ABSTRACT
Pseudomonas aeruginosa (PA) and Aspergillus fumigatus (AF) chronically colonize the airways of patients with cystic fibrosis or chronic immunosuppression and mutually affect each other’s pathogenesis. Here, we evaluated IncuCyte time-lapse imaging and NeuroTrackTM (NT) analysis (Wurster et al., 2019, mBio) as a toolbox to study mycelial expansion and morphogenesis of AF during interaction with PA. Co-incubation of AF with supernatant filtrates of wild-type (WT) PA strains strongly inhibited hyphal growth and branching. Consonant with prior metabolic studies, pyoverdine-deficient PA mutants had significantly attenuated inhibitory capacity. Accordingly, purified PA products pyoverdine and pyocyanin suppressed mycelial expansion of AF in a concentration-dependent way. Using fluorescence-guided tracking of GFP-AF293 mycelia during co-culture with live WT PA cells, we found significant inoculum-dependent mycelial growth inhibition and robust precision of the NT algorithm. Collectively, our experiments position IncuCyte NT as an efficient platform for longitudinal analysis of fungal growth and morphogenesis during bacterial co-infection.
Introduction
Polymicrobial infections have increasingly become a focus of interest in infectious diseases research as inter-kingdom interplay of pathogens can mutually affect their virulence, susceptibility to antimicrobial therapy, and interactions with host immune surveillance [Citation1]. In particular, Aspergillus fumigatus (AF) and Pseudomonas aeruginosa (PA) frequently co-colonize the airways of patients with cystic fibrosis, chronic bronchiectasis, or chronically immunocompromised patients and compete for nutrients in their ecological niche [Citation2]. The mutually antagonistic relationship of AF and PA is, to a large extent, mediated by extracellular PA products, including toxins and siderophores [Citation2]. Specifically, PA metabolites with iron-binding capacity, such as pyoverdine or pyocyanin, inhibit growth, biofilm formation, and metabolism of AF [Citation2–Citation4].
Whereas most of these studies used conventional measurements such as growth inhibition or changes in metabolism [Citation2], there is a need for robust and efficient experimental systems that specifically assess morphogenesis, a key feature of fungal virulence [Citation5], in the context of bacterial interactions. Previous work revealed that dirhamnolipids released by PA induce ultrastructural modifications of AF hyphae, resulting in altered mycelial morphology and reduced growth of AF [Citation6]. However, currently available modalities to study mycelial expansion, hyphal morphology, and biofilm formation, such as fluorescence or electron microscopy [Citation6–Citation8] and spectroscopy techniques [Citation9], have low throughput and are time-intensive. In addition, most currently employed assays for inter-kingdom interaction studies [Citation2–Citation4,Citation6–Citation9] present endpoint analyses that are not suitable for efficient longitudinal monitoring.
Inspired by morphological and functional similarities of neuronal networks and fungal mycelia [Citation10], we have recently proposed the IncuCyte time-lapse microscopy platform and its NeuroTrackTM (NT) image processing module as an efficient and reliable tool to study the expansion kinetics and viability of mycelial trees [Citation11]. In this study, we hypothesized that NT analysis could present an appealing platform to quantitatively analyze mycelial morphology and expansion in the context of intermicrobial interaction. To that end, we validated the NT technology for the assessment of PA and AF interplay and implemented fluorescence-guided real-time tracking of AF mycelia in the presence of live PA cells.
Materials & methods
Fungal strains and culture
Conidia of A. fumigatus AF10 and a GFP-expressing AF293 strain were collected from mature colonies grown on yeast extract agar for 48–72 h at 37 °C. Conidia were passed through a 40 µm cell strainer, washed twice in sterile saline, and counted with a hemocytometer.
Bacterial culture and generation of filtrates
Planktonic culture filtrates of wild-type (WT) PA strains and PA14 mutants (Table S1) were prepared as detailed previously [Citation12]. Briefly, PA was quantitated, suspended in RPMI 1640 medium (Sigma-Aldrich) either supplemented or not with 25 µM FeCl3, and grown for 24 h at 37°C. Bacterial cultures were filtered (0.22 μm) after the growth period to remove bacterial cells and debris.
Co-culture set-up
Aliquots of conidial suspensions (200 conidia in 100 µl RPMI medium) were dispensed per well of a 96-well flat-bottom plate. RPMI medium (100 µl, control), purified PA metabolites pyoverdine (1–10 µM) or pyocyanin (50–500 µM) (both: Sigma-Aldrich), filtrates of WT PA strains and siderophore loss-of-function mutants, or viable PA cells were added. For live cell co-culture, PA/AF ratios of 0.01 to 100 were tested in 10-fold serial dilutions.
IncuCyte imaging
Well plates were incubated and imaged for 18 h at 37 °C in the IncuCyte ZOOM HD/2CLR time lapse microscopy system (Sartorius) equipped with an IncuCyte ZOOM 10 x PLAN FLUOR objective (Sartorius) as previously described [Citation11]. Phase images were acquired hourly for all experiments. For live cell co-culture using AF293-GFP, green fluorescence images were obtained hourly with an acquisition time of 400 milliseconds. NT image processing algorithms were applied using our published parameters for A. fumigatus and AF293-GFP [Citation11].
Data analysis
Raw data for NT endpoints “neurite (hyphal) length” and “branch points” were exported to Microsoft Excel and further processed using GraphPad Prism v8. For each well and NT endpoint, the area-under-the-curve (AUC) was determined, considering the first 18 h of culture. AUCs were normalized to a medium or “AF only” control (= 1.0). Unless indicated otherwise, 3 or 4 replicates with independently prepared bacterial and fungal inocula and/or filtrates were assessed. Significance tests and levels are specified in the figure legends. Coefficients of variation (CVs) for relative AUCs values were calculated by dividing standard deviations by arithmetic means.
Results
To validate the IncuCyte NT technology for studies of AF-PA interplay, we tested the differential inhibitory impact of well-characterized WT PA strains and PA mutants [Citation2] on the expansion and morphogenesis of AF mycelia. Compared to the RPMI control, co-incubation of AF10 with culture filtrates of WT PA strains PA01 and PA14 strongly inhibited hyphal growth and branching (). Mean relative AUC values for NT endpoints in cultures of AF conidia exposed to filtrates of either WT PA strain were 0.05 for hyphal length and 0.02 for branch points (, p < 0.001). In line with prior metabolic studies [Citation2], the inhibitory effect was largely attenuated when using supernatants of a pyoverdine-deficient PA mutant (PA14 pvdD−) or a pyoverdine-pyochelin double PA mutant (PA14 pvdD−pchE−) (p < 0.001), whereas pyochelin deficiency alone (PA14 pchE−) had no impact on the inhibitory capacity of PA ().
Figure 1. Comparative inhibitory activity of culture filtrates of WT and siderophore-deficient P. aeruginosa strains on mycelial expansion of A. fumigatus.
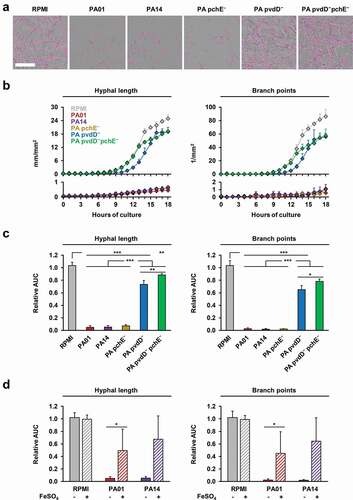
Furthermore, we compared mycelial growth inhibition by WT PA filtrates depending on iron availability, as iron is a source of competition between AF and PA in their in vivo environment [Citation2]. FeCl3 supplementation of the RPMI control did not affect the NT endpoints of AF (relative AUC, 0.99–1.02, ), suggesting that baseline iron levels in our growth medium were not a limiting factor for mycelial expansion. In contrast, iron supplementation considerably attenuated the inhibitory effect of PA culture filtrates on AF growth and morphogenesis, as indicated by an increase in relative hyphal length and branch point AUC values from 0.05 to 0.49–0.68 (p = 0.03–0.07) and 0.02 to 0.45–0.64 (p = 0.03–0.08), respectively ().
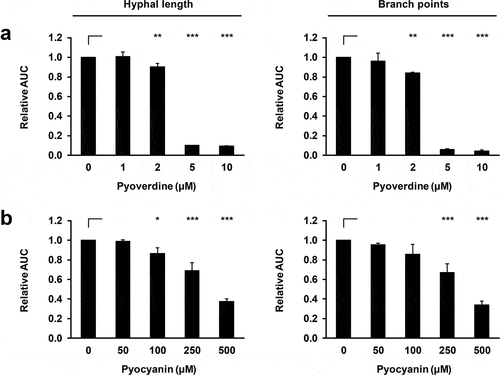
To underscore the critical role of iron in our NT assay, we exposed AF to different concentrations of purified iron-sequestering PA products pyoverdine and pyocyanin that are known inhibitors of AF growth and metabolism [Citation2–Citation4,Citation13]. Pyoverdine at concentrations between 2 and 5 µM resulted in a sharp inhibition of hyphal growth (p < 0.001, ). Consonant with an earlier report [Citation13], gradually increasing inhibitory activity of pyocyanin across a broader range of concentrations (50–500 µM) was seen ().
Figure 2. P. aeruginosa products pyoverdine and pyocyanin concentration-dependently inhibit mycelial expansion and branching of A. fumigatus.
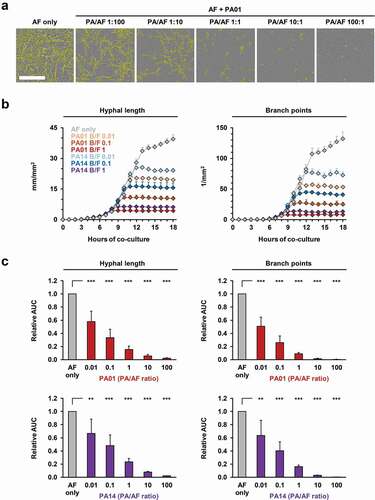
To facilitate real-time image analysis of mycelial proliferation and morphogenesis during co-culture with different concentrations of viable PA cells, we used a GFP-expressing AF293 strain and a fluorescence-based NT algorithm. Cells of either PA WT strain suppressed both hyphal growth and branch point formation in an inoculum-dependent fashion (, Figure S1A). Although initial inhibition of AF morphogenesis at low PA inoculums was rather moderate, a time-dependent increase in inhibition was seen, with a plateau reached after 8–10 h of co-culture (, Figure S1B), whereas PA cells continued to proliferate (Movie S1). Even at PA/AF ratios as low as 0.01, a 33–49% reduction in mycelial elongation and hyphal branching was seen. Relative AUCs further dropped to 0.09–0.23 at a 1:1 ratio (p < 0.01) and a 100:1 PA/AF ratio completely abolished fungal germination (p < 0.001, ). No interference of bacterial cells or auto-fluorescence with the image analysis algorithm and no deviations in autofocus accuracy were observed (Movie S1). Median intra-assay CVs for hyphal length and branching in bacterial co-culture were 7.8% and 11.8%, respectively, and inter-replicate CVs were even lower (Table S2).
Figure 3. Fluorescence-based NT analysis facilitates efficient longitudinal tracking of inoculum-dependent inhibition of A. fumigatus growth and morphogenesis during co-culture with live P. aeruginosa cells.
Discussion
Efficient techniques for longitudinal visualization and analysis of morphological endpoints could expand our understanding of the complex inter-kingdom relationships of fungal and bacterial pathogens competing for the same ecological niches. To that end, we applied the IncuCyte NT live imaging and analysis platform to AF co-cultures with purified PA products, filtrates of PA mutants, and viable PA cells. NT analysis has a number of crucial advantages compared with conventional endpoint assays of fungal proliferation, including an improved longitudinal resolution of phenotypic parameters, capacity to assess multiple well plates concurrently, and a high degree of automation based on pre-programmed routines, resulting in decreased hands-on time [Citation11,Citation14].
To evaluate the suitability of the platform for bacterial and fungal co-culture studies, we first verified the system’s capacity to recapitulate known inhibitory effects of siderophore-deficient PA mutants and purified iron-binding PA metabolites on AF mycelial formation. Iron is a central nutrient for AF [Citation15], and the antifungal activity of PA has been tightly linked to the production of iron-sequestering metabolites, interfering with fungal iron availability [Citation2–Citation4]. The PA siderophore pyoverdine has been suggested as the key inhibitory molecule, especially in low iron environments [Citation2,Citation3], while more recent studies have stressed the dominance of pyocyanin as the main PA competitive molecule when iron is plentiful [Citation16]. Our data suggest that NT analysis has excellent statistical power to detect the differential inhibitory activity of PA mutants and concentration-dependent effects of purified PA products. Therefore, the NT technology would be appealing as an efficient screening assay for bacterial metabolites or mutants interfering with fungal morphogenesis and expansion.
Despite the explosive growth of PA in RPMI medium resulting in considerable turbidity of the wells at high PA/AF ratios, no changes to the processing algorithms for fungal mono-culture or immune cell challenge [Citation11] were needed to maintain high accuracy of NT analysis during co-culture with viable PA cells. Furthermore, no interference of bacterial auto-fluorescence with the image analysis algorithm was encountered. This observation is consonant with a previous report that excitation and emission spectrums of PA, but also other bacterial pathogens of potential interest for future studies (e.g. Escherichia coli or Staphylococcus aureus), do not overlap with the IncuCyte fluorescence channels [Citation17]. Technical variation of NT endpoints in bacterial co-cultures slightly exceeded that in fungal monocultures, likely due to the additional variation introduced by the bacterial seeding step and potential asynchrony of inhibitory PA effects on AF mycelia. However, the reproducibility of NT endpoints was well within recommended ranges for microbiological cellular bioassays [Citation18]. Importantly, reproducibility remained excellent even at very low PA inocula. Therefore, our titratable system can avoid or delay overgrowth of mycelia by PA cells during biofilm formation, a common issue of experimental in vitro systems for co-culture studies [Citation19].
In contrast to NT analysis, most metabolic methods to track fungal proliferation are severely confounded by the presence of viable bacteria. For example, widely used colorimetric tetrazolium-based assays to quantify metabolically active fungal biomass require the use of bacterial filtrates [Citation3] or extensive post-processing of mycelia after recovery from bacterial co-culture [Citation20], as residuals of viable bacteria would metabolize tetrazolium [Citation21] and cause false-positive results.
As with all assays, there are limitations of NT analysis to consider. Differential activity of PA mutants against preformed AF biofilms and planktonic cultures has been described [Citation3,Citation7]. While IncuCyte NT assessment is powerful to longitudinally study germination and early mycelium formation in planktonic cells, the method performs poorly in resolving morphological features of preformed biofilms as the autofocus-guided image acquisition procedure does not provide control of the focusing depth by the user [Citation11]. Another limitation inherent to the GFP-based live cell co-culture approach is the need for sufficiently bright and stable fluorescent labeling approaches and/or availability of fluorescent fungal mutants, a circumstance that could complicate the assessment of clinical mold isolates. In addition, the effects described pertain directly to intermicrobial interactions in liquid media, mimicking, for example, what would occur in bronchial secretions. However, the dynamics and effects of mutations and microbial products might be different in studies of microbes on solid substrates, such as agar, mimicking epithelial or endothelial surfaces. As a specific limitation of the present study, the experiments were performed under normoxic conditions, whereas hypoxia is a common feature in a cystic fibrosis microenvironment, affecting growth characteristics and inter-microbial interactions of the studied pathogens [Citation22]. Future studies with an IncuCyte system placed in oxygen-controlled environments could, therefore, present an exciting research direction.
In summary, the IncuCyte NT technology provides a new, powerful toolbox for intermicrobial interaction studies, facilitating real-time analysis of key morphological endpoints during live cell co-culture with unparalleled longitudinal resolution and robust technical precision. Furthermore, employing a commercially available platform and published processing algorithms [Citation11], NT analysis could contribute to improved standardization of inter-microbial co-culture experiments across different laboratories. Paired with its capacity to accurately track antifungal drug effects in a spectrum of clinically relevant molds and yeasts [Citation11], NT analysis could have important translational implications for studies of antifungal drug activity. Of note, Briard and colleagues showed that quorum-sensing controlled PA rhamnolipids have synergistic activity with azole antifungals against AF [Citation6]. Providing a rapid and reliable platform to test these interdependencies during live cell co-culture, IncuCyte NT could refine drug screening studies in inter-kingdom infection settings such as mixed fungal and bacterial pneumonia in cancer patients or poly-microbial wound infections.
Acknowledgment
DPK acknowledges the Texas 4000 Distinguished Professorship for Cancer Research and the NIH-NCI Cancer Center CORE Support grant no. 16672. DAS has received support from the Flatley Foundation, grant CIMR no. 3770.
Supplemental Material
Download PDF (874.8 KB)Disclosure statement
DPK reports research support from Astellas Pharma and honoraria for lectures from Merck & Co, Gilead, and United Medical. He has served as a consultant for Astellas Pharma, Cidara, Pulmocide, and Mayne Pharma. The Stevens laboratory has received research grants from Astellas Pharma, Riptide, Aridis, and Valley Fever Solutions. DAS has consulted for Riptide, Pulmatrix, Mayne, and the US Department of Justice, and received honoraria for lectures from Pfizer China. The authors are not affiliated with Sartorius or the IncuCyte® brand and have not received any support with the exception of general consumer services.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Arvanitis M, Mylonakis E. Fungal-bacterial interactions and their relevance in health. Cell Microbiol. 2015;17(10):1442–1446.
- Sass G, Nazik H, Penner J, et al. Aspergillus-Pseudomonas interaction, relevant to competition in airways. Med Mycol. 2019;57(Supplement_2):S228–32.
- Sass G, Nazik H, Penner J, et al. Studies of Pseudomonas aeruginosa mutants indicate pyoverdine as the central factor in inhibition of Aspergillus fumigatus biofilm. J Bacteriol. 2017;200(1):e00345–17.
- Briard B, Bomme P, Lechner BE, et al. Pseudomonas aeruginosa manipulates redox and iron homeostasis of its microbiota partner Aspergillus fumigatus via phenazines. Sci Rep. 2015;5:8220.
- Rooney PJ, Klein BS. Linking fungal morphogenesis with virulence. Cell Microbiol. 2002;4(3):127–137.
- Briard B, Rasoldier V, Bomme P, et al. Dirhamnolipids secreted from Pseudomonas aeruginosa modify anjpegungal susceptibility of Aspergillus fumigatus by inhibiting β1,3 glucan synthase activity. Isme J. 2017;11(7):1578–1591.
- Shirazi F, Ferreira J, Stevens DA, et al. Biofilm filtrates of Pseudomonas aeruginosa strains isolated from cystic fibrosis patients inhibit preformed Aspergillus fumigatus biofilms via apoptosis. PLoS One. 2016;11(3):e0150155.
- Joubert LM, Ferreira JA, Stevens DA, et al. Visualization of Aspergillus fumigatus biofilms with scanning electron microscopy and variable pressure-scanning electron microscopy: a comparison of processing techniques. J Microbiol Methods. 2017;132:46–55.
- Reichhardt C, Ferreira JA, Joubert LM, et al. Analysis of the Aspergillus fumigatus biofilm extracellular matrix by solid-state nuclear magnetic resonance spectroscopy. Eukaryot Cell. 2015;14:1064–1072.
- Etxebeste O, Espeso EA. Neurons show the path: tip-to-nucleus communication in filamentous fungal development and pathogenesis. FEMS Microbiol Rev. 2016;40(5):610–624.
- Wurster S, Kumaresan PR, Albert ND, et al. Live monitoring and analysis of fungal growth, viability, and mycelial morphology using the incucyte neurotrack processing module. mBio. 2019;10(3):e00673–19.
- Ferreira JA, Penner JC, Moss RB, et al. Inhibition of Aspergillus fumigatus and its biofilm by Pseudomonas aeruginosa is dependent on the source, phenotype and growth conditions of the bacterium. PLoS One. 2015;10:e0134692.
- Kerr JR, Taylor GW, Rutman A, et al. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J Clin Pathol. 1999;52(5):385–387.
- Gelles JD, Chipuk JE. Robust high-throughput kinetic analysis of apoptosis with real-time high-content live-cell imaging. Cell Death Dis. 2016;7(12):e2493.
- Matthaiou EI, Sass G, Stevens DA, et al. Iron: an essential nutrient for Aspergillus fumigatus and a fulcrum for pathogenesis. Curr Opin Infect Dis. 2018;31(6):506–511.
- Sass G, Nazik H, Chatterjee P, et al. In high iron environment, pyocyanin is a major anti-fungal molecule; differences in prototype P. aeruginosa strains. Med Mycol. 2020, in press. DOI:https://doi.org/10.1093/mmy/myaa066
- Molyneux PM, Kilvington S, Wakefield MJ, et al. Autofluorescence signatures of seven pathogens: preliminary in vitro investigations of a potential diagnostic for acanthamoeba keratitis. Cornea. 2015;34(12):1588–1592.
- Shurtleff AC, Bloomfield HA, Mort S, et al. Validation of the filovirus plaque assay for use in preclinical studies. Viruses. 2016;8(4):113.
- Briard B, Mislin GLA, Latgé JP, et al. Interactions between Aspergillus fumigatus and pulmonary bacteria: current state of the field, new data, and future perspective. J Fungi (Basel). 2019;5(2):48.
- Patel N, Oudemans PV, Hillman BI, et al. Use of the tetrazolium salt MTT to measure cell viability effects of the bacterial antagonist Lysobacter enzymogenes on the filamentous fungus Cryphonectria parasitica. Antonie Van Leeuwenhoek. 2013;103(6):1271–1280.
- Roy R, Tiwari M, Donelli G, et al. Strategies for combating bacterial biofilms: a focus on anti-biofilm agents and their mechanisms of action. Virulence. 2018;9(1):522–554.
- Anand R, Clemons KV, Stevens DA. Effect of anaerobiasis or hypoxia on Pseudomonas aeruginosa inhibition of Aspergillus fumigatus biofilm. Arch Microbiol. 2017;199(6):881–890.
