ABSTRACT
Aeromonas dhakensis is an emerging human pathogen which causes fast and severe infections worldwide. Under the gradual pressure of lacking useful antibiotics, finding a new strategy against A. dhakensis infection is urgent. To understand its pathogenesis, we created an A. dhakensis AAK1 mini-Tn10 transposon library to study the mechanism of A. dhakensis infection. By using a Caenorhabditis elegans model, we established a screening platform for the purpose of identifying attenuated mutants. The uvrY mutant, which conferred the most attenuated toxicity toward C. elegans, was identified. The uvrY mutant was also less virulent in C2C12 fibroblast and mice models, in line with in vitro results. To further elucidate the mechanism of UvrY in controlling the toxicity in A. dhakensis, we conducted a transcriptomic analysis. The RNAseq results showed that the expression of a unique hemolysin ahh1 and other virulence factors were regulated by UvrY. Complementation of Ahh1, one of the most important virulence factors, rescued the pore-formation phenotype of uvrY mutant in C. elegans; however, complementation of ahh1 endogenous promoter-driven ahh1 could not produce Ahh1 and rescue the virulence in the uvrY mutant. These findings suggest that UvrY is required for the expression of Ahh1 in A. dhakensis. Taken together, our results suggested that UvrY controls several different virulence factors and is required for the full virulence of A. dhakensis. The two-component regulator UvrY therefore a potential therapeutic target which is worthy of further study.
Introduction
Aeromonas dhakensis, also called A. hydrophila sub. dhakensis, is an endemic pathogen causes a variety of human diseases, including gastroenteritis, wound infections, septicemia, respiratory infections, hepatobiliary infections, and urinary tract infections worldwide [Citation1–Citation4]. The reported mortality rate among patients with A. dhakensis extra-intestinal infection varies from 25.5% to 37.5%, which is much higher than those infected by other Aeromonas species (ex. 0% to 14.0% in A. veronii infection) [Citation5,Citation6]. Previous studies have reported that A. dhakensis carries an array of virulence factors and exhibits the most potent toxicity to human blood cell lines among the tested Aeromonas species [Citation6–Citation9]. With the higher mortality and the abundance of virulence genes, A. dhakensis has become an extremely important pathogenic species. However, the whole picture of A. dhakensis pathogenesis remains unclear. Moreover, the regulation of virulence is not understood due to a lack of a comprehensive study.
UvrY has been known as the response regulator of the BarA-UvrY two-component system and is thought to function as a transcription factor in bacteria [Citation10,Citation11]. A. dhakensis uvrY has orthologs in many other species, such as uvrY in E. coli, gacA in Pseudomonas, varA in Vibrio and sirA in Salmonella [Citation12–Citation14]. Previous studies have reported that UvrY or its orthologues repress glycolysis and enhance gluconeogenesis with the aid of small RNAs csrB and csrC by inhibiting the translational initiator csrA [Citation14,Citation15]. Studies have also revealed that UvrY contributes to the activation of bacterial toxicity in many bacterial pathogens [Citation12,Citation13,Citation16–Citation18]. However, the route and regulon through which UvrY affects bacterial toxicity remain unclear.
In this study, we highlighted the contribution of UvrY to A. dhakensis infection and discovered that UvrY is responsible for the virulence through regulating an array of virulence factors in A. dhakensis. Of the virulence factors regulated by UvrY, pore-forming hemolysin Ahh1 contributes to the virulence of A. dhakensis. Taken together, our results suggested that the pathogenic role of UvrY is worth verifying in order to develop novel therapy against UvrY and UvrY-related virulent factors.
Materials and methods
Bacteria strains and culture conditions
All the bacteria strains and plasmids used in this study are listed in . Aeromonas dhakensis AAK1, Staphylococcus aureus subsp. aureus ATCC 29213, Bacillus subtilis 3610, and Escherichia coli strains OP50, DH5α SM10 λπ and S17-1 λπ were cultured in a Luria-Bertani (LB) broth at 37°C for 16 hours. A. dhakensis AAK1 deletion and transposon mutants were cultured with 100 μg/ml kanamycin (Kan), and complement strains were cultured with 50 μg/ml chloramphenicol (Cm) in LB broth at 37°C for 16 hours. E. coli SM10 λπ pBSL180 strain was cultured with 50 μg/ml Kan in LB broth at 37°C for 16 hours.
Table 1. Summary of the characteristics of 31 mutant strains
Table 2. Bacteria strains and plasmids used in this study
Worm strains and culture conditions
The C. elegans wild-type Bristol N2 strain and PD4793 mIs10[myo-2p::GFP + pes-10p::GFP + gut-promoter::GFP] worms were provided by the Caenorhabditis Genetics Center (CGC), which is supported by the National Institutes of Health – Office of Research Infrastructure Programs (P40 OD010440). The animals were maintained on Nematode Growth Medium (NGM) with E. coli strain OP50 as the normal food source.
A. dhakensis mini-Tn10 transposon library
The library was built through conjugation with pBSL180, which contains a mini-Tn10 transposon and kanamycin resistant gene nptII. Overall, 500 μl of A. dhakensis AAK1 and E. coli SM10 λπ pBSL180 were cultured overnight and mixed gently. After centrifuging at 12,000 x g for 3 minutes, the supernatant was removed, and the bacteria pellet was washed 3 times with 1 ml of a conjugation buffer (10 mM MgSO4). The washed bacteria mixture was spread evenly on a 0.45 μm membrane filter. The filter was incubated continuously at 37°C on an LB plate for 5 hours, and then on an LB plate with 1 mM IPTG for 1 hour, and finally on an LB plate for 1 hour. Bacteria were collected from the filter through washing with 10 ml deionized sterile water and 50 μl of the bacteria were cultivated on Aeromonas Agar (LAB167, LAB MTM) with 100 μg/ml Kan at 37°C for 16 to 18 hours. Generally, there were 150 colonies growing on each plate. Each colony was picked and cultured in a well with 100 μl LB and 100 μg/ml Kan in a 96-well plate (353,072, Falcon) at 37°C for 16 to 18 hours. The 96-well plates were stored at −80°C after adding 100 μl LB with 30% glycerol to each well and mixing carefully.
Liquid toxicity (LT) screen
By using 96 solid pin multi-blot replicators, each 96-well plate in the transposon library was triplicated to another 96-well plate with 180 μl LB in each well and kept at 37°C for 16 to 18 hours. After centrifuging at 4,000 x g for 15 minutes, the supernatant was removed, and the bacteria pellet was resuspended with 200 μl S medium combined with approximately 20 to 30 synchronized PD4793 larva 4 (L4) stage worms. The plates were incubated at 25°C for 60 hours with constant shaking at 80 x g. The survival rate and GFP expression rate of the worms in each well were measured.
Arbitrary PCR
The arbitrary primed PCR was used to identify the insertion site of the mini-Tn10 transposon of A. dhakensis transposon mutants [Citation19,Citation20]. Briefly, a single colony PCR with an arbitrary primer (Arb1 or Arb6) and a transposon primer (nptR) were used for a first-round PCR. The PCR program was set as follows: 95°C for 10 min, followed by six cycles at 95 oC for 30 s, 30 oC for 30 s, and 72 oC for 2 min; 30 cycles at 95 oC for 30 s, 45 oC for 30 s, and 72 oC for 2 min, and finally, at 72 oC for 3 min. One-tenth of the cleaned-up PCR product was then used as the DNA template for a second-round PCR with the Arb2 primer and a nested transposon primer (nptR1). The PCR program was set as follows: 95 oC for 1 min, followed by 30 cycles at 95 oC for 30 s, 55 oC for 30 s, and 72 oC for 2.5 min, and finally, then 72 oC for an additional 4 min. The second-round PCR products were electrophoresed, and bands over 1,000 bp were gel-extracted and sequenced using Arb2 and nptR1 primers. The primers mentioned above are listed as follows:
Arb1: GGCCACGCGTCGACTAGTACNNNNNNNNNNGATAT
Arb6: GGCCACGCGTCGACTAGTACNNNNNNNNNNACGCC
Arb2: GGCCACGCGTCGACTAGTAC
nptR: GCATTGCATCAGCCATGATGG
nptR1: CATCAGAGCAGCCGATTGTCTG
Swimming and swarming motility
LB with 0.3% Bacto agar (Difco Laboratories) was used to determine swimming motility, and LB with 0.5% Bacto agar was used to measure the swarming motility of all mutants [Citation21,Citation22]. The plates were incubated at 37°C overnight, and motility was measured by examining the distance bacteria migrated from the center toward the periphery of the plate.
Auxotroph test
M9 minimal media was used to incubate all mutants for the auxotroph test [Citation23]. Briefly, a colony of mutants was picked into at least 6 wells of a 96-well plate. Each well contained 200 μL M9 media. The plate was incubated at 37°C for 18 hours. The bacterial growth, as determined by the OD600 values of each well, was recorded.
Growth analysis of bacteria
The OD-Monitor C&T (Taitec, Japan) was used to continuously observe the growth of bacteria. Briefly, of each mutant colony was seeded into 5 ml LB broth and incubated at 37°C in the OD-Monitor. The growth of bacteria measured according to the OD600 value was detected every 30 minutes.
Biofilm formation assay
Crystal violet staining was used to detect the expression of biofilm [Citation24]. Briefly, a colony of mutants was picked into at least 6 wells of a 96-well plate. Each well contained 200 μl of LB broth. The plate was incubated at 37°C for 18 hours. After removing the culture media and washing three times with deionized water, 125 μl of a 0.1% crystal violet solution in water was added to each well and left for 15 min at room temperature. After washing three times with deionized water, 125 μl of 30% acetic acid in water was added to each well to solubilize the crystal violet dye. The OD600 values of each well were recorded.
C. elegans survival assay
The procedures followed those of previous studies [Citation25–Citation27]. Synchronized L1 worms were seeded onto an ENG plate with E. coli OP50 and incubated at 20°C for 44 hours. Approximately 50 L4 worms were transferred onto an NG plate spread and cultured with OD600 = 2.0 A. dhakensis 30 μl. Live worms were counted and transferred daily. All the survival assays were conducted at on 20°C.
Deletion mutant of a. dhakensis
To generate deletion mutations, the host cell proteins driving homologous recombination between the allelic exchange vector and the recipient chromosome were used as the deletion mutants of A. dhakensis AAK1 constructed via allelic exchange, using the vector pwf300 containing a suicide gene sacB (levansucrase) and a kanamycin resistant gene nptII as selective markers [Citation28]. Briefly, about 500 bp of the 3ʹ and the 5ʹ regions of target gene were cloned into the left and right site of nptII. The plasmid was electroporated into E. coli S17-1 λπ and transferred into AAK1 through conjugation. The transconjugants that grew on Aeromonas Agar (LAB167, LAB MTM) was then cultured on an LB plate with 6% sucrose and 50 μg/ml Kan at 20°C to allow the levansucrase convert sucrose to form levans and force the consummation of homologous recombination.
Generation of complement strain
Broad host plasmid pBBRMCS1 was used to create the complement strain [Citation29]. In brief, the target gene was cloned into the multiple cloning sites of pBBRMCS1, and the reconstructed plasmid was transferred into E. coli S17-1 λπ. The conjugants were spread on Aeromonas Agar (LAB167, LAB MTM) plates containing 50 μg/ml Cm and 100 μg/ml Kan.
Cell cytotoxicity
The C2C12 mouse fibroblast cells were cultured in a complete medium consisting of Dulbecco’s Modified Eagle’s medium (DMEM, Gibco) and 10% fetal bovine serum (FBS, Invitrogen) at 37°C with 5% CO2 [Citation6]. The C2C12 fibroblast cells were seeded into 24-well plates (5x104 cells/well) and incubated overnight. With a MOI of 20 ratio, refreshed A. dhakensis were added to DMEM without FBS. After 2 hours of infection, the culture medium was examined for the release of lactate dehydrogenase (LDH) using a CytoTox 96 kit (Promega). A 0.1% Triton X-100 solution was used as a positive control, and serum-free Roswell Park Memorial Institute (RPMI) medium was used as a negative control. The cytotoxicity activity was expressed as the mean of triplicate measurements of released LDH levels compared with Triton X-100 exposure (defined as 100% cytotoxicity).
Mice survival
Based on our previous study [Citation6], 6–10 week-old female BALB/c mice weighing 18–22 grams were obtained from the National Laboratory Animal Center. Each mouse was injected intraperitoneally with 100 μl containing various bacterial loads of A. dhakensis. Three mice were tested for each experimental group in each experiment. The mice survival was monitored daily for 3 days. All the animal experiments in this study were carried out in strict accordance with the recommendations set forth in the guidelines for the Committee of Laboratory Care and Use of Animals, developed by National Cheng Kung University. The protocol was ethically approved by the Institutional Animal Care and Use Committees and National Cheng Kung University (Permit No: 107,080).
Serum resistance
The susceptibility of the bacterial isolates to human serum was analyzed as described previously [Citation30]. In brief, twenty-five microliters of the bacterial suspension (about 2 × 106 CFU) were mixed with 75 μl of pooled normal human serum in microtiter plates and then incubated at 37 °C for 3 hours. The test was performed in triplicate and the number of recovered bacteria was determined.
Determination of in vivo bacterial load
Mouse organs were individually homogenized in a suitable volume of PBS using the Micro Tissue Homogenizer (Pierce Chemical Co., Ill., USA). Homogenates were serially diluted in PBS and plated on TSB to determine bacterial numbers. The detection limit was defined as 30 to 300 CFU, as a modification of a previous report [Citation31].
Blood cytokines
Interleukin 1β (IL-1β), monocyte chemoattractant protein-1 (MCP-1/CCL2) and tumor necrosis factor α (TNF-α) were measured using ELISA (R&D Systems, Minneapolis, MN) [Citation31].
RNAseq
Approximately 5 mL of LB broth containing bacteria grown overnight were collected for RNA extraction. RNAs for sequencing were prepared using the Ion Total RNA-Seq Kit v2 (Ion Torrent™) according to the manufacturer’s instructions. Quality and base trimming were performed on Ion OneTouch™ 2 System reads from RNA libraries. The sequencing raw data was mapped according to the reference genome [Citation32]. The normalized count of sequencing reads (RPKM) was determined for each gene. The raw data and normalized results were updated to GEO (accession number: GSE130707).
qRT-PCR
The expression of the UvrY-regulated genes and gyrB (internal control) was measured using quantitative real time-PCR (qRT-PCR,) as described previously [Citation26,Citation33]. Approximately 3 ml of LB broth containing bacteria that had been cultivated overnight were collected for RNA extraction. RNA samples (2 μg for each experimental group) were converted to cDNA via reverse transcription. All qRT-PCRs were carried out using the FastStart Universal SYBR Green Master kit (ROX) according to the manufacturer’s specifications and analyzed on a StepOnePlus Real-Time PCR System. The expression level of each target was collected and compared to the internal control and conduct as a ΔCt value,where the Ct was equal to the number of PCR cycles required to amplify a given gene from a cDNA population. The fold-change values were estimated using the following equation: Fold change = 2[-ΔCt (AAK1)]/2[-ΔCt (OP50)]. The qRT-PCR primers are listed below:
Zn-dependent exopeptidase M28 Forward primer: TACAGCTTCGATCCCAAGGTA
Zn-dependent exopeptidase M28 Reverse primer: CCCTCAGGGTAGCCATCATA
pilus pilN Forward primer: TGTCAAATATAAACCTCCTTCCCT
pilus pilN Reverse primer: CCGTGATGGCCATGAAGATA
pilus pilO Forward primer: CAAGGATCGTCATTCTGGAGTC
pilus pilO Reverse primer: ACCGTCTTGCCGTTGTATTTA
pilus pilQ Forward primer: CTGCAGGAGGTCAAAGTCAA
pilus pilQ Reverse primer: TAGGAGAGCCGATCGGTAAA
exonuclease Forward primer: GTACCGACACCTACTCCTACA
nuclease Reverse primer: AATCCTCGATGCCGATCAC
exotoxin exoA Forward primer: TCATCAGGTTTGCGCCATA
exotoxin exoA Reverse primer: GATGAGATTGTGGACCGGATAG
hemolysin ahh1 Forward primer: GATCTCGCGATGCTCATACTT
hemolysin ahh1 Reverse primer: AGGATTACCGCTTCAGCATC
protease lasA Forward primer: CACTTCTCCCTGCTCTACAAC
protease lasA Reverse primer: TTGCAGTTGTCGTCGTAGTT
chitinase chiA Forward primer: GGTGAACTTGCGACCATAGA
chitinase chiA Reverse primer: CCGCTCAAGGAGAACAACA
collagenase Forward primer: GAAGAGCAGCAGTTCCATCA
collagenase Reverse primer: GGAGGAGTCGAAGATTACCAAC
lipase estA Forward primer: GACCGGCAGCTTCTTCAA
lipase estA Reverse primer: CAGATGGTTCATCCGGTAGTC
lipase chaperone chaP Forward primer: CTGCAGGCTCGCTATGA
lipase chaperone chaP Reverse primer: CTCGTCGAACAGCACCT
aerolysin aeroA Forward primer: GTATCCCAAATAGTGGGCAAGA
aerolysin aeroA Reverse primer: ATATTCCGGCAGGTGATGAAG
gyrase subunit B gyrB Forward primer: CCTGCTGCTAACCTTCTTCTATC
gyrase subunit B gyrB Reverse primer: CTTGCCCTTCTTCACCTTGTA
RAST analysis
The RNAseq results were re-mapping using RAST software available online [Citation34,Citation35]. The virulence genes with expression levels indicating a 10-fold decrease in the uvrY mutant compared to wild type AAK1 and uvrY complement strains were analyzed.
Pore-formation assay
Based on our previous studies [Citation36,Citation37], L4 stage N2 worms were infected with either A. dhakensis or Ahh1-overexpressed E. coli. At each time point, 10 worms were transferred to microcentrifuge tubes with M9 medium mixed with 5 mg/ml serotonin and 6.7 mg/ml propidium iodide dye and incubated for 30 min at room temperature while undergoing shaking. The worms were mounted on slides after being washed twice with M9 medium. The experimental animals were scored positive for pore-formation if at least one of the intestinal cells was stained with propidium iodide. Three independent repeats were performed in each treatment.
Western blot
An analysis of Ahh1 hemolysin production of either A. dhakensis or E. coli was performed using a western blot method as described previously with some modifications [Citation26]. In brief, 3 ml of LB broth containing bacteria cultivated overnight was collected to extract total protein using Bullet Blender® homogenization and precipitating with acetone. Rabbit polyclonal antibody to Ahh1 (Leadgene Biomedical Inc., Tainan, Taiwan) and rabbit monoclonal antibody to RpoB (EPR18704, abcam) were used for detection.
Image
The worms were mounted on a 2% agarose gel with M9 medium on a slide. Two microliters of 1% sodium azide was added to paralyze the worms. The samples were covered with a slip and observed using a DIC or RFP filter from a fluorescent microscope (Nikon Eclipase Ti inverted microscope system) equipped with a CCD Camera (QImaging Retiga-2000 R Fast 1394).
Statistical analysis
All experiments were performed a minimum of three times independently. The survival assay was analyzed using the Kaplan-Meier method, and survival difference was accessed using a log-rank test. The Student’s t-test was used to analyze the statistical results of the two different groups. A one-way ANOVA test was used to analyze the statistical values among the groups for one independent variable. Statistical significance was at P < 0.05. (***: P < 0.001; **: P < 0.01; *: P < 0.05) (Error bars: standard deviation, SD)
Results
A forward genetic screen to identify the A. dhakensis gene involved in the attenuation of A. dhakensis pathogenesis.
We created a mini-Tn10 transposon library of Aeromonas dhakensis AAK1 [Citation32] through conjugation with plasmid pBSL180 in E. coli SM10 ()) [Citation38]. Our transposon library contained 25,809 clones which gave a 99.74% coverage [Citation39] of A. dhakensis AAK1 genome. By using bacteriovorus nematode Caenorhabditis elegans as the model host, we screened the transposon library to isolate mutants with attenuated virulence. The screen scores were measured by the survival of worms and the retained green fluorescence among PD4793 worms after infection in liquid toxicity assay [Citation25,Citation27]. Thirty-one attenuated mutants were selected for further analysis ()). The transposon-inserted genes responsible for the 31 mutants are summarized in . By the Gene Ontology (GO) analysis [Citation40], the mutated genes were categorized based on their previously known function ( and S1 Fig). According to the description of each hit on the PANTHER website, most of them were reported to involve in metabolism and flagellar biosynthesis.
To exclude the genes responsible for mobility, growth and development, phenotypes of the swimming (S2A Fig) [Citation21], swarming (S2B Fig) [Citation21,Citation22], auxotroph (S2C Fig) [Citation41], and growth deficiency (S3 Fig) of these 31 mutants were tested. The biofilm formation ability (S2D Fig) as well as virulence to C. elegans (S4 Fig) were also examined [Citation26,Citation27]. Following the selection protocol, we turned our attention to the two-component system mutant strain uvrY strain for further study ()).
The attenuated virulence of the uvrY mutant was not related to defects in bacterial physiological features
To validate the virulence reduction of the uvrY transposon mutant, we generated an isogenic uvrY mutant of A. dhakensis AAK1 (S5 Fig) and rechecked the characteristics tested as discussed above. Compared to the uvrY transposon mutant, the motility (,)), auxotroph ()), growth ()), biofilm formation ()), and toxicity to C. elegans ()) remained unchanged in the uvrY isogenic mutant. The attenuation of virulence also displayed in the uvrY deletion mutant both in vitro ()) and in vivo ()). The biofilm formation level and toxicity to C. elegans of the uvrY complement strain (ΔuvrY-puvrY) were both rescued (-)). Taken together, these findings suggested that uvrY contributes to the toxicity of A. dhakensis without altering its physiological features.
Two-component system barA-uvrY participates in the regulation of a. dhakensis toxicity.
Previous study of other bacteria has shown that UvrY is a response regulator that is phosphorylated by BarA [Citation42]. To understand the role of BarA and the BarA-UvrY two-component system in the regulation of A. dhakensis toxicity, the correlation between UvrY and BarA was analyzed. In the C. elegans infection model, the virulence of the A. dhakensis barA isogenic mutant was attenuated as the uvrY mutant ()). However, the toxicity of the uvrY complement in the barA mutant (ΔbarA-puvrY) was significantly increased ()). A similar phenomenon was also observed in a cell infection model with C2C12 mouse myoblast (-)). These results suggested that the response regulator UvrY regulates the toxicity of A. dhakensis even without BarA.
Previous studies have found that the UvrY is phosphorylated by BarA at D54 residue [Citation11,Citation42,Citation43]. To determine the importance of D54 residue of UvrY, we generated D54E (constitutively active form) and D54A (inactive form) mutants of UvrY. Interestingly, in both the C. elegans ()) and C2C12 cell models ()), mutation of D54 residue does not alter the toxicity of UvrY in A. dhakensis. These results suggested that BarA is not the only phosphate donor to UvrY, which is similar to previous findings in E. coli [Citation44,Citation45], and the phosphorylation of the D54 residue of UvrY is not essential for the virulence of A. dhakensis.
UvrY regulates the virulence of a. dhakensis in a mouse infection model
To determine if the virulence attenuation of the uvrY mutant is cohesive in C. elegans, in cell, and in mammalian animals, we established an intraperitoneal (IP) infection of A. dhakensis in a mouse model. With the wild type A. dhakensis AAK1 strain, only a few mice survived for more than 24 hours after IP injection with a lower infection dose (5x105 CFU) ()). At a higher infection dose (4x106 CFU) of the wide type strain, the mice were all dead within 24 hours. However, IP infection of the uvrY mutant at a dose of 4 × 106 CFU resulted in no death of mice 72 hours post infection ()). These results suggested that the virulence of uvrY mutant is attenuated in the murine model.
Inflammation is an important immune response against bacterial infection. The levels of inflammatory markers such as TNF-α, IL-6, and MCP-1, is all correlated with the severity of infection [Citation31,Citation46]. By analyzing the concentrations of IL-6 ), MCP-1 ) and TNF-α ) in mice serum, we found that the uvrY mutant did not cause severe inflammation, which occurred with the wild type AAK1 and the uvrY complement strain. We also measured the bacterial load in the blood ), liver ) and spleen ) of infected mice and the results showed the bacterial load of the uvrY mutant was significantly lower than the case for both the wild type AAK1 and uvrY complement strains in all organs. In the serum resistance assay, the wild type AAK1, uvrY mutant and uvrY complement strains all demonstrated serum resistance at similar levels ()). These results suggested that the attenuation of the uvrY mutant is not related to alterations in the structure of the cell envelope. In other words, specific toxins rather than the components of the cell envelope are regulated by UvrY and required for its virulence in A. dhakensis.
UvrY controls an array of virulence factors in A. dhakensis.
The response regulator of a two-component system is usually a transcription factor [Citation47]. To determine whether UvrY regulates the expression of virulence-associated molecules, the transcriptome of wild type strain and uvrY mutants were analyzed. The RNAseq results of uvrY mutant were compared to wild type AAK1 and uvrY complement strains. The genes with more than 2X-fold change were thought to be the potential candidates ()). A total of 120 genes with expression at a lower transcription level in the uvrY mutant as compared to the wild type AAK1 and uvrY complement strains that achieved a 10X-fold difference were enrolled for analysis () and S6 File).
With Gene Ontology (GO) analysis, all 120 genes were sorted according to their already known function ()). More than half of the genes exhibit catalytic activity. Expression of those genes involving in glycolysis and gluconeogenesis, however, is not significantly different between wild type and uvrY mutants. To clarify and highlight the potential route by which uvrY regulates virulence of A. dhakensis, the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) [Citation34,Citation35] was also applied for further analysis. Among the 120 candidate genes, 6.9% of them were found in the Virulence, Disease and Defense subsystem of RAST ()). According to the RAST analysis, the expression level of several well-known virulence factors in Aeromonas were in line with a change in the uvrY expression level, including pilus, protease, exo-toxin, collagenase, chitinase, aerolysin and hemolysin [Citation9]. The RNAseq results of these RNAs ()) were reconfirmed with qRT-PCR ()) and showed a cohesive expression pattern where these RNAs all had lower expression levels in the uvrY mutant as compared to the wild type AAK1 or uvrY complement strains. These findings suggested that uvrY controls the expression of different virulence factors in A. dhakensis.
Hemolysin ahh1, a type of pore-forming toxin, is regulated by UvrY
Clinically, a severe bacterial infection, such as soft tissue necrotizing fasciitis, is usually caused by pore-forming toxins, including hemolysin and aerolysin in Aeromonas, cytolysin in Vibrio and leucocidin in Staphylococcus aureus [Citation48]. To determine if the attenuation of the uvrY mutant contributes to the reduced production of pore-forming toxins, a pore-forming assay with propidium iodide (PI) was conducted using the C. elegans infection model [Citation36,Citation37]. Propidium iodide (PI), a dye with red fluorescence cannot pass through the cell membrane unless the membrane integrity is broken. The intestinal cells of C. elegans were intact when they were fed with normal food source E. coli OP50 -). The retention of PI dye in the gut indicated the absence of pore-formation in the intestine cells. Comparably, the dye was dispersed into the intestinal cells of the nematodes after infection with A. dhakensis -). In line with our prediction, worms fed with the uvrY mutant did not show the pore-formation phenotype -), and by contrast, the worms infected with the uvrY complement strain were similar to the wild type A. dhakensis AAK1 -).
To clarify whether the uvrY-regulated pore-forming toxins contribute to the virulence of A. dhakensis, isogenic mutants of hemolysin (ahh1), aerolysin (aeroA), and collagenase were generated. Interestingly, only worms infected with the ahh1 mutant failed to demonstrate pore-forming phenotypes, as was also the case for the worms infected with OP50 and the uvrY mutant -). The worms fed with the aeroA mutant and collagenase mutant exhibited obvious pore-formation phenotypes -). The ahh1 complement strain rescued the pore-formation phenotype in C. elegans -).
To link pore-formation pathology and virulence, the standard C. elegans survival assay was conducted ()). The ahh1 as well as the uvrY mutants were less virulent as compared to with the aeroA and collagenase mutants. Although the aeroA and collagenase mutants did show pore-formation phenotypes in C. elegans, these two virulence factors still exhibited toxicity to a lesser degree in C. elegans. In summary, hemolysin ahh1, being one of the most potent virulence factors in A. dhakensis, is regulated by uvrY and has the characteristics of a pore-forming toxin.
UvrY is required for the expression of hemolysin Ahh1.
To determine whether UvrY is necessary for the expression of Ahh1, the Ahh1 complement strain under a uvrY deletion background was generated. Interestingly, the virulence of the ahh1-promoter-driven Ahh1 complement strain under the uvrY deletion background [AAK1 ΔuvrY-pahh1(pahh1)] was not reversed. In contrast, the virulence of the Ahh1 complement strain under the ahh1 deletion background [AAK1 Δahh1-pahh1(pahh1)] was reversed in the C. elegans model ()). The toxicity of the ΔuvrY-pahh1(pahh1) strain was reduced, similar to the uvrY deletion mutant and the ahh1 deletion mutant. However, the strain with complementation of a constitutively active house-keeping gene gyrB promotor-driven Ahh1 strain in the uvrY deletion mutant [AAK1 ΔuvrY-pahh1(pgyrB)] increased the toxicity ). Over-expression of Ahh1 in a nonpathogenic E. coli DH5α showed an increase in toxicity ). Similar to AAK1 ), a higher proportion of worms infected with AAK1 Δahh1-pahh1(pahh1) strain showed a pore-formation phenotype (-)), in contrast to a lower proportion in the AAK1 ΔuvrY-pahh1(pahh1) strain (-)). The pore-formation rate was also increased in the DH5αpahh1(pgyrB) strain as compared to DH5α (, and ). The pore-formation ratio was correlated to the virulence in terms of inverse survival in C. elegans.
To reconfirm the increase in toxicity was due to an effective Ahh1 complementation, a western blot analysis was performed on Ahh1. With a polyclonal anti-Ahh1 antibody, Ahh1 proteins were detected only in the wild type AAK1, AAK1 ΔuvrY-pahh1(pgyrB), and DH5αpahh1(pgyrB) (). It is thus suggested that the toxicity of bacteria results from the expression of Ahh1. Taken together, UvrY controls the virulence of hemolysin Ahh1 and is required for the full expression of virulence in A. dhakensis.
Discussion
In this research, we generated a transposon mutation library of A. dhakensis and surveyed the A. dhakensis mutants with attenuated virulence in C. elegans. We identified UvrY and discovered that UvrY regulates the expression of hemolysin Ahh1 as well as other virulence factors in A. dhakensis. The pore-formation caused by Ahh1 is a vital pathogenesis of A. dhakensis infection in C. elegans ().
In our mutant library, mutation of genes involved in flagella biosynthesis are abundant. These hits present reduced virulence in C. elegans and suggest that flagella biosynthesis is also important for the virulence of A. dhakensis. Flagella is a key mobility structure responsible for chemotaxis and biofilm formation [Citation49,Citation50]. Mutation of flagella may cause a decrease in attachment to the cell surface and reduce the infection efficiency in hosts.
We identified UvrY as a key component in the control of the full virulence of A. dhakensis. The similarity of the UvrY homologue among different bacteria genera is shown in S7A Fig. Using a RAST analysis, the UvrY of A. dhakensis AAK1 and its homologue in other bacteria genera were demonstrated, as shown in S7B Fig and S7 C Fig. It is believed that the homologue of UvrY plays a similar role in regulating virulence, such as in E. coli, Salmonella, Pseudomonas, and Vibrio [Citation43]. Since UvrY or UvrY -like regulators control a broad range of virulence factors in pathogenic bacteria, further studies are warranted to figure out the exact mechanism mediated by UvrY.
As a member of a two-component system, the upstream sensor kinase which phosphorylates UvrY is also thought to play a role in determining the virulence of A. dhakensis. The most well-known sensor kinase of UvrY is BarA. Studies have found that BarA phosphorylates UvrY D54 residue [Citation11,Citation42,Citation43]. In this study, however, complementation of UvrY D54E (constitutively active form) and D54A (inactive form) both rescued the virulence (,)). It suggests that the phosphorylation of D54 residue is not essential for UvrY-associated virulence. Possible interpretations for this finding include (1) D54 of UvrY is phosphorylated by BarA, however, is not essential for UvrY-associated toxicity; (2) Beside BarA, there is another phosphate donor that phosphorylates UvrY at non-D54 residue.
A. dhakensis barA mutant is as attenuated as uvrY mutant (,)), However, the increased toxicity of ΔbarA-puvrY in (,)) suggested that without the phosphorylation by BarA, overexpressed UvrY is able to activate the downstream signaling. This result is similar to previous finding that UvrY may be phosphorylated by other molecule, such as acetyl-phosphate [Citation44,Citation45]. Interestingly, barA mutant which showed attenuated virulence still maintained a copy of uvrY gene. These evidences suggest that the attenuated virulence of barA mutant may come from suppressed expression of downstream signaling which is regulated by different mechanism. Base on this, we tested the expression of Ahh1 in barA mutant (S8 Fig). The result showed that barA mutant still produced pro-Ahh1, a pro-form of Ahh1. In contrast, expression of pro-Ahh1 and Ahh1 is largely deficient in uvrY mutant (). In a brief summary, the roles of BarA, UvrY, BarA-UvrY two-component system, are complex then as expected. Further studies for identification of other factors which play a role in regulating virulence are needed.
Based on the transcriptome results, UvrY is believed to control the expression of several virulent genes. However, how can UvrY regulate these downstream virulence genes is still unclear. As a response regulator in a two-component system, UvrY is thought to be a transcription factor. It is possible that UvrY participates in the transcription of these virulence directly. However, the possibility also exists that UvrY affects the expression of some downstream virulent genes indirectly. For example, UvrY transcribes small RNA csrB, csrC [Citation14,Citation15], rsmY, and rsmZ [Citation51–Citation53], and these small RNAs bind to translation initiator CsrA to suppress the function of CsrA, which is an RNA binding protein influencing post-transcriptional regulation and causing changes in translation [Citation54]. The consensus binding sequences of UvrY at the promoter region of small RNAs is not clearly understood [Citation11,Citation53,Citation55]. In addition, the sequences of these potential binding regions are not cohesive. The lack of consistent binding sequences increases the possibility that UvrY regulates a broad range of RNAs that subsequently activate the expression of downstream virulent genes.
In our case, UvrY was required for the expression of hemolysin ahh1. We therefore provide some proposals which explain how UvrY regulate ahh1 (1) UvrY binds to the ahh1 promoter and transcribes ahh1 directly. (2) CsrA, the downstream of UvrY, affects the expression of ahh. (3) Other unknown components, which are regulated by UvrY, bind to either the promoter of ahh1 or the mRNA of ahh1. We screened transcription factors which were affected by UvrY, and identified NarL and PdhR as potential transcription regulators of ahh1 based on RNAseq analysis. Since NarL and PdhR have been suggested to be negative transcription regulators in previous studies [Citation56,Citation57], the transcription levels of narL and pdhR are increased as expected in the RNAseq profiles of uvrY mutant (S6 file), and are supposed to repress ahh1expression consequently. However, further research to prove this hypothesis is warranted.
Although UvrY regulates many virulence factors, the complementation of Ahh1 to the uvrY mutant rescues full virulence. In contrast, the well-known aerolysin, an important pore-forming toxin of Aeromonas in the cell and mouse models [Citation58,Citation59], is not associated with A. dhakensis virulence in C. elegans. This phenomenon may be explained by the fact that aerolysin is not an effective pore-forming toxin to C. elegans. Similarly, collagenase may contribute to the limited virulence of A. dhakensis toxicity in C. elegans. As has been previously found [Citation60], it is possible that collagenase is not the central virulence factor of A. dhakensis. Due to the virulent traits of Ahh1, further study to the determine its pathogenesis is necessary.
Because UvrY is prevalent in a variety of bacteria genera and acts as a key regulator of many virulence factors, UvrY is a potential therapeutic target of antimicrobial agents. Previous studies have shown that two-component signaling systems are potential drug targets when developing new antimicrobial agents [Citation61]. Therapy with novel agents inhibiting uvrY expression along with traditional antibiotics may lead to additional benefits for patients suffering from severe sepsis.
Figure 1. Identification of genes participating in the virulence of A. dhakensis. (a) Diagram of the establishment of the A. dhakensis AAK1 mini-Tn10 transposon library. (b) Screen of transposon mutants with attenuated virulence. (c) GO analysis of candidate genes based on molecular function. (d) Identification of uvrY as a target gene. (***P < 0.001, **P < 0.01 and *P < 0.05)
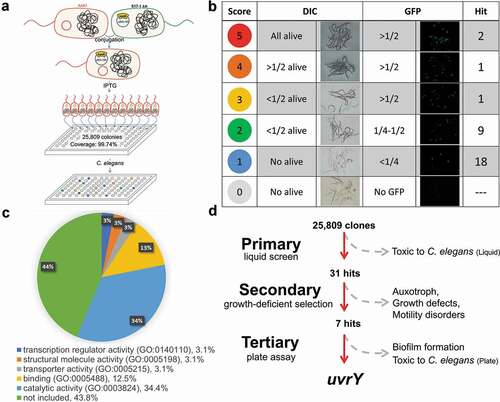
Figure 2. UvrY dominates the virulence without altering the physiological functions of Aeromonas dhakensis in terms of (a) swimming, (b) swarming, (c) auxotroph, (d) growth curve, (e) biofilm formation, and (f) virulence to C.elegans. s. aureus ATCC 29213 is a negative control strain for swimming. B. subtilis 3610 is a positive control strain for swarming. E. coli OP50 is a negative control strain for auxotroph, growth curve and biofilm formation. (***P < 0.001, **P < 0.01 and *P < 0.05)
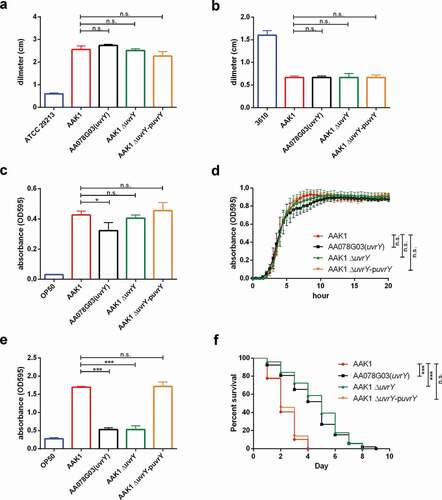
Figure 3. The BarA-UvrY two-component system controls the toxicity of A. dhakensis AAK1. the virulence of the barA mutant is similar to the uuvrY mutant in (a) C. elegans and (b) the C2C12 mouse myoblast cell line. complementation of uvrY rescues the toxicity not only in the uvry mutant but also in the bara mutant in (c) C. elegans and (d) the C2C12 mouse myoblast cell line. complementation of both UvrY D54E (constitutively active form) and UvrY D54A (inactive form) reverse the toxicity in (e) C. elegans and (f) the C2C12 mouse myoblast cell line. (***p < 0.001, **p < 0.01 and *p < 0.05)
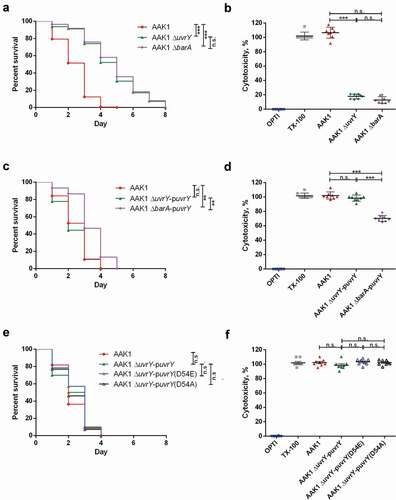
Figure 4. UvrY is required for A. dhakensis virulence in a mouse infection model. (a) Survival of mice infected with various doses of wild type A. dhakensis AAK1. (b) Survival of mice infected with the wild type A. dhakensis AAK1, uvrY mutant, and uvrY complement strains at high doses (4x106 CFU). The levels of cytokine (c) IL-6, (d) MCP-1, and (e) TNF-α in mice infected with the wild type A. dhakensis AAK1, uvrY mutant and uvrY complement strains. Bacterial load in the (f) blood, (g) liver, and (h) spleen of mice infected with A. dhakensis AAK1, uvrY mutant, and the uvrY complement strain. (i) Differences in serum resistance among the A. dhakensis AAK1, uvrY mutant, and the uvrY complement strains were not significant. (***P < 0.001, **P < 0.01 and *P < 0.05)
Figure 5. Several virulence factors are regulated by UvrY in A. dhakensis. The transcriptome of UvrY was analyzed using an RNAseq analysis. (a) Scatter diagram of the fold change of in gene transcriptional levels in the uvrY mutant as compared to the wild type AAK1 (FCA) and in the uvrY mutant as compared to the uvrY complement (FCC), where each dot represents a transcript. The red dots are the transcripts with more than a two-fold increase in expression levels in the uvrY mutant as compared to the control strains. The blue dots are transcripts with more than a two-fold decrease in the expression levels in the uvrY mutant as compared to the control strains. (b) Venn diagram of the blue dots shown in Fig 5A. There are 120, 271, and 699 genes associated with the intersection of FCA10X and FCC10X, FCA5X and FCC5X, and FCA2X and FCC2X, respectively. (c) GO analysis of 120 candidate genes based on molecular function. (d) Category analysis of the 120 candidate genes using a RAST analysis. (e) Genes in the “Virulence, Disease and Defense” category shown in Fig 5D. (f) qRT-PCR of all genes belonging the “Virulence, Disease and Defense” category shown in Fig 5D
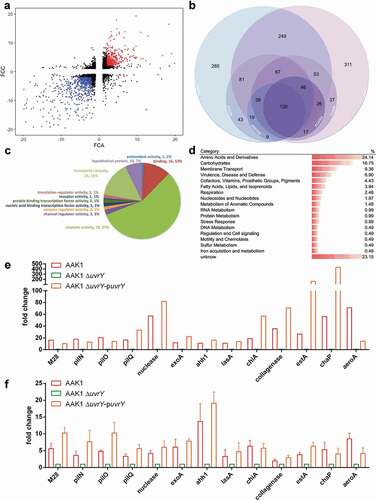
Figure 6. Hemolysin (Ahh1), a pore-forming toxin is regulated by UvrY in A. dhakensis. The pore-forming phenotype in the intestine of C. elegans after infection with (a) E. coli OP50, (b) A. dhakensis AAK1, (c) the A. dhakensis AAK1 uvrY deletion mutant, (d) the A. dhakensis AAK1 uvrY complement, (e) the A. dhakensis AAK1 hemolysin ahh1 deletion mutant, (f) the A. dhakensis AAK1 ahh1 complement, (g) the A. dhakensis AAK1 aerolysin aeroA deletion mutant, and (h) the A. dhakensis AAK1 collagenase deletion mutant. The quantification of pore formation is summarized in (i). (j) Survival assay of C. elegans infected with A. dhakensis mutants. The scale bars in (A) to (H) are all 30 μm (***P < 0.001, **P < 0.01 and *P < 0.05)
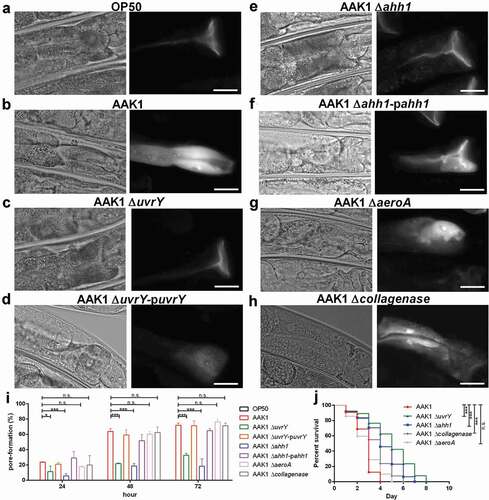
Figure 7. UvrY is required for the expression of hemolysin Ahh1 in A. dhakensis. (a) Survival assay of C. elegans infected with A. dhakensis mutants. Complementation of ahh1-promoter-driven ahh1 did not rescue the toxicity of the uvrY mutant. (b) A. dhakensis mutants with complementation of gyrB-promoter-driven ahh1 recovered the toxicity in the C. elegans survival assay. (c) C. elegans infected with Ahh1 over-expressed bacteria had lower survival rates. The pore-formation phenotype in the intestine of C. elegans infected with (d) A. dhakensis AAK1 (e) the ahh1-promoter-driven ahh1 complement to uvrY deletion mutant (f) the gyrB-promoter-driven ahh1 complement to uvrY deletion mutant (g) E. coli DH5α and (h) Ahh1 over-expressed E. coli DH5α. The quantification of pore-formation is summarized in (i) and (j). (k) Western blot of Ahh1 in the cell lysates of different strains. The scale bars in (D) to (H) are all 30 μm. (***P < 0.001, **P < 0.01 and *P < 0.05)
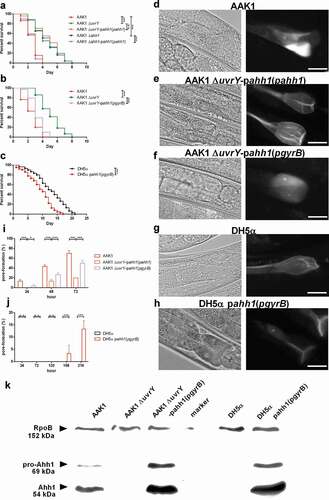
Figure 8. Schematic diagram of UvrY-mediated pathogenesis in Aeromonas dhakensis. Activation of UvrY is induced by BarA or other unknown molecules. UvrY regulates the expression of Ahh1, the most potent toxin as well as other virulence factors in A. dhakensis. Ahh1 results in pore-formation in cells and cause severe infection in hosts
Author contributions
YWC designed the experiments, performed the experiments and wrote the manuscript. WHY, YCS, STW and CJK conducted the experiments. HJT, YCC and CCC performed the mouse experiments. JWC conducted NGS analysis. HYS performed transposon library. CRW and WCK provided bacterial strains. CSC designed the experiments and supervised the project. PLC designed the experiments, supervised the project and wrote the manuscript.
Supplemental Material
Download Zip (9.8 MB)Acknowledgments
The NGS and mapping of RNAseq were performed using the system provided by the Bioinformatics Core at National Cheng Kung University, supported by the Ministry of Science and Technology, Taiwan. We would like to thank for providing experimental space and facilities by the Diagnostic Microbiology and Antimicrobial Resistance Laboratory, National Cheng Kung University Hospital, College of Medicine, National Cheng Kung University, Tainan, Taiwan. We also thank Savana Moore, who is a native speaker of English and an expert in editing science research for the Foreign Language Center at National Cheng Kung University.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Wu CJ, Chen PL, Tang HJ, et al. Incidence of aeromonas bacteremia in Southern Taiwan: vibrio and salmonella bacteremia as comparators. J Microbiol Immunol Infect. 2014;47(2):145–148. PubMed PMID: 23063268.
- Janda JM, Abbott SL. The genus aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev. 2010;231:35–73. PubMed PMID: 20065325; PubMed Central PMCID: PMCPMC2806660.
- Chen PL, Wu CJ, Chen CS, et al. A comparative study of clinical aeromonas dhakensis and aeromonas hydrophila isolates in southern Taiwan: A. dhakensis is more predominant and virulent. Clin Microbiol Infect. 2014;20(7):O428–34. Epub 2013/ 11/19.. PubMed PMID: 24237662.
- Parker JL, Shaw JG. Aeromonas spp. clinical microbiology and disease. J Infect. 2011;622:109–118. PubMed PMID: 21163298.
- Chen PL, Lamy B, Ko WC. Aeromonas dhakensis, an increasingly recognized human pathogen. Front Microbiol. 2016;7:793. Epub 2016/06/16. PubMed PMID: 27303382; PubMed Central PMCID: PMCPMC4882333. .
- Chen PL, Wu CJ, Tsai PJ, et al. Virulence diversity among bacteremic Aeromonas isolates: ex vivo, animal, and clinical evidences. PLoS One. 2014;9(11):e111213. Epub 2014/ 11/07.. PubMed PMID: 25375798; PubMed Central PMCID: PMCPMC4222899.
- Morinaga Y, Yanagihara K, Eugenin FLL, et al. Identification error of aeromonas aquariorum: a causative agent of septicemia. Diagn Microbiol Infect Dis. 2013;76(1):106–109. Epub 2013/ 03/07.. PubMed PMID: 23461831.
- Tomas JM. The main aeromonas pathogenic factors. ISRN Microbiol. 2012;2012:256261. PubMed PMID: 23724321; PubMed Central PMCID: PMCPMC3658858.
- Rasmussen-Ivey CR, Figueras MJ, McGarey D, et al. Virulence factors of aeromonas hydrophila: in the Wake of reclassification. Front Microbiol. 2016;7:1337. Epub 2016/ 09/10.. PubMed PMID: 27610107; PubMed Central PMCID: PMCPMC4997093. .
- Pernestig AK, Georgellis D, Romeo T, et al. The Escherichia coli BarA-UvrY two-component system is needed for efficient switching between glycolytic and gluconeogenic carbon sources. J Bacteriol. 2003;185(3):843–853. PubMed PMID: 12533459; PubMed Central PMCID: PMCPMC142795..
- Zere TR, Vakulskas CA, Leng Y, et al. genomic targets and features of bara-uvry (-sira) signal transduction systems. PLoS One. 2015;10(12):e0145035. Epub 2015/ 12/18.. PubMed PMID: 26673755; PubMed Central PMCID: PMCPMC4682653.
- Goodier RI, Ahmer BM. SirA orthologs affect both motility and virulence. J Bacteriol. 2001;1837:2249–2258. PubMed PMID: 11244064; PubMed Central PMCID: PMCPMC95131.
- Herren CD, Mitra A, Palaniyandi SK, et al. The bara-uvry two-component system regulates virulence in avian pathogenic escherichia coli O78: K80:H9. Infect Immun. 2006;748:4900–4909. PubMed PMID: 16861679; PubMed Central PMCID: PMCPMC1539585.
- Seyll E, Van Melderen L. The ribonucleoprotein csr network. Int J Mol Sci. 2013;1411:22117–22131. PubMed PMID: 24217225; PubMed Central PMCID: PMCPMC3856055.
- Gore AL, Payne SM. CsrA and cra influence shigella flexneri pathogenesis. Infect Immun. 2010;7811:4674–4682. PubMed PMID: 20713625; PubMed Central PMCID: PMCPMC2976347.
- Teplitski M, Goodier RI, Ahmer BM. Pathways leading from bara/sira to motility and virulence gene expression in salmonella. J Bacteriol. 2003;185(24):7257–7265. PubMed PMID: 14645287; PubMed Central PMCID: PMCPMC296259..
- Gauthier JD, Jones MK, Thiaville P, et al. Role of gaca in virulence of vibrio vulnificus. Microbiology. 2010;156(Pt 12):3722–3733. PubMed PMID: 20817642.
- Parkins MD, Ceri H, Storey DG. Pseudomonas aeruginosa gaca, a factor in multihost virulence, is also essential for biofilm formation. Mol Microbiol. 2001;40(5):1215–1226. PubMed PMID: 11401724..
- Boddicker JD, Anderson RA, Jagnow J, et al. Signature-tagged mutagenesis of klebsiella pneumoniae to identify genes that influence biofilm formation on extracellular matrix material. Infect Immun. 2006;74(8):4590–4597. Epub 2006/07/25. PubMed PMID: 16861646; PubMed Central PMCID: PMCPMC1539622.
- Das S, Noe JC, Paik S, et al. An improved arbitrary primed PCR method for rapid characterization of transposon insertion sites. J Microbiol Methods. 2005;63(1):89–94. Epub 2005/09/15. PubMed PMID: 16157212.
- Murray TS, Kazmierczak BI. FlhF is required for swimming and swarming in pseudomonas aeruginosa. J Bacteriol. 2006;188(19):6995–7004. Epub 2006/09/19. PubMed PMID: 16980502; PubMed Central PMCID: PMCPMC1595508.
- Kearns DB, Losick R. Swarming motility in undomesticated Bacillus subtilis. Mol Microbiol. 2003;49(3):581–590. Epub 2003/07/17.PubMed PMID: 12864845..
- Stiernagle T. Maintenance of C. elegans. WormBook. 2006;1–11. Epub 2007/ 12/01. PubMed PMID: 18050451; PubMed Central PMCID: PMCPMC4781397. Doi:https://doi.org/10.1895/wormbook.1.101.1.
- O’Toole GA. Microtiter dish biofilm formation assay. J Vis Exp. 2011;(47): Epub 2011/02/11. PubMed PMID: 21307833; PubMed Central PMCID: PMCPMC3182663. Doi:https://doi.org/10.3791/2437.
- Chen PL, Chen YW, Ou CC, et al. A disease model of muscle necrosis caused by aeromonas dhakensis infection in caenorhabditis elegans. Front Microbiol. 2016;7:2058. Epub 2017/ 01/20.. PubMed PMID: 28101079; PubMed Central PMCID: PMCPMC5209350. .
- Chen YW, Ko WC, Chen CS, et al. RIOK-1 Is a suppressor of the p38 mapk innate immune pathway in caenorhabditis elegans. Front Immunol. 2018;9:774. Epub 2018/05/03. PubMed PMID: 29719537; PubMed Central PMCID: PMCPMC5913292. .
- Chen YW, Ko WC, Chen CS, et al. Evaluating virulence and pathogenesis of aeromonas infection in a caenorhabditis elegans model. J Vis Exp. 2018;(142): Epub 2019/ 01/08. PubMed PMID: 30614490. doi: https://doi.org/10.3791/58768.
- Hmelo LR, Borlee BR, Almblad H, et al. Precision-engineering the pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc. 2015;10(11):1820–1841. Epub 2015/10/23.. PubMed PMID: 26492139; PubMed Central PMCID: PMCPMC4862005.
- Kovach ME, Elzer PH, Hill DS, et al. et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166(1):175–176. Epub 1995/12/01. PubMed PMID: 8529885. .
- Ku YH, Chuang YC, Chen CC, et al. Klebsiella pneumoniae Isolates from meningitis: epidemiology, virulence and antibiotic resistance. Sci Rep. 2017;7(1):6634. Epub 2017/07/28. PubMed PMID: 28747788; PubMed Central PMCID: PMCPMC5529541.
- Pollitt EJG, Szkuta PT, Burns N, et al. Staphylococcus aureus infection dynamics. PLoS Pathog. 2018;14(6):e1007112. Epub 2018/06/15. PubMed PMID: 29902272; PubMed Central PMCID: PMCPMC6019756.
- Wu CJ, Wang HC, Chen CS, et al. Genome sequence of a novel human pathogen, aeromonas aquariorum. J Bacteriol. 2012;194(15):4114–4115. Epub 2012/07/21. PubMed PMID: 22815437; PubMed Central PMCID: PMCPMC3416550.
- Kuo CJ, Wang ST, Lin CM, et al. A multi-omic analysis reveals the role of fumarate in regulating the virulence of enterohemorrhagic Escherichia coli. Cell Death Dis. 2018;9(3):381. Epub 2018/03/09. PubMed PMID: 29515100; PubMed Central PMCID: PMCPMC5841434.
- Aziz RK, Bartels D, Best AA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9(1):75. Epub 2008/ 02/12. PubMed PMID: 18261238; PubMed Central PMCID: PMCPMC2265698.
- Overbeek R, Olson R, Pusch GD, et al. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 2014;42(Database issue):D206–14. Epub 2013/ 12/03. PubMed PMID: 24293654; PubMed Central PMCID: PMCPMC3965101.
- Los FC, Kao CY, Smitham J, et al. RAB-5- and RAB-11-dependent vesicle-trafficking pathways are required for plasma membrane repair after attack by bacterial pore-forming toxin. Cell Host Microbe. 2011;9(2):147–157. Epub 2011/02/16. PubMed PMID: 21320697; PubMed Central PMCID: PMCPMC3057397.
- Chen HD, Kao CY, Liu BY, et al. HLH-30/TFEB-mediated autophagy functions in a cell-autonomous manner for epithelium intrinsic cellular defense against bacterial pore-forming toxin in C. elegans. Autophagy. Epub 2016/ 11/23. PubMed PMID: 27875098; PubMed Central PMCID: PMCPMC5324838. 2017;13(2):371–385. .
- Alexeyev MF, Shokolenko IN. Mini-Tn10 transposon derivatives for insertion mutagenesis and gene delivery into the chromosome of gram-negative bacteria. Gene. Epub 1995/07/04. PubMed PMID: 7628717. 1995;160(1):59–62. .
- Perez-Ortin JE, Lí Del Olmo M, Matallana E, et al. Making your own gene library. Biochemical Education. 1997;25(4):237–242.
- Mi H, Muruganujan A, Ebert D, et al. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47(D1):D419–D26. Epub 2018/ 11/09. PubMed PMID: 30407594; PubMed Central PMCID: PMCPMC6323939.
- Bertels F, Merker H, Kost C. Design and characterization of auxotrophy-based amino acid biosensors. PLoS One. 2012;7(7):e41349. Epub 2012/07/26. PubMed PMID: 22829942; PubMed Central PMCID: PMCPMC3400592.
- Zuber S, Carruthers F, Keel C, et al. GacS sensor domains pertinent to the regulation of exoproduct formation and to the biocontrol potential of pseudomonas fluorescens CHA0. Mol Plant Microbe Interact. 2003;16(7):634–644. Epub 2003/07/10. PubMed PMID: 12848429.
- Pernestig AK, Melefors O, Georgellis D. Identification of uvry as the cognate response regulator for the bara sensor kinase in escherichia coli. J Biol Chem. 2001;276(1):225–231. Epub 2000/ 10/07. PubMed PMID: 11022030.
- Tomenius H, Pernestig AK, Mendez-Catala CF, et al. Genetic and functional characterization of the escherichia coli bara-uvry two-component system: point mutations in the hamp linker of the bara sensor give a dominant-negative phenotype. J Bacteriol. 2005;187(21):7317–7324. Epub 2005/ 10/21. PubMed PMID: 16237014; PubMed Central PMCID: PMCPMC1273000.
- Camacho MI, Alvarez AF, Chavez RG, et al. Effects of the global regulator csra on the bara/uvry two-component signaling system. J Bacteriol. 2015;197(5):983–991. Epub 2014/12/24. PubMed PMID: 25535275; PubMed Central PMCID: PMCPMC4325108.
- Holub M, Lawrence DA, Andersen N, et al. Cytokines and chemokines as biomarkers of community-acquired bacterial infection. Mediators Inflamm. 2013;2013:190145. Epub 2013/05/22.. PubMed PMID: 23690657; PubMed Central PMCID: PMCPMC3652189. .
- Szurmant H, White RA, Hoch JA. Sensor complexes regulating two-component signal transduction. Curr Opin Struct Biol. 2007;17(6):706–715. Epub 2007/10/05. PubMed PMID: 17913492; PubMed Central PMCID: PMCPMC2175030.
- Los FC, Randis TM, Aroian RV, et al. Role of pore-forming toxins in bacterial infectious diseases. Microbiol Mol Biol Rev. 2013;77(2):173–207.
- Santos PG, Santos PA, Bello AR, et al. Association of aeromonas caviae polar and lateral flagella with biofilm formation. Lett Appl Microbiol. 2011;52(1):49–55. Epub 2010/ 12/02. PubMed PMID: 21118278.
- Stecher B, Hapfelmeier S, Muller C, et al. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar typhimurium colitis in streptomycin-pretreated mice. Infect Immun. 2004;72(7):4138–4150. Epub 2004/06/24. PubMed PMID: 15213159; PubMed Central PMCID: PMCPMC427403.
- Brencic A, McFarland KA, McManus HR, et al. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol Microbiol. 2009;73(3):434–445. Epub 2009/07/16. PubMed PMID: 19602144; PubMed Central PMCID: PMCPMC2761719.
- Chambonnier G, Roux L, Redelberger D, et al. The hybrid histidine kinase lads forms a multicomponent signal transduction system with the gacs/gaca two-component system in pseudomonas aeruginosa. PLoS Genet. 2016;12(5):e1006032. Epub 2016/05/14. PubMed PMID: 27176226; PubMed Central PMCID: PMCPMC4866733.
- Humair B, Wackwitz B, Haas D. GacA-controlled activation of promoters for small RNA genes in Pseudomonas fluorescens. Appl Environ Microbiol. 2010;76(5):1497–1506. Epub 2010/01/06. PubMed PMID: 20048056; PubMed Central PMCID: PMCPMC2832403.
- Potts AH, Vakulskas CA, Pannuri A, et al. Global role of the bacterial post-transcriptional regulator CsrA revealed by integrated transcriptomics. Nat Commun. 2017;8(1):1596. Epub 2017/11/19. https://doi.org/10.1038/s41467-017-01613-1. PubMed PMID: 29150605; PubMed Central PMCID: PMCPMC5694010.
- Teplitski M, Al-Agely A, Ahmer BM. Contribution of the SirA regulon to biofilm formation in Salmonella enterica serovar typhimurium. Microbiology. 2006;152(Pt 11):3411–3424. Epub 2006/11/01. PubMed PMID: 17074910.
- Bearson SM, Albrecht JA, Gunsalus RP. Oxygen and nitrate-dependent regulation of dmsABC operon expression in Escherichia coli: sites for Fnr and NarL protein interactions. BMC Microbiol. 2002;2(1):13. Epub 2002/ 06/25.PubMed PMID: 12079504; PubMed Central PMCID: PMCPMC116602..
- Maeda S, Shimizu K, Kihira C, et al. Pyruvate dehydrogenase complex regulator (PdhR) gene deletion boosts glucose metabolism in Escherichia coli under oxygen-limited culture conditions. J Biosci Bioeng. 2017;123(4):437–443. Epub 2016/ 12/23. PubMed PMID: 28007420.
- Szczesny P, Iacovache I, Muszewska A, et al. Extending the aerolysin family: from bacteria to vertebrates. PLoS One. 2011;6(6):e20349. Epub 2011/ 06/21. PubMed PMID: 21687664; PubMed Central PMCID: PMCPMC3110756.
- McCoy AJ, Koizumi Y, Higa N, et al. Differential regulation of caspase-1 activation via NLRP3/NLRC4 inflammasomes mediated by aerolysin and type III secretion system during Aeromonas veronii infection. J Immunol. 2010;185(11):7077–7084. Epub 2010/11/03. PubMed PMID: 21037094.
- Duarte AS, Correia A, Esteves AC. Bacterial collagenases - A review. Crit Rev Microbiol. 2016;42(1):106–126. Epub 2014/ 04/24. PubMed PMID: 24754251.
- Shor E, Chauhan N, Goldman WE. A case for two-component signaling systems as antifungal drug targets. PLoS Pathog. 2015;11(2):e1004632. Epub 2015/02/28. PubMed PMID: 25723524; PubMed Central PMCID: PMCPMC4344368.
- Maier W, Adilov B, Regenass M, et al. A neuromedin U receptor acts with the sensory system to modulate food type-dependent effects on C. elegans lifespan. PLoS Biol. 2010;8(5):e1000376. Epub 2010/06/04. PubMed PMID: 20520844; PubMed Central PMCID: PMCPMC2876044 subject matter of a patent application (PCT/EP2008/061541).
- Pollitt EJG, Diggle SP. Defining motility in the Staphylococci. Cell Mol Life Sci. 2017;74(16):2943–2958. Epub 2017/04/06.. PubMed PMID: 28378043; PubMed Central PMCID: PMCPMC5501909.
