ABSTRACT
In contrast to mammalia, fungi are able to synthesize the branched-chain amino acid leucine de novo. Recently, the transcription factor LeuB has been shown to cross-regulate leucine biosynthesis, nitrogen metabolism and iron homeostasis in Aspergillus fumigatus, the most common human mold pathogen. Moreover, the leucine biosynthetic pathway intermediate α-isopropylmalate (α-IPM) has previously been shown to posttranslationally activate LeuB homologs in S. cerevisiae and A. nidulans. Here, we demonstrate that in A. fumigatus inactivation of both leucine biosynthetic enzymes α-IPM synthase (LeuC), which disrupts α-IPM synthesis, and α-IPM isomerase (LeuA), which causes cellular α-IPM accumulation, results in leucine auxotrophy. However, compared to lack of LeuA, lack of LeuC resulted in increased leucine dependence, a growth defect during iron starvation and decreased expression of LeuB-regulated genes including genes involved in iron acquisition. Lack of either LeuA or LeuC decreased virulence in an insect infection model, and inactivation of LeuC rendered A. fumigatus avirulent in a pulmonary aspergillosis mouse model. Taken together, we demonstrate that the lack of two leucine biosynthetic enzymes, LeuA and LeuC, results in significant phenotypic consequences indicating that the regulator LeuB is activated by α-IPM in A. fumigatus and that the leucine biosynthetic pathway is an attractive target for the development of antifungal drugs.
Introduction
Aspergillus fumigatus is a ubiquitous saprophytic mold and, at the same time, the most common mold pathogen in humans causing severe invasive diseases, termed aspergillosis, particularly in patients with a suppressed immune system [Citation1]. Due to the increasing number of people with impaired immunological defense because of medication or underlying diseases, the number of patients suffering from opportunistic fungal infections has risen dramatically over the last 20 years [Citation2,Citation3]. Treatment of invasive aspergillosis (IA) displays various limitations due to increasing resistance, toxicity and the limited spectrum of currently used antifungal substances [Citation4]. Together with the limitations in diagnosing IA, this results in high mortality rates of between 30% and 90% depending on the patient cohort [Citation5]. To address this problem, new modes of antifungal therapy are needed. In this regard, fungal pathways that are both essential for fungal growth and/or virulence and absent in mammals are of particular interest. Consequently, pathways for biosynthesis of amino acids that are not produced by humans are attractive candidates for antifungal development.
The branched-chain amino acids (BCAA) leucine, isoleucine, and valine are synthesized by bacteria, archaebacteria and eukaryotes such as plants and fungi. In contrast, mammalia do not have the capacity to synthesize BCAAs and satisfy their needs by uptake from the diet. BCAA biosynthesis has three common steps until the production of 2-ketoisovalerate and subsequently splits in the synthesis of valine, leucine, and isoleucine. In Saccharomyces cerevisiae [Citation6], leucine-specific biosynthesis starts with the conversion of 2-ketoisovalerate to α-isopropylmalate (α-IPM) by the α-IPM synthase LeuC (S. cerevisiae contains two paralogs Leu4p and Leu9p). α-IPM is then further processed by the α-IPM isomerase (Leu1p), the β-isopropylmalate (β-IPM) dehydrogenase (Leu2p), and the branched-chain amino acid aminotransferase (Bat2p). Leucine biosynthesis is feedback regulated via inhibition of Leu4p enzyme activity by leucine. Moreover, in S. cerevisiae the pathway intermediate α-IPM has been shown to posttranslationally activate the transcription factor Leu3p, which activates several steps in leucine biosynthesis as well as the NADP-dependent glutamate dehydrogenase Gdh1p involved in nitrogen metabolism [Citation6]. Likewise, the Leu3p orthologue LeuB transcriptionally activates genes involved in leucine biosynthesis as well as the glutamate dehydrogenase-encoding gene gdhA upon posttranslational activation by α-IPM, in Aspergillus nidulans [Citation7]. Recent studies revealed that LeuB additionally regulates iron acquisition in A. fumigatus via transcriptional activation of the genes encoding the iron regulator HapX as well as siderophore biosynthetic enzymes [Citation8]. HapX is the major regulator for adaptation to iron starvation in A. fumigatus and stimulates the expression of genes involved in iron uptake, including the siderophore system, and represses genes involved in iron consuming pathways, like the tricarboxylic acid (TCA) cycle, respiration or heme biosynthesis [Citation9]. The link to iron homeostasis might be explained by the fact that two BCAA biosynthetic enzymes, Ilv3 and LeuA, require iron-sulfur clusters as prosthetic groups, i.e. BCAA biosynthesis is iron-dependent and requires increased iron accumulation. Moreover, HapX has been shown to transcriptionally repress LeuA and Ilv3A during iron starvation [Citation10]. An overview of leucine-specific biosynthesis in S. cerevisiae is shown in Figure S1 and the final proposed steps in A. fumigatus, supported by BLAST searches, are shown in .
Figure 1. Overview of leucine biosynthesis in A. fumigatus. As shown in S. cerevisiae [Citation6], 2-ketoisovalerate, a common step of the biosynthesis of valine and leucine, is converted by LeuC (α-IPM synthase) to α-IPM. LeuA (α-IPM isomerase) produces β-IPM, which is subsequently converted by Leu2A (dehydrogenase), spontaneous decarboxylation and Bat2 (BCAA aminotransferase) to leucine. The biosynthesis is feedback-inhibited via blocking of LeuC enzymatic activity by the end-product leucine [Citation6]. The intermediate α-IPM was shown to posttranslationally activate LeuB in S. cerevisiae and A. nidulans [Citation6,Citation7]. The results of this study indicate that α-IPM also activates LeuB in A. fumigatus including the regulation of BCAA encoding genes, gdhA, and genes involved in adaptation to iron starvation.
![Figure 1. Overview of leucine biosynthesis in A. fumigatus. As shown in S. cerevisiae [Citation6], 2-ketoisovalerate, a common step of the biosynthesis of valine and leucine, is converted by LeuC (α-IPM synthase) to α-IPM. LeuA (α-IPM isomerase) produces β-IPM, which is subsequently converted by Leu2A (dehydrogenase), spontaneous decarboxylation and Bat2 (BCAA aminotransferase) to leucine. The biosynthesis is feedback-inhibited via blocking of LeuC enzymatic activity by the end-product leucine [Citation6]. The intermediate α-IPM was shown to posttranslationally activate LeuB in S. cerevisiae and A. nidulans [Citation6,Citation7]. The results of this study indicate that α-IPM also activates LeuB in A. fumigatus including the regulation of BCAA encoding genes, gdhA, and genes involved in adaptation to iron starvation.](/cms/asset/6e0c122e-d995-4a5e-a367-805b5c705baf/kvir_a_1682760_f0001_oc.jpg)
The importance of the early steps in BCAA biosynthesis for virulence has been studied in different fungi. Inactivation of the first common step in BCAA biosynthesis, the acetolactate synthase (termed Ilv2), resulted in decreased virulence in Cryptococcus neoformans and Candida albicans as well as reduced in vivo persistence of S. cerevisiae [Citation11–Citation13]. Moreover, the third common step, the dihydroxyacid dehydratase was shown to play a crucial role in virulence of A. fumigatus, whereby this mold possesses four potential paralogs, termed Ilv3A, Ilv3B, Ilv3C and Ilv3D [Citation14].
In this study, we investigated the role of the leucine biosynthesis in iron homeostasis and virulence of A. fumigatus via deletion of the genes encoding the biosynthetic enzymes LeuC and LeuA, and analyzed the potential regulatory role of α-IPM in A. fumigatus.
Materials and methods
Media & growth conditions
A. fumigatus strains were grown at 37°C on complete medium (2 g/L peptone and 1g/L yeast extract), on blood agar (containing 25% blood) or on/in Aspergillus minimal medium [Citation15], containing 20 mM glutamine (nitrogen source), 1% (w/v) glucose (carbon source) and 30 µM FeSO4. For iron starvation conditions, the addition of FeSO4 was omitted, and for harsher iron starvation conditions on solid media, BPS (bathophenanthrolinedisulfonic acid disodium salt; Sigma-Aldrich, Vienna, Austria), a ferrous iron chelator, was added. All other supplements are described in the respective experiments. In liquid cultures, 106 spores/mL medium were inoculated; and on solid media 103 spores were point-inoculated.
A. fumigatus strains, deletion of leuA and leuC and reconstitution of ∆leuA and ∆leuC
To generate the mutant strains lacking the leucine biosynthetic enzymes LeuA (AFUB_027020) or LeuC (AFUB_014560), the bipartite marker technique was used [Citation16]. The fragments of the incomplete but overlapping hygromycin (hph) resistance cassette were fused to 1 kb of the 5ʹ- and 3ʹ- flanking region of leuA or leuC, respectively, and transformed into the A. fumigatus wildtype (wt) strain A1160P+, resulting in the mutant strains ∆leuA and ∆leuC. The flanking regions and the hph resistance cassette were PCR-amplified from genomic A1160P+ DNA or the plasmid pAN7.1 using the primer pairs leuA5ʹFW+/leuA5ʹRV+, leuA3ʹFW+/leuA3ʹRV+ and leuAhphFW/leuAhphRV for leuA and TO3/TO4, leuC3ʹFW+/leuC3ʹRV+ and TO15/leuChphRV for leuC, respectively. Together with the pUC19L vector (containing an ampicillin resistance cassette), the fragments were assembled with the GeneArt Seamless PLUS Cloning and Assembly Kit (Thermo Fisher Scientific, Vienna, Austria) resulting in the plasmids p∆leuA and p∆leuC. Deletion constructs for transformation were amplified with the primers leuA5ʹFW/ohph14 (2.3 kb) and leuA3ʹRV/ohph15 (2.5 kb) or TO14/ohph14 (2.3 kb) and leuC3ʹRV/ohph15 (2.5 kb). Protoplasts were transformed with the two split marker fragments for each gene and the mutant strains were selected on minimal media containing 0.1 mg/mL hygromycin B (Calbiochem).
To reconstitute leuA and leuC in the deletion mutants, fragments containing a complete leuA (4.4 kb) or leuC (4.3 kb) copy were amplified from genomic A1160P+ DNA with the primer pairs TO52/TO53 for leuA and TO54/TO55 for leuC, respectively, and subsequent to gel purification, the fragments were digested with HindIII and NdeI (leuA) or NotI and SpeI (leuC). The plasmid pSK275 carrying the pyrithiamine resistance cassette (ptrA) was digested with the respective enzymes and ligated with the leuA or leuC fragment, resulting in the plasmids pleuArec and pleuCrec. These plasmids were linearized by digestion with AvrII (pleuArec) or HpaI (pleuCrec) to promote homologous recombination in the 5ʹ-flanking regions of leuA or leuC and integrated into the deletion locus of the corresponding leucine biosynthetic enzyme by protoplast transformation. The reconstituted strains were selected with 0.1 µg/mL pyrithiamine hydrobromide and for leucine prototrophy. The correct integration of all constructs was verified by Southern blot analysis (data not shown). An overview of the deletion strategy is shown in Figure S2. All strains used in this study are listed in Table S1. Primers used for genetic manipulation are shown in Table S2.
RNA isolation and northern blot analysis
Fungal strains were cultured in the liquid minimal medium under iron starvation conditions at 37°C and 200 rpm with or without the addition of 5 mM leucine as described in the Figure legends. RNA isolation was performed using TRI Reagent® (Sigma-Aldrich, Vienna, Austria) following the manufacturer’s manual. Ten micrograms of RNA (if not indicated otherwise) were loaded on 2.2 M formaldehyde agarose gels and blotted after electrophoresis onto Amersham Hybond N membranes (GE Healthcare, Vienna, Austria). RNA was detected with DIG-labeled probes, amplified by PCR. Primers used for these probes are shown in Table S2.
Southern blot analysis
Genomic DNA was isolated according to Sambrook et al. [Citation17]. To confirm the correct integration of the genetic manipulations, the DNA was digested withBamHI (∆leuA, leuArec, wt) or PvuII (∆leuC, leuCrec, wt), separated on an agarose gels and blotted onto Amersham Hybond N membranes (GE Healthcare, Vienna, Austria). The different fragment sizes were detected with DIG-labeled probes of the 5ʹ- and 3ʹ-flanks of the corresponding gene. Primers used for these probes are shown in Table S2.
Quantification of siderophore production
Extracellular siderophores were extracted from culture supernatants and detection was performed as described previously [Citation10].
Virulence assay in Galleria mellonella
Virulence studies in the G. mellonella infection model were performed according to Fallon et al. [Citation18]. Larvae of the greater wax-moth G. mellonella (SAGIP, Italy) were kept at 18°C in the dark before use to prevent pupation. Randomly selected larvae, weighing between 0.3 and 0.4 g, were injected through one of the hind pro-legs with 107 spores of A. fumigatus wt, ∆leuA, leuArec, ∆leuC, or leuCrec in 20 µL insect physiological saline (IPS) into the hemocoel. As controls, untreated larvae and larvae injected with IPS were used. All larvae were incubated at 30°C in the dark, to circumvent temperature-related immune response of the larvae as shown previously [Citation19], and viability was monitored for 6 days post infection. Experiments were repeated three times with groups of 20 larvae per sample each time. Survival data representing the average survival rates of all experiments (total of 60 larvae per sample) were evaluated using Kaplan-Meier curves and analyzed with the log-rank (Mantel-Cox) test utilizing GraphPad Prism 7.00 software. Differences were considered significant at p ≤ 0.05.
Virulence testing in a pulmonary mouse model
A non-neutropenic mouse model was used for virulence testing. Therefore, six-week-old female ICR mice (10 per group) were immunocompromised by subcutaneous injection of 300 mg/kg body weight cortisone acetate on day −3, 0, 3, 7 and 11 of infection. For pulmonary infection, a suspension of 5 × 105 spores of wt, ∆leuC or leuCrec in 20 µL PBS plus 0.2% Tween 20 was applied intranasally (10 µL in each mouse nostril). Survival was observed for 21 days post infection and analysis of the survival data was performed by Kaplan-Meier survival analysis. Lung fungal burden was assessed by infecting immunocompromised mice (n = 5) as described above, sacrificing them 48 h postinfection, homogenizing the lungs and plating dilutions on YAG plates for CFU (colony forming unit) enumeration. Three mice in the wt group and two in the leuCrec group were found moribund before completion of the 48 h infection and were therefore not included in the analysis. Histology was performed with gomori methenamine silver stain (GMS, stains fungal elements black) or haematoxylin and eosin stain (H&E, stains host-cell nuclei purple, cytosol pink).
The Ministry of Health (MOH) Animal Welfare Committee, Israel, ethically approved these experiments.
Results
Lack of leuC results in higher leucine requirement for growth compared to lack of leuA
Homologues of leucine biosynthetic and regulatory enzymes in A. fumigatus were identified by BLAST analysis (Table S3). In contrast to S. cerevisiae, which contains two α-IPM synthase homologs (termed Leu4 and Leu9), A. fumigatus possesses only a single enzyme, termed LeuC (AFUB_014560); the α-IPM isomerase (in S. cerevisiae LEU1p) was termed LeuA (AFUB_027020), the β-isopropylmalate dehydrogenase (in S. cerevisiae Leu2p) was termed Leu2A (AFUB_015310) and the homolog of the transcription factor Leu3p is termed LeuB (AFUB_020530).
To investigate the role of leucine biosynthesis in A. fumigatus, we generated two mutant strains lacking either the α-IPM isomerase LeuA (strain ∆leuA) or the α-IPM synthase LeuC (strain ∆leuC) by replacing the coding region of the respective gene with the hygromycin resistance cassette. A single copy of the respective gene was reintegrated in the same locus yielding the strains leuArec and leuCrec to ensure that the phenotypic consequences observed are indeed due to the gene deletions. Correct genetic integration was confirmed by Southern blot analysis.
Growth analysis of all strains on plates ()) revealed leucine auxotrophy of both the ∆leuA and ∆leuC strain, which was cured by the reintegration of the respective gene resulting in leuArec and leuCrec strains. Remarkably, the ∆leuC mutant required a significantly higher leucine supplementation for growth compared to the ∆leuA mutant, i.e. on minimal medium plate cultures, ∆leuA was able to grow with 1 mM leucine supplementation while the ∆leuC mutant required ≥5 mM ()). Nine millimolar leucine supplementation did improve the growth of the ∆leuC mutant but did not fully reconstitute wt-like growth. Supplementation with the leucine biosynthesis intermediates α-IPM or β-IPM did not rescue growth of the ∆leuA or ∆leuC mutants (Figure S3), which indicates that α-IPM or β-IPM are not taken up efficiently by A. fumigatus. As leucine biosynthesis is dependent on iron and as the leucine biosynthesis regulator LeuB was recently found to be involved in the regulation of iron acquisition [Citation8], we examined the effect of iron availability on the growth of the two leucine auxotrophic strains. Supplementation with different iron concentrations or the presence of the ferrous iron-specific chelator BPS, which generates iron starvation [Citation20], did not significantly affect the leucine requirement of both mutants on plates ()). On complete medium containing yeast extract and peptone, which both contain leucine, both mutant strains showed wt-like growth ()). In contrast, on blood agar plates, neither ∆leuA nor ∆leuC was able to grow ()).
Figure 2. Strains lacking LeuA or LeuC display leucine auxotrophy and ∆leuC requires higher leucine supplementation for growth. Growth of ∆leuA and ∆leuC compared to wt and the reconstituted strains was analyzed on minimal medium (a) complete medium (b) or 25% blood agar (c). Generally, 103 spores were point-inoculated on the respective solid growth medium and incubated at 37°C. Minimal medium contained different concentrations of ferrous iron (0 mM, 0.03 mM, or 4 mM), the ferrous iron-specific chelator BPS, and/or leucine (0 mM, 1 mM, 2 mM, 5 mM, or 9 mM) as indicated. 0.03 mM iron reflects the standard minimal medium. Growth was evaluated after incubation for 48 h on minimal medium, 24 h on complete medium and 36 h on blood agar. Representatives of at least three biological replicates are shown.
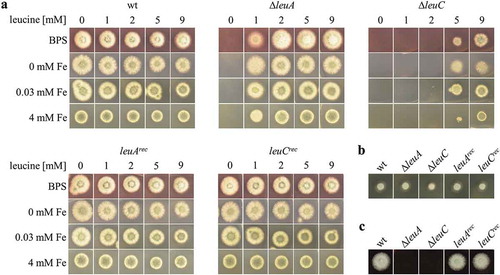
Taken together, the deletion of either leuA or leuC causes leucine auxotrophy, demonstrating that there are no other genes able to compensate for their loss. The difference in the degree of leucine requirement for growth between the two leucine auxotrophic strains indicates a regulatory role of α-IPM, which is missing in ∆leuC but accumulating in ∆leuA ().
Lack of leuC impairs growth particularly during iron starvation
Similar to plate growth assays, the ∆leuC mutant required significantly higher leucine supplementation for growth compared to the ∆leuA mutant in liquid growth conditions ()), i.e. under standard iron sufficient conditions (30 µM FeSO4) with 1 mM leucine supplementation, ∆leuA reached about 50% of the wt biomass, while ∆leuC did not grow. In contrast, growth of wt, leuArec, and leuCrec in liquid cultures was not affected by leucine supplementation, neither under iron starvation nor under iron sufficient conditions ()).
Figure 3. Lack of LeuC results in decreased growth and TAFC production in iron-limited liquid cultures. 106 spores/mL of the respective strains were cultured in 100 mL liquid minimal medium containing 0, 1, 2, 5 or 9 mM of leucine with or without addition of 0.03 mM of FeSO4 at 37°C for 30 h. (a) For the wt, the absolute biomass is displayed, while for the deletion and reconstituted strains, the biomass is normalized to the wt biomass under the same conditions. (b) TAFC content of the culture supernatants was normalized to the respective biomass. TAFC production of the wt is shown normalized to that without leucine supplementation, while TAFC production of the deletion and reconstituted mutant strains is shown normalized to the wt production at the same leucine concentration. The data show the mean ± standard deviation of three biological replicates.
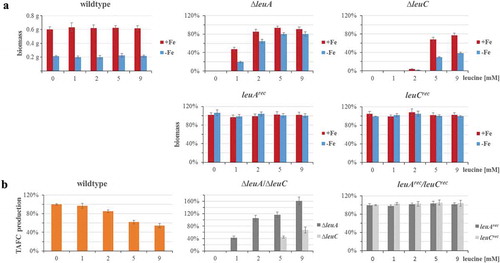
Iron limitation reduced biomass formation of wt and reconstituted mutant strains to about 33% compared to iron sufficiency. With 9 mM leucine supplementation, the ∆leuA mutant produced 90% of the wt biomass during iron sufficiency and 80% of the wt biomass during iron starvation. In comparison, with 9 mM leucine supplementation, the ∆leuC mutant produced 78% of the wt biomass during iron sufficiency, but only 39% of the wt biomass during iron starvation. In other words, with 9 mM leucine supplementation, ∆leuC produced 87% of the ∆leuA biomass during iron sufficiency but only 49% of the ∆leuA biomass during iron starvation, which indicates a particular growth defect during iron starvation.
Siderophore production is repressed by leucine supplementation and lack of leuC
Production of the extracellular siderophore triacetylfusarinine C (TAFC) was decreased in wt in a leucine concentration-dependent manner with 9 mM leucine reducing TAFC production to 54% of that without leucine supplementation ()).
The ∆leuA strain displayed wt-like TAFC production at 2 mM and 5 mM leucine and exceeded the wt by 1.6-fold at the same leucine supplementation of 9 mM leucine ()). In contrast, the ∆leuC mutant strain displayed significantly reduced TAFC production, i.e. compared to wt, TAFC production was decreased to 45% and 68% with 5 mM and 9 mM leucine supplementation, respectively. Taken together, the ∆leuC mutant displayed significantly decreased (>50%) TAFC production compared to the ∆leuA strain ()). The reconstituted mutant strains displayed wt-like TAFC production, confirming that these effects are indeed caused by the gene deletions.
Lack of leuC but not of leuA impairs activation of leuB target genes involved in leucine biosynthesis, nitrogen metabolism, and iron acquisition
To assess the potential impact of α-IPM on LeuB-mediated regulation [Citation8], we analyzed the transcript levels of leuB, and recently identified LeuB target genes [Citation8] in the wt, ∆leuA, and ∆leuC strains grown under iron starvation conditions with 5 mM leucine supplementation by Northern analysis ()). Compared to LeuA inactivation, LeuC inactivation resulted in strongly decreased transcript levels of leuB, leu2A, gdhA, mirB (encoding a transporter for uptake of ferric TAFC), sidA (encoding a siderophore biosynthetic gene), and hapX (encoding a regulator for adaptation to iron starvation). Compared to wt, lack of LeuA resulted in increased transcript levels of these genes, except hapX, also including leuC, which is not expressed in ∆leuC due to the deletion of the gene. The reintegration of the leuC gene in ∆leuC (strain leuCrec) cured the defect in gene regulation (Figure S4).
Figure 4. Transcript levels of leuB and LeuB target genes are upregulated in ∆leuA but downregulated in ∆leuC compared to wt (a) and downregulated by leucine supplementation in wt (b). (a) For Northern blot analysis, fungal strains were cultured under iron starvation conditions with 5 mM leucine supplementation. The wt strain was grown for 16 h, ∆leuA for 20 h, and ∆leuC for 28 h to compensate for the different growth rate and to reach the same biomass. (b) To analyze potential autoregulation of LeuB, the wt strain was cultured for 16 h under iron starvation prior the addition of leucine to a final concentration of 5 mM. For Northern analysis, samples were taken at 0, 15 and 30 min after leucine addition.
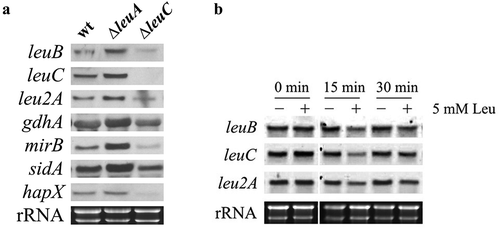
The downregulation of leuB in ∆leuC compared to ∆leuA indicates transcriptional autoregulation of LeuB ()). This autoregulation is further supported by the fact that supplementation of liquid culture with leucine, which inhibits the enzymatic activity of LeuC (), to a final concentration of 5 mM resulted in downregulation of leuB, leu2A, and leuC within 15 min ()).
Lack of leuA or leuC attenuates virulence in the Galleria mellonella infection model
To investigate the influence of leucine biosynthesis on A. fumigatus infections, we compared the virulence potential of wt, ∆leuA and ∆leuC in the G. mellonella wax-moth infection model. Lack of LeuA or LeuC resulted in significantly higher survival rates (p < 0.0001) of the larvae compared to wt over the examined period of 6 days (,)). While all larvae infected with wt spores died on day 3, only 10% of ∆leuA and ∆leuC infected larvae were dead. By the end of the experiment (day 6) survival rates of ΔleuA and ΔleuC still were 60% and 80%, respectively. Untouched larvae showed 100% survival (data not shown) and reintegration of the deleted genes (strains leuArec and leuCrec) cured the virulence defect (,)). Taken together, these data indicate a crucial role for leucine biosynthesis in the virulence of A. fumigatus.
Figure 5. ∆leuA and ∆leuC mutants display attenuated virulence in Galleria mellonella and ∆leuC is avirulent in a pulmonary aspergillosis mouse model. In G. mellonella, ∆leuA (a) and ∆leuC (b) showed significantly increased survival rates compared to wt (Log-Rank test, p < 0.0001), while survival of the reconstituted strains (leuArec and leuCrec) was statistically indifferent to the wt (p = 0.80 and 0.56, respectively). The curves display the average survival of three independent experiments (in total 60 larvae per sample). (c) In the mouse model (10 mice per strain), the leuC deletion strain was avirulent compared to the wt and the reconstituted strain (p < 0.0001). (d) Mouse lung fungal burden is greatly reduced in mice infected with the LeuC-deletion strain compared to the wt (p < 0.0001) and reconstituted strain (p < 0.008). (e) Histological analysis of infected lungs stained with GMS (gomori methenamine silver, stains fungi black, upper panel) shows large foci containing numerous invasive hyphae in mice infected with the wt and reconstituted strain. In contrast, only small infrequent aggregates of swollen conidia were seen in lung sections of mice infected with the ∆leuC strain (black arrow, upper panel on the right). Staining of lung cells with H&E shows granulocyte accumulation around the wt and leuCrec A. fumigatus strains, but not the ∆leuC strain (lower panel).
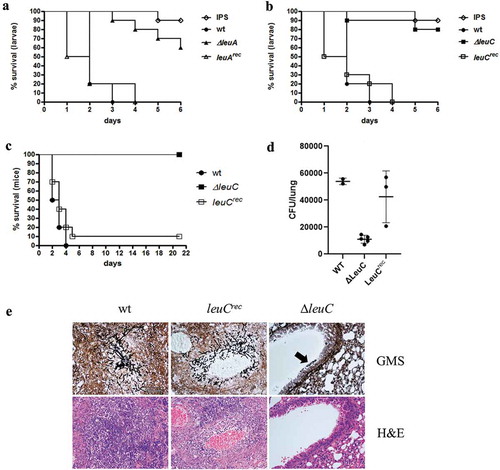
Lack of leuC results in attenuated virulence in a pulmonary aspergillosis mouse model
LeuA has a homolog in humans, the aconitase 2, while LeuC shows no significant similarity to human proteins. Moreover, LeuC-deletion resulted in a more severe growth defect compared to the deletion of LeuA. Therefore, only the ∆leuC mutant strain was used to assess the role of leucine biosynthesis in a murine pulmonary aspergillosis model. Mice infected with wt or leuCrec displayed high mortality, resulting in survival rates of 0% (wt) and 10% (leuCrec). In contrast, the leuC deletion strain demonstrated total avirulence (p < 0.0001) compared to the wt and the reconstituted strain with all the mice surviving over the examined period of 21 days ()). The lung fungal burden of mice infected with the ∆leuC mutant strain was significantly reduced in comparison to those of mice infected with wt (p < 0.0001) and leuCrec(p < 0.008) strains ()). Histological examination of mouse lungs showed greatly reduced fungal growth and inflammatory cell influx for mice infected with the ∆leuC mutant strain compared to those infected with the wt or leuCrec strains ()).
Discussion
The BCAAs leucine, valine, and isoleucine are produced exclusively by bacteria, archaebacteria and lower eukaryotes including plants and fungi [Citation6]. Mammals lack these biosynthetic pathways and consequently have to satisfy the need for these essential amino acids by nutritional uptake. This makes the BCAA biosynthetic pathway an attractive target for new antifungal agents unless the concentration of the amino acids in the host niche is high enough to cure the auxotrophy caused by inhibition of the targeted enzyme. Compounds targeting the first step of BCAA biosynthesis, the acetolactate synthase (termed Ilv2 in S. cerevisiae), are already used as herbicides [Citation21]. Moreover, the deletion of the gene encoding this enzyme decreased in vivo persistence of S. cerevisiae [Citation11] and attenuated virulence of C. neoformans and C. albicans in murine models [Citation12,Citation13]. However, antifungal activity of currently available Ilv2 inhibitors, such as triazolopyrimidine-sulfonamide compounds, can be bypassed through supplementation with BCAA or serum, which limits their application for treatment of mammalian infections [Citation22]. Deletion of the third BCAA biosynthesis step, the dihydroxyacid dehydratase (termed Ilv3A), was shown to attenuate virulence in A. fumigatus [Citation14]. Interestingly, the latter study revealed that the growth defect of dihydroxyacid dehydratase-lacking strains can be restored by valine and isoleucine supplementation, while leucine supplementation had a negative influence on the growth [Citation14]. Focusing on leucine biosynthesis alone, lack of the LeuA homolog Leu1 was shown to attenuate virulence in a pulmonary mouse model of C. neoformans [Citation23]. Interestingly, lack of the Leu1 homolog was also found to increase abundance of two mitochondrial iron-sulfur cluster proteins (aconitase and the iron-sulfur biosynthetic enzyme Nfu1) during iron starvation, but not iron sufficiency, but the mechanism behind this was unclear. In contrast, leucine auxotrophy did not affect in vivo persistence of Candida glabrata or C. albicans, respectively [Citation24,Citation25].
To analyze exclusively the role of leucine biosynthesis in virulence, we focused on two enzymes, LeuA and LeuC (in contrast to A. fumigatus, S. cerevisiae employs two homologs termed Leu4 and Leu9). BLAST search revealed that mammals have a LeuA homologous protein, termed aconitase (CAG38805.1; E-value = 9e-26 with 25% identity in 484 amino acids) but lack LeuC homologous proteins.
In this study, we found that lack of either LeuA or LeuC causes leucine auxotrophy in A. fumigatus. Lack of either LeuA or LeuB blocked growth on blood agar plates indicating that the leucine content of blood is too low to support the growth of these mutant strains. The latter is in agreement with the reported blood leucine concentration of 0.10–0.14 mM in humans [Citation26].
Remarkably, inactivation of LeuC and LeuA, respectively, resulted in significant differences, i.e. compared to lack of LeuA, lack of LeuC resulted in (i) significantly higher leucine requirement for growth, (ii) decreased growth rate during iron starvation, (iii) decreased siderophore production, and (iv) decreased expression of the gene encoding the NADP-dependent glutamate dehydrogenase, genes involved in leucine biosynthesis, as well as of genes involved in siderophore-mediated iron acquisition.
These data revealed that the inactivation of different steps in the leucine biosynthetic pathway has different phenotypic consequences. The difference between the two mutant strains is that inactivation of LeuC results in the lack of α-IPM, while this pathway intermediate accumulates in LeuA lacking cells. Apart from leucine biosynthesis, other α-IPM-dependent enzymatic pathways are not known. In S. cerevisiae and A. nidulans, LeuB homologs have been shown to be posttranslationally activated by the leucine biosynthesis intermediate α-IPM, leading to transcriptional activation of LeuB target genes involved in nitrogen metabolism and leucine biosynthesis [Citation6,Citation7]. Recently, LeuB was found to transcriptionally regulate not only leucine biosynthesis and glutamate dehydrogenase but also iron acquisition in A. fumigatus and is consequently important for adaptation to iron starvation [Citation8]. The comparison of mutants lacking either LeuA or LeuC offers the possibility to study the regulatory role of α-IPM, as this leucine pathway intermediate is missing in ∆leuC but accumulating in ∆leuA strains. Inactivation of LeuC had similar consequences as lack of LeuB: a particular growth defect during iron starvation, decreased siderophore production and the decreased expression of the gene encoding glutamate dehydrogenase, genes involved in leucine biosynthesis and genes involved in siderophore-mediated iron acquisition. Taken together, the current data indicate that α-IPM is important for the activation of all currently known functions of LeuB in A. fumigatus. The function of the leucine biosynthetic pathway in adaptation to iron starvation is further underlined by the finding that leucine supplementation, which blocks LeuB activity, decreased siderophore production in A. fumigatus wt. Consequently, this study reveals for the first time the role of α-IPM in fungal iron homeostasis. The decreased expression of genes involved in siderophore metabolism is in agreement with the reduced siderophore production and explains the reduced growth of the ∆leuC compared to the ∆leuA mutant strain during iron starvation. The increased leucine requirement for growth of the ∆leuC compared to the ∆leuA mutant strain might indicate that α-IPM-mediated activation of LeuB is required for transcriptional activation of leucine uptake. The leucine permeases of A. fumigatus have not been identified yet. BLASTp (https://blast.ncbi.nlm.nih.gov/Blast.cgi) searches revealed that the A. fumigatus amino acid permease-encoding gene gap1 (AFUB_089830) is the A. fumigatus amino acid permease with the highest degree of similarity to both S. cerevisiae Gap1 (57% identity) and S. cerevisiae Bap2 (41% identity), which mediate uptake of leucine in this yeast [Citation27]. The similar gap1 transcript levels in wt, leuA, and leuC mutants suggest that the growth defect of the leuC mutant compared to the leuA mutant might be due to differences in intracellular leucine handling rather than to leucine uptake.
Northern analysis revealed downregulation of leuB in ∆leuC compared to ∆leuA, which indicates the transcriptional autoregulation of LeuB. This is supported by the fact that LeuB was previously found to bind to the promoter of its encoding gene in vivo [Citation8] and that we found here that leucine supplementation, which inhibits the enzymatic activity of LeuC and thereby activity of LeuB, results in downregulation of leuB, leu2A, and leuC within 15 min.
In the Galleria mellonella infection model, lack of either LeuA or LeuC attenuated virulence of A. fumigatus. Consequently, for insect virulence, the defect in leucine biosynthesis appears to be a major reason for the defective virulence. The fact that lack of LeuC caused higher attenuation compared to lack of LeuA is in agreement with the differences in the growth pattern of the respective mutant strains.
In a murine pulmonary aspergillosis model, lack of LeuC resulted in avirulence. This might be the consequence of lacking leucine biosynthesis, the malfunction in adaptation to iron starvation, which has been shown to be important for virulence [Citation10,Citation20], or a combination. In this system, we tested only the virulence of the LeuC-deletion mutant for the following reasons: LeuC-deletion caused a more severe growth defect, particular during iron starvation, which is a known feature of the host niche [Citation28]. Moreover, LeuC lacks mammalian homologs, while LeuA displays significant similarity to aconitase 2. Taken together, LeuC appears to be a more promising target for developing novel antifungal therapies.
In summary, our study shows that (i) inactivation of different components of the same pathway can have significantly different phenotypic consequences, (ii) LeuC may be an attractive target for the development of novel antifungals, and (iii) α-IPM has a regulatory role in A. fumigatus similar to S. cerevisiae and A. nidulans, whereby a role in fungal iron homeostasis is shown for the first time here.
Abbreviations
| α-IPM | = | α-isopropylmalate |
| β-IPM | = | β-isopropylmalate |
| BCAA | = | branched chain amino acid |
| BPS | = | bathophenanthrolinedisulfonic acid disodium salt |
| IA | = | invasive aspergillosis |
| IPS | = | insect physiological saline |
| TAFC | = | triacetylfusarinine C |
| wt | = | wildtype |
Supplemental Material
Download Zip (796.5 KB)Acknowledgments
We are grateful to Carmen Kandelbauer for technical assistance.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Tekaia F, Latge JP. Aspergillus fumigatus: saprophyte or pathogen? Curr Opin Microbiol. 2005;8:385–392.
- Sanguinetti M, Posteraro B, Beigelman-Aubry C, et al. Diagnosis and treatment of invasive fungal infections: looking ahead. J Antimicrob Chemother. 2019;74:ii27–ii37.
- Brown GD, Denning DW, Gow NA, et al. Hidden killers: human fungal infections. Sci Transl Med. 2012;4:165rv13.
- Denning DW, Hope WW. Therapy for fungal diseases: opportunities and priorities. Trends Microbiol. 2010;18:195–204.
- Brakhage AA, Langfelder K. Menacing mold: the molecular biology of Aspergillus fumigatus. Annu Rev Microbiol. 2002;56:433–455.
- Kohlhaw GB. Leucine biosynthesis in fungi: entering metabolism through the back door. Microbiol Mol Biol Rev. 2003;67: 1–15. table of contents.
- Downes DJ, Davis MA, Kreutzberger SD, et al. Regulation of the NADP-glutamate dehydrogenase gene gdhA in Aspergillus nidulans by the Zn(II)2Cys6 transcription factor LeuB. Microbiology. 2013;159:2467–2480.
- Long N, Orasch T, Zhang S, et al. The Zn2Cys6-type transcription factor LeuB cross-links regulation of leucine biosynthesis and iron acquisition in Aspergillus fumigatus. PLoS Genet. 2018;14:e1007762.
- Haas H, Eisendle M, Turgeon BG. Siderophores in fungal physiology and virulence. Annu Rev Phytopathol. 2008;46:149–187.
- Schrettl M, Beckmann N, Varga J, et al. HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog. 2010;6:e1001124.
- Kingsbury JM, Goldstein AL, McCusker JH. Role of nitrogen and carbon transport, regulation, and metabolism genes for Saccharomyces cerevisiae survival in vivo. Eukaryot Cell. 2006;5:816–824.
- Kingsbury JM, McCusker JH. Cytocidal amino acid starvation of Saccharomyces cerevisiae and Candida albicans acetolactate synthase (ilv2{Delta}) mutants is influenced by the carbon source and rapamycin. Microbiology (Reading, England). 2010;156:929–939.
- Kingsbury JM, Yang Z, Ganous TM, et al. Cryptococcus neoformans Ilv2p confers resistance to sulfometuron methyl and is required for survival at 37 degrees C and in vivo. Microbiology (Reading, England). 2004;150:1547–1558.
- Oliver JD, Kaye SJ, Tuckwell D, et al. The Aspergillus fumigatus dihydroxyacid dehydratase Ilv3A/IlvC is required for full virulence. PloS One. 2012;7:e43559.
- Pontecorvo G, Roper JA, Hemmons LM, et al. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238.
- Nielsen ML, Albertsen L, Lettier G, et al. Efficient PCR-based gene targeting with a recyclable marker for Aspergillus nidulans. Fungal Genet Biol. 2006;43:54–64.
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. New York: Cold Spring Harbor Laboratory Press; 1989.
- Fallon JP, Troy N, Kavanagh K. Pre-exposure of Galleria mellonella larvae to different doses of Aspergillus fumigatus conidia causes differential activation of cellular and humoral immune responses. Virulence. 2011;2:413–421.
- Mowlds P, Kavanagh K. Effect of pre-incubation temperature on susceptibility of Galleria mellonella larvae to infection by Candida albicans. Mycopathologia. 2008;165:5–12.
- Schrettl M, Bignell E, Kragl C, et al. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J Exp Med. 2004;200:1213–1219.
- Tan S, Evans R, Singh B. Herbicidal inhibitors of amino acid biosynthesis and herbicide-tolerant crops. Amino Acids. 2006;30:195–204.
- Richie DL, Thompson KV, Studer C, et al. Identification and evaluation of novel acetolactate synthase inhibitors as antifungal agents. Antimicrob Agents Chemother. 2013;57:2272–2280.
- Do E, Hu G, Caza M, et al. Leu1 plays a role in iron metabolism and is required for virulence in Cryptococcus neoformans. Fungal Genet Biol. 2015;75:11–19.
- Kirsch DR, Whitney RR. Pathogenicity of Candida albicans auxotrophic mutants in experimental infections. Infect Immun. 1991;59:3297–3300.
- Jacobsen ID, Brunke S, Seider K, et al. Candida glabrata persistence in mice does not depend on host immunosuppression and is unaffected by fungal amino acid auxotrophy. Infect Immun. 2010;78:1066–1077.
- Badoud F, Lam KP, DiBattista A, et al. Serum and adipose tissue amino acid homeostasis in the metabolically healthy obese. J Proteome Res. 2014;13:3455–3466.
- Didion T, Grauslund M, Kielland-Brandt MC, et al. Amino acids induce expression of BAP2, a branched-chain amino acid permease gene in Saccharomyces cerevisiae. J Bacteriol. 1996;178:2025–2029.
- McDonagh A, Fedorova ND, Crabtree J, et al. Sub-telomere directed gene expression during initiation of invasive aspergillosis. PLoS Pathog. 2008;4:e1000154.
- Fraczek MG, Bromley M, Buied A, et al. The cdr1B efflux transporter is associated with non-cyp51a-mediated itraconazole resistance in Aspergillus fumigatus. J Antimicrob Chemother. 2013;68:1486–1496.
