ABSTRACT
This study investigates the twitching ability of 28 clinical and five environmental strains of S. maltophilia grown under iron-depleted condition through in-silico, phenotypic and proteomics approaches. Rapid Annotations using Subsystem Technology (RAST) analysis revealed the presence of 21 targets of type IV pilus shared across S. maltophilia strains K279a, R551-3, D457 and JV3. The macroscopic twitching assay showed that only clinical isolates produced a zone of twitching with a mean of 22.00 mm under normal and 25.00 mm under iron-depleted conditions. (p = 0.002). Environmental isolates did not show any significant twitching activity in both conditions tested. Isobaric Tags for Relative and Absolute Quantification (ITRAQ) analysis showed altered expression of twitching motility protein PilT (99.08-fold change), flagellar biosynthesis protein FliC (20.14-fold change), and fimbrial protein (0.70-fold change) in response to iron-depleted condition. Most of the strains that have the ability to twitch under the normal condition, exhibit enhanced twitching during iron limitation.
Introduction
The prevalence of nosocomial infections caused by a Gram-negative opportunistic pathogen, Stenotrophomonas maltophilia, has remarkably increased in recent years [Citation1–Citation7]. S. maltophilia is ubiquitously widespread in the water, soil, plants, animals, and foods [Citation8,Citation9]. In hospital settings, the pathogen has been isolated from various medical devices, disinfectant solutions, and found as part of endogenous flora of healthcare workers [Citation10–Citation13]. These serve as the source of transmission among patient who is immunocompromised, with indwelling medical devices, admitted to intensive care unit (ICU), on broad-spectrum antibiotics, prolonged hospitalization and exposed mucocutaneous barrier. The potential transmission of S. maltophilia is mostly contributed by the intrinsic resistance to multiple antibiotics [Citation14–Citation17] and the presence of virulence factors [Citation18–Citation21] in establishing its pathogenesis.
In general, pathogens express their virulence factors such as exotoxins, extracellular enzymes, biofilm formation, quorum sensing, etc. in order to access new niches [Citation22]. In such circumstances, motility plays an important role that allows the bacteria to migrate into a favorable niche and evade intracellular host. Motile bacteria demonstrate its ability to move due to the expression of multiple genes under sophisticated regulatory control, in response to environmental conditions [Citation23,Citation24]. Bacteria can sense a wide range of environmental stimuli such as osmolarity, pH, oxygen tension, temperature, nutrient availability, and consequently adapting their morphology and physiology for survival. Among all these factors, iron depletion causes a reduced production of cellular components, metabolic/enzyme activity and its products [Citation25]. In such condition, pathogens must scavenge available iron sources within the host’s cells to thrive this stressful environment.
Flagella is known to be the functional appendage of bacteria motility. The flagella filaments of S. maltophilia are composed of a 38-kDa subunit, SMFliC, and appeared to be similar to Serratia marcescens (78.6%), Escherichia coli, Proteus mirabilis, Shigella sonnei (71.4%), and Pseudomonas aeruginosa (57.2%) [Citation26]. Numerous studies have reported the correlation between S. maltophilia’s motility and biofilm formation on abiotic surfaces, and its capability of invading host epithelial cells under normal nutritional condition [Citation18,Citation27–Citation30]. Iron depletion in S. maltophilia was found to be associated with biofilm formation, extracellular polymeric substances (EPS) production, oxidative stress response, outer membrane proteins (OMPs) regulation, quorum sensing, siderophore production, and expression of iron acquisition systems [Citation31–Citation34]. However, the influence of low iron concentration on motility, especially twitching activity in S. maltophilia remains unclear. The motility-associated proteins that are altered during iron depletion also require future investigation. Therefore, this study is aimed at investigating the twitching motility in S. maltophilia through in-silico, phenotypic and proteomic approaches.
Materials and methods
Bacterial strains
A total of 28 clinical isolates (referred to as SM in ) including CS17 (clinical invasive) and CS24 (clinical noninvasive) as reference strains obtained from the laboratory culture collections (Department of Medical Microbiology and Parasitology, Universiti Putra Malaysia, Serdang, Selangor, Malaysia) were used in this study. Five environmental isolates, LMG959, LMG10871, LMG10879, LMG11104, and LMG11108; purchased from Belgian Coordinated Collections of Microorganisms (BCCM) (Laboratorium voor Microbiologie, Universiteit Gent, Belgium) were also studied. The isolates were incubated aerobically for 24 hours at 37°C for clinical and 30°C for environmental isolates [Citation18,Citation21,Citation33].
Table 1. S. maltophiia strains used in this study and their average zone of twitching under normal and iron-depleted conditions
In-silico analysis for targets of twitching motility
The complete genome sequences of four S. maltophilia strains, K279a, R551-3, D457 and JV3 (refer to ) were downloaded from the National Center for Biotechnology Information (NCBI, Bethesda, MD, USA) Genbank (www.ncbi.nlm.nih.gov/genome/browse/). The genomes were annotated by Rapid Annotations using Subsystem Technology (RAST) server (http://rast.nmpdr.org/) [Citation35]. Comparative genomics in SEED viewer (Genome Viewer) was used to confirm the identification and conservation of twitching motility genes within S. maltophilia genome sequences [Citation36].
Bacterial culture under iron-depleted and normal conditions
An iron-depleted condition was achieved by adding an iron chelator, 100 µM 2,2ʹ-dipyridyl (DIP) (Sigma Aldrich, Darmstadt, Germany) to LB broth (LB-DIP), while the normal condition was achieved by inoculating the suspension into usual LB broth (without DIP). The tubes were incubated aerobically (37°C for clinical and 30°C for environmental isolates) for 48 hours in an incubator shaker (Model IKA® KS 4000 i control, IKA® Works (Asia) Sdn Bhd, Selangor, Malaysia) at 200 rpm to ensure stationary phase bacterial growth [Citation31,Citation37]. The resulting bacterial growths were adjusted to 0.2 with an Eppendorf BioPhotometer Plus (Hamburg, Germany) at an optical density (OD) of 600 nm.
Twitching motility assay
P. aeruginosa ATCC27853 was used as a positive control as it exhibits twitching motility. Twitching motility was assayed by the modified subsurface agar method previously described [Citation38], with some modifications. A well was made in the center of the twitching agar plate, and 2 µl aliquot of the standardized suspension was inoculated into the well [Citation39]. The zone of twitching was visualized 96 hours after aerobic incubation (37°C for clinical and 30°C for environmental isolates). All assays were carried out at least in triplicate for each of the culture conditions.
Protein extraction and identification by Isobaric Tags for Relative and Absolute Quantification (ITRAQ)
CS17 and LMG959 strains under both normal and iron-depleted conditions were subjected to ITRAQ analysis. Total protein was extracted from the bacterial supernatant by the trichloroacetic acid (TCA) method. The extracted proteins were quantitated using RC-DC protein assay (Bio-Rad, California, US) as per manufacturer instructions. The ITRAQ assay was outsourced to Proteomic International, Perth, WA, Australia. The ITRAQ assay was performed independently using two biological replicas for each strain at two different conditions in a two 4-plex design. The protein sequences for S. maltophilia were obtained from UniProt database using ProteinPilotTM 4.5 software (AB SCIEX, USA). All mass spectrometry-based proteomics data were deposited in the ProteomeXchange Consortium database (http://proteomecentral.proteomexchange.org) via PRoteomics IDEntifications (PRIDE) with the dataset identifier PXD004370.
Statistical analysis
The twitching motility data were analyzed using IBM SPSS version 24.0. Wilcoxon signed-rank test (related samples, within-group) was used to compare twitching motility under normal versus iron-depleted conditions, respective of clinical and environmental strains. Furthermore, the twitching motility between clinical and environmental isolates under two growth conditions was analyzed using the Mann-Whitney test (unrelated samples, different group). Lastly, the differences in the strains that showed twitching positive and negative strains were analyzed. The p-values <0.05 is considered statistically significant for all the analysis. For proteomic analysis, an average protein ratio and p-values, which indicate significant differential expression was calculated by ProteinPilotTM 4.5 software (AB SCIEX, USA).
Results
Targets of twitching motility in S. maltophilia
Targeted in-silico analysis of the four complete genomes of S. maltophilia (see ), revealed the presence of shared type IV pilus subsystem across all the strains. This subsystem is categorized under “membrane transport” and subcategorized under “protein and nucleoprotein secretion system, type IV”. The 21 targets and functional roles obtained from RAST server are listed in .
Table 2. Functional roles of type IV pilus associated with twitching motility and their abbreviations obtained from RAST server
Twitching motility in S. maltophilia
The zone of twitching measured after 96 hours is tabulated in . Typical subsurface twitching motility agar interpretation is shown in ). The zone of twitching motility on twitching agar plates on normal and iron-depleted conditions for P. aeruginosa, SM45 and LMG10879 are shown in ). P. aeruginosa exhibited an average of 67 mm and 71 mm in normal and iron-depleted conditions respectively. Among the 33 S. maltophilia tested, 19 clinical isolates and one environmental isolate exhibited twitching motility in both conditions. The clinical isolates produced a zone of twitching with a mean of 22.00 mm under normal and 25.00 mm under iron-depleted conditions. The statistical analysis showed p < 0.05 (0.002), inferring significant difference between a zone of twitching under normal and iron-depleted conditions among 28 clinical strains tested. ) shows box-and-whisker plot (boxplot) for clinical isolates under normal and iron-depleted conditions.
Figure 1. (a) Typical subsurface twitching motility agar interpretation. A colony on the surface of agar around the inoculation point (top colony) and a visible halo or hazy zone of bacteria that have twitched across the plate between the bottom of the agar and the Petri plate (interstitial colony). (b) A representative of the zone of twitching motility on twitching agar plates on normal and iron-depleted conditions. P. aeruginosa ATCC27853 (positive control), SM45 (clinical isolate) and LMG10879 (environmental isolate). The plates were flooded with TM developer solution. The zone of twitching motility was marked, and 1 cm line was drawn onto the plates. (c) Box-and-whisker plot for clinical isolates under normal and iron-depleted conditions. The twitching zone under normal condition (min: 13 mm, median: 22 mm, max: 45 mm), while iron-depleted condition (min:14 mm, median: 25 mm, max: 50 mm)
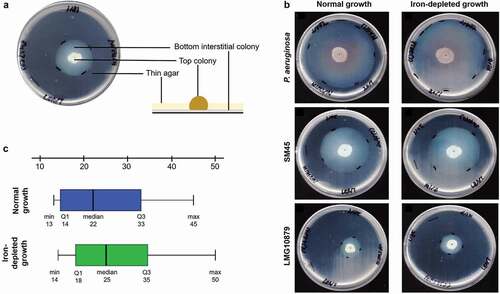
On the other hand, the only environmental strain, LMG10879, although twitched in the normal condition did not show any increase in zone size under iron-depleted condition (23.00 mm in both conditions), thereby no significant difference was found (p = 1.000). Furthermore, there was no significant difference in twitching motility between clinical and environmental isolates under normal and iron-depleted conditions, with p = 0.104 and p = 0.083, respectively. Those strains (n = 20) that exhibited positive twitching activity, there was no significant difference for all isolates under normal and iron-depleted conditions, with p = 1.000 and p = 0.900 respectively. While, strains (n = 13) that exhibited negative twitching activity, there was no significant difference for all isolates under normal and iron-depleted conditions, with p = 1.000 and p = 1.000 respectively.
Motility-associated proteins of S. maltophilia under iron-depleted
A total of 687 differentially expressed proteins were detected, of which 151 were commonly found to be present in both replicates. Among the 687 proteins detected, a total of 122 proteins (58 up-regulated and 19 down-regulated in CS17 and 36 up-regulated and seven down-regulated in LMG959) were seen with altered expression in response to iron-depleted condition (data not shown). The motility-associated proteins that were detected under normal versus iron-depleted conditions for both CS17 and LMG959 are shown in . The ITRAQ analysis showed that, only targets for CS17 was found significantly different between normal and iron-depleted conditions, including: (1) Flagellar biosynthesis protein FliC, S. maltophilia EPM1, 20.14-fold change, p = 0.0065; (2) Flagellar biosynthesis protein FliC, S. maltophilia D457, 1.54-fold change, p = 0.0046; (3) Fimbrial protein, S. maltophilia EPM1, 0.70-fold change, p = 0.0147; and (4) twitching motility protein PilT, S. maltophilia 5BA-I-2, 99.08-fold change, p = 0.0156.
Table 3. Differentially expressed motility-associated proteins identified via ITRAQ assay for CS17 and LMG959 isolates under iron-depleted condition
Discussion
Our in-silico analysis revealed that adhesion and motility factors such as pili and fimbriae were found in all S. maltophilia genomes (see ). A previous study reported that, S. maltophilia isolates K279a and SKK35 (clinical strains), R551-3 (environmental strain), SKA14 (seawater strain), and RA8 (wastewater strain) were found to harbor genes involved in pili and fimbriae formation, including: (1) Twitching motility proteins (pilU, pilT, pilE, pilI, pilJ, pilH, pilG, cheAW); (2) Fimbrial proteins (smf1); (3) Pilus assembly proteins (pilZ, pilW, pilV, pilX, pilY, pilE, pilF, pilQ, pilP, pilO, pilN, pilM); and (4) Twitching motility proteins, pilus assembly proteins (pilS) [Citation40]. Notably, both environmental and clinical strains of S. maltophilia have been found to contain genes encoding for twitching motility. In such circumstances, S. maltophilia shows the dual nature (i.e., commensal and true pathogen) [Citation34], and it is utmost essential to treat it as health hazard even when isolated from environment [Citation41].
P. aeruginosa exhibited active twitching motility at the agar-plate interface, with radial rates of expansion approaching 1 mm/h, resulting in large but fine twitching zones approaching 20 to 30 mm in diameter after overnight growth [Citation42]. In our study, nearly similar zone sizes were found, as, within four days of incubation, an average of 67 mm and 71 mm in normal and iron-depleted conditions respectively. These results were in accordance with several earlier studies which showed enhanced twitching motility for P. aeruginosa under reduced iron concentration [Citation43–Citation45]. In the present study, a variance of twitching pattern was observed, where only 60.6% exhibited the twitching motility. This variability could be attributed to the source of the isolates, and a larger zone of twitching was observed for isolates obtained from blood [Citation28]. In our study, five clinical isolates from blood exhibited larger zone of twitching under iron-depleted compared to normal conditions (20.00 mm for CS17, 25.00 mm for SM24, 33.00 mm for SM27, 36.00 mm for SM40, 35.00 mm for SM41) (see ), but not statistically significant (p = 0.136). In such circumstances, high disease severity and mortality were associated with hospital-acquired S. maltophilia bacteremia in comparison with community [Citation46–Citation50].
In contrast, some studies also reported the absence of twitching motility among S. maltophilia [Citation39,Citation43]. Aforementioned, only 19 clinical isolates formed a hazy zone with different diameter of the zone, suggestive that not all strains are capable of twitching motility. Regarding environmental isolates, a study reported that LMG959 showed a zone of twitching under normal growth condition [Citation27], but we did not observe in our study. Instead, LMG10879 exhibited twitching with an average zone of 23.00 mm in both normal and iron-depleted conditions. Environmental strains exhibit twitching activity and may not necessarily iron-dependent, thus deemed further investigation to identify other factors.
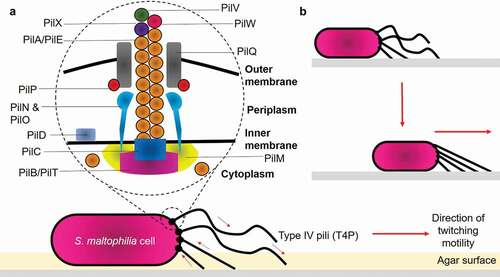
The ITRAQ analysis showed that, CS17 was found significantly to fimbrial-related protein. (A0A0U5I9J4, 0.70- fold change, p = 0.0147). The expression of pili/fimbriae is crucial for adhesion and biofilm formation [Citation26]. Particularly, the Smf-1 fimbrial operon in S. maltophilia mediates adherence at early stages of biofilm formation [Citation51]. Smf-1 is a peritrichous semiflexible fimbria of 5 to 7 nm under electron microscopy and only formed at 37°C, suggesting the reason why there was no significant change-fold observed in LMG959, as this strain was grown at 30°C. The flagellar biosynthesis protein FliC (A0A0X3QSM8, 20.14- fold change, p = 0.0065; A0A2Y9U6E5, 1.54-fold change, p = 0.0046) is relatively important, as it mediates the attachment of bacteria to epithelial cells, and initiate host’s infection process [Citation52]. Interestingly, the twitching motility protein PilT (A0A501PPD4, p = 0.0156) was found the highest (99.08-fold change) in S. maltophilia. Twitching motility was proposed to be mediated by type IV pili (T4P) located at one or both poles of the cells. Structurally, the pili are 5 to 7 nm in diameter and composed of small protein subunit called PilA (also known as pilin). T4P is capable of binding to a variety of surfaces such as inert surfaces, bacterial or eukaryotic cells, thereby exhibiting contact and promote colonization via a specific mechanism [Citation42,Citation53]. A model for S. maltophilia type IV pilus can reasonably be proposed based on RAST server and previous other studies on P. aeruginosa [Citation54–Citation56] as shown in ). It was elucidated that functional retractile pili are the mechanical basis for twitching motility. The removal of PilA from the base of assembled pilus fiber, followed by shortening of the pilus, pulls the cell closer to the site of attachment ()) [Citation54,Citation57–Citation59].
Figure 2. Structure of type IV pili and mechanisms of bacterial twitching motility. (a) Structurally, T4P is composed of: (1) Major pilin PilA/PilE; (2) Secretin PilQ; (3) Alignment proteins PilM, PilN, PilO, PilP; (4) Retraction ATPase PilT; (5) Assembly ATPase PilB; and (6) Minor pilins PilX, PilW, PilV. (b) Pilus retraction results in the forward movement of the cell across the surface. Arrows indicate the direction of pilus retraction/extension
Twitching motility may facilitate the bacteria to spread in the infected tissue, particularly in patients with the iron deficit, and promote tissue colonization to initiate its pathogenesis. Hence, further studies are warranted to determine why not all S. maltophilia strains have the ability to twitch (i.e., environmental isolates) and also to understand the biology behind increased twitching during reduced iron availability in an in-vivo model.
Acknowledgments
This research was funded by the Ministry of Higher Education, Malaysia through Fundamental Research Grant Scheme [04-01-14-53FR] and Universiti Putra Malaysia through Geran Putra—Inisiatif Putra Siswazah (IPS) [GP-IPS/2016/9478200].
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The datasets generated and/or analysed during the current study are available in the ProteomeXchange Consortium database (http://proteomecentral.proteomexchange.org) via PRoteomics IDEntifications (PRIDE) with the dataset identifier PXD004370.
Additional information
Funding
References
- Tian L, Sun Z, Zhang Z. Antimicrobial resistance of pathogens causing nosocomial bloodstream infection in Hubei Province, China, from 2014 to 2016: a multicenter retrospective study. BMC Public Health. 2018;18:1121.
- Matta R, Hallit S, Hallit R, et al. Epidemiology and microbiological profile comparison between community and hospital acquired infections: A multicenter retrospective study in Lebanon. J Infect Public Health. 2018;11:405–411.
- Tang CQ, Li JQ, Shou BM, et al. Epidemiology and outcomes of bloodstream infections in 177 severe burn patients from an industrial disaster: a multicentre retrospective study. Clin Microbiol Infect. 2018;24:199.e1-199.e7.
- Rosenthal VD, Desse J, Maurizi DM, et al. Impact of the International Nosocomial Infection Control Consortium’s multidimensional approach on rates of ventilator-associated pneumonia in 14 intensive care units in 11 hospitals of 5 cities within Argentina. Am J Infect Control. 2018;46:674–679.
- Montravers P, Tubach F, Lescot T, et al.; For the DURAPOP Trial Group. Short-course antibiotic therapy for critically ill patients treated for postoperative intra-abdominal infection: the DURAPOP randomised clinical trial. Intensive Care Med. 2018;44:300–310.
- Velázquez-Acosta C, Zarco-Márquez S, Jiménez-Andrade MC, et al. Stenotrophomonas maltophilia bacteremia and pneumonia at a tertiary-care oncology center: a review of 16 years. Support Care Cancer. 2018;26:1953–1960.
- Kuo S-H, Lin W-R, Lin J-Y, et al. The epidemiology, antibiograms and predictors of mortality among critically-ill patients with central line-associated bloodstream infections. J Microbiol Immunol Infect. 2018;51:401–410.
- Brooke JS. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev. 2012;25:2–41.
- Ryan RP, Monchy S, Cardinale M, et al. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nature Rev Microbiol. 2009;7:514–525.
- Looney WJ, Narita M, Mühlemann K. Stenotrophomonas maltophilia: an emerging opportunist human pathogen. Lancet Infect Dis. 2009;9:312–323.
- Abbott IJ, Slavin MA, Turnidge JD, et al. Stenotrophomonas maltophilia: emerging disease patterns and challenges for treatment. Expert Rev Anti Infect Ther. 2011;9:471–488.
- Nyč O, Matějková J. Stenotrophomonas maltophilia: significant contemporary hospital pathogen — review. Folia Microbiol (Praha). 2010;55:286–294.
- Senol E. Stenotrophomonas maltophilia: the significance and role as a nosocomial pathogen. J Hosp Infect. 2004;57:1–7.
- Crossman LC, Gould VC, Dow JM, et al. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 2008;9:R74.
- Zhang L, Li X-Z, Poole K. Multiple antibiotic resistance in Stenotrophomonas maltophilia: involvement of a multidrug efflux system. Antimicrob Agents Chemother. 2000;44:287–293.
- Sánchez MB. Antibiotic resistance in the opportunistic pathogen Stenotrophomonas maltophilia. Front Microbiol [Internet]. 2015 [cited 2019 May 17];6. Available from: http://journal.frontiersin.org/Article/https://doi.org/10.3389/fmicb.2015.00658/abstract
- Sanchez MB, Hernandez A, Martinez JL. Stenotrophomonas maltophilia drug resistance. Future Microbiol. 2009;4:655–660.
- Di Bonaventura G, Prosseda G, Del Chierico F, et al. Molecular characterization of virulence determinants of Stenotrophomonas Maltophilia strains isolated from patients affected by cystic fibrosis. Int J Immunopathol Pharmacol. 2007;20:529–537.
- Figueirêdo PMS, Furumura MT, Santos AM, et al. Cytotoxic activity of clinical Stenotrophomonas maltophilia. Lett Appl Microbiol. 2006;43:443–449.
- Chhibber S, Gupta A, Sharan R, et al. Putative virulence characteristics of Stenotrophomonas maltophilia: a study on clinical isolates. World J Microbiol Biotechnol. 2008;24:2819–2825.
- Nicoletti M, Iacobino A, Prosseda G, et al. Stenotrophomonas maltophilia strains from cystic fibrosis patients: genomic variability and molecular characterization of some virulence determinants. Int J Med Microbiol. 2011;301:34–43.
- Rohmer L, Hocquet D, Miller SI. Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 2011;19:341–348.
- Casadevall A, Pirofski L. Virulence factors and their mechanisms of action: the view from a damage–response framework. J Water Health. 2009;7:S2–18.
- Josenhans C, Suerbaum S. The role of motility as a virulence factor in bacteria. Int J Med Microbiol. 2002;291:605–614.
- Kalidasan V, Joseph N, Kumar S, et al. The ‘Checkmate’ for iron between human host and invading bacteria: chess game analogy. Indian J Microbiol. 2018;58:257–267.
- de Oliveira-garcia D, Dall’Agnol M, Rosales M, et al. Characterization of flagella produced by clinical strains of Stenotrophomonas maltophilia. Emerging Infect Dis. 2002;8:918–923.
- Thomas R, Hamat RA, Neela V. Extracellular enzyme profiling of Stenotrophomonas maltophilia clinical isolates. Virulence. 2014;5:326–330.
- Passerini De Rossi B, Calenda M, Vay C, et al. Biofilm formation by Stenotrophomonas maltophilia isolates from device-associated nosocomial infections. Revista Argentina De Microbiología. 2007;39:204–212.
- Pompilio A, Piccolomini R, Picciani C, et al. Factors associated with adherence to and biofilm formation on polystyrene by Stenotrophomonas maltophilia : the role of cell surface hydrophobicity and motility. FEMS Microbiol Lett. 2008;287:41–47.
- Pompilio A, Pomponio S, Crocetta V, et al. Phenotypic and genotypic characterization of Stenotrophomonas maltophilia isolates from patients with cystic fibrosis: genome diversity, biofilm formation, and virulence. BMC Microbiol. 2011;11:159.
- Garcia CA, Rossi BPD, Alcaraz E, et al. Siderophores of Stenotrophomonas maltophilia: detection and determination of their chemical nature. Revista Argentina De Microbiología. 2012;44:150–154.
- García CA, Alcaraz ES, Franco MA, et al. Iron is a signal for Stenotrophomonas maltophilia biofilm formation, oxidative stress response, OMPs expression, and virulence. Front Microbiol [Internet]. 2015 [cited 2019 May 17];6. Available from: http://journal.frontiersin.org/Article/https://doi.org/10.3389/fmicb.2015.00926/abstract
- Kalidasan V, Azman A, Joseph N, et al. Putative iron acquisition systems in Stenotrophomonas maltophilia. Molecules. 2018;23:2048.
- Kalidasan V, Joseph N, Kumar S, et al. Iron and virulence in Stenotrophomonas Maltophilia: all we know so far. Front Cell Infect Microbiol [Internet]. 2018 [cited 2019 Apr 30];8. Available from: https://www.frontiersin.org/article/https://doi.org/10.3389/fcimb.2018.00401/full
- Aziz RK, Bartels D, Best AA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75.
- Overbeek R, Olson R, Pusch GD, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014;42:D206–14.
- Navarro Llorens JM, Tormo A, Martínez-García E. Stationary phase in gram-negative bacteria. FEMS Microbiol Rev. 2010;34:476–495.
- Turnbull L, Whitchurch CB. Motility assay: twitching MOTILITY [Internet]. In: Filloux A, Ramos J-L, editors. Pseudomonas methods and protocols. New York: Springer New York; 2014 [cited 2019 May 17]:73–86. Available from: http://link.springer.com/https://doi.org/10.1007/978-1-4939-0473-0_9
- Jimenez PN, Koch G, Papaioannou E, et al. Role of PvdQ in Pseudomonas aeruginosa virulence under iron-limiting conditions. Microbiology. 2010;156:49–59.
- Adamek M, Linke B, Schwartz T. Virulence genes in clinical and environmental Stenotrophomas maltophilia isolates: A genome sequencing and gene expression approach. Microb Pathog. 2014;67–68:20–30.
- Adegoke AA, Stenström TA, Okoh AI. Stenotrophomonas maltophilia as an emerging ubiquitous pathogen: looking beyond contemporary antibiotic therapy. Front Microbiol [Internet]. 2017 [cited 2019 Jun 6];8. Available from: http://journal.frontiersin.org/article/https://doi.org/10.3389/fmicb.2017.02276/full
- Mattick JS. Type IV Pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314.
- Patriquin GM, Banin E, Gilmour C, et al. Influence of quorum sensing and iron on twitching motility and biofilm formation in Pseudomonas aeruginosa. J Bacteriol. 2008;190:662–671.
- Singh PK. Iron sequestration by human lactoferrin stimulates P. aeruginosa surface motility and blocks biofilm formation. BioMetals. 2004;17:267–270.
- Singh PK, Parsek MR, Greenberg EP, et al. A component of innate immunity prevents bacterial biofilm development. Nature. 2002;417:552–555.
- Garazi M, Singer C, Tai J, et al. Bloodstream infections caused by Stenotrophomonas maltophilia: a seven-year review. J Hosp Infect. 2012;81:114–118.
- Cho S-Y, Lee D-G, Choi S-M, et al. Stenotrophomonas maltophilia bloodstream infection in patients with hematologic malignancies: a retrospective study and in vitro activities of antimicrobial combinations. BMC Infect Dis [Internet] 2015 [cited 2019 May 18];15. Available from: http://bmcinfectdis.biomedcentral.com/articles/https://doi.org/10.1186/s12879-015-0801-7
- Chang Y-T, Lin C-Y, Lu P-L, et al. Stenotrophomonas maltophilia bloodstream infection: comparison between community-onset and hospital-acquired infections. J Microbiol Immunol Infect. 2014;47:28–35.
- Micozzi A, Venditti M, Monaco M, et al. Bacteremia due to Stenotrophomonas maltophilia in patients with hematologic malignancies. Clinl Infect Dis. 2000;31:705–711.
- Kagen J, Zaoutis TE, McGowan KL, et al. Bloodstream infection caused by Stenotrophomonas maltophilia in children. Pediatr Infect Dis J. 2007;26:508–512.
- de Oliveira-garcia D, Dall’Agnol M, Rosales M, et al. Fimbriae and adherence of Stenotrophomonas maltophilia to epithelial cells and to abiotic surfaces. Cell Microbiol. 2003;5:625–636.
- Chhibber S, Zgair AK. Involvement of Stenotrophomonas maltophilia Flagellin in bacterial adhesion to airway biotic surfaces: an in vitro study. Am J Biomed Sci. 2009;1:188–195.
- O’Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304.
- Burrows LL. Pseudomonas aeruginosa twitching motility: type IV Pili in action. Annu Rev Microbiol. 2012;66:493–520.
- Craig L, Forest KT, Maier B. Type IV pili: dynamics, biophysics and functional consequences. Nature Reviews Microbiology [Internet]. 2019 [cited 2019 Jun 7]. Available from: http://www.nature.com/articles/s41579-019-0195-4
- Leighton TL, Buensuceso RNC, Howell PL, et al. Biogenesis of Pseudomonas aeruginosa type IV pili and regulation of their function: pseudomonas aeruginosa type IV pili. Environ Microbiol. 2015;17:4148–4163.
- Kearns DB. A field guide to bacterial swarming motility. Nature Rev Microbiol. 2010;8:634–644.
- Wall D, Kaiser D. Type IV pili and cell motility. Mol Microbiol. 1999;32:01–10.
- Kaiser D. Bacterial motility: how do pili pull? Curr Biol. 2000;10:R777–80.
