ABSTRACT
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) outbreak resulted in 5,993,317 confirmed cases worldwide with 365,394 confirmed deaths (as of May 29th, 2020, WHO). The molecular mechanism of virus infection and spread in the body is not yet disclosed, but studies on other betacoronaviruses show that, upon cell infection, these viruses inhibit macroautophagy/autophagy flux and cause the accumulation of autophagosomes. No drug has yet been approved for the treatment of SARS-CoV-2 infection; however, preclinical investigations suggested repurposing of several FDA-approved drugs for clinical trials. Half of these drugs are modulators of the autophagy pathway. Unexpectedly, instead of acting by directly antagonizing the effects of viruses, these drugs appear to function by suppressing autophagy flux. Based on the established cross-talk between autophagy and apoptosis, we speculate that over-accumulation of autophagosomes activates an apoptotic pathway that results in apoptotic death of the infected cells and disrupts the virus replication cycle. However, administration of the suggested drugs are associated with severe adverse effects due to their off-target accumulation. Nanoparticle targeting of autophagy at the sites of interest could be a powerful tool to efficiently overcome SARS-CoV-2 infection while avoiding the common adverse effects of these drugs.
Viruses recruit cellular machinery and pathways, such as autophagy, for their replication and spread [Citation1, Citation2]. Autophagy is a part of the cell stress response that works as a quality control mechanism for cells by removing and degrading malfunctioning proteins, damaged organelles, and invasive microbes [Citation1,Citation3]. Macroautophagy, hereafter autophagy, is initiated via the formation of a double-membrane structure (termed a phagophore). The phagophore engulfs the substrates that are targeted for ultimate degradation, and sequesters them within an autophagosome. The mature autophagosome merges with a lysosome to generate an autolysosome where the engulfed material will be degraded [Citation1,Citation4].
Hijacking of cellular autophagy mechanisms has been reported for several viruses. For example, measles virus/MeV induces autophagy through the engagement of CD46; human immunodeficiency virus type 1/HIV-1 envelope glycoproteins gp120 and gp41 induce autophagy in uninfected CD4+ T cells and initiate HIV-1 entry with subsequent T cell apoptosis and immunodeficiency; Chikungunya virus/CHIKV triggers autophagy via an endoplasmic reticulum and oxidative stress pathway [Citation5]; Macacine alphaherpesvirus 1/MCHV, and murine gammaherpesvirus (MHV) 68/MHV-68 inhibit autophagy by blocking phagophore formation [Citation5]; Picornaviruses, coxsackie virus and coronaviruses utilize autophagy to promote their replication [Citation5]. Although these viruses hijack cellular autophagy pathways in favor of their replication and transcription, for other viruses autophagy restricts the viral infection by degrading engulfed viruses in a process called virophagy [Citation5].
The SARS-CoV-2 global outbreak, responsible for coronavirus disease 2019 (COVID-19) [Citation6,Citation7], belongs to the betacoronavirus (βCoV) genus. This genus also includes SARS-CoV, Middle East respiratory syndrome-coronavirus (MERS-CoV) and MHV [Citation8]. βCoV are positive-sense RNA viruses [Citation9]. Among them, MHV has been used as a prototype for βCoV in biological investigations. βCoV utilize double-membrane vesicles (DMVs), which are similar to autophagosomes, for their replication [Citation10]. Using MHV-infected delayed brain tumor/DBT cells, Prentice, and co-workers were the first to show the replication of βCoV inside DMVs [Citation11]. They also showed that βCoV induce ATG5-dependent autophagy [Citation11]. Another study confirmed βCoV induction of ATG5-dependent autophagosome formation via their NSP6 (non-structural protein 6) in MHV-infected VERO cells [Citation12]. Similarly, viral membrane-anchored papain-like protease/PLpro-TM polyprotein produced by both SARS-CoV and MERS-CoV induces the formation of autophagosomes, but inhibits their maturation, preventing the generation of autolysosomes as shown in three different human cell lines [Citation13]. In line with these reports, a recent study, using ATG5 wild-type and ATG5 knockout Vero B4 cells, reported that MERS-CoV infection suppresses autophagy flux by inhibiting the fusion step [Citation14]. In contrast, few studies reported a βCoV infection which is independent of autophagy induction mechanisms [Citation15,Citation16]. For example, Reggiori and co-workers confirmed that replication and release of βCoV are independent of autophagy [Citation15]. However, they showed that the virus utilizes DMVs coated with non-lipidated microtubule-associated protein 1 light chain 3 (LC3)-I for replication. To the best of our knowledge, no similar experiments have been conducted using SARS-CoV-2. However, an evolutionary analysis on SARS-CoV-2 genome sequences of 351 clinical samples revealed mutations in NSP6, a protein that has an inducing effect on autophagosome formation [Citation17]. This finding infers an interaction of SARS-CoV-2 cell infection and autophagy ().
Figure 1. Modulation of the autophagy pathway by coronaviruses and proposal of novel smart drug-loaded nanoparticles to target this pathway to combat COVID-19. Schematic shows how coronaviruses interact with autophagy. The NSP6 protein of SARS and MHV induces the formation of autophagosomes but confines their expansion and blocks their maturation into autolysosomes. A similar effect is observed by PLpro-TM of SARS. Human CoVs (HCoVs) via their NSPs, and MHV induce the formation of LC3-I-coated DMVs needed for viral RNA transcription and replication. MERS decreases the level of BECN1 (beclin 1) and blocks fusion of autophagosomes with lysosomes. Chloroquine/hydroxychloroquine, emtricitabine/tenofovir, interferon alfa-2b, lopinavir/ritonavir and ruxolitinib, which are all under clinical trial for treatment of SARS-CoV-2, induce autophagosome accumulation by blocking their maturation into autolysosomes. Thus, designing nanoparticles for the targeted delivery of these drug to avoid their off-target effects will provide safe and effective powerful tools to combat COVID-19. ATG14: autophagy related 14; DMV: double-membrane vesicles; EDEMosome: LC3-I-positive endoplasmic reticulum-derived vesicles exporting short-lived ERAD regulators; ER: endoplasmic reticulum; LC3-I: processed MAP1LC3; LC3-II: lipidated MAP1LC3; MERS: Middle East respiratory syndrome; MHV: murine gammaherpes virus; NSP6: non-structural protein 6; PIK3C3/VPS34: phosphatidylinositol 3-kinase catalytic subunit type 3; PIK3R4/VPS15: phosphoinositide-3-kinase regulatory subunit 4; PtdIns3 K: class III phosphatidylinositol 3-kinase; PLpro-TM: membrane-anchored papain-like protease; SARS: severe acute respiratory syndrome; ULK1 complex: unc-51 like autophagy activating kinase 1.
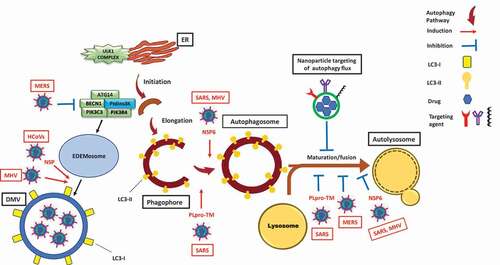
COVID-19 is associated with common symptoms such as fever and shortness of breath. These symptoms could progress to an acute respiratory distress syndrome/ARDS that leads to lung failure, the most common reason of death [Citation18]. To date, there is no clinically approved drug to prevent or cure COVID-19. Repurposing of FDA-approved drugs was associated with promising outcomes and resulted in ongoing clinical trials for 12 drugs tested against COVID-19, based on a recent WHO report [Citation19]. Several potential drug candidates are autophagy modulators (). Surprisingly, almost all of these autophagy modulators do not appear to act by directly antagonizing the effect of βCoVs. Instead, they inhibit autophagy flux in a similar fashion to the effect of βCoVs (). Therefore, we suggest that the beneficial effect of these drugs is possibly due to the over-accumulation of autophagosomes that can potentially induce apoptotic cell death of virally infected cells and disrupt the virus replication cycle, similar to what we observed in our recent study [Citation20].
Table 1. Drugs under clinical trials against SARS-CoV-2 infection based on the World Health Organization report [Citation19], their autophagy-related mechanism of action, and their severe side-effects.
It is very important to consider the unfolded protein response (UPR), an important intracellular pathway that is activated as a response to the accumulation of unfolded proteins in the endoplasmic reticulum (ER) with regard to viral infection. The UPR is usually activated during coronavirus infection because virus replication requires excessive protein biosynthesis and folding to provide sources for viral proteins, and use of the ER membrane for the formation of DMVs [Citation21,Citation22]. Furthermore, the UPR and autophagy are interconnected, and induction of the UPR could potentially facilitate or promote autophagy [Citation4,Citation23,Citation24]. Therefore, SARS-CoV-2 infection could possibly induce autophagy via UPR induction in the cells.
As depicted in , all of the indicated drugs have severe adverse effects and limited patient tolerance. This is attributed to the off-target effects of these drugs upon systemic administration [Citation25]. For instance, chloroquine/CQ has some potential as an effective therapy for COVID-19 based on preliminary clinical trial findings [Citation26], but is associated with retinopathy, neuromyopathy, nephropathy, and cardiomyopathy that makes it difficult to tolerate [Citation27,Citation28].
The body of literature pointing to the mutual effect of SARS-CoV-2 infection and autophagy, in addition to the fact that 58% of the drugs under clinical trials for COVID-19 are autophagy modulators [Citation26], emphasize the need for research in the area of autophagy for the fight against COVID-19. It is very important to consider that the drugs in modulate other mechanisms than auto-phagy to decrease SARS-CoV-2 infection. As an example, chloroquine/hydroxychloroquine has anti-inflammatory effects and might be involved in controlling a SARS-CoV-2-induced cytokine storm [Citation29], endocytosis of the virus [Citation30], and regulation of the SARS-COV-2 receptor, ACE2 (angiotensin I converting enzyme 2) [Citation29]. Some of these effects, including regulation of the cytokine storm, and endocytosis of the virus are indirectly regulated by auto-phagy [Citation30].
Therefore, we recommend two main research targets for scientists who are investigating the interconnection of viral infection and autophagy:
Mechanistic understanding of the intracellular trafficking and replication of SARS-CoV-2.
Developing effective therapies that are specific to SARS-CoV-2 and the autophagy pathway.
Successful implementation of an autophagy modulator as a safe and efficacious therapy for COVID-19 requires a carrier to deliver it to the site of action (infected cells) and mitigate off-target effects. Applications of nanotechnology in medicine (called nanomedicine), have introduced the use of nanoparticles for targeting active sites and avoiding off-target accumulation. This is based on the unique physical properties of nanoparticles, which affect their bioavailability and circulation time. Decorating the nanoparticles with ligands directed to specific cell targets amplifies nanoparticle specificity [Citation31,Citation32]. Other advantages offered by nanoparticles include their ability to cross biological barriers [Citation33], improved bioavailability of poorly soluble drugs (based on the large surface-area-to-volume ratio of nanoparticles compared to large particles) [Citation34] and tunability of nanoparticle surface charge and chemistry to further control interactions with cells and barriers [Citation33,Citation35]. Recently, nanoparticles were shown to modulate auto-phagy, and have been exploited for overcoming obstacles encountered with autophagy modulators [Citation36]. Several nanoparticle-based products are approved or under evaluation for the treatment of viral infections, including Inflexal V® (Crucell, Berna Biotech), and PegIntron® (Merck) [Citation37]. Therefore, nanotechnology has a great potential for contributing significantly to the fight against COVID-19 by developing effective therapies that can selectively block the replication of the virus in target cells [Citation38].
Further, SARS-CoV-2 could be considered as natural spherical nanoparticles (60- to 140-nm size range). Therefore, mechanisms established for nanoparticle interaction with target cells and subcellular organelles, could be used to enhance our understanding of cell binding and intracellular trafficking mechanisms of the virus [Citation39]. We strongly recommend cross-disciplinary collaborations between autophagy and nanotechnology communities in order to accelerate the discovery of potential drug candidates and the translation of these discoveries into clinically-approved COVID-19 therapies that are both effective and safe.
Abbreviations
Disclosure statement
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Choi Y, Bowman JW, Jung JU. Autophagy during viral infection - a double-edged sword. Nat Rev Microbiol. 2018;16(6):341–354.
- Yeganeh B, Ghavami S, Rahim MN, et al. Autophagy activation is required for influenza A virus-induced apoptosis and replication. Biochim Biophys Acta Mol Cell Res. 2018;1865(2):364–378.
- Ghavami S, Mutawe MM, Schaafsma D, et al. Geranylgeranyl transferase 1 modulates autophagy and apoptosis in human airway smooth muscle. Am J Physiol Lung Cell Mol Physiol. 2012;302(4):L420–428.
- Mehrbod P, Ande SR, Alizadeh J, et al. The roles of apoptosis, autophagy and unfolded protein response in arbovirus, influenza virus, and HIV infections. Virulence. 2019;10(1):376–413.
- Mao J, Lin E, He L, et al. Autophagy and Viral Infection. Adv Exp Med Biol. 2019;1209:55–78.
- Habtemariam S, Nabavi SF, Ghavami S, et al. Possible use of the mucolytic drug, bromhexine hydrochloride, as a prophylactic agent against SARS-CoV-2 infection based on its action on the transmembrane serine protease 2. Pharmacol Res. 2020;157:104853.
- Moradian N, Ochs HD, Sedikies C, et al. The urgent need for integrated science to fight COVID-19 pandemic and beyond. J Transl Med. 2020;18(1):205.
- Rehman SU, Shafique L, Ihsan A, et al. Evolutionary trajectory for the emergence of novel coronavirus SARS-CoV-2. Pathogens. 2020;9(3):240.
- Mousavizadeh L, Ghasemi S. Genotype and phenotype of COVID-19: their roles in pathogenesis. J Microbiol Immunol Infect. 2020. doi:https://doi.org/10.1016/j.jmii.2020.03.022
- Fung TS, Liu DX. Human coronavirus: host-pathogen interaction. Annu Rev Microbiol. 2019;73:529–557.
- Prentice E, Jerome WG, Yoshimori T, et al. Coronavirus replication complex formation utilizes components of cellular autophagy. J Biol Chem. 2004;279(11):10136–10141.
- Cottam EM, Whelband MC, Wileman T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014;10(8):1426–1441.
- Chen X, Wang K, Xing Y, et al. Coronavirus membrane-associated papain-like proteases induce autophagy through interacting with Beclin1 to negatively regulate antiviral innate immunity. Protein Cell. 2014;5(12):912–927.
- Gassen NC, Niemeyer D, Muth D, et al. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection. Nat Commun. 2019;10(1):5770.
- Reggiori F, Monastyrska I, Verheije MH, et al. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7(6):500–508.
- Zhao Z, Thackray LB, Miller BC, et al. Coronavirus replication does not require the autophagy gene ATG5. Autophagy. 2007;3(6):581–585.
- Benvenuto D, Angeletti S, Giovanetti M, et al. Evolutionary analysis of SARS-CoV-2: how mutation of non-structural protein 6 (NSP6) could affect viral autophagy. J Infect. 2020;81(1):e24-e27.
- Du Y, Tu L, Zhu P, et al. Clinical features of 85 fatal cases of COVID-19 from Wuhan: a retrospective observational study. Am J Respir Crit Care Med. 2020;201(11):1372–1379.
- WHO; 2020. Table of therapeutics against COVID-19 as 21st March 2020 WHO. Available from: https://www.who.int/blueprint/priority-diseases/key-action/Table_of_therapeutics_Appendix_17022020.pdf?ua=1
- Shojaei S, Koleini N, Samiei E, et al. Simvastatin increases temozolomide-induced cell death by targeting the fusion of autophagosomes and lysosomes. Febs J. 2020;287(5):1005–1034.
- Fung TS, Huang M, Liu DX. Coronavirus-induced ER stress response and its involvement in regulation of coronavirus–host interactions. Virus Res. 2014;194:110–123.
- Fung TS, Liu DX. Coronavirus infection, ER stress, apoptosis and innate immunity. Front Microbiol. 2014;5:296.
- Ghavami S, Sharma P, Yeganeh B, et al. Airway mesenchymal cell death by mevalonate cascade inhibition: integration of autophagy, unfolded protein response and apoptosis focusing on Bcl2 family proteins. Biochim Biophys Acta. 2014;1843(7):1259–1271.
- Ghavami S, Yeganeh B, Zeki AA, et al. Autophagy and the unfolded protein response promote profibrotic effects of TGF-beta1 in human lung fibroblasts. Am J Physiol Lung Cell Mol Physiol. 2018;314(3):L493–L504.
- Pestka S. The interferons: 50 years after their discovery, there is much more to learn. J Biol Chem. 2007;282(28):20047–20051.
- Nabirotchkin S, Peluffo AE, Bouaziz J, et al. Focusing on the unfolded protein response and autophagy related pathways to reposition common approved drugs against COVID-19, MDPI Priprint, 2020
- Edelstein CL, Venkatachalam MA, Dong Z. Autophagy inhibition by chloroquine and hydroxychloroquine could adversely affect AKI and other organ injury in critically ill patients with COVID-19. Kidney Int. 2020. doi:https://doi.org/10.1016/j.kint.2020.05.001
- Pereira BB. Challenges and cares to promote rational use of chloroquine and hydroxychloroquine in the management of coronavirus disease 2020 (COVID-19) pandemic: a timely review. J Toxicol Environ Health B Crit Rev. 2020;18;23(4):177-181. doi:https://doi.org/10.1080/10937404.2020.1752340
- Pahan P, Pahan K. Smooth or risky revisit of an old malaria drug for COVID-19? J Neuroimmune Pharmacol. 2020;15(2):174–180.
- Yang N, Shen HM. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int J Biol Sci. 2020;16(10):1724–1731.
- Bertrand N, Wu J, Xu X, et al. Cancer nanotechnology: the impact of passive and active targeting in the era of modern cancer biology. Adv Drug Deliv Rev. 2014;66:2–25.
- Labouta HI, Menina S, Kochut A, et al. Bacteriomimetic invasin-functionalized nanocarriers for intracellular delivery. J Control Release. 2015;220(Part A):414–424.
- Labouta HI, El-Khordagui LK, Kraus T, et al. Mechanism and determinants of nanoparticle penetration through human skin. Nanoscale. 2011;3(12):4989–4999.
- Castoldi A, Herr C, Niederstraßer J, et al. Calcifediol-loaded liposomes for local treatment of pulmonary bacterial infections. Eur J Pharm Biopharm. 2017;118:62–67.
- Labouta HI, Gomez-Garcia MJ, Sarsons CD, et al. Surface-grafted polyethylene glycol conformation impacts the transport of PEG-functionalized liposomes through a tumour extracellular matrix model. RSC Adv. 2018;8(14):7697–7708.
- Cordani M, Somoza Á. Targeting autophagy using metallic nanoparticles: a promising strategy for cancer treatment. Cell Mol Life Sci. 2019;76(7):1215–1242.
- Singh L, Kruger HG, Maguire GEM, et al. The role of nanotechnology in the treatment of viral infections. Ther Adv Infect Dis. 2017;4(4):105–131.
- Chan WCW. Nano research for COVID-19. ACS Nano. 2020;14(4):3719–3720.
- Hu TY, Frieman M, Wolfram J. Insights from nanomedicine into chloroquine efficacy against COVID-19. Nat Nanotechnol. 2020;15(4):247–249.
- Mauthe M, Orhon I, Rocchi C, et al. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14(8):1435–1455.
- Schrezenmeier E, Dorner T. Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat Rev Rheumatol. 2020;16(3):155–166.
- Kyrmizi I, Gresnigt MS, Akoumianaki T, et al. Corticosteroids block autophagy protein recruitment in Aspergillus fumigatus phagosomes via targeting dectin-1/Syk kinase signaling. J Immunol. 2013;191(3):1287–1299.
- Langhammer A, Forsmo S, Syversen U. Long-term therapy in COPD: any evidence of adverse effect on bone? Int J Chron Obstruct Pulmon Dis. 2009;4:365–380.
- Rodriguez M, Lapierre J, Ojha CR, et al. Morphine counteracts the antiviral effect of antiretroviral drugs and causes upregulation of p62/SQSTM1 and histone-modifying enzymes in HIV-infected astrocytes. J Neurovirol. 2019;25(2):263–274.
- Tripathi A, Thangaraj A, Chivero ET, et al. Antiretroviral-mediated microglial activation involves dysregulated autophagy and lysosomal dysfunction. Cells. 2019;8(10):1168.
- Stravitz RT, Shiffman ML, Kimmel M, et al. Substitution of tenofovir/emtricitabine for Hepatitis B immune globulin prevents recurrence of Hepatitis B after liver transplantation. Liver Int. 2012;32(7):1138–1145.
- Zhao J, Wang ML, Li Z, et al. Interferon-alpha-2b induces autophagy in hepatocellular carcinoma cells through Beclin1 pathway. Cancer Biol Med. 2014;11(1):64–68.
- Hejny C, Sternberg P, Lawson DH, et al. Retinopathy associated with high-dose interferon alfa-2b therapy. Am J Ophthalmol. 2001;131(6):782–787.
- Zha BS, Wan X, Zhang X, et al. HIV protease inhibitors disrupt lipid metabolism by activating endoplasmic reticulum stress and inhibiting autophagy activity in adipocytes. PLoS One. 2013;8(3):e59514.
- Kim MJ, Kim SW, Chang HH, et al. Comparison of antiretroviral regimens: adverse effects and tolerability failure that cause regimen switching. Infect Chemother. 2015;47(4):231–238.
- Ishida S, Akiyama H, Umezawa Y, et al. Mechanisms for mTORC1 activation and synergistic induction of apoptosis by ruxolitinib and BH3 mimetics or autophagy inhibitors in JAK2-V617F-expressing leukemic cells including newly established PVTL-2. Oncotarget. 2018;9(42):26834–26851.
- Kusoglu A, Bagca BG, Ozates Ay NP, et al. Ruxolitinib regulates the autophagy machinery in multiple myeloma cells. Anticancer Agents Med Chem. 2020;20. DOI:https://doi.org/10.2174/1871520620666200218105159
- Alimam S, McLornan D, Harrison C. The use of JAK inhibitors for low-risk myelofibrosis. Expert Rev Hematol. 2015;8(5):551–553.
