ABSTRACT
The twin-arginine translocation (Tat) system is involved in a variety of important bacterial physiological processes. Conserved among bacteria and crucial for virulence, the Tat system is deemed as a promising anti-microbial drug target. However, the mechanism of how the Tat system functions in bacterial pathogenesis has not been fully understood. In this study, we showed that the Tat system was critical for the virulence of an extra-intestinal pathogenic E. coli (ExPEC) strain PCN033. A total of 20 Tat-related mutant strains were constructed, and competitive infection assays were performed to evaluate the relative virulence of these mutants. The results demonstrated that several Tat substrate mutants, including the ΔsufI, ΔamiAΔamiC double mutant as well as each single mutant, ΔyahJ, ΔcueO, and ΔnapG, were significantly outcompeted by the WT strain, among which the ΔsufI and ΔamiAΔamiC strains showed the lowest competitive index (CI) value. Results of individual mouse infection assay, in vitro cell adhesion assay, whole blood bactericidal assay, and serum bactericidal assay further confirmed the virulence attenuation phenotype of the ΔsufI and ΔamiAΔamiC strains. Moreover, the two mutants displayed chained morphology in the log phase resembling the Δtat and were defective in stress response. Our results suggest that the Tat system and its dependent cell division proteins SufI, AmiA, and AmiC play critical roles during ExPEC pathogenesis.
Introduction
Protein translocation and secretion are critical for bacterial survival, environmental adaption, and pathogenesis [Citation1,Citation2]. So far, a variety of protein secretion systems have been discovered in bacteria, among which the twin-arginine translocation (Tat) system is a unique one that facilitates folded proteins to be inserted into or translocated across the cytoplasmic membrane [Citation2–5]. The Tat system is present in most bacteria, plant chloroplasts, but absent in mammalian cells. It is composed of a core membrane protein TatC and one or two TatA-like proteins [Citation6]. The proteins exported through the Tat pathway encompass an SRRxFLK motif at their N-terminus [Citation7]. In E. coli, over 30 proteins have been verified or predicted to be transported through the Tat pathway, which are distributed in diverse physiological pathways [Citation5,Citation8].
Due to the diversity of Tat substrate proteins, Tat system disruption causes pleiotropic defects, affecting bacterial growth, cell division, motility, biofilm formation, iron acquisition, stress response, etc [Citation9,Citation10]. Moreover, the crucial role of the Tat system in pathogenesis has been reported in several important bacterial pathogens, including Pseudomonas aeruginosa [Citation11], E. coli O157 [Citation12], Salmonella enterica serovar Typhimurium [Citation13,Citation14], Citrobacter freundii [Citation15], and Yersinia pseudotuberculosis [Citation16]. Efforts have been made by several research groups to unravel how the Tat system affects bacterial virulence. Envelop defects are previously proposed to be the major cause of virulence attenuation in S. Typhimurium [Citation17]. However, a recent study shows that it is the cell division defects under stress conditions in the gut that contribute to the in vivo fitness decrease of the tat mutant of S. Typhimurium [Citation18]. Besides, the Tat system has been suggested to affect the efficiency of type III secretion in a plant pathogen P. syringae [Citation19]. However, it is not the case in S. Typhimurium [Citation17]. Therefore, the role of the Tat system during bacterial pathogenesis needs further investigation.
Extra-intestinal pathogenic Escherichia coli (ExPEC) is one of the most important pathogens causing neonatal meningitis, urinary tract infections, and sepsis in humans [Citation20]. Recently, ExPEC has also been frequently isolated from livestock which are believed as important reservoirs causing human infections [Citation21–23]. A substantial amount of ExPEC isolates of animal origin were reported that possessed high-level antimicrobial resistance and were highly virulent, and some isolates share similar virulence factors with those of human origin [Citation24–26]. Therefore, ExPEC is becoming a potential threat to food safety as well as public health. Understanding the pathogenesis of the ExPEC is of great significance.
In this study, we reveal that the Tat system is critical for the virulence of ExPEC. By constructing Tat-related mutants and performing competitive infection assays, we showed that the Tat-dependent cell division proteins SufI, AmiA, and AmiC are the key Tat substrate proteins accounting for the virulence attenuation of the tat mutant. Further analysis suggests that ΔsufI and ΔamiAΔamiC displayed severe cell division defect in the log phase and their growth was compromised under different stress conditions.
Materials and methods
Bacterial strains and cell culture conditions
All strains used in this study are listed in . The ExPEC strain PCN033 was isolated from the brain of a diseased pig with meningitis as described previously [Citation27,Citation28], which exhibited meningitis and high virulence in the mouse infection model [Citation29,Citation30]. E. coli strain χ7213 is a diaminopimelic acid (DAP) autotrophic strain used for delivering plasmid into ExPEC PCN033 through transconjugation [Citation31]. E. coli DH5α λpir was used as the host strain for the propagation of pRE112 [Citation32] or its derived plasmids. E. coli DH5α was used as the host strain for routine cloning. The E. coli χ7213 is grown in LB supplemented with 50 μg/mL of DAP. Chloramphenicol and apramycin were used at a final concentration of 50 μg/ml. PK-15 (pig kidney epithelial cell) and BHK-21 (derived from baby hamster kidney) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, Carlsbad, CA, USA) supplemented with 10% heat-inactivated fetal bovine serum in a 37°C incubator with 5% CO2.
Table 1. Strains used in this study.
Construction of plasmids and mutant strains
All primers and plasmids used in this study are listed in Table S1 and Table S2, respectively. Plasmids were constructed by seamless cloning using the ClonExpress® MultiS One Step Cloning Kit (Cat# C113, Vazyme Biotech Co., Ltd, Nanjing, China). All the mutant strains were constructed as described previously [Citation30]. Briefly, E. coli χ7213 competent cells were transformed with a pRE112 derived plasmid, which served as the donor strain for transconjugation. Cells of donor strain and recipient strain were mixed with a ratio of 10:1 and dripped onto a sterile filter membrane disc (Φ 0.45 μm) which was then placed on LB agar plate containing 50 μg/ml of DAP followed by incubation at 37°C for 5 hours. The bacterial cells were washed off from the disc and the cells plated onto LB agar containing chloramphenicol followed by overnight growth at 37°C. The colonies were picked and single exchanged strains were identified by PCR. The double exchanged mutants were then screened by using LB agar plate containing 10% sucrose and PCR identification of the presence of the target gene.
Growth assay
To measure bacterial growth in liquid medium, overnight-grown cell culture was diluted in LB medium giving an initial OD600nm of 0.01 and grown at 37°C with shaking at 200 round/min (rpm). OD600nm was recorded at each time point using a spectrometer. When necessary, cell culture at certain time point was taken, diluted in LB, and plated onto LB plate for viable cell counting. When doing growth assay on agar plates, bacterial cells at the mid-log phase were diluted in LB to give an identical OD600nm value, which were subject to 10-fold dilution, and 3 μL of cells of each indicated strain were spotted onto each specific LB agar plate. When measuring bacterial resistance against porcine β-defensin 2 (PDB2), 5 × 103 CFU of bacterial cells were mixed with different concentrations of synthetic porcine β defensin at 37°C for 1 hour, and the samples were plated onto LB plate and the viable cells were counted. The plates were then grown at each indicated temperature. The assay was performed in triplicate.
Morphological analysis
Gram staining and fluorescence imaging were used to analyze bacterial morphology. For Gram staining, cells of each strain were grown in LB to the log phase, washed three times with PBS, stained with Gram staining reagents according to the regular procedure, and observed with an optical microscope. For fluorescence imaging, each strain was transformed with pQE80Apra-GFP plasmid which constitutively expresses green fluorescent protein. The cells were grown in LB and harvested at each indicated time point. The cells were washed three times with PBS and imaged using a fluorescence confocal microscope.
Bacterial swimming assay
The swimming assay was performed as previously described [Citation18]. Briefly, 5 μL of cells of each indicated strain at the mid-log phase (OD600nm at 0.6) were spotted onto agar plate containing 10 g/L tryptone, 5 g/L yeast extract, 5 g/L NaCl, 0.5% glucose (w/v), and 0.45% agar (w/v). The plate was photographed after incubation at 37°C for 8 h.
Animal infection experiments
All animal experiments were approved by the Laboratory Animal Monitoring Committee of Huazhong Agricultural University and performed according to the recommendations in the Guide for the Care and Use of Laboratory Animals of Hubei Province, China. Female Kunming mice were purchased from the Experimental Animal Center, Huazhong Agricultural University, Wuhan, China. When evaluating the virulence of an individual ExPEC strain, bacterial cells were grown to the mid-log phase, pelleted, washed with sterile saline, and diluted in sterile saline to get an appropriate amount of viable cells which were then used to inject mouse intraperitoneally. The survival rate of the mice was recorded every 24 hours post-infection for 7 days. When calculating the in vivo bacterial loads, the mice were euthanized at each indicated time points and the organs were collected, weighed, homogenized in sterile saline, and plated on to LB for cell counting. Competitive infection assay, an accurate and sensitive approach to determine relative virulence, was used to determine whether the mutant was attenuated compared with the WT strain [Citation17,Citation34–36]. In the competitive infection assay, a similar amount of viable cells in the mid-log phase of the WT strain and each Tat-related mutant were mixed and used to inject mouse intraperitoneally. The mice were euthanized at each indicated time point, and the number of the WT strain and mutant strain in each organ were counted after plating on LB plate with or without chloramphenicol, respectively. The competitive index (CI) was calculated by dividing the ratio of the mutant cells to the WT cells recovered from the tissues by the ratio of the mutant cells to the WT cells in the injection mixture [output (CFUmutant/CFUWT)/Input (CFUmutant/CFUWT)]. When yagT, nrfC, hyaA and ydhX, which were located within operons, were in-frame substituted with Cmr coding sequence, the strains did not show chloramphenicol resistance. To calculate the competitive index for these strains, the numbers of the WT strain and the mutant strain in the recovered colonies were determined by using PCR.
In vitro cell adhesion assay, whole blood bactericidal assay, and serum bactericidal assay
PK-15 and BHK-21 cells were used to test the adhesion of the WT and the mutant strains as previously described [Citation30]. Briefly, bacterial cells grown to mid-log phase were harvested and washed three times with DMEM. PK-15 cells and BHK-21 cells grown in 6-well plates were infected with cells of each indicated bacterial strain with a ratio of 10:1 followed by incubation at 37°C with 5% CO2 for 2 hours. The cells were then washed five times with PBS and lysed with sterile water. The input bacterial cells and the cell lysates were then diluted and plated onto LB plates for bacterial enumeration. The adhesion rate of the WT strain was set as 100%. Whole blood bactericidal assay was performed as previously described [Citation37]. Briefly, 450 μL heparinized mouse whole blood were mixed with 50 μL bacterial cells of each indicated strain grown to mid-log phase (approximately 108 CFU/mL) and incubated at 37°C for 1 hour. The samples were then diluted and plated onto LB plates for bacterial enumeration. Serum bactericidal assay was performed as previously described [Citation30,Citation38]. Briefly, 100 μL bacterial cells of each indicated strain grown to mid-log phase (approximately 108 CFU/mL) were mixed with 100 μL of normal mouse serum (NS), or serum inactivated at 56°C for 30 min (IS) at 37°C, for 1 hour. The initial input samples and the incubated samples were then diluted and plated onto LB plates for bacterial enumeration. The survival rate was calculated as (CFUrecovered/CFUinput) × 100.
Statistical analysis
Statistical analysis was performed using GraphPad Prism (version 5) software. The Student’s t-test was used to calculate the differences between two groups. Error bars in the graphs represent the standard deviations of the means.
Results
The Tat system is essential for the virulence of ExPEC
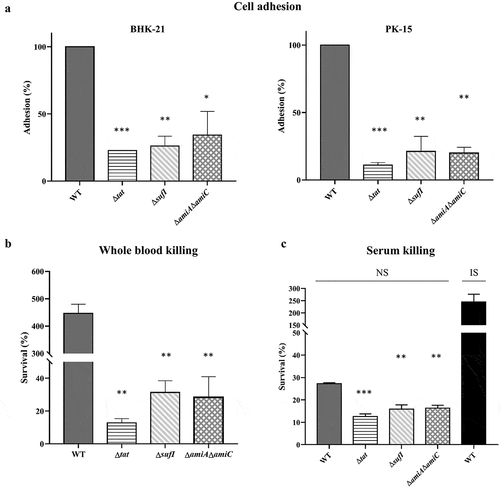
In the genome of ExPEC strain PCN033, an operon encoding TatA, TatB and TatC was present. To test whether the Tat system was functional, we constructed a tatABC deletion mutant (Δtat) and its complement strain (CΔtat). As shown in , the Δtat formed chained morphology in contrast to the wild-type (WT) and the complement strains, which was consistent with the observation reported in previous studies [Citation39,Citation40]. This suggested that the Tat system was functional in the ExPEC PCN033 strain. The growth assay showed that the tat mutant exhibited similar growth to WT strain (). Next, a mouse infection assay was performed to test whether the deletion of the Tat system had any effect on the virulence of the ExPEC strain. Mice were intraperitoneally injected with 6 × 106 CFU of WT and Δtat strains, respectively. It was shown that the survival rate of the mice infected with Δtat strain kept 80% (4/5), while those infected with WT strain fell to 0 (0/5) within 5 days post-infection (). The ability of the WT and Δtat cells to survive within mice was further tested. Mice were intraperitoneally injected with 6.7 × 105 cells (a non-lethal dose) of WT and Δtat strain, respectively. As shown in , the bacterial loads of the Δtat strain in the brain, lung, spleen, and blood decreased drastically within 36 hours post-infection, and the cells were almost completely cleared at the end of the experiment. In contrast, a very high level of bacterial loads of WT strain were still observed in each organ at 36 hours post-infection (). The adhesion assay result further showed that the ability of the Δtat strain to adhere to host cells was dramatically decreased compared with the WT strain (). In the whole blood bactericidal assay, it was shown that, instead of being killed, the WT strain could even grow in mouse blood. In contrast, the number of viable bacteria of the Δtat strain significantly dropped after incubation in whole blood (). The ability of the Δtat strain to survive in serum was also significantly lower than that of the WT strain (). These results strongly suggested that the Tat system was essential for the virulence of ExPEC.
Figure 1. Morphological, growth and virulence characterization of the parental ExPEC PCN033 strain and the tat mutant (Δtat).
Tat substrates prediction and mutants construction
It is the Tat-exported substrate proteins, instead of the Tat system itself, that function in bacterial pathogenesis. The Tat system recognizes its substrate proteins via the N-terminally located signal sequence containing an SRRxFLK motif [Citation5,Citation7]. So, we performed a bioinformatics prediction by using TatP 1.0 server (http://www.cbs.dtu.dk/services/TatP/)[Citation41] to search for Tat substrate proteins encoded in the genome of ExPEC PCN033 strain. As listed in , a total of 25 Tat substrates were found. We next constructed a series of Tat-related mutants. To facilitate in vivo competitive infection assay, we used a chloramphenicol resistance cassette to replace the target genes that are not present in the middle of an operon. As NapA, DmsA, TorA, TorZ, FdnG, FdoG, and YedY have been reported to be functional only when cofactor molybdenum (Mo) is incorporated with and have an overlapping function in respiration, single deletion of each of these genes may not have an obvious phenotype [Citation42–46]. Therefore, a ΔmoaA mutant was constructed which was deficient in Mo cofactor biogenesis, in which the Tat substrates containing Mo as cofactor were functionally disrupted [Citation36,Citation47]. AmiA and AmiC are two amidases that are both exported through the Tat pathway and have overlapping functions [Citation39]. Therefore, a double deletion mutant ΔamiAΔamiC was also constructed.
Table 2. Predicted Tat substrates encoded in ExPEC PCN033 genome.
Table 3. Competitive index (n = 5).
Competitive infection assay to assess the in vivo fitness of Tat-related mutants
To identify which Tat substrate protein accounted for the virulence attenuation of the tat mutant, we performed a series of competitive animal infection assays. As shown in , consistent with the result that disputing the Tat system significantly attenuated the virulence, the competitive infection assay also showed that the WT strain significantly outcompeted the Δtat strain (CI = 0.00568, p < 0.0001). Seven out of the 20 mutants, including ΔsufI, ΔamiA ΔamiC, ΔamiA, ΔamiC, ΔcueO, ΔyahJ, and ΔnapG, showed a CI less than 1 with statistical significance, indicating virulence attenuation compared with the WT strain. Among them ΔsufI displayed the lowest virulence, however, was not as avirulent as the Δtat strain. Double deletion of amiA and amiC genes resulted in the second most attenuated mutant which showed a lower CI value than each single amidase mutant. Individual deletion of yahJ, encoding an uncharacterized protein, and cueO, encoding a blue copper oxidase, also led to somewhat virulence attenuation. The remaining mutants, including 10 Tat substrate mutants and the ΔmoaA mutant, did not show significant virulence attenuation.
SufI as well as AmiA and AmiC play an important role in pathogenesis of ExPEC
As ΔsufI and ΔamiA ΔamiC showed the lowest CI value in the competitive infection assay, individual infection experiment was carried out to further assess their virulence. Three groups of mice were intraperitoneally injected with 9.4 × 105 CFU of WT, ΔsufI, ΔamiAΔamiC strains, respectively. As shown in , at 24 hours post-infection, a very high level of WT bacteria was present in each indicated organs. In contrast, both ΔsufI and ΔamiAΔamiC strains were rapidly cleared in vivo. Growth assay showed that ΔsufI and ΔamiA ΔamiC strains exhibited a slightly slower growth rate than the WT strain during the log phase, but reached a similar cell density with the WT strain in the stationary phase (). We further compared the ability of cell adhesion, and the resistance to whole blood and serum killing between the WT and the sufI and ΔamiAΔamiC strains. As shown in , the abilities of adhesion to PK-15 and BHK-21 cells of the ΔsufI and ΔamiAΔamiC strains were significantly lower than that of the WT strain, but were comparable to that of the Δtat. The whole blood bactericidal and serum bactericidal assays further revealed that the ΔsufI and ΔamiAΔamiC strains were more vulnerable in blood and serum than the WT strain (). These results further confirmed that SufI, and the two amidases play an important role in the pathogenesis of ExPEC.
Figure 2. Colonization of WT, ΔsufI, and ΔamiAΔamiC strains in mouse.
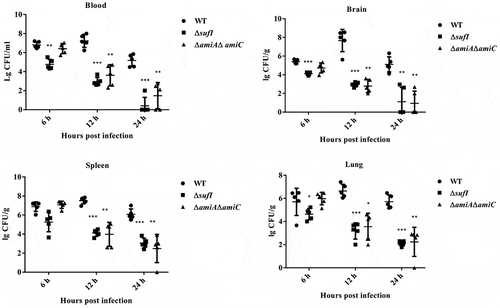
Figure 3. In vitro cell adhesion, and whole blood and serum bactericidal assays. (a). In vitro cell adhesion.
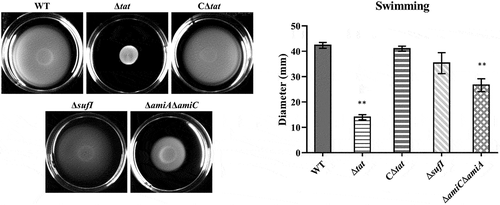
Motility was disrupted in the ΔamiAΔamiC strain but not in the ΔsufI
By performing a bacterial swimming assay, we found that the deletion of the Tat system caused severe defects in the motility of ExPEC (). As the motility-related functions have been recognized as a critical virulence factor [Citation48–51], we next tested whether the motility phenotype of the ΔsufI and ΔamiA ΔamiC mutants was deficient as well, thus causing virulence attenuation. As shown in , the swimming phenotype was significantly affected in the ΔamiA ΔamiC strain, although was not completely disrupted as that of the Δtat strain. However, it was not disturbed in the ΔsufI strain. These results suggest that at least the virulence attenuation of the ΔsufI strain was not due to motility defect.
Figure 4. Motility assay.
Δsufi and ΔamiA ΔamiC displayed severe cell division defect in the log growth phase
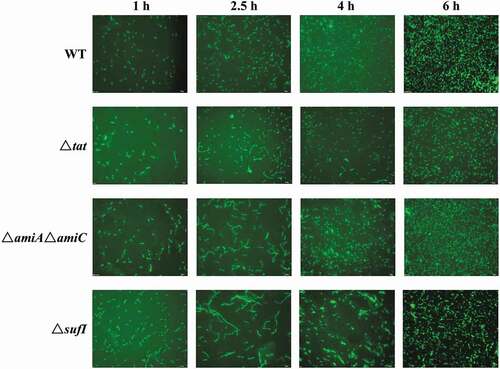
As SufI and the Tat-dependent amidases have been reported to be involved in cell division [Citation52–Citation54], the morphology of the mutant strains was analyzed. As shown in , at 1 hour of growth, all the strains showed normal cell morphology. However, at the early log phase (2.5 hours of growth), in contrast to the wild-type strain that still displayed normal separated cells, all the Δtat, ΔsufI and ΔamiA ΔamiC strains formed chained morphology suggesting abnormal cell division, among which the ΔsufI showed the most severe cell division defect. However, this chained morphology almost disappeared at 6 hours of growth. These results suggest that SufI, AmiA, and AmiC are critical for cell division, especially during the log phase.
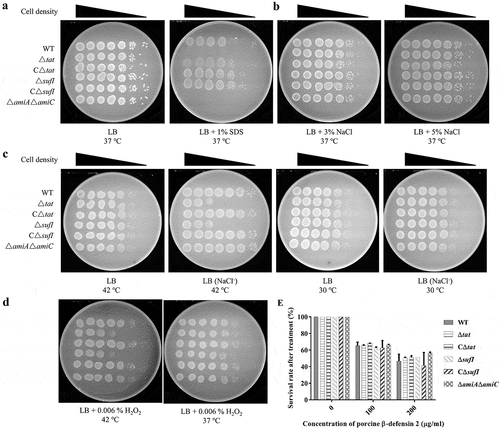
Δsufi and ΔamiA ΔamiC were defective in stress response
SDS sensitivity as an indicator of bacterial envelop integrity has been widely used, by which approach the Δtat strain is shown to have a defective envelop [Citation17,Citation55,Citation56]. Therefore, we tested whether the above mutants have a similar defect. As shown in , consistent with previous reports, both the Δtat and ΔamiA ΔamiC showed high sensitivity to SDS, while the ΔsufI was as resistant as the WT strain. The growth of the strains in stress conditions was further investigated. Different stress conditions were generated, including high and low osmotic stresses, high-temperature stress, H2O2 induced oxidative stress, and porcine β-defensin 2 (PDB2) mediated antimicrobial stress. As shown in , all of Δtat, ΔsufI and ΔamiA ΔamiC were able to grow in the medium containing up to 5% NaCl. However, ΔsufI exhibited impaired growth in LB medium without NaCl at 42°C but not at 30°C (). Furthermore, ΔsufI as well as Δtat were sensitive to H2O2, while this sensitivity was only observed at 42°C, but not at 37°C (). None of Δtat, ΔsufI and ΔamiA ΔamiC showed a different resistance to PDB2 ().
Figure 6. Stress response assay. (a). SDS resistance.
Discussion
In several important bacterial pathogens, the Tat system has been reported to be crucial for pathogenesis [Citation8,Citation14,Citation17,Citation35]. Consistently, our results also revealed that the deletion of the Tat system significantly attenuated the virulence of extra-intestinal pathogenic E. coli (ExPEC). In some pathogens, for example, Legionella pneumophila and Pseudomonas aeruginosa, the Tat system is involved in the secretion of exotoxins so as to contribute to virulence [Citation57,Citation58]. However, in the genome of the ExPEC PCN033 strain used in this study, we did not find any obvious virulence factors encompassing a Tat signal peptide through bioinformatics analysis.
In order to reveal how the Tat system affects bacterial pathogenesis, by constructing a total of 20 Tat-related mutants and performing competitive infection experiments, we demonstrated that several Tat substrate mutants exhibited significant virulence attenuation. Among these mutants, ΔsufI and ΔamiAΔamiC showed the lowest CI value indicating the most severe virulence decrease. Similar findings have also been reported previously. In Y. pseudotuberculosis, several Tat substrates are identified critical to cause systematic infection, among which ΔsufI shows the largest attenuation [Citation35]. In Citrobacter freundii, it has also been shown that sufI mutation results in a very significant in vivo fitness defect [Citation15]. A recent study in S. Typhimurium demonstrates that the deletion of the two Tat-dependent amidase encoding genes leads to significant in vivo fitness attenuation, mainly due to cell division defect especially in the inflamed gut where stress conditions including high osmolarity and antimicrobial peptides are present [Citation18]. However, a previous study which was also carried out in S. Typhimurium shows that neither the mutant devoid of sufI nor that lacking amidases alone, but the triple deletion mutant, display a defective virulence phenotype similar as the tat mutant, in which they propose that the virulence attenuation is primarily due to envelop defects [Citation17].
Bacterial pathogens encounter a variety of stresses during infection, including antimicrobial peptides secreted by epithelial cells [Citation59], hyperosmolarity [Citation60], and ROS- or iNOS-mediated oxidative stress [Citation61,Citation62]. The ability of bacterial stress response contributes much to its in vivo fitness. In this study, ΔamiAΔamiC and ΔsufI both showed significant virulence attenuation. SufI and the Tat-exported amidases are both involved in cell division of E. coli. sufI serves as a genetic suppressor of ftsI which encodes an essential cell division protein and has been shown to function under stress conditions [Citation53,Citation54,Citation63]. AmiA and AmiC are N-acetylmuramyl-L-alanine amidases involved in cell wall remodeling [Citation52]. Consistent with their critical roles in cell division, our data demonstrate that the two mutant strains exhibited abnormal morphology in the log growth phase. It is worth noting that although AmiA and AmiC are paralogs functioning in cell septation and separation, they contribute differently to virulence as the two single mutants gave different CI values in the competitive infection assay. This has also been observed in a previous study [Citation17]. Actually, it has been reported that the subcellular localization of AmiA and AmiC differs markedly and they play different roles in cleaving the glycan chains of peptidoglycan [Citation64,Citation65]. Additionally, both of the ΔamiAΔamiC and ΔsufI mutants are defective in stress response. Therefore, it is possible that the virulence attenuation in ExPEC caused by the disruption of the Tat substrates may be attributed to cell division defect-mediated compromise in stress response.
Bacterial motility has been recognized as an important virulence factor [Citation48–Citation51]. In consistence with the data reported in previous studies, our results also reveal that the disruption of the Tat system severely compromises bacterial motility. Therefore, we tested whether ΔsufI and ΔamiAΔamiC possessed a similar non-motile phenotype as the Δtat mutant, which may as a result account for the virulence attenuation. Our results showed that the motility was deficient in the ΔamiAΔamiC strain, but not the ΔsufI. A similar motility defect in the ΔamiAΔamiC strain has been also revealed in S. Typhimurium [Citation17]. However, considering flagella expression is down-regulated during S. Typhimurium infection, the motility defect is not believed as the cause of virulence attenuation of the ΔamiAΔamiC strain [Citation17]. In ExPEC, our previous study has shown that the deletion of a flagella biosynthesis gene, flgD, causes significant virulence attenuation, indicating that motility is crucial for the virulence [Citation51]. Therefore, whether and to which extent the motility defect can explain the virulence attenuation of the ΔamiAΔamiC needs further investigations.
ΔcueO mutant was also significantly outcompeted by the WT strain in the competitive infection assay. cueO encodes a copper oxidase involved in bacterial copper homeostasis [Citation66,Citation67]. Although copper is an essential metal ion functioning as a cofactor of specific bacterial enzymes, too much copper is toxic. Copper is proposed to catalyze the production of hydroxyl radicals causing oxidative damage [Citation68]. Excessive copper is also reported to interfere with disulfide bond formation in the periplasm of bacteria [Citation69]. It has been shown that the copper level in the microenvironments within the host increases dramatically during bacterial infection [Citation70]. Therefore, the ability of bacterial pathogens to tolerate high concentration copper may contribute to the pathogenicity. CueO itself is a copper-containing protein, therefore may contribute to copper efflux during its export through the Tat pathway which as a result decreases the copper concentration in the cytoplasm. Meanwhile, CueO, as an oxidase, is able to catalyze the toxic and membrane-permeable Cu(I) to the less toxic and less permeable Cu(II) [Citation67,Citation71]. Therefore, CueO may increase the copper tolerance of E. coli which contributes its survival in vivo.
Iron acquisition is another important physiological process linked to bacterial pathogenesis [Citation10,Citation72,Citation73]. Bacteria have evolved different strategies to acquire iron, among which the Efe system is an iron transporter responsible for ferrous iron uptake [Citation74]. The Efe system encompasses a membrane protein EfeU and two periplasmic proteins EfeO and EfeB which both bear a Tat signal peptide [Citation74]. The Efe system, or its homologous systems, has been revealed to be required for the virulence of several important bacterial pathogens, including Brucella abortus, Burkholderia [Citation75,Citation76]. In the genome of E. coli K-12 MG1655 strain, there is a frameshift within the efe operon leading to the inactivation of this iron transporting function, and restoring this function by replacing the native efeUOB operon with an in-frame one increased the growth of E. coli under iron-restricted conditions [Citation77]. However, in the genome of ExPEC PCN033, the efeUOB is intact without any frameshift, suggesting that it may be functional. Therefore, it is interesting to test whether the EfeUOB transporter is important for the pathogenesis of ExPEC. However, our results showed that ΔefeOB had a comparable virulence to the WT strain, indicating the EfeO and EfeB are not critical for the virulence and therefore not responsible for the virulence attenuation of the tat mutant.
A large proportion of the Tat substrates are co-factor containing redox enzymes, including trimethylamine N-oxide (TMAO) reductase TorA, dimethyl sulfoxide (DSMO) reductase DmsD, which mainly functions in electron transport chains in bacterial respiration [Citation10]. In some pathogens, these redox enzymes have been shown to be crucial for virulence. In Actinobacillus pleuropneumoniae, the ΔdmsA mutant was significantly attenuated in a pig infection model [Citation78]. In Vibrio cholera, deletion of torD, which encodes a chaperone protein required for the maturation of TorA, resulted in decrease in cholera toxin production and virulence, which was similar as the tat mutant [Citation79]. In the genome of ExPEC PCN033 strain, seven Tat substrates, NapA, DmsA, TorA, TorZ, FdnG, FdoG, and YedY, are reported to be molybdenum-containing enzymes [Citation45]. To assess the role of these enzymes in ExPEC virulence, instead of constructing every individual mutant, we deleted the moaA gene, which is responsible for molybdenum incorporation, therefore inactivates all of the above Tat substrates [Citation44,Citation47]. However, the animal infection experiment results showed that ΔmoaA did not outcompete the WT strain, indicating no obvious virulence attenuation. Therefore, the molybdenum-containing Tat substrate proteins are not involved in the virulence attenuation.
In conclusion, our results demonstrate that the Tat system is critical for virulence of ExPEC, and SufI as well as AmiA and AmiC, are the key Tat substrates responsible for the virulence attenuation.
Supplemental Material
Download MS Word (17 KB)Supplemental Material
Download MS Word (23.6 KB)Disclosure statement
No potential conflicts of interest were disclosed.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Tseng TT, Tyler BM, Setubal JC. Protein secretion systems in bacterial-host associations, and their description in the gene ontology. BMC Microbiol. 2009;9(Suppl 1):S2.
- Green ER, Mecsas J. Bacterial secretion systems: an overview. Microbiol Spectr. 2016;4.
- Bhoite S, van Gerven N, Chapman MR, et al. Curli biogenesis: bacterial amyloid assembly by the type VIII secretion pathway. EcoSal Plus. 2019;8.
- Lauber F, Deme JC, Lea SM, et al. Type 9 secretion system structures reveal a new protein transport mechanism. Nature. 2018;564:77.
- Palmer T, Berks BC. The twin-arginine translocation (Tat) protein export pathway. Nature Rev Microbiol. 2012;10:483–496.
- Berks BC. The twin-arginine protein translocation pathway. Annu Rev Biochem. 2015;84:843–864.
- Berks BC. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol. 1996;22:393–404.
- Tullman-Ercek D, DeLisa MP, Kawarasaki Y, et al. Export pathway selectivity of Escherichia coli twin arginine translocation signal peptides. J Biol Chem. 2007;282:8309–8316.
- Berks BC, Palmer T, Sargent F. The Tat protein translocation pathway and its role in microbial physiology. Adv Microb Physiol. 2003;47:187–254.
- Palmer T, Sargent F, Berks BC. The Tat protein export pathway. EcoSal Plus. 2010;4.
- Ochsner UA, Snyder A, Vasil AI, et al. Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc Natl Acad Sci U S A. 2002;99:8312–8317.
- Pradel N, Ye C, Livrelli V, et al. Contribution of the twin arginine translocation system to the virulence of enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2003;71:4908–4916.
- Reynolds MM, Bogomolnaya L, Guo J, et al. Abrogation of the twin arginine transport system in Salmonella enterica serovar Typhimurium leads to colonization defects during infection. PloS One. 2011;6:e15800.
- Urrutia IM, Sabag A, Valenzuela C, et al. Contribution of the twin-arginine translocation system to the intracellular survival of salmonella typhimurium in dictyostelium discoideum. Front Microbiol. 2018;9:3001.
- Anderson MT, Mitchell LA, Zhao L, et al. Citrobacter freundii fitness during bloodstream infection. Sci Rep. 2018;8:11792.
- Lavander M, Ericsson SK, Broms JE, et al. The twin arginine translocation system is essential for virulence of Yersinia pseudotuberculosis. Infect Immun. 2006;74:1768–1776.
- Craig M, Sadik AY, Golubeva YA, et al. Twin-arginine translocation system (tat) mutants of Salmonella are attenuated due to envelope defects, not respiratory defects. Mol Microbiol. 2013;89:887–902.
- Fujimoto M, Goto R, Hirota R, et al. Tat-exported peptidoglycan amidase-dependent cell division contributes to Salmonella Typhimurium fitness in the inflamed gut. PLoS Pathog. 2018;14:e1007391.
- Bronstein PA, Marrichi M, Cartinhour S, et al. Identification of a twin-arginine translocation system in Pseudomonas syringae pv. tomato DC3000 and its contribution to pathogenicity and fitness. J Bacteriol. 2005;187:8450–8461.
- Biran D, Ron EZ. Extraintestinal pathogenic Escherichia coli. Curr Top Microbiol Immunol. 2018;416:149–161.
- Liu CM, Stegger M, Aziz M, et al. Escherichia coli ST131-H22 as a foodborne uropathogen. mBio. 2018;9.
- Bergeron CR, Prussing C, Boerlin P, et al. Chicken as reservoir for extraintestinal pathogenic Escherichia coli in humans, Canada. Emerg Infect Dis. 2012;18:415–421.
- Maluta RP, Logue CM, Casas MR, et al. Overlapped sequence types (STs) and serogroups of avian pathogenic (APEC) and human extra-intestinal pathogenic (ExPEC) Escherichia coli isolated in Brazil. PloS One. 2014;9:e105016.
- Nandanwar N, Janssen T, Kuhl M, et al. Extraintestinal pathogenic Escherichia coli (ExPEC) of human and avian origin belonging to sequence type complex 95 (STC95) portray indistinguishable virulence features. Int J Med Microbiol IJMM. 2014;304:835–842.
- Ge X Z, Jiang J, Pan Z, et al. Comparative genomic analysis shows that avian pathogenic Escherichia coli isolate IMT5155 (O2: K1:H5;ST complex 95, ST140) shares close relationship with ST95 APEC O1: k1and human ExPEC O18: k1strains. PloS One. 2014;9:e112048.
- Gao J, Duan X, Li X, et al. Emerging of a highly pathogenic and multi-drug resistant strain of Escherichia coli causing an outbreak of colibacillosis in chickens. Infect Genet Evol. 2018;65:392–398.
- Tan C, Xu Z, Zheng H, et al. Genome sequence of a porcine extraintestinal pathogenic Escherichia coli strain. J Bacteriol. 2011;193:5038.
- Liu C, Zheng H, Yang M, et al. Genome analysis and in vivo virulence of porcine extraintestinal pathogenic Escherichia coli strain PCN033. BMC Genomics. 2015;16:717.
- Tan C, Tang X, Zhang X, et al. Serotypes and virulence genes of extraintestinal pathogenic Escherichia coli isolates from diseased pigs in China. Vet J. 2012;192:483–488.
- Zong B, Liu W, Zhang Y, et al. Effect of kpsM on the virulence of porcine extraintestinal pathogenic Escherichia coli. FEMS Microbiol Lett. 2016;363.
- Roland K, Curtiss R 3rd, Sizemore D. Construction and evaluation of a delta cya delta crp Salmonella typhimurium strain expressing avian pathogenic Escherichia coli O78 LPS as a vaccine to prevent airsacculitis in chickens. Avian Dis. 1999;43:429–441.
- Edwards RA, Keller LH, Schifferli DM. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene. 1998;207:149–157.
- Platt R, Drescher C, Park SK, et al. Genetic system for reversible integration of DNA constructs and lacZ gene fusions into the Escherichia coli chromosome. Plasmid. 2000;43:12–23.
- Macho AP, Rufian JS, Ruiz-Albert J, et al. Competitive index: mixed infection-based virulence assays for genetic analysis in pseudomonas syringae-plant interactions. Methods Mol Biol. 2016;1363:209–217.
- Avican U, Doruk T, Ostberg Y, et al. The Tat substrate sufi is critical for the ability of yersinia pseudotuberculosis to cause systemic infection. Infect Immun. 2017;85.
- Hughes ER, Winter MG, Duerkop BA, et al. Microbial respiration and formate oxidation as metabolic signatures of inflammation-associated dysbiosis. Cell Host Microbe. 2017;21:208–219.
- Zong B, Zhang Y, Wang X, et al. Characterization of multiple type-VI secretion system (T6SS) VgrG proteins in the pathogenicity and antibacterial activity of porcine extra-intestinal pathogenic Escherichia coli. Virulence. 2019;10:118–132.
- Wang H, Liu L, Cao Q, et al. Haemophilus parasuis alpha-2,3-sialyltransferase-mediated lipooligosaccharide sialylation contributes to bacterial pathogenicity. Virulence. 2018;9:1247–1262.
- Ize B, Stanley NR, Buchanan G, et al. Role of the Escherichia coli Tat pathway in outer membrane integrity. Mol Microbiol. 2003;48:1183–1193.
- Alcock F, Stansfeld PJ, Basit H, et al. Assembling the Tat protein translocase. Elife. 2016;5.
- Bendtsen JD, Nielsen H, Widdick D, et al. Prediction of twin-arginine signal peptides. BMC Bioinformatics. 2005;6:167.
- Cheng VW, Weiner JH. S- and N-oxide reductases. EcoSal Plus. 2007;2.
- Cole JA, Richardson DJ. Respiration of nitrate and nitrite. EcoSal Plus. 2008;3.
- Magalon A, Mendel RR. Biosynthesis and insertion of the molybdenum cofactor. EcoSal Plus. 2008;3.
- Leimkuhler S, Iobbi-Nivol C. Bacterial molybdoenzymes: old enzymes for new purposes. FEMS Microbiol Rev. 2016;40:1–18.
- Pinske C, Sawers RG. Anaerobic formate and hydrogen metabolism. EcoSal Plus. 2016;7.
- Rivers SL, McNairn E, Blasco F, et al. Molecular genetic analysis of the moa operon of Escherichia coli K-12 required for molybdenum cofactor biosynthesis. Mol Microbiol. 1993;8:1071–1081.
- Chaban B, Hughes HV, Beeby M. The flagellum in bacterial pathogens: for motility and a whole lot more. Semin Cell Dev Biol. 2015;46:91–103.
- Erhardt M. Strategies to block bacterial pathogenesis by interference with motility and chemotaxis. Curr Top Microbiol Immunol. 2016;398:185–205.
- Weller-Stuart T, Toth I, De Maayer P, et al. Swimming and twitching motility are essential for attachment and virulence of Pantoea ananatis in onion seedlings. Mol Plant Pathol. 2017;18:734–745.
- Liu F, Fu J, Liu C, et al. Characterization and distinction of two flagellar systems in extraintestinal pathogenic Escherichia coli PCN033. Microbiol Res. 2017;196:69–79.
- Heidrich C, Templin MF, Ursinus A, et al. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol Microbiol. 2001;41:167–178.
- Samaluru H, SaiSree L, Reddy M. Role of SufI (FtsP) in cell division of Escherichia coli: evidence for its involvement in stabilizing the assembly of the divisome. J Bacteriol. 2007;189:8044–8052.
- Tarry M, Arends SJ, Roversi P, et al. The Escherichia coli cell division protein and model Tat substrate SufI (FtsP) localizes to the septal ring and has a multicopper oxidase-like structure. J Mol Biol. 2009;386:504–519.
- Huang Q, Alcock F, Kneuper H, et al. A signal sequence suppressor mutant that stabilizes an assembled state of the twin arginine translocase. Proc Natl Acad Sci U S A. 2017;114:E1958–E67.
- Huang Q, Palmer T. Signal peptide hydrophobicity modulates interaction with the twin-arginine translocase. mBio. 2017;8.
- Voulhoux R, Ball G, Ize B, et al. Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. Embo J. 2001;20:6735–6741.
- Rossier O, Cianciotto NP. The Legionella pneumophila tatB gene facilitates secretion of phospholipase C, growth under iron-limiting conditions, and intracellular infection. Infect Immun. 2005;73:2020–2032.
- Xia X, Cheng L, Zhang S, et al. The role of natural antimicrobial peptides during infection and chronic inflammation. Antonie Van Leeuwenhoek. 2018;111:5–26.
- Henderson AG, Ehre C, Button B, et al. Cystic fibrosis airway secretions exhibit mucin hyperconcentration and increased osmotic pressure. J Clin Invest. 2014;124:3047–3060.
- Dryden M. Reactive oxygen species: a novel antimicrobial. Int J Antimicrob Agents. 2018;51:299–303.
- Fang FC, Vazquez-Torres A. Reactive nitrogen species in host-bacterial interactions. Curr Opin Immunol. 2019;60:96–102.
- Spratt BG. Temperature-sensitive cell division mutants of Escherichia coli with thermolabile penicillin-binding proteins. J Bacteriol. 1977;131:293–305.
- Bernhardt TG, de Boer PA. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol Microbiol. 2003;48:1171–1182.
- Priyadarshini R, Popham DL, Young KD. Daughter cell separation by penicillin-binding proteins and peptidoglycan amidases in Escherichia coli. J Bacteriol. 2006;188:5345–5355.
- Grass G, Rensing C. CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochem Biophys Res Commun. 2001;286:902–908.
- Djoko KY, Chong LX, Wedd AG, et al. Reaction mechanisms of the multicopper oxidase CueO from Escherichia coli support its functional role as a cuprous oxidase. J Am Chem Soc. 2010;132:2005–2015.
- Liochev SI, Fridovich I. The Haber-Weiss cycle – 70 years later: an alternative view. Redox Rep. 2002;7:55–57.
- Hiniker A, Collet JF, Bardwell JC. Copper stress causes an in vivo requirement for the Escherichia coli disulfide isomerase DsbC. J Biol Chem. 2005;280:33785–33791.
- Wagner D, Maser J, Lai B, et al. Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell’s endosomal system. J Iimmunol. 2005;174:1491–1500.
- Outten FW, Huffman DL, Hale JA, et al. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J Biol Chem. 2001;276:30670–30677.
- Sheldon JR, Laakso HA, Heinrichs DE. Iron acquisition strategies of bacterial pathogens. Microbiol Spectr. 2016;4.
- Lemos ML, Balado M. Iron uptake mechanisms as key virulence factors in bacterial fish pathogens. J Appl Microbiol. 2020;129:104–115.
- Grosse C, Scherer J, Koch D, et al. A new ferrous iron-uptake transporter, EfeU (YcdN), from Escherichia coli. Mol Microbiol. 2006;62:120–131.
- Elhassanny AE, Anderson ES, Menscher EA, et al. The ferrous iron transporter FtrABCD is required for the virulence of Brucella abortus 2308 in mice. Mol Microbiol. 2013;88:1070–1082.
- Mathew A, Eberl L, Carlier AL. A novel siderophore-independent strategy of iron uptake in the genus Burkholderia. Mol Microbiol. 2014;91:805–820.
- Cao J, Woodhall MR, Alvarez J, et al. EfeUOB (YcdNOB) is a tripartite, acid-induced and CpxAR-regulated, low-pH Fe2+ transporter that is cryptic in Escherichia coli K-12 but functional in E. coli O157: H7. Mol Microbiol. 2007;65:857–875.
- Baltes N, Hennig-Pauka I, Jacobsen I, et al. Identification of dimethyl sulfoxide reductase in Actinobacillus pleuropneumoniae and its role in infection. Infect Immun. 2003;71:6784–6792.
- Lee KM, Park Y, Bari W, et al. Activation of cholera toxin production by anaerobic respiration of trimethylamine N-oxide in Vibrio cholerae. J Biol Chem. 2012;287:39742–39752.