilm producing Serratia strain [Citation100] served as a positive control for biofilm formation (* p < 0.05, **p < 0.01, ***p < 0.001 compared to empty vector using one-way ANOVA).
![Figure 8. Sha, Vat, and Tsh promote biofilm formation.Clones of E. coli fim-negative strain ORN172 expressing SPATE proteins were grown at different temperatures (25°C, 30°C, 37°C, and 42°C) in polystyrene plate wells for 48 h and then stained with crystal violet. Remaining crystal violet after washing with acetone was measured as absorbance at 595 nm. Data are the means of three independent experiments, and error bars represent standard errors of the means. Empty vector (pBCsk+) was used as a negative control, and a string biofilm producing Serratia strain [Citation100] served as a positive control for biofilm formation (* p < 0.05, **p < 0.01, ***p < 0.001 compared to empty vector using one-way ANOVA).](/cms/asset/0f1aa0c8-7bbb-4fb0-8adb-c4f36e0262d9/kvir_a_1624102_f0008_oc.jpg)
Assessment of the cytopathic effect of 5 different SPATEs on bladder cells
Since some SPATEs produce cytopathic activity on host cells, we assessed the cytopathic effect of extracts of supernatants of the different SPATEs as well as the supernatant of wild-type strain QT598 and SPATE-free Δ5ATs mutant, on the human bladder 5637 cell line. Incubation of SPATEs from concentrated filtered supernatant () of strain QT598 with bladder cells triggered a cytopathic effect, characterized by the dissolution of cytoplasm and enlargement of the nucleus (), after 5 h of incubation. After 12 h, the majority of the cells were affected and showed similar morphological changes. These phenotypes were absent upon the treatment with the supernatant of the Δ5AT SPATE-free mutant or with culture media alone.
Figure 9. All five different SPATEs from strain QT598 induce cytopathic effects.
a. Concentrated supernatants (30 μg of protein per well) from wild type QT598 as well SPATE free Δ5ATs were incubated with human bladder cell line 5637 for 5 h and 12 h. Cytopathic cells (arrowheads) were found in the cells treated with QT598 supernatant while there were no morphological changes in the cells treated with supernatant of Δ5ATs. b. Concentrated supernatants of E. coli BL21 pBCsk+ (30 μg of protein per well) clones overexpressing different SPATEs were incubated with monolayers of 5637 bladder cell lines for 12 h at 37°C. TagB, TagC, Sha, and Vat showed more cytopathic effect (arrowheads) compared to Tsh and empty vector. c. LDH release by 5637 cells after incubation with culture filtrates of different clones (30 μg of protein per well) expressing SPATEs or from supernatants from wild-type E. coli strain QT598 and Δ5ATs mutant derivatives with 5637 human bladder cells at 37°C for 5 h and 12 h. Empty vector (pBCsk+) was used as a negative control. Lysis solution was added as a positive control for maximum LDH release. Data are the means of three independent experiments, and error bars represent the standard errors of the means (* p < 0.05, **p < 0.01, ***p < 0.001 vs empty vector using one-way ANOVA). Bar represents 50 µm.
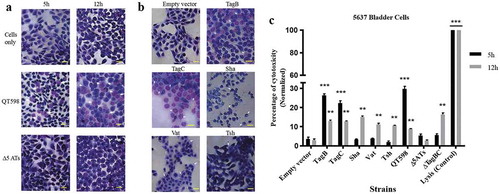
Further, we assessed the cytopathic effect of individual SPATEs that were from recombinant clones. Following 12 h of interaction, cells incubated with TagB or TagC proteins elicited distinct cytopathic changes including dissolution of cytoplasm, nuclear enlargement and vacuolation in the nucleus, cells treated with Sha were rounded, and Vat-treated cells showed numerous vacuoles within the cytoplasm (). By contrast, cells exposed to Tsh did not show distinct morphological changes. These results suggest that TagB, TagC, Sha and Vat proteases demonstrate cytopathic effects that alter bladder cell morphology, whereas Tsh was less cytopathic to this cell line. We further investigated the cytopathic effect of these SPATEs by measuring the release of lactate dehydrogenase (LDH) after 5 h and 12 h following exposure to supernatant extracts. The release of LDH after 5 h was demonstrated only following exposure to either TagB or TagC (). Interestingly, although LDH was not detected from samples exposed to Vat, Tsh and Sha after 5 h (), some LDH release was observed after 12 h of exposure, suggesting these SPATEs may demonstrate a delayed cytotoxic effect. Hence, all 5 SPATEs elicited some cytotoxicity that corresponded with cytotoxic effects that were observed in cells following Giemsa staining. Of further interest, the concentrated supernatant filtrates from ExPEC strain QT598 showed early toxicity comparable to the concentrated supernatant filtrates from tagB or tagC expressing clones. However, loss of tagB and tagC resulted in only a late onset cytotoxic effect at 12 h, and loss of all 5 SPATEs abrogated LDH release at either early or late time points (). Taken together, these results indicate TagB and TagC can mediate an early (5 h) cytotoxicity, whereas Vat, Tsh, and Sha mediate late onset (12 h) cytotoxicity, and that these 5 SPATES collectively mediate the overall cytotoxic effects of ExPEC QT598 on bladder epithelial cells.
Cumulative role of SPATEs for colonization during urinary infection in mice
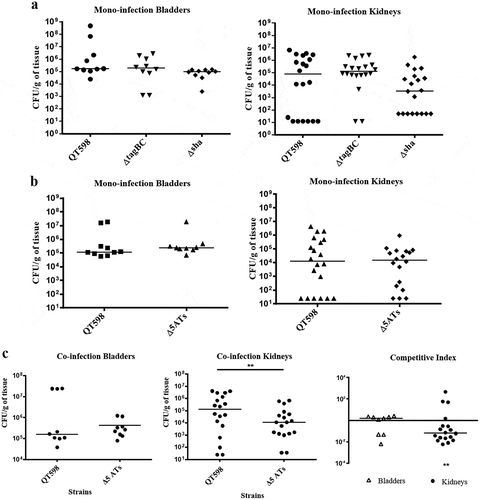
In order to determine the potential role of SPATEs during UTI, we tested isogenic mutants with deletions of the tagB, tagC, and sha genes in a murine transurethral infection model. We observed no significant differences in colonization of the wild-type compared to tagB, tagC or sha knockout mutants (). To determine the collective role of the SPATEs in UTI for QT598, we deleted all five SPATE encoding genes and did transurethral infections. We again observed no significant difference in the colonization of kidneys or bladder following single-strain infections. By contrast, when a co-infection model was used (1 mouse died after 24 h), the Δ5ATs mutant was significantly outcompeted by the wild-type by more than 10-fold in kidneys (p = 0.0037) ()
Figure 10. Role of SPATEs for E. coli QT598 in the murine model of ascending UTI.
CBA/J mice were challenged transurethrally with QT598 and isogenic strains ΔtagBC, Δsha, or Δ5ATs (wherein all 5 SPATE genes are inactivated). Mice were euthanized after 48 h, and bladder and kidneys were harvested for colony counts. a. Single-strain infections to compare wild-type strain QT598 to ΔtagBC, Δsha mutants. There were no significant differences in bacterial numbers in either the bladders or kidneys. b. Single-strain infections to compare wild-type strain QT598 to the Δ5ATs mutant. Similarly, there were no significant differences in colonization observed. c. Co-infection experiments between the QT598Δlac and the Δ5ATs mutant. The Δ5ATs mutant colonized the bladder at similar levels to the wild-type (Δlac) strain; however, the Δ5ATs strain was outcompeted in the kidneys by over 10-fold (Data are means ± standard errors of the means of 10 mice (* p < 0.05, ** p < 0.01, Mann–Whitney Test).
Sha gene expression is upregulated during infection in the mouse bladder
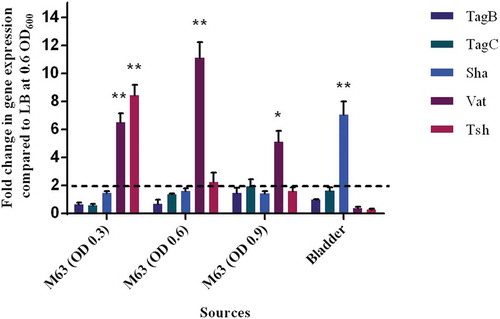
The level of expression of SPATE encoding genes from samples grown in different culture conditions as well as from infected mouse bladders was determined. All 5 SPATE genes were expressed in LB medium as well as in minimal M63-glycerol medium. Interestingly, the vat gene was upregulated by 10-fold in the minimal medium compared to LB. In bladder samples from infected mice, all the SPATE genes were detected and expressed during infection. Interestingly, the sha gene was upregulated 6-fold in the bladder (). However, expression levels of the four other SPATE genes were not significantly different in bladders when compared to expression during culture in LB.
Figure 11. Differential expression of some SPATE genes occurs in vitro and in mouse bladder.
qRT-PCR analysis of SPATE gene transcription from QT598 strain grown in different conditions. Growth in rich medium (LB) to OD600 of 0.6 was used as a standard and compared to growth in M63 minimal medium (with glycerol as carbon) at different growth phases (OD600 of 0.3, 0.6. and 0.9). RNAs were also extracted from infected mouse bladder. Transcription of tsh and vat genes were significantly increased in minimal medium. Further, the sha gene was shown to be significantly upregulated in the mouse bladder. (* p < 0.05, ** p < 0.01, error bars indicate standard deviations, Student t-test). Expression of other SPATE genes was tested under similar conditions. See methods section for details concerning the calculation of gene expression levels. Expressions of other SPATE genes were similar under all conditions tested. The dashed line corresponds to the cutoff for a significant difference in expression.
Discussion
Some pathogenic E. coli produce multiple SPATEs, and this may provide a greater capacity to infect different host species or tissues. Having a combination of SPATEs may also allow a reserve of functions that may include both specific and redundant functions which may importantly be differentially regulated during infection or colonization. UPEC strain CFT073 has 10 autotransporter proteins including 3 SPATEs: Sat, Vat and PicU [Citation41]. Shigella flexneri also contains 3 SPATEs: SepA, Pic and SigA [Citation37,Citation72,Citation73]. Similarly, Citrobacter rodentium, a model for A/E lesions of EPEC and Enterohemorrhagic E. coli (EHEC) also produces 3 SPATEs [Citation74]. The ExPEC strain QT598 we investigated in this report produces a total of 5 SPATEs including three previously uncharacterized members: Sha, TagB, and TagC. Each of the 5 SPATEs demonstrated individual as well as shared properties or functions. Sha, located on a ColV-type plasmid (pEC598), is a class 2 SPATE, and it is closely related to Tsh and Vat proteins. We found this new AT has a modified cleavage site lacking twin asparagine (NN) sites between the passenger domain and the β-barrel. This absence of a typical cleavage site was also seen in rpeA (Rabbit-specific enteropathogenic Escherichia coli (REPEC) plasmid-encoded autotransporter) [Citation75], indicating that the passenger domain is not cleaved from the outer membrane. However, in the case of Sha when cloned in BL21, a band was detected in polyacrylamide gel indicating that the modified cleavage site contributes to the separation of the passenger domain from the β-barrel.
TagB and TagC were found on a genomic island between the conserved E. coli genes yjdI and yjdK. Prevalence of the SPATE sequences among UPEC and APEC demonstrated that tagB and tagC sequences were present in at least 10% of the UPEC strains and that these genes were largely associated with strains belonging to phylogenetic group B2. Interestingly, genomic islands harboring tagB and tagC are also present in the genomes of numerous multi-resistant clinical isolates including members of the E. coli ST131 pandemic clone such as E. coli JJ1877 [Citation76] and many other CTX-producing clinical isolates from urinary tract infections or sepsis (Supplemental Table 2). Further investigation into the potential role of these newly identified SPATE toxins for the virulence of such human ExPEC is therefore warranted.
Although tagB and tagC were less prevalent in APEC, the strains carrying those genes were mostly O1 strains belonging to phylogenetic group B2 and were all isolated from diseased turkeys. Although sha was only present in 1% of UPEC, it was present in 20% of APEC strains. As with Tsh, Sha is plasmid-encoded and these ColV-type plasmids are present in nearly all APEC but are less common in UPEC [Citation77]. Similar phenotypes were observed for the Tsh, Vat, and Sha autotransporters, including hemagglutination, adherence, protease activity, biofilm formation, and cytopathic effects. As such, the role of these three SPATEs may also be cumulative for some APEC strains.
We assessed the cumulative role of SPATEs in APEC strain QT598 with cell cytotoxicity assays and infection experiments in the murine UTI model. It has been shown that certain APEC strains are highly similar to human ExPEC and can belong to the same clonal groups and contain similar virulence gene profiles [Citation2,Citation6,Citation9,Citation11,Citation12,Citation14,Citation24,Citation25]. Previous reports have also tested APEC strains in the mouse UTI model [Citation78–Citation81]. QT598 is a serogroup O1 strain, which is a common serogroup of both APEC and human ExPEC strains, and is clonally related to some human ExPEC, further supporting verification of the role of SPATEs for this strain in a UTI model. QT598 was initially isolated from a young turkey poult and was virulent by subcutaneous infection of 1-day-old chicks. We initially tested QT598 strain in a 3-week-old chicken air sac inoculation model. However, the strain only caused very limited disease and was rapidly cleared in this model. It is possible that the strain is only able to infect very young chicks or it may be more specific for infection of turkeys than chickens. It will be of interest to investigate this in the future.
By cloning each of the SPATE encoding genes in E. coli K-12, specific activities of individual SPATEs could be determined. Protease cleavage using oligopeptides demonstrated that Sha, as well as Tsh and Vat, demonstrated elastase-like activity, whereas TagB and TagC demonstrated trypsin-like activity, similar to the EspC autotransporter (). Oligopeptide degradation was also observed in strain QT598 with TagB and TagC being required for trypsin-like activity, whereas the Class 2 SPATEs contributed to Elastase-like activity. As such, the combination of these SPATEs provides an expanded spectrum of protease activity. However, adding a serine protease inhibitor (PMSF) to the supernatant of the SPATEs neutralized their effect on the cleavage of these oligopeptides. Further, when the catalytic site of the ATs was mutated (serine was replaced by alanine) proteolytic activity was absent, indicating that the serine protease motif is important for the activity of these SPATEs (Supplemental Fig. S3). Previously, Tsh was shown to be proteolytic to substrates including mucin and factor V, whereas EspC could cleave other proteins such as pepsin and spectrin [Citation62]. This suggests that the combination of SPATEs produced by QT598 can target multiple substrates. It will be of interest to more specifically investigate cleavage of different host substrates by the newly characterized SPATEs Sha, TagB, and TagC to try to identify their mechanisms of interaction with host cells. Adherence to host cell lines was also increased by the production of SPATEs. In particular, TagB, TagC, and Sha increased adherence to both human and avian epithelial cells, whereas Tsh only increased adherence to bladder cells, and Vat only increased adherence to kidney cells (). Interestingly, in addition to promoting general adherence to host cells, TagB, TagC, and Sha also mediated bacterial aggregation, suggesting a self-associating phenotype similar to the AIDA-1 autotransporter (). Although Tsh and Vat were less effective at general adherence to different epithelial cells, these SPATEs as well as Sha were effective hemagglutinins for erythrocytes of a variety of animal species and also demonstrated increased biofilm formation, whereas production of TagB and TagC only conferred limited hemagglutination activity () and no increase in biofilm formation (). It is interesting to note that some class 2 SPATEs have been shown to recognize a variety of glycans on leukocyte surfaces [Citation82]. As similar carbohydrates may be present on erythrocyte surfaces, it is not surprising that Sha as well as Tsh and Vat autotransporters demonstrated extensive hemagglutination activity for a variety of erythrocytes. It will be of interest to further determine if these SPATEs can also recognize glycosylated surface receptors on either human or avian leukocytes that may alter host immune function.
Notably, some phenotypes such as autoaggregation, hemagglutination, and biofilm formation were only present when the genes were expressed in high-copy vectors but were absent in the clinical strain. Cloning in a higher copy vector provides a means to constitutively express proteins in vitro, whereas these proteins or systems may not always be expressed when in a single copy or on a native plasmid in the clinical strain under in vitro growth conditions. Also, in the wild-type clinical strain, there can be a great deal of redundancy of function with multiple proteins (both the autotransporters and other outer membrane proteins and fimbrial adhesins that may similarly mediate hemagglutination to different erythrocytes or adherence to host cells).
Interestingly all of the SPATEs demonstrated cytopathic effects on epithelial cells. However, the class 2 SPATEs, Vat, Tsh, and Sha, only caused delayed cell death after 12 h exposure, compared to the TagB and TagC SPATEs that demonstrated cytotoxicity within 5 h of interaction (). Importantly, loss of all 5 of the SPATE encoding genes from QT598 was required to abrogate the cytopathic effect (). However, the “slow acting” cytopathic effect may be due to a less directed or non-specific internalization such as pinocytosis for some of the SPATEs and since this effect only occurs after long-term 12-h exposure it may indicate that the cytotoxic capacity of these SPATEs may be limited and that the roles of such SPATEs may be more specifically linked to other protease functions such as mucinase activity or immunomodulatory roles as has been proposed as the main function of some of the Class-2 SPATEs. Similarly, a slow effect was seen in the case of EspC, when it was internalized slowly into the cells by pinocytosis, and no receptor was required for this process [Citation83]. Further experiments are in progress to elucidate the cytopathic effects of Sha, TagB, and TagC.
Of further note, loss of all 5 SPATEs resulted in decreased fitness in the kidneys of infected mice (), whereas loss of individual SPATEs did not have any reduction in virulence or fitness. This suggests that collectively these SPATEs can provide a selective advantage during kidney infection for QT598 in the murine UTI model. The levels of expression of the 5 SPATEs were different depending on the growth conditions – all 5 SPATEs were expressed in LB broth, mass spectrometry results have confirmed that Tsh (48% of coverage) and Vat (22% of coverage) proteins were highly expressed compared to the other SPATE proteins. In minimal M63-glycerol medium, the vat gene was upregulated by 10-fold compared to LB, and the sha gene was upregulated six-fold in infected bladder compared to culture in LB. These results indicate that SPATE-encoding genes can be subjected to environmental changes that influence their regulation. However, other than the vat gene [Citation58], there is very limited information concerning identification of factors that regulate the expression of different SPATEs.
In conclusion, we investigated the role of three new SPATEs in an APEC strain, TagB, TagC, and Sha, which were present in some APEC and UPEC strains. These SPATEs may confer fitness and capabilities to infect multiple hosts. Our findings highlight the potential role of these proteins in virulence and show they significantly contribute to autoaggregation, hemagglutination, adherence to epithelial cell lines and also exhibit cytopathic effects. Furthermore, we have shown, in combination, all five ATs contribute to fitness and colonization of QT598 during urinary tract infection in the mouse model. In future studies, it will be of interest to determine mechanisms of action of the TagB, TagC, and Sha autotransporters, identify specific targets, and determine the effects of these SPATEs on host immune responses.
Materials and methods
Strains, media, and PCR screening
Strains and plasmids are listed in . The 697 E. coli strains isolated from urinary tract infections were clinical isolates from Guadeloupe. Strains were collected by laboratories or hospitals over a period of 17 months and included community or nosocomial urinary tract infection isolates [Citation61]. The 299 APEC strains were previously described elsewhere [Citation22]. MT156 (QT598) is an APEC serogroup O1 strain that was isolated from the liver of a young turkey poult [Citation53]. E. coli DH5α, ORN172, and BL21 were used for gene cloning and phenotypic tests. Bacteria were grown at 37°C on solid or liquid Luria-Bertani medium (Alpha Bioscience, Baltimore, MD, USA) supplemented with the appropriate antibiotics when required at concentrations of 100µg/ml ampicillin, 30 µg/ml chloramphenicol, or 50 µg/ml of kanamycin. For mice infection studies, QT598 and mutant derivatives were grown in brain heart infusion broth (Alpha Bioscience). M63-glycerol minimal medium contained the following per liter: 5.3 g KH2PO4, 13.9 g K2HPO4 · 3H2O, and 2.0 g (NH4)2SO4. The pH was adjusted to 7.5 with KOH, and the medium was supplemented with 1 mM MgSO4, 1 mM CaCl2, 1 mM thiamine, and 0.6% (wt/vol) glycerol.
Multiplex PCR was performed to determine the prevalence of different ATs within clinical isolates using primers listed in Supplemental Table 1. PCR amplification was done in a 25 µl reaction mixture containing 10 mM of primers, 1X of Taq FroggaMix (FroggaBio, Toronto, ON, Canada), DNA templates and deionized water when necessary. To detect sat, tsh, tagB and tagC genes, the reaction mixture was placed in a thermocycler (Eppendorf, Mississauga, ON, Canada) and set for initial denaturation at 95°C for 2 min followed by 30 cycle of denaturation at 95°C for 30 sec, annealing at 58°C for 40 sec and extension at 72°C for 30 sec. A final extension step was added at 72°C for 5 min. In order to detect sha gene, PCR was set for initial denaturation at 95°C for 2 min, 35 cycle of denaturation (95°C, 30 sec), annealing (56°C, 40 sec) and extension (72°C, 1 min), and a final extension step at 72°C for 5 min. To detect vat gene, PCR was set for initial denaturation at 95°C for 2 min, 30 cycle of denaturation (95°C, 30 sec), annealing (60°C, 40 sec) and extension (72°C, 30 sec), and a final extension step at 72°C for 5 min. The phylogenetic group of strains was determined by multiplexed PCR as described [Citation84]. Amplified samples were separated by electrophoresis on 0.8% agarose gel stained with gel stain (Civic Bioscience, Beloeil, QC, Canada) and DNA was visualized using Gel Doc (Syngene Chemi Genius, Frederick, MD, USA) at 400 nm.
DNA and genetic manipulations
Plasmid DNA was extracted using the EZ DNA Miniprep kit (Bio Basic, Markham, ON, Canada). PCR products and DNA were purified using the EZ-10 Spin Column PCR Product Purification Kit (Bio Basic). DNA for SPATE-encoding genes was amplified using Q5 High Fidelity-DNA polymerase (New England Biolabs, Whitby, ON, Canada).
Bioinformatic analysis
Presence of AT sequences in silico was determined in E. coli genomes available from the NCBI database by Blast analyses using the tagB, tagC and sha sequences from the QT598 genome. Phylogenetic analyses of the predicted full-length passenger domain sequence of each SPATE were performed by Clustal W, and the phylogenetic tree was constructed using PhyML/bootstrapping in MEGA6 [Citation85]. The Conserved Domain Database (CDD) and SignalP were used to predict the three domains of the AT proteins [Citation86,Citation87]. The I-TASSER server and chimera were used for three-dimensional (3D) structure [Citation65,Citation66] visualization of the predicted Sha AT protein, and the Protein Data Bank (PDB) server was used to obtain the predicted 3-D structure of the Sha protein for comparison to Hbp [Citation88]. Amino acid sequence comparisons were performed using Clone Manager Suite 7 (SciED, Denver, CO, USA) and online BLAST programs available from the National Center for Biotechnology Information (NCBI).
Construction of plasmids
AT-encoding genes were amplified by PCR using specific primers (listed in Supplemental Table 2). The tagB, tagC, sha, vat, and tsh genes were amplified from QT598. The espC gene was amplified from EPEC strain E2348/69 (Accession number AF297061) [Citation60]. To clone tagB, tagC, espC, sha, tsh, and vat, PCR products contained 15 bp extensions homologous to the pBCsk+ multi-cloning site. Linearized pBCsk+ digested with XhoI and BamHI was used to clone inserts by fusion reaction with the Quick-fusion cloning kit (Biotool, Houston, TX, USA). All the recombinant plasmids (pIJ548, pIJ549, pIJ550, pIJ551, pIJ552, pIJ553) were first transformed into E. coli DH5α then the plasmids were extracted using a Miniprep kit according to the manufacturer’s recommendations (Bio Basic) and transformed into E. coli BL21 for protein expression into culture supernatants and into the fim-negative E. coli strain ORN172 for other phenotypic assays.
Mutagenesis of SPATE-encoding genes
Mutants were generated using the lambda red recombinase method [Citation89]. The tagBC genes were first replaced with a kanamycin resistance cassette, from plasmid pKD13 with knock out primers: 2094 and 2095 (Supplemental Table 1). PCR products were electroporated into QT598 containing the lambda red recombinase-expressing plasmid pKD46. Deletion of the tagB and tagC was confirmed by PCR with screening primers (Supplemental Table 1). The kanamycin resistance cassette, flanked by FLP recombination target (FRT) sequences, was removed by the introduction of plasmid pCP20 expressing the FLP recombinase [Citation90]. Then, the vat gene was replaced by a chloramphenicol resistance cassette amplified from pKD4 in the background QT598ΔtagBC::FRT (QT5187) using the same approach. Similarly, kanamycin resistance cassette amplified from plasmid pKD3 was used to replace the sha gene in QT598 ΔtagBC Δvat::cat (QT5188). Finally, the tsh gene was replaced with a tetracycline-resistance cassette by allelic exchange as detailed in [Citation22] in QT598 ΔtagBC Δvat::cat Δsha::kan (QT5189) generating the five SPATE genes, Δ5ATs mutant (QT598 ΔtagBC Δvat::cat Δsha::kan Δtsh::tetAR(B)) (QT5182).
Protein preparation and analysis
Culture supernatants, from LB broth cultures of E. coli BL21 expressing AT proteins were centrifuged at 7500 × g for 15 min at 4°C. Supernatants were filtered through 0.22 μm filters and concentrated through 50 kDa Amicon filters (Millipore Sigma, St. Louis, MO, USA). Protein concentrations were determined using the Pierce Coomassie Plus Assay Reagent kit (Thermo Fisher Scientific, St. Laurent, QC, Canada) and proteins were visualized on SDS-PAGE by Coomassie blue as well as silver staining and identified based on the protein markers (10–200 kDa) (Bio Basic). To identify the proteins, bands were excised from denatured gels. Protein digestion by the trypsin, peptide labeling, and mass spectrometry analyses was performed by the proteomics platform of the Institut de Recherche en Immunologie et en Cancérologie (IRIC) of the Université de Montréal (Montréal, QC, Canada). The data were visualized and analyzed using Scaffold 4 Proteomics software.
Oligopeptide cleavage assays
Synthetic peptide cleavage was performed as previously described in [Citation91] with slight modifications. Briefly, 5 µg/ml of each SPATES was added to 200 μl of three different pNA-conjugated oligopeptides: N-Succinyl-Ala-Ala-Ala-p-nitroanilide, N-Benzoyl-L-arginine 4-nitroanilide and N-succinyl-ala-ala-pro-phe-p-nitroanilide (Millipore Sigma) at 1 mM concentration in a buffer containing 0.2 M NaCl, 0.01 mMZnSO4, 0.1 M MOPS (3-(N-morpholino) propanesulfonic acid), pH 7.3 were incubated at 37°C for 3 h and absorbance readings were determined at 410 nm. Readings were normalized to the maximum absorbance of positive control. All reactions were performed in triplicate.
Autoaggregation
The autoaggregation test was carried out as described before [Citation92]. Briefly, overnight cultures were adjusted to an OD600 of 1.5. A volume of 10 ml of each culture was placed in sterile 20 ml glass tubes. Tubes were then vortexed for 5 s then left at 4°C for 3 h. Samples 1 cm from the top were taken, and the percentage of change in OD600 was used as the value for autoaggregation.
Hemagglutination tests
Hemagglutination in microtiter plates was performed as described by [Citation93] with some modifications. Human, chicken, turkey, pig, bovine, canine, rabbit, horse and sheep red blood cells (RBCs) were washed and resuspended in PBS at a final concentration of 3%. The E. coli fim-negative K-12 strain ORN172 expressing different SPATEs was grown overnight at 37°C in LB medium, harvested and adjusted to an optical density (O.D.600) of 60. Suspensions were serially diluted in 96-well round bottom plates containing 20 µl of PBS mixed with 20 µl of 3% red blood cells and incubated for 30 min at 4°C.
Biofilm assay
Biofilm formation on polystyrene surfaces was assessed in 96-well plates as previously described [Citation67]. Strains were grown at various temperatures (25°C, 30°C, 37°C, and 42°C) for 48 h under static conditions in LB medium. Wells were washed and stained with 0.1% crystal violet (Millipore Sigma) for 15 min, then 200 µl of ethanol-acetone (80:20) solution was added, followed by measuring at an optical density at OD595.
Adherence assays
The 5637 human bladder cells (ATCC HTB-9), human embryonic kidney cells HEK-293 (ATCC® CRL-1573™), and the avian fibroblast cell line (CEC-32) were used to determine adherence of E. coli ORN172 expressing different SPATEs as described before [Citation94]. The 5637 cells were maintained in RPMI 1640 (Thermo Fisher Scientific) supplemented with 10% heat-inactivated fetal bovine serum (FBS) at 37°C in 5% CO2, and 2 × 105 cells/well were seeded into 24-well cell culture plates. Cell lines were washed twice with phosphate-buffered saline (pH 7.2) and then incubated at a multiplicity of infection (MOI) of 10 at 37°C for 2 h in RPMI 1640 medium with 10% FBS. Non-adherent bacteria were removed by washing with PBS three times. Cells were then exposed to 1% Triton X-100 for 5 min, and serial dilutions were plated on LB agar plus an antibiotic. For HEK-293 cells, Eagle’s Minimum Essential Medium supplemented with 10% of FBS was used. For CEC-32 cells Dulbecco’s Modified Eagle’s medium supplemented with 10% of FBS was used. The adherence assays were done in triplicate for each sample.
Determination of cytopathic effects
The 5637 cells were maintained in RPMI 1640 medium (Thermo Fisher Scientific) supplemented with 10% heat-inactivated FBS at 37°C in 5% CO2, and 2 × 105 cells/well were seeded into eight-well chamber slides (Thermo Fisher Scientific) and allowed to grow to 75% confluence. Thirty μg/ml of each SPATE (final concentration) was added directly to monolayers and incubated for 5 h or 12 h at 37°C with 5% CO2. Cells were then washed twice with PBS (phosphate-buffered saline), fixed with 70% methanol, and stained with Giemsa stain.
Lactate dehydrogenase (LDH) release assay
Culture supernatants were incubated with monolayers of 5637 cells in RPMI 1640 supplemented with 10% heat-inactivated FBS at 37°C in 5% CO2 for up to 12 h. Release of LDH was quantified by CytoTox 96® Non-Radioactive Cytotoxicity Assay kit (Promega, Madison, WI, USA) at 5 h and 12 h. A lysis solution (provided in the kit) was added to the non-infected cells to generate the maximum LDH release control from lysed cells.
Ascending urinary tract infection in mice
For single strain infections with the wild-type parent QT598 and isogenic SPATE mutants QT598ΔtagBC and QT598Δsha, 25 μl (109 CFU) were tested in an ascending UTI model adapted from [Citation95] with 10 mice in each group. Similarly, a murine ascending UTI model with 10 mice in each group was used for co-infection, in which a virulent ΔlacZYA derivative of QT598 was co-infected with the Δ5ATs strain, a QT598-derivative lacking all 5 SPATE-encoding genes. Twenty-five μl (109 CFU) of a mixed culture containing equal amounts of each strain were inoculated through a catheter in six-week-old CBA/J female mice. Mice were euthanized after 48 h, and bladders and kidneys were harvested aseptically, homogenized, diluted and plated on MacConkey agar plates. Bladder samples were frozen at −80°C in TRIzol® reagent (Thermo Fisher Scientific) for RNA extraction.
qRT-PCR analysis of SPATE gene expression in vivo and in vitro
Expression of the 5 SPATE-encoding genes was determined after growth in LB medium, minimal M63 medium, and from infected mouse bladders. Total RNAs from bacterial samples were extracted in the EZ-10 Spin Column Total RNA Miniprep Kit (Bio Basic) as described elsewhere [Citation96]. Briefly, to extract RNA from infected bladder, samples were homogenized with TRIzol® reagent (Thermo Fisher Scientific), centrifuged for 30 sec at 12,000 × g, the supernatant was incubated with ethanol (95–100%) and transferred into Zymo-Spin™ IICR Column to extract RNA using Direct-zol RNA Miniprep kit (Zymo Research, Irvine, CA, USA) according to the manufacturer’s recommendations. All RNA samples were treated with TURBO DNase (Thermo Fisher Scientific). PCR (35 cycles) was used to verify DNA contamination. cDNAs were synthesized by using the IscriptTM Reverse transcription supermix according to the manufacturer’s protocol (Bio-Rad Life Science, Mississauga, ON, Canada). The RNA polymerase sigma factor rpoD gene was used as a housekeeping control. Primers designed to amplify rpoD, tagB, tagC, vat, tsh, and sha (Supplemental Table 1) were used. For each sample, 50 ng of cDNA and 100 nM concentrations of each primer set were mixed with 10 μl of SsoFast Evagreen supermix (Bio-Rad Life Science) per well. Assays were performed in triplicate on a Corbett Rotorgene (Thermo Fisher Scientific) instrument. All data were normalized to rpoD expression levels. Melting-curve analysis was verified to differentiate accumulation of Evagreen-bound DNA and determine that signal was gene-amplification specific and not due to the primer-dimer formation. The data were analyzed by the 2−ΔΔCT method [Citation97].
Statistical analyses
Experimental data were expressed as a mean ± standard error of the mean (SEM) in each group. The means of groups were combined and analyzed by a two-tailed Student t-test for pairwise comparisons and analysis of variance (ANOVA) to compare means of more than two populations. For mouse infection experiments, the Mann–Whitney test was used to compare the samples by pairs, and the Kruskal–Wallis test was used to compare groups. A P value of <0.05 was considered statistically significant. All data were analyzed with the Graph Pad Prism 7 software (GraphPad Software, San Diego, CA, USA).
Ethics statement
The protocol for mice urinary tract infection was approved by the animal ethics evaluation committee – Comité Institutionel de Protection des Animaux (CIPA No 1608–02) of the INRS-Institut Armand-Frappier.
Nucleotide accession numbers
Sections of the genome of E. coli strain QT598 containing the five different SPATE-encoding genes were submitted to NCBI Genbank from the analysis of whole-genome survey sequence. The corresponding accession numbers are tsh (MH899683), sha (MH899684), vat (MH899682), and tagB and tagC (MH899681) genetic regions.
Supplemental Material
Download Zip (4.9 MB)Acknowledgments
We thank E. Bonneil, manager at the proteomics platform of Université de Montréal for assistance with mass spectrometry and peptide analysis and Y. Lopez de Los Santos for assistance for 3-D model prediction comparison of autotransporters. Funding for this work was supported by NSERC Canada Discovery Grants 2014-06622 and 2019-06642 (C.M.D.) and a collaborative ACIP grant from the Institut Pasteur International Network (to A.T., S.G.-R., and C.M.D).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Croxen MA, Finlay BB. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol. 2010;8(1):26–38.
- Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nature Rev Microbiol. 2004;2(2):123.
- Bauchart P, Germon P, Bree A, et al. Pathogenomic comparison of human extraintestinal and avian pathogenic Escherichia coli–search for factors involved in host specificity or zoonotic potential. Microb Pathog. 2010;49(3):105–115.
- Bélanger L, Garenaux A, Harel J, et al. Escherichia coli from animal reservoirs as a potential source of human extraintestinal pathogenic E. coli. FEMS Immunol Med Microbiol. 2011;62(1):1–10.
- Clermont O, Olier M, Hoede C, et al. Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infect Genet Evol. 2011;11(3):654–662.
- Ewers C, Li G, Wilking H, et al. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they?. Int J Med Microbiol. 2007;297(3):163–176.
- Moulin-Schouleur M, Schouler C, Tailliez P, et al. Common virulence factors and genetic relationships between O18: K1:H7 Escherichia coli isolates of human and avian origin. J Clin Microbiol. 2006;44(10):3484–3492.
- Nandanwar N, Janssen T, Kuhl M, et al. Extraintestinal pathogenic Escherichia coli (ExPEC) of human and avian origin belonging to sequence type complex 95 (STC95) portray indistinguishable virulence features. Int J Med Microbiol. 2014;304(7):835–842.
- Johnson JR, Russo TA. Extraintestinal pathogenic Escherichia coli:“the other bad E.coli”. J Lab Clin Med. 2002;139(3):155–162.
- Guabiraba R, Schouler C. Avian colibacillosis: still many black holes. FEMS Microbiol Lett. 2015;362(15):fnv118.
- Dho-Moulin M, Fairbrother JM. Avian pathogenic Escherichia coli (APEC). Vet Res. 1999;30(2–3):299–316.
- Dziva F, Stevens MP. Colibacillosis in poultry: unravelling the molecular basis of virulence of avian pathogenic Escherichia coli in their natural hosts. Avian Pathol. 2008;37(4):355–366.
- Dziva F, Hauser H, Connor TR, et al. Sequencing and functional annotation of avian pathogenic Escherichia coli serogroup O78 strains reveal the evolution of E.coli lineages pathogenic for poultry via distinct mechanisms. Infect Immun. 2013;81(3):838–849.
- Johnson TJ, Kariyawasam S, Wannemuehler Y, et al. The genome sequence of avian pathogenic Escherichia coli strain O1: K1: H7 shares strong similarities with human extraintestinal pathogenic E.coli genomes. J Bacteriol. 2007;189(8):3228–3236.
- Johnson TJ, Nolan LK. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol Mol Biol Rev. 2009;73(4):750–774.
- Johnson TJ, Wannemuehler YM, Johnson SJ, et al. Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl Environ Microbiol. 2007;73(6):1976–1983.
- Mellata M, Touchman JW, Curtiss R. Full sequence and comparative analysis of the plasmid pAPEC-1 of avian pathogenic E.coli chi7122 (O78: K80:H9). PLoS One. 2009;4(1):e4232.
- Peigne C, Bidet P, Mahjoub-Messai F, et al. The plasmid of Escherichia coli strain S88 (O45: K1:H7) that causes neonatal meningitis is closely related to avian pathogenic E.coli plasmids and is associated with high-level bacteremia in a neonatal rat meningitis model. Infect Immun. 2009;77(6):2272–2284.
- Cordoni G, Woodward MJ, Wu H, et al. Comparative genomics of European avian pathogenic E.coli (APEC). BMC Genomics. 2016;17(1):960.
- Leimbach A, Hacker J, Dobrindt U. E.coli as an all-rounder: the thin line between commensalism and pathogenicity. Curr Top Microbiol Immunol. 2013;358:3–32.
- Mokady D, Gophna U, Ron EZ. Extensive gene diversity in septicemic Escherichia colistrains. J Clin Microbiol. 2005;43(1):66–73.
- Dozois CM, Dho-Moulin M, Bree A, et al. Relationship between the Tsh autotransporter and pathogenicity of avian Escherichia coli and localization and analysis of the Tsh genetic region. Infect Immun. 2000;68(7):4145–4154.
- Dozois CM, Daigle F, Curtiss R 3rd. Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc Natl Acad Sci U S A. 2003;100(1):247–252.
- Johnson TJ, Johnson SJ, Nolan LK. Complete DNA sequence of a ColBM plasmid from avian pathogenic Escherichia coli suggests that it evolved from closely related ColV virulence plasmids. J Bacteriol. 2006;188(16):5975–5983.
- Krishnan S, Chang AC, Hodges J, et al. Serotype O18 avian pathogenic and neonatal meningitis Escherichia coli strains employ similar pathogenic strategies for the onset of meningitis. Virulence. 2015;6(8):777–786.
- Johnson TJ, Wannemuehler Y, Johnson SJ, et al. Comparison of extraintestinal pathogenic Escherichia coli strains from human and avian sources reveals a mixed subset representing potential zoonotic pathogens. Appl Environ Microbiol. 2008;74(22):7043–7050.
- Mellata M. Human and avian extraintestinal pathogenic Escherichia coli: infections, zoonotic risks, and antibiotic resistance trends. Foodborne Pathog Dis. 2013;10(11):916–932.
- Rodriguez-Siek KE, Giddings CW, Doetkott C, et al. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology. 2005;151(Pt 6):2097–2110.
- Henderson IR, Navarro-Garcia F, Nataro JP. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 1998;6(9):370–378.
- Klemm P, Vejborg RM, Sherlock O. Self-associating autotransporters, SAATs: functional and structural similarities. Int J Med Microbiol. 2006;296(4–5):187–195.
- Wells TJ, Totsika M, Schembri MA. Autotransporters of Escherichia coli: a sequence-based characterization. Microbiology. 2010;156(8):2459–2469.
- Ruiz-Perez F, Nataro JP. Bacterial serine proteases secreted by the autotransporter pathway: classification, specificity, and role in virulence. Cell Mol Life Sci. 2014;71(5):745–770.
- Albenne C, Ieva R. Job contenders: roles of the beta-barrel assembly machinery and the translocation and assembly module in autotransporter secretion. Mol Microbiol. 2017;106(4):505–517.
- Bernstein HD. Looks can be deceiving: recent insights into the mechanism of protein secretion by the autotransporter pathway. Mol Microbiol. 2015;97(2):205–215.
- Henderson IR, Navarro-Garcia F, Desvaux M, et al. Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev. 2004;68(4):692–744.
- Otto BR, Van Dooren SJ, Nuijens JH, et al. Characterization of a hemoglobin protease secreted by the pathogenic Escherichia coli strain EB1. J Exp Med. 1998;188(6):1091–1103.
- Henderson IR, Czeczulin J, Eslava C, et al. Characterization of Pic, a Secreted Protease of Shigella flexneri and Enteroaggregative Escherichia coli. Infect Immun. 1999;67(11):5587–5596.
- Brunder W, Schmidt H, Karch H. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157: H7 cleaves human coagulation factor V. Mol Microbiol. 1997;24(4):767–778.
- Eslava C, Navarro-García F, Czeczulin JR, et al. Pet, an autotransporter enterotoxin from enteroaggregative Escherichia coli. Infect Immun. 1998;66(7):3155–3163.
- Guyer DM, Henderson IR, Nataro JP, et al. Identification of sat, an autotransporter toxin produced by uropathogenic Escherichia coli. Mol Microbiol. 2000;38(1):53–66.
- Parham NJ, Srinivasan U, Desvaux M, et al. PicU, a second serine protease autotransporter of uropathogenic Escherichia coli. FEMS Microbiol Lett. 2004;230(1):73–83.
- Stein M, Kenny B, Stein MA, et al. Characterization of EspC, a 110-kilodalton protein secreted by enteropathogenic Escherichia coli which is homologous to members of the immunoglobulin A protease-like family of secreted proteins. J Bacteriol. 1996;178(22):6546–6554.
- Subashchandrabose S, Smith SN, Spurbeck RR, et al. Genome-wide detection of fitness genes in uropathogenic Escherichia coli during systemic infection. PLoS Pathog. 2013;9(12):e1003788.
- Guyer DM, Radulovic S, Jones F-E, et al. Sat, the secreted autotransporter toxin of uropathogenic Escherichia coli, is a vacuolating cytotoxin for bladder and kidney epithelial cells. Infect Immun. 2002;70(8):4539–4546.
- Parreira VR, Gyles CL. A novel pathogenicity island integrated adjacent to the thrW tRNA gene of avian pathogenic Escherichia coli encodes a vacuolating autotransporter toxin. Infect Immun. 2003;71(9):5087–5096.
- Parham NJ, Pollard SJ, Desvaux M, et al. Distribution of the serine protease autotransporters of the Enterobacteriaceae among extraintestinal clinical isolates of Escherichia coli. J Clin Microbiol. 2005;43(8):4076–4082.
- Restieri C, Garriss G, Locas MC, et al. Autotransporter-encoding sequences are phylogenetically distributed among Escherichia coli clinical isolates and reference strains. Appl Environ Microbiol. 2007;73(5):1553–1562.
- Spurbeck RR, Dinh PC Jr., Walk ST, et al. Escherichia coli isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect Immun. 2012;80(12):4115–4122.
- Provence DL, Curtiss R. Isolation and characterization of a gene involved in hemagglutination by an avian pathogenic Escherichia coli strain. Infect Immun. 1994;62(4):1369–1380.
- Cyoia PS, Rodrigues GR, Nishio EK, et al. Presence of virulence genes and pathogenicity islands in extraintestinal pathogenic Escherichia coli isolates from Brazil. J Infect Dev Ctries. 2015;9(10):1068–1075.
- Maluta RP, Logue CM, Casas MR, et al. Overlapped sequence types (STs) and serogroups of avian pathogenic (APEC) and human extra-intestinal pathogenic (ExPEC) Escherichia coli isolated in Brazil. PLoS One. 2014;9(8):e105016.
- Otto BR, van Dooren SJ, Dozois CM, et al. Escherichia coli hemoglobin protease autotransporter contributes to synergistic abscess formation and heme-dependent growth of Bacteroides fragilis. Infect Immun. 2002;70(1):5–10.
- Marc D, Dho-Moulin M. Analysis of the fim cluster of an avian O2 strain of Escherichia coli: serogroup-specific sites within fimA and nucleotide sequence of fimI. J Med Microbiol. 1996;44(6):444–452.
- Dozois CM, Fairbrother JM, Harel J, et al. pap-and pil-related DNA sequences and other virulence determinants associated with Escherichia coli isolated from septicemic chickens and turkeys. Infect Immun. 1992;60(7):2648–2656.
- Dobrindt U, Agerer F, Michaelis K, et al. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J Bacteriol. 2003;185(6):1831–1840.
- Wang X, Wei L, Wang B, et al. Complete genome sequence and characterization of avian pathogenic Escherichia coli field isolate ACN001. Stand Genomic Sci. 2016;11:13.
- Kostakioti M, Stathopoulos C. Role of the alpha-helical linker of the C-terminal translocator in the biogenesis of the serine protease subfamily of autotransporters. Infect Immun. 2006;74(9):4961–4969.
- Nichols KB, Totsika M, Moriel DG, et al. Molecular characterization of the vacuolating autotransporter Toxin in Uropathogenic Escherichia coli. J Bacteriol. 2016;198(10):1487–1498.
- Sandt CH, Hill CW. Four Different Genes Responsible for Nonimmune Immunoglobulin-Binding Activities within a Single Strain of Escherichia coli. Infect Immun. 2000;68(4):2205–2214.
- Mellies JL, Navarro-Garcia F, Okeke I, et al. espC pathogenicity island of enteropathogenic Escherichia coli encodes an enterotoxin. Infect Immun. 2001;69(1):315–324.
- Guyomard-Rabenirina S, Malespine J, Ducat C, et al. Temporal trends and risks factors for antimicrobial resistant Enterobacteriaceae urinary isolates from outpatients in Guadeloupe. BMC Microbiol. 2016;16(1):121.
- Dutta PR, Cappello R, Navarro-García F, et al. Functional comparison of serine protease autotransporters of Enterobacteriaceae. Infect Immun. 2002;70(12):7105–7113.
- Stathopoulos C, Provence DL, Curtiss R. Characterization of the Avian Pathogenic Escherichia coliHemagglutinin Tsh, a Member of the Immunoglobulin A Protease-Type Family of Autotransporters. Infect Immun. 1999;67(2):772–781.
- Gutierrez D, Pardo M, Montero D, et al. TleA, a Tsh-like autotransporter identified in a human enterotoxigenic Escherichia coli strain. Infect Immun. 2015;83(5):1893–1903.
- Yang J, Yan R, Roy A, et al. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12(1):7.
- Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612.
- Genevaux P, Muller S, Bauda P. A rapid screening procedure to identify mini-Tn10 insertion mutants of Escherichia coli K-12 with altered adhesion properties. FEMS Microbiol Lett. 1996;142(1):27–30.
- Kostakioti M, Stathopoulos C. Functional analysis of the Tsh autotransporter from an avian pathogenic Escherichia coli strain. Infect Immun. 2004;72(10):5548–5554.
- Sherlock O, Vejborg RM, Klemm P. The TibA adhesin/invasin from enterotoxigenic Escherichia coli is self recognizing and induces bacterial aggregation and biofilm formation. Infect Immun. 2005;73(4):1954–1963.
- Hasman H, Chakraborty T, Klemm P. Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J Bacteriol. 1999;181(16):4834–4841.
- Benz I, Schmidt MA. Cloning and expression of an adhesin (AIDA-I) involved in diffuse adherence of enteropathogenic Escherichia coli. Infect Immun. 1989;57(5):1506–1511.
- Al-Hasani K, Henderson IR, Sakellaris H, et al. The sigA gene which is borne on the she pathogenicity island of Shigella flexneri 2a encodes an exported cytopathic protease involved in intestinal fluid accumulation. Infect Immun. 2000;68(5):2457–2463.
- Benjelloun‐Touimi Z, Sansonetti PJ, Parsot C. SepA, the major extracellular protein of Shigella flexneri: autonomous secretion and involvement in tissue invasion. Mol Microbiol. 1995;17(1):123–135.
- Vijayakumar V, Santiago A, Smith R, et al. Role of class 1 serine protease autotransporter in the pathogenesis of Citrobacter rodentium colitis. Infect Immun. 2014;82(6):2626–2636.
- Leyton DL, Adams LM, Kelly M, et al. Contribution of a novel gene, rpeA, encoding a putative autotransporter adhesin to intestinal colonization by rabbit-specific enteropathogenic Escherichia coli. Infect Immun. 2007;75(9):4664–4669.
- Johnson TJ, Aziz M, Liu CM, et al. Complete genome sequence of a CTX-M-15-producing Escherichia coli strain from the H30Rx subclone of sequence type 131 from a patient with recurrent urinary tract infections, closely related to a lethal urosepsis isolate from the patient’s sister. Genome Announc. 2016;4(3):e00334–16.
- Johnson TJ, Siek KE, Johnson SJ, et al. DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. J Bacteriol. 2006;188(2):745–758.
- Jakobsen L, Hammerum AM, Frimodt-Møller N. Virulence of Escherichia coli B2 isolates from meat and animals in a murine model of ascending urinary tract infection (UTI): evidence that UTI is a zoonosis. J Clin Microbiol. 2010;48(8):2978–2980.
- Mellata M, Johnson J, Curtiss III R. Escherichia coliisolates from commercial chicken meat and eggs cause sepsis, meningitis and urinary tract infection in rodent models of human infections. Zoonoses Public Health. 2018;65(1):103–113.
- Pavanelo DB, Houle S, Matter LB, et al. The periplasmic trehalase affects type 1 fimbriae production and virulence of the extraintestinal pathogenic E.coli strain MT78. Infect Immun. 2018;IAI:00241.
- Skyberg JA, Johnson TJ, Johnson JR, et al. Acquisition of avian pathogenic Escherichia coli plasmids by a commensal E.coli isolate enhances its abilities to kill chicken embryos, grow in human urine, and colonize the murine kidney. Infect Immun. 2006;74(11):6287–6292.
- Ruiz-Perez F, Wahid R, Faherty CS, et al. Serine protease autotransporters from Shigella flexneri and pathogenic Escherichia coli target a broad range of leukocyte glycoproteins. Proc Nat Acad Sci. 2011;108(31):12881–12886.
- Vidal JE, Navarro-García F. Efficient translocation of EspC into epithelial cells depends on enteropathogenic Escherichia coli and host cell contact. Infect Immun. 2006;74(4):2293–2303.
- Clermont O, Christenson JK, Denamur E, et al. The Clermont Escherichia coli phylo‐typing method revisited: improvement of specificity and detection of new phylo‐groups. Environ Microbiol Rep. 2013;5(1):58–65.
- Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729.
- Bendtsen JD, Nielsen H, von Heijne G, et al. Improved prediction of signal peptides: signalP 3.0. J Mol Biol. 2004;340(4):783–795.
- Marchler-Bauer A, Lu S, Anderson JB, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2010;39(suppl_1):D225–D9.
- Kouranov A, Xie L, de la Cruz J, et al. The RCSB PDB information portal for structural genomics. Nucleic Acids Res. 2006;34(suppl_1):D302–D5.
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Nat Acad Sci. 2000;97(12):6640–6645.
- Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: Tc R and Km R cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158(1):9–14.
- Benjelloun-Touimi Z, Tahar MS, Montecucco C, et al. SepA, the 110 kDa protein secreted by Shigella flexneri: two-domain structure and proteolytic activity. Microbiology. 1998;144(7):1815–1822.
- Charbonneau M-È, Berthiaume F, Mourez M. Proteolytic processing is not essential for multiple functions of the Escherichia coli autotransporter adhesin involved in diffuse adherence (AIDA-I). J Bacteriol. 2006;188(24):8504–8512.
- Provence D, Curtiss R. Role of crl in avian pathogenic Escherichia coli: a knockout mutation of crl does not affect hemagglutination activity, fibronectin binding, or curli production. Infect Immun. 1992;60(11):4460–4467.
- Matter LB, Barbieri NL, Nordhoff M, et al. Avian pathogenic Escherichia coli MT78 invades chicken fibroblasts. Vet Microbiol. 2011;148(1):51–59.
- Hagberg L, Engberg I, Freter R, et al. Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect Immun. 1983;40(1):273–283.
- Porcheron G, Habib R, Houle S, et al. The small RNA RyhB contributes to siderophore production and virulence of uropathogenic Escherichia coli. Infect Immun. 2014;82(12):5056–5068.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–408.
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425.
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Bioinformatics. 1992;8(3):275–282.
- Labbate M, Queck SY, Koh KS, et al. Quorum sensing-controlled biofilm development in Serratia liquefaciens MG1. J Bacteriol. 2004;186(3):692–698.
- Woodall LD, Russell PW, Harris SL, et al. Rapid, synchronous, and stable induction of type 1 piliation in Escherichia coli by using a chromosomal lacUV5 promoter. J Bacteriol. 1993;175(9):2770–2778.
