ABSTRACT
Fimbriae play an important role in adhesion and are therefore essential for the interaction of bacteria with the environments they encounter. Most of them are expressed in vivo but not in vitro, thus making difficult the full characterization of these fimbriae. Here, we characterized the silencing of plasmid-encoded fimbriae (Pef) expression, encoded by the pef operon, in the worldwide pathogen Salmonella Typhimurium. We demonstrated that the nucleoid-associated proteins H-NS and Hha, and their respective paralogs StpA and YdgT, negatively regulate at pH 5.1 and pH 7.1 the transcription of the pef operon. Two promoters, PpefB and PpefA, direct the transcription of this operon. All the nucleoid-associated proteins silence the PpefB promoter and H-NS also targets the PpefA promoter. While Hha and YdgT are mainly considered as acting primarily through H-NS to modulate gene transcription, our results strongly suggest that Hha and YdgT silence pef transcription at acidic pH either by interacting with StpA or independently of H-NS and StpA. We also confirmed the previously described post-transcriptional repression of Pef fimbriae by CsrA titration via the fim mRNA and CsrB and CsrC sRNA. Finally, among all these regulators, H-NS clearly appeared as the major repressor of Pef expression. These results open new avenues of research to better characterize the regulation of these bacterial adhesive proteins and to clarify their role in the virulence of pathogens.
Introduction
Fimbriae are filamentous appendages present on the surface of bacteria which play an important role in the virulence of pathogens. They mediate adhesion to target host cells and to many surfaces encountered by bacteria in their host but also in the environment [Citation1–Citation3]. Salmonella Typhimurium genome harbors 13 fimbrial operons: fim, agf, bcf, sti, sth, lpf, sef, stc, stb, stj, std, stf and pef [Citation4]. All of them have been visualized after heterologous expression in E. coli [Citation5] and evidence for their expression by Salmonella in animals and/or plants has been observed [Citation6–Citation10]. By contrast, in vitro, S. Typhimurium is only able to assemble Type 1 fimbriae encoded by the fim operon in standard laboratory growth conditions [Citation11–Citation13]. Altogether, these data are in agreement with a tight regulation of fimbrial expression characterized by a strong in vitro repression that can be relieved by the environmental conditions encountered by the bacteria. However, little is known about the physiological environments allowing the expression of fimbriae and the regulatory mechanisms involved.
Plasmid-encoded fimbriae (Pef) of Salmonella are thin (2–5 nm diameter) flexible fimbriae belonging to the κ-fimbriae clade characterized by the absence of a tip adhesin [Citation6,Citation14,Citation15]. Pef fimbriae are among the fimbriae whose expression has been observed in animals. Indeed, a low expression of these fimbriae has been identified by flow cytometry after recovery of infected bovine ligated ileal loops [Citation8] and a seroconversion has also been observed after mice or chicken inoculation with S. Typhimurium or S. Enteritidis respectively [Citation7,Citation16]. Moreover, pef mutants of S. Typhimurium were shown to be attenuated in their ability to form mature biofilms on a chicken intestinal tissue [Citation17], and also in their colonization of chick, pig, cattle and in a mouse model of long-term systemic persistence [Citation18,Citation19]. Pef fimbriae have also been shown to be necessary for fluid accumulation in the infant mouse [Citation6]. Conversely, Pef fimbriae are not expressed by Salmonella in standard laboratory culture conditions, i.e. growth in rich medium at pH 7.0 with or without agitation [Citation7,Citation20–Citation22], except when the genes involved in the biosynthesis of these fimbriae are overexpressed [Citation6,Citation21]. The only culture conditions, known to date, allowing the expression of Pef fimbriae by Salmonella are cultures in LB broth buffered at pH 5.1 without agitation [Citation21].
Pef fimbriae biogenesis depends on the pefBACDorf5orf6 operon, also called pef operon, located on the virulence plasmid of S. Typhimurium [Citation14]. This operon is also present on the virulence plasmid of S. Enteritidis, S. Bovismorbificans, S. Paratyphi C and S. Choleraesuis but orf5 and orf6 are not present in all these serotypes. This operon encodes the major Pef fimbriae subunit PefA, the usher protein PefC required for the assembly of the fimbriae and the PefD periplasmic chaperone for PefA. pefB, orf5 and orf6 are predicted to encode a regulatory protein and two outer membrane proteins respectively. Two promoters located upstream of pefB and pefA seem to drive the transcription of this operon [Citation6,Citation23].
Only two papers describe the regulatory mechanisms of Pef fimbriae expression. In 2000, Nicholson and Low [Citation21] described how DNA adenine methylase (Dam) and the leucine-responsive regulatory protein (Lrp) modulate GATC sites methylation upstream of pefB, the first open reading frame (ORF) of the pef operon, and consequently positively regulate the transcription of this operon. More recently, Sterzenbach et al. [Citation23] characterized how the 5ʹ untranslated region of the fim mRNA, encoding Type I fimbriae in Salmonella, cooperates with the two small untranslated RNA CsrB and CsrC to prevent Pef fimbriae expression by sequestering the CsrA protein required to stabilize the pefACDorf5orf6 mRNA. In addition to this latter mechanism, an analysis of the literature led us to postulate that the H-NS, Hha and YdgT nucleoid-associated proteins (NAPs) could also be involved in the repression of Pef fimbriae expression in vitro. First, a Δhns mutant was shown to produce more PefA protein than its parental strain [Citation21,Citation22], and then two large transcriptomic studies using Salmonella microarrays suggested that the pef operon was negatively regulated by the H-NS, Hha and YdgT NAPs [Citation24,Citation25]. Although the three regulators seem thus involved in Pef fimbriae negative regulation, the mechanisms beneath this control remained unknown.
The nucleoprotein H-NS is known to repress the transcription of many genes in Gram-negative enteric bacteria, including virulence genes acquired by horizontal transfer, by directly binding DNA targeted regions in an oligomerized state [Citation26]. In S. Typhimurium, almost 10% of the ORFs present on the chromosome or on the virulence plasmid are repressed by H-NS, especially the major virulence genes including most pathogenicity islands [Citation24]. H-NS has a paralog, the nucleoprotein StpA, that is present in Salmonella and other enteric bacteria. The StpA and H-NS proteins share 52% identity and display common properties, especially a capacity to bind AT-rich regions found in curved DNA [Citation27]. Moreover, they are able to form heterodimers due to the similarity of their dimerization domains [Citation28]. StpA was shown to regulate 5% of the Salmonella genome and most of the StpA regulon is part of the H-NS-dependent genes [Citation29]. Hha and its paralog YdgT belong to the Hha/YmoA family of nucleoproteins whose members are known to interact with other NAPs but do not have a DNA binding domain [Citation30]. These two NAPs exhibit 34% identity and 71% similarity. Hha nucleoprotein has been shown to be a negative regulator for many virulence genes like the enterocyte effacement locus in enterohemorrhagic E. coli [Citation31], the Salmonella pathogenicity islands 1 (SPI-1) and 2 (SPI-2) [Citation32,Citation33], and the α-hemolysin genes hlyCABD in E. coli [Citation34]. YdgT seems to act as a support or backup molecule for Hha contributing to the negative regulation of SPI-2 expression in Salmonella and the hly operon in E. coli for example [Citation33,Citation35,Citation36]. To date, the regulatory mechanisms involving Hha and YdgT are not completely understood. Indeed, in 2013, Ali et al. [Citation37] proposed a model where H-NS and Hha nucleoproteins cooperate to repress gene transcription. In this model, H-NS in its oligomerized form binds to the target DNA and interacts with Hha that increases H-NS DNA-binding thanks to positively charged residues on its surface and thus improves H-NS silencing. However, a repression of the transcription of 120 genes by Hha independently of H-NS/StpA has been observed by Solórzano et al. and a direct interaction of Hha with the rtsA promoter region has been described [Citation38,Citation39].
Deciphering the regulatory mechanisms of fimbriae expression is important to fully understand the role of these adhesive structures. The objective of our study was thus to demonstrate the repressive effect of H-NS, StpA, Hha and YdgT nucleoproteins on Pef fimbriae expression in S. Typhimurium and to evaluate their respective importance in Pef repression compared to the repression exerted by the sequestration of CsrA via the 5ʹUTR of the fim mRNA and CsrB/CsrC sRNA.
Results
The absence of CsrA sequestration does not allow Pef expression by S. Typhimurium after culture under standing condition at pH 5.1
Nicholson and Low [Citation21] previously observed expression of Pef fimbriae after culture of S. Typhimurium in LB broth at pH 5.1 without agitation. More recently, Sterzenbach et al. [Citation23] described the first mechanism of Pef fimbriae repression in vitro but in a slightly different culture condition, i.e. in LB buffered to pH 5.5 containing a sterile pipette tip, as a tip was previously shown to enhance Pef fimbriae production in vitro [Citation22]. As a first step of our study, we analyzed the importance of this repression mechanism in the culture conditions used by Nicholson and Low as a tip is not an environmental condition encountered by Salmonella. As a control, we repeated the experiments of Sterzenbach et al. [Citation23]. For this purpose, we constructed a double ΔfimA-FΔsirA mutant of S. Typhimurium 14028 strain and performed immunoblots against PefA protein, the major Pef fimbriae subunit. Indeed, in the repression mechanism described by Sterzenbach et al. [Citation23], CsrA is required to stabilize the pefACDorf5orf6 mRNA. However, in vitro, the 5ʹUTR of the fimAICDHF transcript cooperates with the CsrB and CsrC sRNA, which are positively regulated by SirA, to antagonize CsrA activity leading to an absence of Pef expression in a wild-type strain (see model ). Consequently, a double ΔfimA-FΔsirA mutant has a marked increase in PefA expression which is related to active CsrA availability in this double mutant that potentiate pefACDorf5orf6 mRNA translation. shows that PefA production was increased in ΔfimA-FΔsirA mutants when bacteria were grown statically in conditions similar to that of Sterzenbach et al. [Citation23], i.e. our control conditions. By contrast, we did not observe this increase when bacteria were grown statically in TSB buffered to pH 5.1, the culture conditions used by Nicholson and Low [Citation21] ().
Figure 1. Absence of CsrA sequestration by the 5ʹUTR of fim mRNA and CsrB/C allows PefA expression but only in specific culture conditions.
Western blot against PefA or Hsp60 (loading control) proteins. S. Typhimurium 14028 wild-type and ΔfimA-FΔsirA mutants were grown statically at 37°C in TSB-MES pH 5.1 (a) in 5 mL of culture medium put in a 50 mL conical tube containing a sterile 200 μL pipet tip or (b) in 20 mL of culture medium put in a 100 mL flask until stationary phase. For PefA results, the time of membrane exposure necessary to obtain the signal is mentioned.
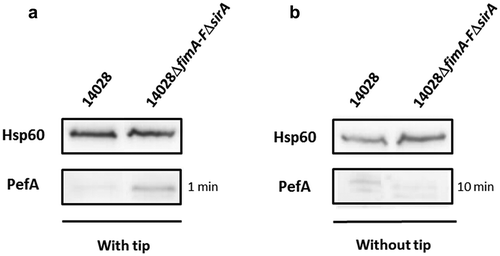
These results are in accordance with a role of the 5ʹUTR of the fimAICDHF transcript and CsrB and CsrC sRNA in preventing PefA protein production via the sequestration of the CsrA protein. However, they also suggest that this repression depends on the culture conditions used. For the next experiments, as Pef fimbriae have been shown to be expressed statically at pH 5.1 by Nicholson and Low [Citation21], which is a relevant environmental condition encountered by Salmonella, we decided to continue our study on the repression of Pef fimbriae expression in vitro in this culture condition. We only kept the culture conditions used by Sterzenbach et al. [Citation23] and Nuccio et al. [Citation22] when we tested an impact of CsrA.
H-NS strongly represses Pef fimbriae expression
The next step of our study was to confirm the repression of Pef fimbriae expression by H-NS. Indeed, as stated in the introduction, several results in the literature suggested a transcriptional repression of the pef operon by H-NS [Citation21,Citation24,Citation40]. We first performed RT-PCR experiments on S. Typhimurium strains grown statically at 37°C in TSB medium buffered to pH 5.1. We chose to measure the mRNA level of pefB, pefA, pefC and orf5 ORFs which are distributed along the pef operon. Moreover, as secondary mutations can occur in Δhns mutants [Citation41], at least 3 independent Δhns mutants were tested in these experiments but also in all the following experiments. A very low level of transcripts was detected in the wild-type strain compared to Δhns mutants for all the ORFs tested. Complementation of the Δhns mutants with pAChns restored the wild-type phenotype (). These results demonstrate that H-NS silences pef operon transcription.
Figure 2. H-NS strongly represses pef mRNA level and PefA protein expression in standing cultures at pH 5.1.
S. Typhimurium 14028 wild-type, Δhns::kan mutants and the complemented strains were grown at 37°C in TSB-MES pH 5.1 under standing conditions. At stationary phase, cells were harvested for RT(reverse transcription)-PCR and Western blot analyses. At least three independent Δhns::kan mutants were tested and the experiment was repeated at least twice. (a) Estimation of pefB, pefA, pefC and orf5 mRNA levels by RT-PCR. PCR were performed on mRNA samples without RT (-) or with RT (+). A representative experiment is presented. (b) Western blot against PefA or Hsp60 (loading control) proteins. For the PefA results, the time of membrane exposure necessary to obtain the signal is mentioned. A typical result obtained with three different Δhns::kan mutants (1 to 3) and three different complemented mutants (a to c) is shown.
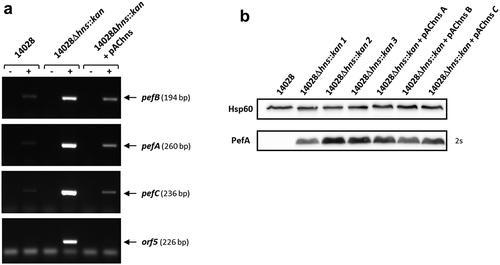
Immunoblots were then performed to check whether this transcriptional regulation had an impact on PefA protein level. As expected, Western blots revealed a greater amount of PefA protein in Δhns mutants compared to the wild-type strain (). It should also be noted that, whatever the samples we tested and the blots we performed, this signal was reproducibly detected after an exposure time of the blots of maximum 2 s for the Δhns mutants while an exposure of about 1 minute was always required to observe PefA expression in ΔfimA-FΔsirA mutants (). This demonstrates a considerably stronger repression of PefA protein expression by H-NS than by the combined action of the 5ʹUTR of the fimAICDHF mRNA and CsrB/CsrC. For unknown reasons and contrary to RT-PCR and β-galactosidase experiments (this paper, see below) and to our previous experiments [Citation42], complementation of the Δhns mutants gave no convincing complementation of PefA protein expression in Western blot experiments (). Therefore, all further immunoblot analyses on Δhns mutants in this study, will not include the complemented strain. Altogether, these results demonstrate that H-NS strongly silences Pef fimbriae expression.
H-NS exerts its repression activity mostly on the PpefB promoter
In order to better characterize the mechanism of pef operon regulation by H-NS, we tried to identify the promoter region(s) regulated by this nucleoprotein. So far, two promoters that initiate pef operon transcription have been described. Putative −10 and −35 RNA polymerase binding sites have been identified upstream of the pefB start codon by Nicholson and Low [Citation21]. These boxes correspond to the transcriptional start site (TSS) recently identified at eight bases downstream of the end of the predicted −10 region and 62 bases upstream of the pefB start codon using differential RNA-seq (J. Hinton, personal communication). The second promoter located upstream of pefA was recently characterized. The TSS of this promoter is located 76 bases upstream of the pefA start codon [Citation23].
Two plasmid-based transcriptional fusions were constructed in the low copy number promoter-probe vector pQF50Cm [Citation43,Citation44] with DNA fragments containing the promoter region upstream of pefB (PpefB) or upstream of pefA (PpefA) and introduced into the wild-type S. Typhimurium 14028 strain. Then Δhns::kan mutation was introduced by transduction in these strains and several mutants harboring each transcriptional fusion were selected for β-galactosidase experiments (). Experiments were performed after bacterial culture at 37°C in TSB-MES pH 5.1 under standing conditions. No significant transcription of the lacZ reporter gene was detected with the empty vector pQF50Cm in this culture condition (data not shown). Both pQF-PpefB and pQF-PpefA transcriptional fusions exhibited a weak or no β-galactosidase activity in the wild-type strain, 17 ± 1.8 and 0.66 ± 0.13 Miller units respectively. By contrast, a high increase of β-galactosidase activity was observed for the pQF-PpefB fusion in the Δhns::kan mutants (565 ± 34 Miller units) and pAChns plasmid was shown to efficiently restore the wild-type phenotype. These experiments demonstrate that H-NS acts as a strong silencer of the PpefB promoter (). Similar results were obtained with the pQF-PpefA::lacZ fusion. The β-galactosidase activities of the pQF-PpefA fusion remained low in Δhns mutants (26.5 ± 2.4 Miller Units), but they were nevertheless 40 times higher in Δhns mutants than in the wild-type strain. As CsrA was shown to induce PefA expression by binding to the 5ʹUTR of the pefACDorf5orf6 transcript [Citation23], we hypothesized that the impact of H-NS on pQF-PpefA fusion could be masked due to the unavailability of active CsrA protein to stabilize lacZ mRNA in our experiments. To test this hypothesis, we measured the β-galactosidase activity of this fusion in ΔhnsΔfimA-FΔsirA mutants. No increase of β-galactosidase activities of the pQF-PpefA fusion was observed in ΔhnsΔfimA-FΔsirA triple mutants compared to Δhns mutants neither after bacterial growth in TSB-MES pH 5.1 under standing conditions (data not shown) nor after growth in Sterzenbach et al. culture conditions [Citation23] (). These results demonstrate that the low β-galactosidase activity of the pQF-PpefA fusion in the Δhns mutants is not due to CsrA sequestration by fim operon and CsrB and CsrC sRNA. Together, these results are in favor of a regulation of PpefA promoter by H-NS, even if our experiments were probably not performed in the best environmental conditions for observing this regulation.
Figure 3. The promoter upstream of pefB is the main target of H-NS.
(a) Diagram representing the PpefB and PpefA promoters of the S. Typhimurium pef operon and the DNA fragments cloned upstream of the lacZ gene used as a transcriptional reporter. (b and c) β–galactosidase activities expressed by pQF-PpefA or pQF-PpefB transcriptional fusions in S. Typhimurium 14028 wild-type strain or its derivatives after growth until stationary phase at 37°C in TSB-MES pH 5.1 under standing conditions without a sterile pipette tip (b) and in presence of a sterile pipette tip (c). Aliquots were collected for measurements of the OD600nm and β–galactosidase activity. β-galactosidase activities are expressed in Miller units. Average values (± standard error of the mean) of activity units were calculated based on at least three independent assays performed in duplicates for each strain. For Δhns::kan strains, several mutants were tested in each assay and results presented are the mean of the data obtained for all these mutants. (d) Western blot against PefA or Hsp60 proteins (loading control) prepared from S. Typhimurium strains grown at 37°C until stationary phase in TSB-MES pH 5.1 under standing conditions. For the PefA results, the time of membrane exposure necessary to obtain the signal is mentioned. At least three independent Δhns::kan and Δhns::kanΔPpefB mutants were tested and a representative result is shown.
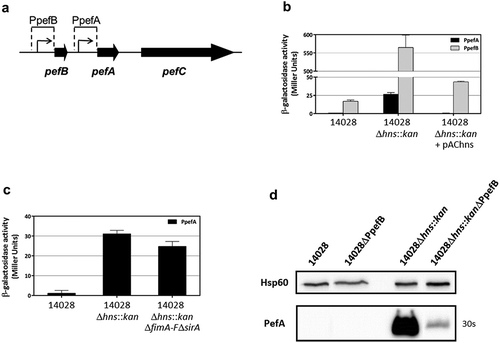
To confirm the predominant role of the PpefB promoter in the regulation of PefA expression by H-NS, we deleted the PpefB promoter and performed immunoblot analyses. A decrease of PefA expression was observed in the ΔhnsΔPpefB mutants compared to the Δhns mutant strains, demonstrating that the silencing of PefA by H-NS is essentially mediated by the PpefB promoter in our culture conditions (). Altogether, these results demonstrate that the main target of H-NS involved in the negative regulation of Pef fimbriae expression is the promoter region located upstream of pefB at least when bacteria are cultured in TSB-MES pH 5.1 under standing conditions. However, we have to consider that PpefB is a stronger promoter than PpefA in our culture conditions. We therefore cannot exclude a predominant role of the PpefA promoter in the regulation of Pef fimbriae by H-NS in different culture conditions.
Hha and YdgT negatively regulate PefA expression but at a lower level than H-NS
In addition to H-NS, the nucleoproteins Hha and YdgT have also been suggested to be involved in the repression of Pef fimbriae expression in S. Typhimurium [Citation25]. To test this hypothesis, single Δhha or ΔydgT mutants and a double ΔhhaΔydgT mutant were constructed. PefA expression was detected only in the double mutant by immunoblot analyses and neither in the wild-type strain nor in the single mutants (). The wild-type phenotype was completely restored by complementation of the double ΔhhaΔydgT mutant with the pAChha-ydgT plasmid. These results were confirmed by RT-PCR experiments on the double ΔhhaΔydgT mutant which exhibited an increase of pefB, pefA, pefC and orf5 mRNA level compared to the wild-type or the complemented strain (). These results demonstrate that Hha and YdgT negatively regulate PefA expression and that the simultaneous absence of Hha and YdgT is necessary to relieve this repression.
Figure 4. Hha and YdgT negatively regulate PefA expression by acting on the pefB promoter.
S. Typhimurium 14028 and its derivative strains were grown statically at 37°C in TSB-MES pH 5.1. At stationary phase, cells were harvested for Western blot analysis, RT-PCR and β-galactosidase activity measurement. At least three independent assays were performed for each experiment. (a) Western blot against PefA or Hsp60 proteins (loading control) prepared from S. Typhimurium wild-type, Δhha and ΔydgT single mutants, ΔhhaΔydgT double mutant and its complemented strain. For PefA results, the time of membrane exposure necessary to obtain the signal is mentioned. (b) Estimation of pefB, pefA, pefC and orf5 mRNA levels by RT-PCR. PCR were performed on mRNA samples without RT (-) or with RT (+). (c) β–galactosidase activities expressed by pQF-PpefA and pQF-PpefB transcriptional fusions in S. Typhimurium wild-type, ΔhhaΔydgT double mutant and its complemented strain. β-galactosidase activities are expressed in Miller units. Average values (± standard error of the mean) of activity are presented. (d) Western blot against PefA or Hsp60 proteins (loading control) prepared from S. Typhimurium wild-type, ΔPpefB, ΔhhaΔydgT double mutant and the ΔhhaΔydgTΔPpefB triple mutant. For PefA results, the time of membrane exposure necessary to obtain the signal is mentioned.
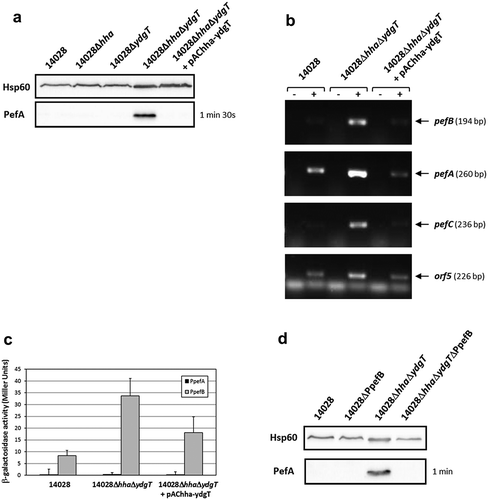
Determination of the promoter(s) targeted by Hha and YdgT was performed using the transcriptional fusions pQF-PpefA and pQF-PpefB and after bacterial culture at 37°C in TSB-MES pH 5.1 under standing conditions. No transcriptional activity was observed for the PpefA promoter whatever the strain tested, while a low β-galactosidase activity (34 ± 7.4 Miller units), partially complemented by pAChha-ydgT, was detected for the PpefB promoter in the double ΔhhaΔydgT mutant (). Moreover, deletion of the PpefB promoter in the ΔhhaΔydgT mutant abolished the expression of PefA protein as shown by immunoblot analysis (). Taken together, these results demonstrate that Hha and YdgT negatively regulate PefA expression by acting, as for H-NS, on the PpefB promoter. However, this repression was much weaker than the one exerted by H-NS as the transcriptional fusion PpefB::lacZ displayed a reporter gene expression about 17-fold lower in a double ΔhhaΔydgT mutant than in a Δhns mutant. Moreover, acquisition times used for detection of PefA protein on Western blots were always much higher for the double ΔhhaΔydgT mutant than for the Δhns mutant (in the minute range versus in the second range respectively).
Higher PefA expression is observed at pH 7.1 than pH 5.1 in Δhns or ΔhhaΔydgT or ΔfimA-FΔsirA mutants
All our previous experiments were performed in culture conditions described in the literature as optimal for the expression of Pef fimbriae, i.e. under standing conditions at pH 5.1 [Citation21]. However, during an experiment designed to study the expression of another target of the Hha/YdgT regulon, we observed an expression of PefA after bacterial culture at pH 7.1 suggesting that an expression of this protein can be detected in culture conditions other than pH 5.1 without agitation. We, thus, decided to study the impact of agitation and pH on pef expression in Δhns, ΔhhaΔydgT and ΔfimA-FΔsirA mutants. Bacteria were grown at pH 5.1 or 7.1 with or without shaking. PefA was detected by immunoblot in Δhns mutants in all the conditions tested but at a higher level in standing cultures than after bacterial growth under shaking conditions ()). Moreover, the signal detected for PefA was always more intense at pH 7.1 than at pH 5.1. The measure of the transcriptional activity of the PpefB promoter in Δhns mutants in these four culture conditions confirmed these results. Standing cultures allowed a higher expression of lacZ reporter gene than shaking cultures (). Moreover, in agreement with immunoblot results, the β-galactosidase activities measured for pQF-PpefB fusion (1287 ± 60 Miller units for standing culture and 198 ± 19 Miller units for shaking culture) were always higher at pH 7.1 than at pH 5.1 (565 ± 34 Miller units for standing culture, 72 ± 8 Miller units for shaking culture). The wild-type strain displayed weak β-galactosidase activities, the highest being also those measured in standing conditions at pH 7.1 (40 ± 1.7 Miller units).
Figure 5. Higher PefA expression at pH 7.1 than pH 5.1 in Δhns, ΔhhaΔydgT or ΔfimA-FΔsirA mutants.
S. Typhimurium 14028 and isogenic mutants were grown at 37°C in TSB-MES pH 5.1 or pH 7.1 under standing or shaking conditions. At stationary phase, cells were harvested for Western blot analysis and β-galactosidase activity measurement. (a) Western blot against PefA or Hsp60 proteins (loading control) prepared from wild-type strain and several Δhns::kan mutants. A typical result is presented for each condition tested. For PefA results, the time of membrane exposure necessary to obtain the signal is mentioned. (b) β–galactosidase activities expressed by the transcriptional fusion pQF-PpefB in S. Typhimurium 14028 wild-type, Δhns::kan mutant and its complemented strain. Activities are expressed in Miller units. Average values (± standard error of the mean) of activity were calculated based on at least three independent assays. For Δhns::kan strain, several mutants were tested in each assay. (c) Western blot against PefA or Hsp60 proteins (loading control) prepared from wild-type strain and ΔhhaΔydgT double mutant. For the PefA results, the time of membrane exposure necessary to obtain the signal is mentioned. (d) β–galactosidase activity expressed by the transcriptional fusion pQF-PpefB in S. Typhimurium 14028 wild-type, ΔhhaΔydgT double mutant and its complemented strain. Activities are expressed in Miller units. Average values (± standard error of the mean) of activity were calculated based on at least three independent assays. (e) Western blot against PefA or Hsp60 proteins (loading control) prepared from wild-type strain and ΔfimA-FΔsirA double mutant. Bacteria were grown with or without a sterile pipette tip added in the culture medium to mimick the conditions used by Sterzenbach et al. who demonstrated the role of fim and sirA in the regulation of Pef expression [Citation23]. For the PefA results, the time of membrane exposure necessary to obtain the signal is mentioned.
![Figure 5. Higher PefA expression at pH 7.1 than pH 5.1 in Δhns, ΔhhaΔydgT or ΔfimA-FΔsirA mutants.S. Typhimurium 14028 and isogenic mutants were grown at 37°C in TSB-MES pH 5.1 or pH 7.1 under standing or shaking conditions. At stationary phase, cells were harvested for Western blot analysis and β-galactosidase activity measurement. (a) Western blot against PefA or Hsp60 proteins (loading control) prepared from wild-type strain and several Δhns::kan mutants. A typical result is presented for each condition tested. For PefA results, the time of membrane exposure necessary to obtain the signal is mentioned. (b) β–galactosidase activities expressed by the transcriptional fusion pQF-PpefB in S. Typhimurium 14028 wild-type, Δhns::kan mutant and its complemented strain. Activities are expressed in Miller units. Average values (± standard error of the mean) of activity were calculated based on at least three independent assays. For Δhns::kan strain, several mutants were tested in each assay. (c) Western blot against PefA or Hsp60 proteins (loading control) prepared from wild-type strain and ΔhhaΔydgT double mutant. For the PefA results, the time of membrane exposure necessary to obtain the signal is mentioned. (d) β–galactosidase activity expressed by the transcriptional fusion pQF-PpefB in S. Typhimurium 14028 wild-type, ΔhhaΔydgT double mutant and its complemented strain. Activities are expressed in Miller units. Average values (± standard error of the mean) of activity were calculated based on at least three independent assays. (e) Western blot against PefA or Hsp60 proteins (loading control) prepared from wild-type strain and ΔfimA-FΔsirA double mutant. Bacteria were grown with or without a sterile pipette tip added in the culture medium to mimick the conditions used by Sterzenbach et al. who demonstrated the role of fim and sirA in the regulation of Pef expression [Citation23]. For the PefA results, the time of membrane exposure necessary to obtain the signal is mentioned.](/cms/asset/8058c9c5-e5e8-4b81-96ce-0a87f5775275/kvir_a_1682752_f0005_b.gif)
Similar experiments were carried out on the double ΔhhaΔydgT mutant and the results were generally consistent with those obtained for the Δhns mutants. More precisely, PefA was never detected in shaking cultures even after an immunoblot exposure of several minutes whereas, in standing cultures, PefA was more expressed at pH 7.1 than pH 5.1 ( and data not shown). Moreover, as previously observed at pH 5.1 in standing cultures, PefA expression was always lower in the ΔhhaΔydgT double mutant than in Δhns mutants, confirming the highest repression activity of H-NS compared to Hha and YdgT. β-galactosidase experiments on the PpefB promoter in the ΔhhaΔydgT double mutant gave results fully consistent with immunoblot analyses except when bacteria were cultured at pH 7.1 in shaking conditions (). In this latter condition, the highest β-galactosidase activity was detected in the ΔhhaΔydgT mutant for the pQF-PpefB fusion (107 ± 11 Miller units) whereas PefA protein was not detected in this culture condition (,)).
For ΔfimA-FΔsirA mutants, the highest expression of PefA protein was detected after bacterial culture under standing conditions at pH 7.1 and with a tip. Moreover, at this pH, PefA was detected even when bacteria were statically cultivated without a tip. This was not the case at pH 5.1 (). As for ΔhhaΔydgT mutants, no PefA expression was detected after bacterial culture in shaking conditions (data not shown).
In conclusion, these results confirm the importance of the culture conditions for the expression of Pef fimbriae and demonstrate a repression of Pef expression by H-NS in all the culture conditions tested and by Hha/YdgT and CsrA at least in standing culture.
Deletion of hha and ydgT in an hns mutant represses PefA expression more than in a single hns mutant
The nucleoproteins H-NS and, to a lesser extent, Hha-YdgT, are involved in the silencing of Pef fimbriae expression. Subsequently, one of the key questions is whether Hha-YdgT act in synergy with H-NS as it has been shown in vitro that Hha can improve H-NS binding to target regions [Citation37]. We reasoned that if Hha and YdgT repress pef transcription only through H-NS, then the deletion of hha and ydgT in a Δhns mutant will have no greater impact on pef transcription and PefA expression than the impact of hns deletion. We measured the transcriptional activity of the PpefB promoter region in a Δhns::kanΔhhaΔydgT triple mutant grown in standing cultures at pH 5.1 and pH 7.1, which correspond to the culture conditions in which a repression was observed in Δhns and ΔhhaΔydgT mutants (). At pH 7.1, the activity of the pQF-PpefB transcriptional fusion in the Δhns::kanΔhhaΔydgT triple mutant was about 2-fold higher than in the Δhns mutant (2419 ± 275 versus 1287 ± 60 Miller units) whereas at pH 5.1, the activity in the triple mutant was 4.3-fold higher than in the Δhns mutant (2460 ± 349 versus 565 ± 34 Miller units) () and ). Complementation with pAChns plasmid, contrary to pAChha-ydgT, confirmed that H-NS is the main regulator on this promoter region as the transcriptional activity returned to a very low level in the complemented strain. Then we investigated the expression of PefA in the ΔhnsΔhhaΔydgT triple mutant grown in similar culture conditions. Expression of PefA was increased 4.3-fold in the triple mutant compared to the Δhns single mutant when bacteria were grown at pH 5.1 whereas both the triple and the Δhns mutant displayed a similar level of PefA expression at pH 7.1 (1.5-fold higher in the triple mutant compared to the Δhns mutant) (). These results are consistent with β-galactosidase experiments which showed a greater increase of the PpefB promoter activity in the triple mutant compared to the Δhns mutant at pH 5.1 than pH 7.1 ().
Figure 6. Deletion of hha and ydgT in a Δhns::kan mutant has a more pronounced effect on PefA expression at pH 5.1 than at pH 7.1.
(a) β–galactosidase activity expressed by the transcriptional fusion pQF-PpefB in S. Typhimurium Δhns::kanΔhhaΔydgT triple mutant and its complemented strains after growth in TSB-MES pH 5.1 or pH 7.1 at 37°C under standing conditions. In stationary phase, aliquots were collected for measurements of the OD600nm and β–galactosidase activity. β-galactosidase activities are expressed in Miller units. Average values (± standard error of the mean) of activity units were calculated based on at least three independent assays. For Δhns::kan and Δhns::kanΔhhaΔydgT strains, several mutants were tested in each assay. (b) Western blot against PefA or Hsp60 proteins (loading control) prepared from S. Typhimurium 14028 wild-type, ΔhhaΔydgT, Δhns::kan or Δhns::kanΔhhaΔydgT mutants grown statically at 37°C until stationary phase in TSB-MES pH 5.1 or pH 7.1. Several Δhns::kan mutants and Δhns::kanΔhhaΔydgT triple mutants were tested in each experiment. A typical result is presented for each culture condition tested. For the PefA results, the time of membrane exposure necessary to obtain the signal is mentioned. Relative intensities for PefA bands calculated using normalization with the loading control Hsp60 are noted just under the immunoblot exposed for 4 s and for which no signal saturation was detected.
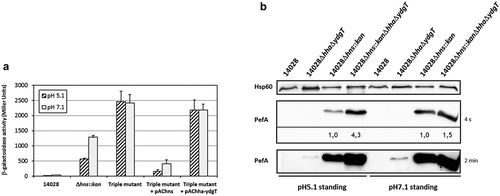
Altogether, these results suggest a negative regulation of PefA expression by Hha-YdgT independently of H-NS in the ΔhnsΔhhaΔydgT triple mutant at pH 5.1. As StpA, the paralog of H-NS, is expressed at high levels in the absence of H-NS [Citation24,Citation40], we tested whether StpA could be responsible for the higher expression of PefA in the triple mutant compared to the Δhns mutant. Unfortunately, we were not able to construct a ΔhnsΔstpA double mutant. As we succeeded in constructing a ΔstpA mutant and in deleting hns in a stpA mutant complemented with stpA, our results strongly suggest that the deletion of both hns and stpA is lethal for Salmonella. Consequently, we cannot rule out a role of StpA in regulating Pef fimbriae expression in an hns background and on its involvement with Hha-YdgT in the higher expression of PefA in the ΔhnsΔhhaΔydgT triple mutant after bacterial culture at pH 5.1.
StpA represses Pef fimbriae expression by acting on the PpefB promoter
While StpA is highly expressed during early and middle exponential phase, expression of StpA is minimal and low during late stationary phase which corresponds to the culture conditions used in our experiments [Citation29,Citation45]. This is why we initially did not test the H-NS paralog. However, when we tried to explain the higher expression of PefA in the ΔhnsΔhhaΔydgT triple mutant at pH 5.1, we surprisingly observed that deletion of stpA in S. Typhimurium 14028 strain led to an increase of PefA expression and decided to study this phenotype. As for Δhns and ΔhhaΔydgT mutants, RT-PCR experiments performed on S. Typhimurium strains grown statically at 37°C in TSB medium buffered to pH 5.1 showed a higher level of transcripts in the ΔstpA mutant than in the wild-type strain for the pefB, pefA, pefC and orf5 ORFs. Complementation of the ΔstpA mutant with pACstpA restored the wild-type phenotype (). Moreover, a greater amount of PefA protein was detected by immunoblots in ΔstpA mutants compared to the wild-type strain or the complemented strain (), thus confirming the RT-PCR results. Intriguingly, the exposure times needed to obtain a signal for ΔstpA mutants were quite variable depending on the mutant used and on the experiment (between a few seconds up to a few minutes) unlike those obtained for Δhns and ΔhhaΔydgT mutants that were highly reproducible. Nevertheless, the exposure times of the blots were always higher than that obtained for Δhns mutants. Then, using the transcriptional fusions pQF-PpefA and pQF-PpefB and by deleting the PpefB promoter in the ΔstpA background, we demonstrated that PpefB is the main promoter targeted by StpA, as previously shown for H-NS and Hha/YdgT. Indeed, a moderate β-galactosidase activity (89 ± 12 Miller units), partially complemented by pACstpA, was detected for the PpefB promoter in the ΔstpA mutant, while no transcriptional activity was observed for the PpefA promoter whatever the strain tested (). Moreover, immunoblot experiments showed that deletion of the PpefB promoter abolished PefA expression in the ΔstpA mutant (). Altogether, these results demonstrate that StpA silences pef operon transcription on the promoter located upstream of pefB when bacteria are cultivated at acidic pH 5.1 until late stationary phase.
Figure 7. StpA silences PefA expression by acting on the pefB promoter.
S. Typhimurium 14028 and its derivative strains were grown statically at 37°C in TSB-MES pH 5.1 (A to D) or at 37°C in TSB-MES pH 5.1 or pH 7.1 under standing or shaking conditions (e and f). At stationary phase, cells were harvested for RT-PCR, Western blot analysis and β-galactosidase activity measurement. At least three independent assays were performed for each experiment. (a) Estimation of pefB, pefA, pefC and orf5 mRNA levels by RT-PCR. PCR were performed on mRNA samples without RT (-) or with RT (+) and prepared from S. Typhimurium wild-type, ΔstpA::kan mutant and its complemented strain. (b) Western blot against PefA or Hsp60 proteins (loading control). For the PefA results, the time of membrane exposure necessary to obtain the signal is mentioned. (c) β–galactosidase activities expressed by pQF-PpefA and pQF-PpefB transcriptional fusions in S. Typhimurium wild-type, ΔstpA::kan mutant and its complemented strain. β-galactosidase activities are expressed in Miller units. Average values (± standard error of the mean) of activity are presented. (d) Western blot against PefA or Hsp60 proteins (loading control) prepared from S. Typhimurium wild-type, ΔPpefB, ΔstpA::kan mutant and the ΔstpA::kanΔPpefB double mutant. For PefA results, the time of membrane exposure necessary to obtain the signal is mentioned. (e) Western blot against PefA or Hsp60 proteins (loading control) prepared from wild-type strain and ΔstpA::kan mutant. For the PefA results, the time of membrane exposure necessary to obtain the signal is mentioned. (f) β–galactosidase activity expressed by the transcriptional fusion pQF-PpefB in S. Typhimurium 14028 wild-type, ΔstpA::kan mutant and its complemented strain. Activities are expressed in Miller units. Average values (± standard error of the mean) of activity were calculated based on at least three independent assays.
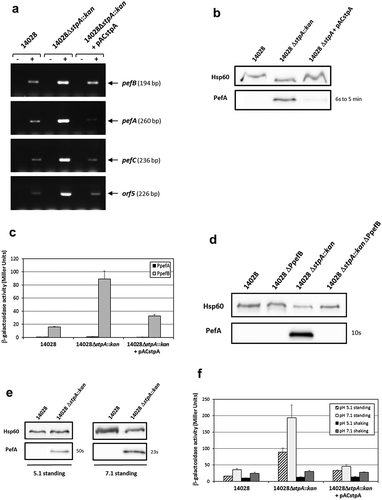
Finally, we tested the impact of the agitation and pH on pef expression in stpA mutants. As observed for the double ΔhhaΔydgT mutant, (i) PefA expression was detected only in standing culture ( and data not shown for shaking cultures), (ii) a higher expression of PefA was detected at pH 7.1 than at pH 5.1 () and (iii) a higher β-galactosidase activity for pQF-PpefB fusion (193 ± 39 Miller units for standing culture and 30 ± 3.1 Miller units for shaking culture) was measured at pH 7.1 than at pH 5.1 (89 ± 12 Miller units for standing culture and 13 ± 0.9 Miller units for shaking culture) (). However, whatever the culture conditions used, the silencing of Pef by StpA was always lower than that of H-NS.
Discussion
Fimbrial expression in vivo but not in vitro suggests a fine-tuned regulation according to the environment encountered by bacteria allowing them to express these complex structures at their surface only when required and thus to optimize virulence against fitness costs. In Salmonella, several genes/proteins have been implicated in the repression of fimbrial expression but the mechanisms of regulation remain unknown [Citation21,Citation46] except in the case of the post-transcriptional control of Pef fimbriae expression via CsrA protein [Citation23]. Analysis of published high throughput transcriptomic data suggested that at least eight of the thirteen fimbrial operons present in the genome of S. Typhimurium, including pef operon, were negatively regulated by H-NS [Citation24,Citation40]. On the other hand, the pef operon is one of the few fimbrial operons which seemed regulated by Hha-YdgT [Citation25]. Our results demonstrate that H-NS strongly silences the pef operon transcription at both the PpefB and the PpefA promoters, the two promoters of the pef operon currently identified. In our culture conditions, i.e. bacterial culture at pH 5.1 and in standing conditions, the main impact occurs on the transcription initiated at the PpefB promoter. The environmental conditions and the regulatory proteins favoring the transcription from the PpefA promoter are currently unknown. We also show that the PpefB promoter, but not the PpefA promoter, is silenced by Hha-YdgT and StpA, the paralog of H-NS, but these nucleoproteins affect pef operon transcription less than H-NS. The use of a double ΔhhaΔydgT mutant was necessary to obtain this phenotype, which is consistent with the already described redundancy of these proteins [Citation25,Citation36,Citation47]. Concerning StpA, our results are the first demonstration of StpA gene silencing after bacterial growth in late stationary phase, i.e. a growth phase where StpA is expressed at a low level in Salmonella but also in other bacteria [Citation45,Citation48,Citation49]. Indeed, Lucchini et al. [Citation29] in Salmonella, but also Müller et al. [Citation50] in E. coli for example, did not observe any altered gene expression in a stpA mutant compared to the wild-type strain in late stationary phase using DNA arrays. Our results cannot be attributed to a reduced expression of H-NS in the stpA mutant as no significant StpA-dependent changes in hns expression was observed at any S. Typhimurium growth time point [Citation29] in accordance with other transcriptomic studies in Shigella flexneri or E. coli [Citation48,Citation50]. pef ORFs seem not to be the only fimbriae related genes regulated by StpA as std, saf, fim, bcf and stf genes are over-expressed in a stpA mutant after Salmonella growth until exponential phase [Citation29]. The absence of the pef operon in the list of the StpA-dependent genes of this study could be due to the culture conditions used which were quite different from those considered optimal for Pef fimbriae expression [Citation21]. In conclusion, in the culture conditions we used, i.e. bacterial culture at 37°C in TSB-MES pH 5.1 under standing conditions, the absence of Pef expression in vitro is mostly due to a strong silencing by H-NS on the PpefB promoter. StpA and Hha/YdgT also repress pef transcription at PpefB promoter. Moreover, Pef expression is also modulated through a weak post-transcriptional mechanism involving the stabilizing effect of CsrA on the pefACDorf5orf6 transcript. A model for in vitro pef regulation summarizing all the results obtained in this study as well as those described for the regulatory protein CsrA is presented .
Figure 8. Model for in vitro pef regulation in S. Typhimurium.
The six ORFs belonging to the pef operon are represented by black arrows. The two promoters identified upstream of pefB and pefA are indicated by red broken arrows. The promoter upstream of pefA is repressed by H-NS and the RNA binding protein CsrA stabilizes the pefACDorf5orf6 transcript. The activity of CsrA is antagonized by the 5ʹ-UTR of the fimA-F transcript and the two small RNAs named CsrB and CsrC [Citation23]. The promoter upstream of pefB is strongly silenced by H-NS and weakly by StpA and Hha-YdgT. The two gray double arrows between H-NS and Hha-YdgT indicate that, at pH 7.1 Hha-YdgT act through H-NS to modulate pef expression, contrary to pH 5.1 where Hha-YdgT seem able to negatively regulate pef expression independently of H-NS. The single dashed double arrow with a question mark connecting StpA and Hha-YdgT signifies a possible interaction at pH 5.1 between these three NAPs to repress pef expression.
![Figure 8. Model for in vitro pef regulation in S. Typhimurium.The six ORFs belonging to the pef operon are represented by black arrows. The two promoters identified upstream of pefB and pefA are indicated by red broken arrows. The promoter upstream of pefA is repressed by H-NS and the RNA binding protein CsrA stabilizes the pefACDorf5orf6 transcript. The activity of CsrA is antagonized by the 5ʹ-UTR of the fimA-F transcript and the two small RNAs named CsrB and CsrC [Citation23]. The promoter upstream of pefB is strongly silenced by H-NS and weakly by StpA and Hha-YdgT. The two gray double arrows between H-NS and Hha-YdgT indicate that, at pH 7.1 Hha-YdgT act through H-NS to modulate pef expression, contrary to pH 5.1 where Hha-YdgT seem able to negatively regulate pef expression independently of H-NS. The single dashed double arrow with a question mark connecting StpA and Hha-YdgT signifies a possible interaction at pH 5.1 between these three NAPs to repress pef expression.](/cms/asset/52d77fcd-d108-4817-b6b4-3c4b2b2a1bb4/kvir_a_1682752_f0008_oc.jpg)
H-NS, StpA, Hha and YdgT silence genes by affecting transcription initiation, elongation and termination. Initiation is blocked by inhibiting the RNA polymerase binding to promoters and by blocking promoter escape [Citation51]. H-NS and StpA form homodimers and interact together to form heterodimers. Both homodimers and heterodimers assemble in large multimers that interact with DNA. Both can also associate with Hha and YdgT [Citation52]. Experimental evidence and a mechanistic model recently described indicate that Hha and YdgT act primarily through H-NS and StpA to modulate gene expression [Citation37,Citation53]. In this model, Hha directly interacts with the N-terminal dimerization domain of H-NS or StpA and interaction of positively charged residues of Hha with DNA facilitates the formation of repressive nucleoprotein-DNA complexes. Recently, Hha-YdgT have been shown to favor H-NS DNA-bridging activity, thanks to conformational changes of H-NS from a close to an open state [Citation54]. On the contrary, few reports show that Hha binds to specific regulatory sequences independently of H-NS, although Hha has no DNA binding domain [Citation38,Citation39,Citation55]. A few other papers report a Hha direct regulation but careful interpretation should be done since, in these studies, Hha protein purification was performed in wild-type E. coli and thus a co-purification of Hha with H-NS and/or StpA could have occurred [Citation56,Citation57]. Our results with the ΔhnsΔhhaΔydgT triple mutant strongly suggest that Hha and YdgT act independently of H-NS to modulate pef expression when bacteria are grown statically at pH 5.1 since an increase of PefA expression and a higher PpefB transcriptional activity have been observed in the triple mutant compared to the Δhns mutant in this culture condition. As Hha-YdgT do not complement the ΔhnsΔhhaΔydgT triple mutant in β-galactosidase assays, this strongly suggest an indirect activity of Hha-YdgT in this mutant. As StpA represses pef operon, the most probable hypothesis is that StpA in conjunction with Hha and YdgT silences pef transcription in the hns mutant. We tried to test whether the higher expression of PefA in the ΔhnsΔhhaΔydgT triple mutant compared to the hns mutant could be due to StpA silencing through Hha/YdgT in the Δhns mutant by constructing a ΔhnsΔstpA mutant. However, we never succeeded in constructing a double ΔhnsΔstpA mutant while we had no problem to transduce a hns::kan mutation in an stpA mutant complemented with stpA. The ΔhnsΔstpA double mutation is thus most probably lethal in Salmonella. In E. coli, ΔhnsΔstpA double mutants have been studied but a severe growth defect has been reported [Citation58–Citation60]. Consequently, we cannot completely rule out the hypothesis that Hha-YdgT act indirectly through another unknown regulatory protein. A recent work identified 120 genes regulated by Hha independently of H-NS/StpA [Citation39] among which two fimbrial genes belonging to sth and stj operons, but not the pef operon. However, bacteria were grown in neutral medium in this study and consequently not in conditions optimal for Pef expression. Further studies are thus necessary to understand the mechanisms used by Hha and YdgT to regulate pef operon transcription.
While the precise mechanism of pef fimbriae regulation by Hha and YdgT remains to be determined, the silencing by H-NS most certainly involves a direct interaction of H-NS with the promoter regions upstream of pefB and pefA since a chromatin immunoprecipitation-on-chip analysis identified an H-NS binding site upstream of both ORFs [Citation24]. Moreover, a search for sequences similar to the H-NS DNA binding motif determined in E. coli [Citation61] allowed us to identify one putative H-NS high-affinity binding site in the promoter regions of both pefB and pefA (). They are located approximately 100 bp upstream of the transcriptional start sites of both promoters. After binding of H-NS, the high AT richness of these regions, especially the one upstream of pefB, will then favor the oligomerization of H-NS along this DNA segment. Future work will include interaction and biophysical studies to verify our hypothesis concerning the ability of Hha-YdgT to regulate the PpefB promoter both through H-NS and/or StpA or independently of these NAPs according to the pH. Electrophoretic mobility shift assays, co-crystallization experiments with H-NS, StpA and/or Hha and their DNA target(s), and amino-acid mutagenesis will help clarify how Hha interacts with DNA according to the H-NS/StpA dependent or independent mechanism.
Figure 9. H-NS predicted sites and AT content of the pef operon.
The upper part of the figure shows the AT percentage along the nucleotide sequence of the pef operon. A schematic diagram of the pef operon, with the H-NS predicted sites symbolized by black triangles, is represented in the lower part of the figure. The AT content was calculated using a window length of 25 bp. Potential H-NS binding sites in pef operon were searched using the Virtual Footprint software [Citation72] and the H-NS position weight matrix from E. coli [Citation61]. Only the sites with a score greater than 5.5 are indicated, while the maximum score for this matrix is 6.73. The experimentally identified PpefB and PpefA promoters are indicated by broken arrows.
![Figure 9. H-NS predicted sites and AT content of the pef operon.The upper part of the figure shows the AT percentage along the nucleotide sequence of the pef operon. A schematic diagram of the pef operon, with the H-NS predicted sites symbolized by black triangles, is represented in the lower part of the figure. The AT content was calculated using a window length of 25 bp. Potential H-NS binding sites in pef operon were searched using the Virtual Footprint software [Citation72] and the H-NS position weight matrix from E. coli [Citation61]. Only the sites with a score greater than 5.5 are indicated, while the maximum score for this matrix is 6.73. The experimentally identified PpefB and PpefA promoters are indicated by broken arrows.](/cms/asset/6060bbef-d1e7-4e92-ab12-ee5b3e384003/kvir_a_1682752_f0009_b.gif)
Adhesion to intestinal epithelium is an essential step for the colonization of the small intestine by Salmonella as bacteria must be in close contact with host cells to successfully invade them. Fimbrial adhesins represent one adhesion system responsible for this initial step of interaction between Salmonella and epithelial cells. As for most of Salmonella fimbriae, standard laboratory culture conditions do not allow the expression of Pef fimbriae. However, a study seeking for culture conditions able to induce Pef biosynthesis showed that maximal expression was obtained after growth of a S. Typhimurium wild-type strain at 37°C in rich acidic medium (LB broth buffered at pH 5.1 with MES) under standing culture conditions for 2 days [Citation21]. In these conditions, Pef fimbriae were readily detected by immunofluorescence analysis in S. Typhimurium, as well as pefA and pefB transcripts. Moreover, PefA protein could be detected at a low level by flow cytometry in S. Typhimurium grown in similar conditions [Citation8]. However, PefA subunit was not detectable by Western blot in S. Typhimurium wild-type strain cultivated in these conditions [Citation7,Citation22]. In agreement with these studies, we were unable to detect PefA by Western blotting in the wild-type strain whatever the growth conditions used. Our results with Δhns and ΔhhaΔydgT mutants also agree with the importance of standing cultures for the expression of Pef fimbriae. In contrast, in our hands, a rich medium buffered at pH 7.1 induced a stronger Pef expression than a rich acidic medium. Indeed, expression of PefA measured by Western blotting and transcriptional activity of the PpefB promoter was higher in Δhns, ΔhhaΔydgT, ΔfimA-FΔsirA or ΔstpA mutants grown statically at 37°C in rich neutral medium (TSB broth buffered at pH 7.1 with MES) for 18 h. These contradictory results regarding optimum pH for Pef expression could be explained by different culture conditions. The main differences were the incubation time which lasted 2 days in the literature and 18 h in our experiments, and/or the presence of a pipette tip in the medium [Citation21,Citation22]. Another hypothesis would be that, in the wild-type strain, H-NS represses Pef expression more strongly at pH 7.1 than pH 5.1. More studies are required to decipher these expression discrepancies according to the culture conditions.
Among the many fimbrial adhesins of S. Typhimurium, the fimbriae Pef are involved in biofilm formation on chicken intestinal epithelium and adhesion to murine small intestine [Citation6,Citation17]. A glycomic study showed that Pef fimbriae specifically binds the Lewis X blood group antigen which is expressed mainly by crypt epithelial cells, suggesting a possible role for Pef fimbriae in adhesion of Salmonella to human crypt epithelium [Citation62]. The growth conditions determined as optimum in our study for expression of PefA in Δhns, ΔstpA or ΔhhaΔydgT mutants can be considered as physiologically relevant since human gastrointestinal pH range from 6.6 in the jejunum to 7.5 in the ileum and oxygen level is low in the intestine [Citation63,Citation64]. Since most fimbrial operons of S. Typhimurium seem negatively regulated by one or several NAPs, this study could be extended to all the fimbrial operons in order to compare their mechanism of negative regulation. Finally, the identification of the transcriptional activator which relieves in vivo the silencing exerted by H-NS, StpA and Hha-YdgT would make it possible to better understand the role of the Pef fimbriae in animals.
Experimental procedures
Bacterial strains and culture conditions
Bacterial strains used in this study are listed in . S. Typhimurium 14028 is a virulent strain isolated from chicken tissues. E. coli MC1061 was used for cloning procedures. Strains were routinely grown in LB (10 g/L bactotryptone, 5 g/L yeast extract, 10 g/L NaCl) or TSB medium (BD Difco, No 211825) at 37°C with agitation. For pef operon transcription and PefA expression studies, S. Typhimurium strains were cultivated under standing or shaking conditions in 20 mL of TSB medium buffered to pH 5.1 or 7.1 with 100 mM MES [2-(N-morpholino)ethanesulfonic acid] and put in a 100 mL flask. When we used culture conditions similar to those of Nuccio et al. and Sterzenbach et al. [Citation22,Citation23], strains were grown in 5 mL TSB medium buffered to pH 5.1 or 7.1 with 100 mM MES in a 50 mL conical tube where a sterile 200 μL pipette tip was added. When necessary, antibiotics were added to the culture medium at the following concentrations: carbenicillin 100 μg/mL, kanamycin 50 μg/mL, chloramphenicol 34 μg/mL.
Table 1. Strains and plasmids used in this study.
Cloning procedures and mutant construction
Plasmids used in this work are listed in . Promoters identified upstream of pefB or pefA were inserted upstream of promoterless lacZ gene in pQF50Cm in order to construct plasmid-based transcriptional fusions (). The fragments were amplified from S. Typhimurium 14028 genomic DNA using Herculase Hotstart DNA Polymerase (Agilent) and primers flanked by SphI and HindIII restriction sites at the 5ʹ and 3ʹ end respectively (Table S1). The resulting PCR products were digested with SphI and HindIII (Promega) and ligated into pQF50Cm cleaved by the same enzymes. All plasmids were validated by DNA sequencing.
For the construction of complementation plasmid pAChha-ydgT, hha and ydgT ORFs were amplified, including their ribosome binding sites (RBS), from S. Typhimurium 14028 genomic DNA using Herculase Hotstart DNA Polymerase (Agilent) and primers flanked by XhoI and ClaI restriction sites for hha, and SmaI and HindIII for ydgT (Table S1). First, the hha PCR product digested with XhoI and ClaI enzymes was ligated into pACYC177 cleaved by the same enzymes yielding the pAChha plasmid. Then, the ydgT PCR product digested with SmaI and HindIII was cloned into pAChha cleaved by the corresponding enzymes to generate pAChha-ydgT vector. The transcription of hha and ydgT in this recombinant plasmid is under the control of the promoter of the Kanr gene of the pACYC177 vector which allows a constitutive expression of Hha and YdgT. In the same way, the complementation plasmid pACstpA was constructed by cloning stpA ORF, including its RBS, into pACYC177 cleaved by XhoI and HindIII. The sequence of pAChha-ydgT and pACstpA inserts were checked by DNA sequencing.
Chromosomal deletions were done in S. Typhimurium 14028 strain by the λ-Red recombinant method using primers stpA-P1/stpA-P2 (stpA deletion), hha-P1/hha-P2 (hha deletion), ydgT-P1/ydgT-P2 (ydgT deletion), sirA-P1/sirAP2 (sirA deletion), FimA-P1/FimF-P2 (fim deletion) as previously described [Citation65] (Table S1). The same protocol was used to delete a 50 bp region encompassing the −10 and −35 boxes of the PpefB promoter. All deletion mutants were checked by PCR, using primers outside of the deletion site, and DNA sequencing. Construction of Δhns::kan, Δhns::kanΔstpA, Δhns::kanΔfimA-FΔsirA, and Δhns::kanΔhhaΔydgT mutants was performed by transduction of the Δhns::kan mutation in the wild-type, the ΔstpA mutant, the ΔfimA-FΔsirA or the ΔhhaΔydgT double mutants with a P22HT105int lysate as previously described [Citation42]. Due to their genetic instability, all mutants containing an hns deletion were freshly prepared by transduction before each experiment and several mutants were studied in each experiment.
Western blot analysis
Total bacterial extracts were prepared as previously described [Citation66]. Bacterial proteins (an equivalent of 108 bacteria/well) were separated by electrophoresis in 15% (wt/vol) SDS-polyacrylamide gel (PAGE) and transferred onto a nitrocellulose membrane. Immunoblotting were performed using either a mouse anti-Hsp60 monoclonal antibody (loading control; 1:6000; Enzo Life Sciences) or a rabbit anti-PefA polyclonal antibody (1:500) [Citation8]. Horseradish peroxidase HRP-conjugated rabbit anti-mouse IgG (1:5000; Dako) or HRP-conjugated goat anti-rabbit IgG (1:5000; Dako) were used as secondary antibodies. Detection was performed by chemiluminescence with Fusion FX7 imaging system (Vilber Lourmat) using the SuperSignal™ West Dura Extended Duration Substrate (Thermo Scientific). Exposure times for PefA detection are indicated on all the blots. For Hsp60 detection, exposure times varied between 100 and 500 ms. At least two independent assays were performed for each experiment.
Reverse transcription-PCR (RT-PCR)
Bacteria were harvested by centrifugation and bacterial RNA were stabilized by a 0.1% SDS, 1% acidic phenol, 19% ethanol mixture [Citation67]. Total RNA were purified using the RNeasy Mini Kit (Qiagen) and genomic DNA was removed using the TURBO DNA-free kit (Ambion) according to manufacturers’ instructions. Then two micrograms of total RNA were reverse transcribed in presence of random hexamers using the AMV Reverse Transcriptase (Promega). PCR amplification mixtures contained 0.5 μM of each primer (Table S1), 200 μM deoxyribonucleoside triphosphate, 1.5 mM MgCl2, 1 U of GoTaq DNA polymerase (Promega) and 1X Green GoTaq Flexi Buffer (Promega). The temperature cycling for the amplification was performed in a Bio-Rad thermocycler (MyCycler) as follows: 1 cycle at 95°C for 3 min; 30 cycles at 95°C for 30 s, 60°C for 30 s and 72°C for 30 s; and finally, 1 cycle at 72°C for 5 min. Reactions without RT were performed to check the absence of DNA contamination in RNA samples. All RT-PCR experiments were duplicated from at least two independent bacterial cultures and RNA extractions.
Measurements of β-galactosidase activity
The measures of the β-galactosidase activities were performed as described by Miller [Citation68]. All assays were performed in triplicate from at least three independent bacterial cultures. β-galactosidase activities are expressed in Miller units.
Supplemental Material
Download MS Word (15.3 KB)Acknowledgments
We would like to thank Andreas Bäumler for the kind gift of anti-PefA serum. We are grateful to Jay Hinton and Sathesh Sivasankaran for sharing unpublished RNA-seq data. We also thank Camille Clamagirand for technical assistance. This work was supported in part by the ERA-NET InfectERA project “Sal host trop” funded by the Agence Nationale pour la Recherche. N.A. was supported by two grants from the Délégation Régionale à la Recherche et à la Technologie du Centre (FEDER) number 1634-32245 and from the Région Centre number 2008-00036085. G.A.H-E. hold a doctoral fellowship from the Ministère de l’Enseignement Supérieur et de la Recherche.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Bäumler AJ, Tsolis RM, Heffron F. Contribution of fimbrial operons to attachment to and invasion of epithelial cell lines by Salmonella typhimurium. Infect Immun. 1996;64:1862–1865. PMID: 8613405.
- Sukupolvi S, Lorenz RG, Gordon JI, et al. Expression of thin aggregative fimbriae promotes interaction of Salmonella typhimurium SR-11 with mouse small intestinal epithelial cells. Infect Immun. 1997;65:5320–5325. PMID: 9393832.
- Wilson RL, Elthon J, Clegg S, et al. Salmonella enterica serovars gallinarum and pullorum expressing Salmonella enterica serovar Typhimurium type 1 fimbriae exhibit increased invasiveness for mammalian cells. Infect Immun. 2000;68:4782–4785. PMID: 10899888.
- McClelland M, Sanderson KE, Spieth J, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. PMID: 11677609.
- Hansmeier N, Miskiewicz K, Elpers L, et al. Functional expression of the entire adhesiome of Salmonella enterica serotype Typhimurium. Sci Rep. 2017;7:10326. PMID: 28871183.
- Bäumler AJ, Tsolis RM, Bowe FA, et al. The pef fimbrial operon of Salmonella typhimurium mediates adhesion to murine small intestine and is necessary for fluid accumulation in the infant mouse. Infect Immun. 1996;64:61–68. PMID: 8557375.
- Humphries A, DeRidder S, Bäumler AJ. Salmonella enterica serotype Typhimurium fimbrial proteins serve as antigens during infection of mice. Infect Immun. 2005;73:5329–5338. PMID: 16113248.
- Humphries AD, Raffatellu M, Winter S, et al. The use of flow cytometry to detect expression of subunits encoded by 11 Salmonella enterica serotype Typhimurium fimbrial operons. Mol Microbiol. 2003;48:1357–1376. PMID: 12787362.
- Łaniewski P, Baek C-H, Roland KL, et al. Analysis of spleen-induced fimbria production in recombinant attenuated Salmonella enterica serovar Typhimurium vaccine strains. MBio. 2017;8:e01189–17. PMID: 28830946.
- Weening EH, Barker JD, Laarakker MC, et al. The Salmonella enterica serotype Typhimurium lpf, bcf, stb, stc, std, and sth fimbrial operons are required for intestinal persistence in mice. Infect Immun. 2005;73:3358–3366. PMID: 15908362.
- Clegg S, Purcell BK, Pruckler J. Characterization of genes encoding type 1 fimbriae of Klebsiella pneumoniae, Salmonella typhimurium, and Serratia marcescens. Infect Immun. 1987;55:281–287. PMID: 2879791.
- Duguid JP, Anderson ES, Campbell I. Fimbriae and adhesive properties in salmonellae. J Pathol Bacteriol. 1966;92:107–138. PMID: 5334095.
- Römling U, Bian Z, Hammar M, et al. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol. 1998;180:722–731. PMID: 9457880.
- Friedrich MJ, Kinsey NE, Vila J, et al. Nucleotide sequence of a 13.9 kb segment of the 90 kb virulence plasmid of Salmonella typhimurium: the presence of fimbrial biosynthetic genes. Mol Microbiol. 1993;8:543–558. PMID: 8100983.
- Nuccio S-P, Bäumler AJ. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol Mol Biol Rev. 2007;71:551–575. PMID: 18063717.
- Woodward MJ, Allen-Vercoe E, Redstone JS. Distribution, gene sequence and expression in vivo of the plasmid encoded fimbrial antigen of Salmonella serotype enteritidis. Epidemiol Infect. 1996;117:17–28. PMID: 8760946.
- Ledeboer NA, Frye JG, McClelland M, et al. Salmonella enterica serovar Typhimurium requires the Lpf, Pef, and Tafi fimbriae for biofilm formation on HEp-2 tissue culture cells and chicken intestinal epithelium. Infect Immun. 2006;74:3156–3169. PMID: 16714543.
- Chaudhuri RR, Morgan E, Peters SE, et al. Comprehensive assignment of roles for Salmonella Typhimurium genes in intestinal colonization of food-producing animals. PLoS Genet. 2013;9:e1003456. PMID: 23637626.
- Lawley TD, Chan K, Thompson LJ, et al. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2006;2:e11. PMID: 16518469.
- Bouwman CW, Kohli M, Killoran A, et al. Characterization of SrgA, a Salmonella enterica serovar Typhimurium virulence plasmid-encoded paralogue of the disulfide oxidoreductase DsbA, essential for biogenesis of plasmid-encoded fimbriae. J Bacteriol. 2003;185:991–1000. PMID: 12533475.
- Nicholson B, Low D. DNA methylation-dependent regulation of Pef expression in Salmonella typhimurium. Mol Microbiol. 2000;35:728–742. PMID: 10692151.
- Nuccio S-P, Chessa D, Weening EH, et al. SIMPLE approach for isolating mutants expressing fimbriae. Appl Environ Microbiol. 2007;73:4455–4462. PMID: 17526787.
- Sterzenbach T, Nguyen KT, Nuccio S-P, et al. A novel CsrA titration mechanism regulates fimbrial gene expression in Salmonella typhimurium. Embo J. 2013;32:2872–2883. PMID: 24056837.
- Navarre WW, Porwollik S, Wang Y, et al. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. PMID: 16763111.
- Vivero A, Baños RC, Mariscotti JF, et al. Modulation of horizontally acquired genes by the Hha-YdgT proteins in Salmonella enterica serovar Typhimurium. J Bacteriol. 2008;190:1152–1156. PMID: 18039769.
- Stoebel DM, Free A, Dorman CJ. Anti-silencing: overcoming H-NS-mediated repression of transcription in gram-negative enteric bacteria. Microbiology. 2008;154:2533–2545. PMID: 18757787.
- Sonnenfield JM, Burns CM, Higgins CF, et al. The nucleoid-associated protein StpA binds curved DNA, has a greater DNA-binding affinity than H-NS and is present in significant levels in hns mutants. Biochimie. 2001;83:243–249. PMID: 11278075.
- Leonard PG, Ono S, Gor J, et al. Investigation of the self-association and hetero-association interactions of H-NS and StpA from Enterobacteria. Mol Microbiol. 2009;73:165–179. PMID: 19508284.
- Lucchini S, McDermott P, Thompson A, et al. The H-NS-like protein StpA represses the RpoS (sigma 38) regulon during exponential growth of Salmonella Typhimurium. Mol Microbiol. 2009;74:1169–1186. PMID: 19843227.
- Madrid C, Balsalobre C, García J, et al. The novel Hha/YmoA family of nucleoid-associated proteins: use of structural mimicry to modulate the activity of the H-NS family of proteins. Mol Microbiol. 2007;63:7–14. PMID: 17116239.
- Sharma VK, Zuerner RL. Role of hha and ler in transcriptional regulation of the esp operon of enterohemorrhagic Escherichia coli O157:H7. J Bacteriol. 2004;186:7290–7301. PMID: 15489441.
- Fahlen TF, Wilson RL, Boddicker JD, et al. Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J Bacteriol. 2001;183:6620–6629. PMID: 11673432.
- Silphaduang U, Mascarenhas M, Karmali M, et al. Repression of intracellular virulence factors in Salmonella by the Hha and YdgT nucleoid-associated proteins. J Bacteriol. 2007;189:3669–3673. PMID: 17307861.
- Nieto JM, Carmona M, Bolland S, et al. The hha gene modulates haemolysin expression in Escherichia coli. Mol Microbiol. 1991;5:1285–1293. PMID: 1956303.
- Coombes BK, Wickham ME, Lowden MJ, et al. Negative regulation of Salmonella pathogenicity island 2 is required for contextual control of virulence during typhoid. Proc Natl Acad Sci U S A. 2005;102:17460–17465. PMID: 16301528.
- Paytubi S, Madrid C, Forns N, et al. YdgT, the Hha paralogue in Escherichia coli, forms heteromeric complexes with H-NS and StpA. Mol Microbiol. 2004;54:251–263. PMID: 15458420.
- Ali SS, Whitney JC, Stevenson J, et al. Structural insights into the regulation of foreign genes in Salmonella by the Hha/H-NS complex. J Biol Chem. 2013;288:13356–13369. PMID: 23515315.
- Olekhnovich IN, Kadner RJ. Role of nucleoid-associated proteins Hha and H-NS in expression of Salmonella enterica activators HilD, HilC, and RtsA required for cell invasion. J Bacteriol. 2007;189:6882–6890. PMID: 17675384.
- Solórzano C, Srikumar S, Canals R, et al. Hha has a defined regulatory role that is not dependent upon H-NS or StpA. Front Microbiol. 2015;6:773. PMID: 26284052.
- Ono S, Goldberg MD, Olsson T, et al. H-NS is a part of a thermally controlled mechanism for bacterial gene regulation. Biochem J. 2005;391:203–213. PMID: 15966862.
- Beraud M, Kolb A, Monteil V, et al. A proteomic analysis reveals differential regulation of the sigma(S)-dependent yciGFE(katN) locus by YncC and H-NS in Salmonella and Escherichia coli K-12. Mol Cell Proteomics. 2010;9:2601–2616. PMID: 20713450.
- Abed N, Grépinet O, Canepa S, et al. Direct regulation of the pefI-srgC operon encoding the Rck invasin by the quorum-sensing regulator SdiA in Salmonella Typhimurium. Mol Microbiol. 2014;94:254–271. PMID: 25080967.
- Farinha MA, Kropinski AM. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J Bacteriol. 1990;172:3496–3499. PMID: 2111810.
- Robbe-Saule V, Schaeffer F, Kowarz L, et al. Relationships between H-NS, sigma S, SpvR and growth phase in the control of spvR, the regulatory gene of the Salmonella plasmid virulence operon. Mol Gen Genet. 1997;256:333–347. PMID: 9393431.
- Kröger C, Colgan A, Srikumar S, et al. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe. 2013;14:683–695. PMID: 24331466.
- Jakomin M, Chessa D, Bäumler AJ, et al. Regulation of the Salmonella enterica std fimbrial operon by DNA adenine methylation, SeqA, and HdfR. J Bacteriol. 2008;190:7406–7413. PMID: 18805972.
- Wallar LE, Bysice AM, Coombes BK. The non-motile phenotype of Salmonella hha ydgT mutants is mediated through PefI-SrgD. BMC Microbiol. 2011;11:141. PMID: 21689395.
- Deighan P, Beloin C, Dorman CJ. Three-way interactions among the Sfh, StpA and H-NS nucleoid-structuring proteins of Shigella flexneri 2a strain 2457T. Mol Microbiol. 2003;48:1401–1416. PMID: 12787365.
- Free A, Dorman CJ. The Escherichia coli stpA gene is transiently expressed during growth in rich medium and is induced in minimal medium and by stress conditions. J Bacteriol. 1997;179:909–918. PMID: 9006049.
- Müller CM, Dobrindt U, Nagy G, et al. Role of histone-like proteins H-NS and StpA in expression of virulence determinants of uropathogenic Escherichia coli. J Bacteriol. 2006;188:5428–5438. PMID: 16855232.
- Landick R, Wade JT. Grainger DC. H-NS and RNA polymerase: a love-hate relationship? Curr Opin Microbiol. 2015;24:53–59. PMID: 25638302.
- Ueda T, Takahashi H, Uyar E, et al. Functions of the Hha and YdgT proteins in transcriptional silencing by the nucleoid proteins, H-NS and StpA, in Escherichia coli. DNA Res. 2013;20:263–271. PMID: 23543115.
- Boudreau BA, Hron DR, Qin L, et al. StpA and Hha stimulate pausing by RNA polymerase by promoting DNA-DNA bridging of H-NS filaments. Nucleic Acids Res. 2018;46:5525–5546. PMID: 29718386.
- van der Valk RA, Vreede J, Qin L, et al. Mechanism of environmentally driven conformational changes that modulate H-NS DNA-bridging activity. Elife. 2017;6:e27369. PMID: 28949292.
- Nieto JM, Madrid C, Miquelay E, et al. Evidence for direct protein-protein interaction between members of the enterobacterial Hha/YmoA and H-NS families of proteins. J Bacteriol. 2002;184:629–635. PMID: 11790731.
- Nieto JM, Madrid C, Prenafeta A, et al. Expression of the hemolysin operon in Escherichia coli is modulated by a nucleoid-protein complex that includes the proteins Hha and H-NS. Mol Gen Genet. 2000;263:349–358. PMID: 10778755.
- Olekhnovich IN, Kadner RJ. Crucial roles of both flanking sequences in silencing of the hilA promoter in Salmonella enterica. J Mol Biol. 2006;357:373–386. PMID: 16443238.
- Bertin P, Hommais F, Krin E, et al. H-NS and H-NS-like proteins in Gram-negative bacteria and their multiple role in the regulation of bacterial metabolism. Biochimie. 2001;83:235–241. PMID: 11278074.
- Sondén B, Uhlin BE. Coordinated and differential expression of histone-like proteins in Escherichia coli: regulation and function of the H-NS analog StpA. Embo J. 1996;15:4970–4980. PMID: 8890170.
- Srinivasan R, Scolari VF, Lagomarsino MC, et al. The genome-scale interplay amongst xenogene silencing, stress response and chromosome architecture in Escherichia coli. Nucleic Acids Res. 2015;43:295–308. PMID: 25429971.
- Lang B, Blot N, Bouffartigues E, et al. High-affinity DNA binding sites for H-NS provide a molecular basis for selective silencing within proteobacterial genomes. Nucleic Acids Res. 2007;35:6330–6337. PMID: 17881364.
- Chessa D, Dorsey CW, Winter M, et al. Binding specificity of Salmonella plasmid-encoded fimbriae assessed by glycomics. J Biol Chem. 2008;283:8118–8124. PMID: 18211897.
- Evans DF, Pye G, Bramley R, et al. Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut. 1988;29:1035–1041. PMID: 3410329.
- He G, Shankar RA, Chzhan M, et al. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc Natl Acad Sci U S A. 1999;96:4586–4591. PMID: 10200306.
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. PMID: 10829079.
- Fardini Y, Chettab K, Grépinet O, et al. The YfgL lipoprotein is essential for type III secretion system expression and virulence of Salmonella enterica serovar enteritidis. Infect Immun. 2007;75:358–370. PMID: 17060472.
- Eriksson S, Lucchini S, Thompson A, et al. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol. 2003;47:103–118. PMID: 12492857.
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. p. 352–355.
- Casadaban MJ, Cohen SN. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. PMID: 6997493.
- Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: tcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. PMID: 7789817.
- Chang ACY, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. PMID: 149110.
- Münch R, Hiller K, Grote A, et al. Virtual footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics. 2005;21:4187–4189. PMID: 16109747.
