ABSTRACT
The type VI secretion system (T6SS) is a new secretion system that is widely distributed among Gram-negative bacteria. The core component hemolysin-coregulated protein (Hcp) can be used as both its structural protein and secretory protein or chaperone protein. Studies on Hcp are important to elucidate the overall virulence mechanism of T6SS. Salmonella typhimurium is an important foodborne pathogen. There are three copies of hcp genes identified in S. Typhimurium 14028s. This study aimed to characterize the functions of the three Hcp family proteins and to elucidate the interactions among them. The hcp gene deletion mutants were constructed by λ Red-based recombination system. Effects of hcp mutation on the pathogenicity of 14028s were studied by bacterial competition assays, Dictyostelium discoideum assays and mouse model. The three Hcp family proteins were found to play different roles. Hcp1 can affect the transcription of rpoS and type 2 flagellar gene and influence the motility of 14028s. It is also involved in the intracellular survival of 14028s in Dictyostelium discoideum; Hcp2 is involved in the early proliferative capacity of 14028s in mice and can prevent its excessive proliferation; Hcp3 did not show direct functions in these assays. Hcp1 can interact with Hcp2 and Hcp3. Deletion of one hcp gene can result in a transcription level variation in the other two hcp genes. Our findings elucidated the functions of the three Hcp family proteins in S.Typhimurium and illustrated that there are interactions between different Hcp proteins. This study will be helpful to fully understand how T6SS actions in an organism.
Introduction
Bacterial pathogens utilize various secretion systems to export toxins or effectors into target cells. There are nine secretion systems discovered so far [Citation1,Citation2]. The type VI secretion system (T6SS) was first described in Vibrio cholerae in 2006 [Citation3]. Studies have shown that T6SS plays important role in the pathogenicity of bacteria [Citation4–6]. Currently, there are three different types of T6SS identified [Citation2]. The second type is only found in Francisella, the third is only found in Bacteroides, and the first type which consists of 13 core genes has been found in most Gram-negative bacteria [Citation2,Citation7]. S.Typhimurium is a common pathogen of foodborne diseases worldwide [Citation8,Citation9]. It can be transmitted through the fecal-oral route, and cause diarrhea or other systemic disease by entering the host’s macrophages or dendritic cells [Citation10,Citation11]. Researches on virulence-related genes are necessary to reveal its invasion mechanism.
It has been reported that in S.Typhimurium, there is a T6SS gene cluster which was found to play important roles in bacterial competition and in the interaction of bacteria with macrophages or hosts [Citation12–18]. The hemolysin-coregulated protein (Hcp) is an important structural protein which can be stacked into a tube of Hcp hexamers to form the tail of T6SS; moreover, it can also play the roles of secretory protein or chaperone protein [Citation19–23]. The secretion of Hcp is the hallmark of a functional T6SS [Citation24]. Understanding the roles of Hcp is critical to explore the function of T6SS. It is interesting that in some bacteria, more than one copy of hcp genes was found. However, even if multiple copies of hcp genes exist, their functions are not redundant. They can take different roles in bacterial competition, interaction with macrophages or involve in the biological phenotypes such as the motility or biofilm formation of bacteria [Citation25–29]. However, it remains a question that how the Hcp family proteins function? Are there interactions among them? In S.Typhimurium 14028s, we found three copies of hcp genes (Additional file 1: Figure S1), two of them (hcp1 and hcp2) are on the Salmonella pathogenic island 6 (SPI-6). The proteins encoded by the two genes are very similar, with only 10 amino acid differences between them [Citation23]. There is also an orphan hcp3 gene not linked with SPI-6 [Citation12]. Hcp3 only shared 26%-27% similarity with Hcp1 and Hcp2 and it is more homologous to the Hcp of S.bongori and Pseudomonas [Citation12]. Previous studies have shown that the expression of Hcp1 is enhanced in bile salt environment and it can interact with amidase Tae4 to exert antibacterial activity [Citation5]. In this study, we constructed hcp mutants and investigated the roles of the three Hcp family proteins by different assays. We further investigated the interactions between different Hcp proteins to elucidate the mechanism behind them.
Results
Effects of hcp mutations on the growth rates, antimicrobial resistance, biofilm formation, and the motility of S.Typhimurium14028s
All mutant strains were confirmed with PCR using specific primers. To investigate whether hcp knock-out would affect the growth of Salmonella, we tested the growth rates of the Δhcp mutants and wild type 14028s in both LB and M9-glucose minimum medium. As shown in ), there were no significant differences among them, indicating that knocking out hcp did not affect the growth of Salmonella. A recent study found that deletion of the T6SS core component can influence the antimicrobial resistance of A. baumannii [Citation30]. In this study, we tested the effects of hcp mutation on the antimicrobial resistance of Salmonella with five antibiotics, ampicillin, gentamicin, streptomycin, naphthalidine, and tetracycline. As shown in , the MIC of the wild type and mutants for the five antibiotics were 16 μg/mL, 2 μg/mL, 32 μg/mL, 4 μg/mL, and 2 μg/mL, respectively. There were no significant differences between wild type and mutant strains. The same result was obtained using the K-B method (Additional file 2: Figure S2). Compared with the wild type strain 14028s, there was a mild decrease tendency in the biofilm formation ability of Δhcp1 and Δhcp2 mutants ()). However, there were no significant differences between them.
Table 1. Effects of hcp mutations on the MICs of antimicrobials
Figure 1. Growth characters in LB (a) and M9-glucose minimum medium (b), biofilm formation ability (c) and motility (d) of wild type and mutant strains. Motility of the wild type and mutant strains was determined by measuring the diameter of the column. Error bars represent the SDs. Significant differences were defined by P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***) compared to the wild type strain 14028s
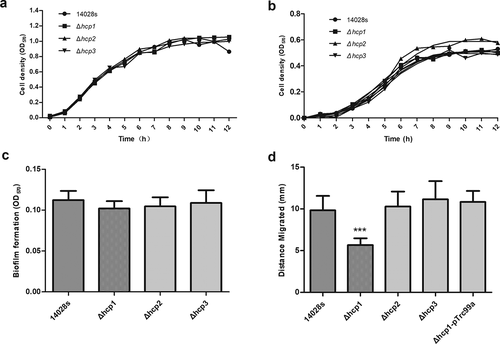
The motility of wild type and mutant strains was examined by measuring the diameter of the circles on the swimming plate. As shown in ), the motility of the Δhcp1 mutant was significantly decreased (P < 0.001). Complementation of hcp1 restored the motility of Δhcp1 mutant. In contrast, deletion of hcp2 and hcp3 did not affect the motility of the strain.
Effects of hcp mutations on the bacterial competition ability
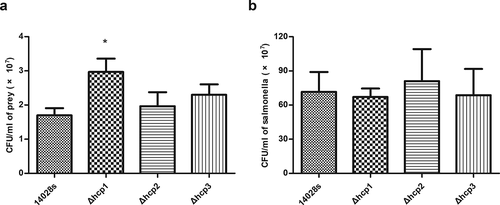
Previous studies revealed that the bacterial competition ability of S.Typhimurium SL1344 to E. coli was significantly reduced after knocking out hcp1, and knocking out hcp2 has no effect on bacterial competition [Citation5]. This study focused on whether hcp3 played a role in bacterial competition. Our results showed that consistent with previous study, mutation of hcp1 significantly affected the bacterial competition ability of 14028s (P < 0.05) and mutation of hcp2 showed no significant effect. The competition ability of Δhcp3 was similar with that of the wild type strain, suggesting that hcp3 was not implicated in Salmonella bacterial competition ()). The CFUs of Salmonella strains after competition showed no significant statistical difference ()).
Figure 2. Results of bacterial competition assay. The surviving prey (E.coli JM109) (a) and the surviving salmonella strains (b) after 12 h co-incubation on LB plate supplemented with 0.05% porcine bile salts were measured by counting CFU. Error bars represent the SDs. Significant differences were defined by P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***) compared to the wild type strain 14028s
Effects of hcp mutations on intracellular survival of Salmonella in D.discoiseum
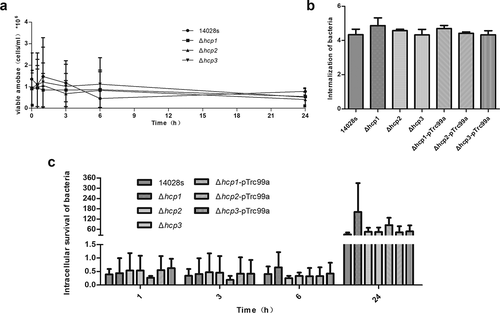
Dictyostelium discoideum is an amoeba model which was widely used for host–pathogen interactions [Citation3]. It was also shown that T6SS was required for S. Typhimurium to survive intracellularly in the social amoeba Dictyostelium discoideum [Citation15]. To determine the roles of the three Hcp family proteins in the intracellular survival of Salmonella, we co-incubated both wild type 14028s and its mutants in D.discoiseum and let D.discoiseum fed on bacteria. There were no significant differences between the number of viable D.discoideum when incubated with the wild type 14028s and hcp mutants ()). So the influence of the number of viable D.discoiseum can be excluded. Then the internalization and intracellular survival of Salmonella in D.discoideum were tested. It was found that all the mutants showed an internalization level similar to that of the wild type14028s ()). For the intracellular survival ability, there were no significant differences for all the strains within 3 h; however, Δhcp1 showed a strong increasing trend at 6 h and 24 h, although there were no statistically significant differences. Complementation of hcp1 can suppress the increasing trend. These results indicated that deletion of hcp1 can result in increased intracellular survival ability in D.discoideum ()).
Figure 3. The population of viable amoebae at each time point when incubated with the wild type and mutant strains (a) were tested by Trypan blue exclusion and counting on a Neubauer chamber. The internalization of the wild type and mutant strains in D.discoideum (b) were calculated as CFUt=0/CFUinoculum and converted negative logarithmically. The intracellular survival of the wild type and mutant strains in D.discoideum (c) were calculated at each time point as CFUt=x/CFUt=0. Error bars represent the SDs. Significant differences were defined by P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***) compared to the wild type strain 14028s
Virulence in mouse model of systemic infection
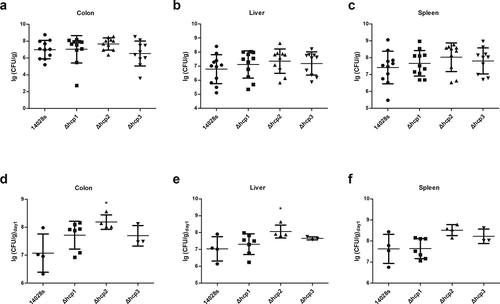
To study the roles of the three Hcp family proteins played in the virulence to mouse model, we challenged BALB/c mice with two different Salmonella levels. At first, we challenged mice with a low level (200 μL bacterial suspensions normalized to 106 CFU/mL) which is not lethal to mice and examined the proliferation of bacteria every day. We found that the weight of mice spleen and the number of the bacteria recovered from the spleen of the Δhcp1 and Δhcp3 group showed no significant difference with wild type 14028s group, in contrast, the Δhcp2 mutant group showed an increasing tendency compared with wild type 14028s group within 2 days, but on day 5 the difference between the two groups disappeared (Additional file 3: Figure S3), suggesting Hcp2 may be involved in the early proliferative capacity of 14028s in mice. To verify this, we challenged mice with a higher level of Salmonella (200 μL bacterial suspensions normalized to 108 CFU/mL). Our results showed that there were no significant differences in the survival rate among different groups (Additional file 4: Figure S4). However, when calculating the number of Salmonella in the mice that died on the first day (within 24 h after inoculation), we found that the Δhcp2 mutant showed a significant higher colonization ability in the colon and liver (p < 0.05), and there was also a higher trend in the spleen (–f)). The other mutants showed no significant differences with the wild type strain. When we examined the number of bacteria on day 5, there were no significant differences between wild type 14028s and the mutants, which is consistent with the results of low-level inoculation (Additional file 3: Figure S3b).
Figure 4. Virulence of the wild type and mutant strains in mouse model of systemic infection were determined by examining the bacteria concentrations in different organs (colon, spleen, and liver)
Interactions of the three Hcp family proteins
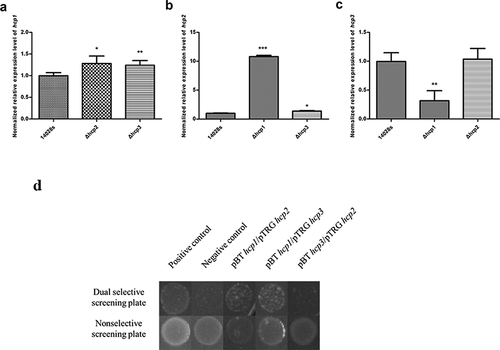
Since there are some similarities among the sequences of the three Hcp family proteins, we tried to see whether there are interactions between them. The hcp transcription levels of the wild type 14028s and hcp mutant strains were tested by qRT-PCR. It was found that the hcp2 transcription level in the Δhcp1 mutant was significantly increased (P < 0.001), and the hcp3 transcription level was significantly decreased (P < 0.01) ()). For the Δhcp2 mutant, the hcp1 transcription level was significantly increased (P < 0.05), while the hcp3 transcription had no difference ()). The hcp1 transcription level of Δhcp3 mutant was significantly increased (P < 0.01), and the hcp2 transcription level was also increased (P < 0.05) ()).We then tested whether there are interactions among the Hcp proteins. The results of bacterial two-hybrid assay indicated that Hcp1 can interact with Hcp2 and Hcp3, while Hcp2 and Hcp3 cannot interact with each other ()).
Figure 5. Interactions of the three Hcps were tested by qRT-PCR and bacteria two-hybrid assay. (a–c) The normalized expression level (2−ΔΔct) of hcps of the wild type and mutant strains. 16S rRNA was used as the internal parameter. Error bars represent the SDs. Significant differences were defined by P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***) compared to the wild type strain 14028s. (d) Bacteria two-hybrid analyses. Different strains that have different plasmid combinations were spotted on dual selective screening plate and nonselective screening plate respectively at 30°C for 24 h. The strains can grow on the dual selective screening plate if the proteins on the plasmids can interact. The experiments were done at least in triplicate, and a representative result is shown
Effects of hcp mutations on the transcription of rpoS, fimH and flagellar genes
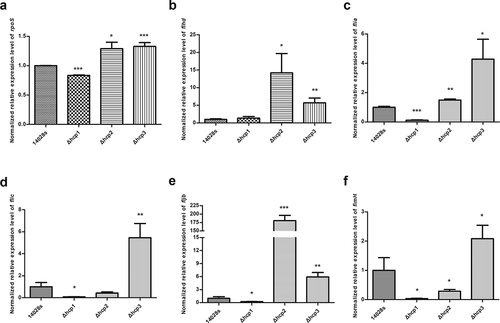
Lastly, we tested the transcription levels of rpoS, fimH and flagellar genes in the wild type 14028s and the hcp mutants. Since our results showed that mutation of hcp1 can influence the motility of Salmonella, we tested the transcription levels of the rpoS, fimH and flagellar genes to elucidate the mechanism behind it. Results () showed that in the Δhcp1 mutant, the transcription of rpoS, flia, fljb, flic, and fimH was significantly decreased. In the Δhcp2 mutant, the transcription of rpoS, flhd, flia and fljb was significantly increased and the transcription of fimH was significantly decreased. In the Δhcp3 mutant, the transcription of rpoS, flia, flhd, fljb, flic, and fimH were increased significantly.
Figure 6. The normalized expression level (2−ΔΔct) of rpoS(a), flagellar genes flhd (b), flia (c), flic (d), fljb (e), and fimH (f) of the wild type and mutant strains were tested by qRT-PCR. 16S rRNA was used as the internal parameter. Error bars represent the SDs. Significant differences were defined by P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***) compared to the wild type strain 14028s
Discussion
In this study, we constructed hcp mutants and tested the effects of hcp mutations on a variety of characters in S.Typhimurium 14028s. Our results showed that hcp mutation had no significant effect on the growth of 14028s. So the subsequent results can exclude the influence that may be caused by growth ability. Previous study showed that mutation of T6SS core genes can influence the antimicrobial resistance in A.baumannii [Citation30]. S.Typhimurium was found to have a high resistant rate to streptomycin, ampicillin, nalidixic acid, chloramphenicol, gentamicin, and tetracycline [Citation31]. In this study, we tested the resistance of the hcp mutants to these antibiotics, and the results showed that hcp mutations of 14028s had no significant effect on the antimicrobial resistance. In Aeromonas hydrophila, it was found that hcp was implicated in the biofilm formation ability [Citation28]. However, in our study, we found that hcp mutation showed no effect on biofilm formation.
The assembly and functional expression of flagella involves three types of genes which were expressed in a cascade. flhD is a representative of type 1 flagellar gene which is necessary for the activation and transcription of type 2 gene promoters. fliA belongs to the type 2 flagellar gene, which encode proteins that constitute the flagellar secretion device and participate in the transcription of the type 3 flagellar genes which participate in the final motility function of bacteria [Citation32]. In this study, we found that deletion of hcp1 led to significant defects in the motility of S.Typhimurium. This can be explained by the data from qRT-PCR, which showed that in Δhcp1 mutant the type 2 flagellar gene fliA and type 3 flagellar genes fliC and fljB all showed significant decreased transcriptions, which may result in reduced synthesis of flagellin and thus influence the motility. Previous studies [Citation33,Citation34] have found that rpoS can regulate the expression of T6SS to help bacteria survive under stress conditions, another study [Citation35] also found that it can be regulated by T6SS. In this study, we found the deletion of hcp can affect the transcription of rpoS. Since rpoS has also been found to be involved in the formation of flagella [Citation33], our study indicated that Hcp can modulate the expression of rpoS to regulate the expression of flagella genes and thus affect the motility of bacteria.
T6SS was required for S. Typhimurium to survive intracellularly in the social amoeba Dictyostelium discoideum [Citation15,Citation36]. However, the mechanism and the effectors associated with this process remain unknown. Some studies revealed that although flagellar mutants are more difficult to invade epithelial cells, mutants have stronger virulence to mice and a faster net growth in macrophages within 4–24 h [Citation32,Citation37]. In addition, in the study of avian pathogenic Escherichia coli (APEC), it was found that deletion of hcp gene significantly affected the expression of fimbriae subunit protein and decreased the adhesion and invasion of cultured cells in vitro [Citation38]. Consistent with these results, our data showed that the transcription of fimbriae gene fimH was significantly decreased in the Δhcp1 and Δhcp2 mutants. And we found that although it was not statistically significant, the internalization level of Δhcp1 and Δhcp2 was lower than that of the wild type strain 14028s. The intracellular survival of Δhcp1 showed a strong increasing trend at 6 h and 24 h. We postulated that the down-regulation of the flagellar gene caused by the deletion of hcp1 makes it impossible for Salmonella to synthesize flagella and thus reduce the metabolic burden, which may be one of the main reasons of increased intracellular survival in amoeba.
In order to survive in a multi-bacterial environment such as the host gut, bacteria need to compete with each other [Citation39]. Sana et al. found that hcp1 can affect the ability of S.Typhimurium to participate in bacterial competition with bile salts, while hcp2 has no effect [Citation5]. The same results were obtained in our experiments. Besides this, we found Δhcp3 mutant had the same competitive ability as the wild type strain. Therefore, Hcp1 is mainly involved in bacteria competition and the other two Hcps showed no direct effects.
By analyzing the transcription levels and protein interactions, we found there were indeed mutual interactions between Hcp proteins. There was a balance between Hcp1 and Hcp2, and they may complement each other due to the similarity in their amino acid sequences. The transcription of hcp3 decreased significantly in the Δhcp1 mutant, we suppose that Hcp3 may mainly assist the function of Hcp1. We hypothesized that hcp3 might function more as a secretory protein. Further studies were needed to verify these assumptions.
In our animal model assay, it was found that the colonization ability of Δhcp2 mutant in the liver and colon increased significantly within 24 h after inoculation. It has been reported that deletion of pcgL and scis can also increase the proliferation of Salmonella in the liver and spleen in mice and in macrophages [Citation40,Citation41]. There is a symbiotic view that bacteria may deliberately reduce their virulence to allow themselves to continue to spread [Citation42]. Gene expression in the host cell will cause a difference in the cellular environment. Some environments will promote cell proliferation, which is conducive to immune evasion and the establishment of niche, while other environments will allow bacteria to reproduce [Citation43]. A study showed that the deletion of T3SS-related genes leads to increased intracellular reproduction and reduced spread [Citation44]. In addition, there is one hypothesis that Salmonella can use the bacteria competition ability mediated by T6SS to clear its niche [Citation39]. We hypothesize that Hcp2 may just play this balance function or elimination mechanism to escape the host’s killing.
In summary, we conducted a comprehensive study and concluded that the three Hcp family proteins play different roles in S.Typhimurium14028s, and the three Hcp family proteins can interact and balance with each other. Hcp1 plays an important role in bacterial competition, long-term intracellular survival in eukaryotic cells, and bacterial motility. These effects may be due to the expression of flagellar genes. Hcp2 mainly contributes to the early stage of bacterial infection in mice. Hcp3 mainly assists the function of Hcp1. We assume that under different conditions, bacteria may selectively use different Hcp proteins to form hexamer pipes, but whether such pipes are a single Hcp stack, or whether two or three Hcp proteins together still need further research. Researches on those virulence-related genes are necessary to reveal the invasion mechanism of the bacteria.
Materials and methods
Bacteria strains and cultural condition
Bacteria and plasmids used in this study were listed in Table S1 (Additional file 5) and Table S2 (Additional file 6). Bacteria were grown in LB broth unless otherwise stated. Antibiotics were added when necessary. The concentrations of antibiotics used in this study were: kanamycin: 50 mg/L; ampicillin: 100 mg/L; chloramphenicol: 25 mg/L; gentamicin: 10 mg/L; tetracycline: 15 mg/L.
The eukaryotic cell model used in this study was Dictyostelium discoideum strain AX4, which was obtained from the Dicty Stock Center and revived with standard protocols [Citation45]. Briefly, solid culture was carried out by plating on SM media or LPB media containing E. coli B/r, and liquid culture was carried out by aseptic culture in HL5 liquid media.
Construction of mutant and complement strains and determination of growth characters
The hcp mutant strains were constructed using the λ Red-based recombination system described in previous study [Citation46]. Briefly, the fragment containing a 56 bp homologous fragment of target gene and kanamycin resistance gene which comes from pKD4 was introduced into 14028s containing pKD46. After recombination, the pKD46 was removed by cultured at 42°C. Then, pCP20 was introduced to obtain a pure gene knockout strain without kanamycin resistance. pTrc99a integrating the target genes were introduced into the mutant strains to produce complement strains.
To verify whether hcp mutation affects the growth ability of 14028s, the growth characters were measured: bacteria were cultured overnight and adjusted to OD600 ~ 1, then diluted 1:100 to fresh LB broth or M9-glucose (0.2%) minimum medium [Citation47] and cultured with shaking, then 200 μL was added to 96-well plate. The growth rates were detected every hour by measuring the value of OD570.
Biofilm assays
Single colonies were picked from LB Agar, transferred to fresh biofilm induction medium YP media (2% glucose, 2% peptone, 1% yeast extract), and cultured overnight at 30°C with shaking. The bacteria solutions were adjusted to OD600 ~ 1 and inoculated in fresh media with a rate of 1:100, and then 200 μL of the culture were added to each well of 96-well plate and incubated at 30°C for 72 h. Then the supernatant and the unattached bacteria were gently removed and washed away with distilled water. The bioflim was stained with 200 μL of 2% crystal violet dye solution for 20 min, and washed three times with water. Then, 200 μL of 95% ethanol was added to dissolve the crystal violet. After 20 min, the biofilm formation ability was determined by measuring the value of OD570.
Antimicrobial susceptibility assays
The antimicrobial susceptibility assays were performed using the MIC broth dilution method for ampicillin, gentamicin, nalidixic acid, streptomycin, and tetracycline. Single colonies of bacteria were inoculated in LB broth and cultured overnight at 37°C, and then the cultures were adjusted to OD600 ~ 0.5, and transferred to fresh MH medium at a ratio of 1:100. At last 100 μL of the culture was added to 96-well plate and cultured at 37°C. After 16–20 h, the OD570 were detected by microplate reader to determine the minimum inhibitory concentration (MICs). In addition, we used K-B method to determine the drug susceptibility of ampicillin, gentamicin, streptomycin, and tetracycline again according to the national clinical test operating protocol.
Motility assays
Single colonies were inoculated in LB broth, and the overnight culture were normalized to OD600 ~ 1; then, 1 μL of the bacteria suspensions were spotted on the center of swimming plate (1% tryptone, 0.25% Sodium chloride, 0.3% agar) respectively. After 18 h incubation, the migrated distances of the colonies were measured.
Bacterial competition assays
Bacterial competition assays were performed according to previous studies [Citation5]. Overnight cultures were inoculated into fresh LB broth at a ratio of 50:1. After 4 h, the bacteria were washed with PBS and re-suspended to a concentration of approximately 1010 CFU/mL. Salmonella and E.coli were mixed at a ratio of 10:1 and then co-cultured on LB agar containing 0.05% porcine bile salts at 37°C for 12 h. The bacteria were washed and re-suspended with PBS, and then the colony-forming units (CFU) were determined by plating serial dilutions on LB agar plates with appropriate antibiotics.
Dictyostelium invasion assay
Dictyostelium invasion assay was performed as described in previous literature with minor modifications [Citation15]. Briefly, the Dictyostelium AX4 was adjusted to a concentration of 5*105 cells/mL, and 2 mL of the suspension was pipetted into a 6-well plate and grown at 22°C for 1 hour to let it adhere to the surface. Then, AX4 was co-incubated with wild type or mutant strains in HL5 media using a multiplicity of infection (MOI) of 100 bacteria/amoeba for 1 h. The Dictyostelium were washed three times with Soerensen buffer and then re-suspended in 2 mL HL5 media (t = 0) and incubated at 22°C for different time points. At the time point 0, 1, 3, 6, 24 h, the infected cells were washed with Soerensen buffer containing 10 mg/L gentamycin. Then the Dictyostelium were washed with Soerensen buffer to remove residual antibiotics, and lysed with 0.2% TritonX-100. The colony-forming units (CFU) were determined by plating serial dilutions on LB agar plates with appropriate antibiotics. The internalization was calculated as CFUt=0/CFUinoculum and expressed as – log10. The survival ability of each time point was calculated as CFUt=x/CFUt=0. Viable amoeba were determined by Trypan blue dying and counted on Neubauer chamber at each time point.
Mice infection assays
Mice infection assays were performed following the Guide for the Care and Use of Laboratory Animals, National Research Council, 1996, and were approved by the ethics committee of Peking University Health Science Center of China. 6–8 week old female BALB/c mice were randomly assigned to five groups and housed under specific pathogen-free condition with sterile water and food. The mice were starved for 16 h before intra-gastrically administered with 200 μL of 5% sodium bicarbonate. After 30 minutes, 200 μL bacterial suspensions normalized to 106 CFU/mL or 108 CFU/mL were inoculated. Once a mouse was dead, it was removed quickly from the cage, dissected and the liver, spleen, and colon were picked for culture. The organs were weighed, homogenized, and cultured on MacConkey agar plate to determine the CFU. On day five, mice were sacrificed with cervical dislocation and their organs were removed for culture to calculate the CFU.
RNA extraction and real-time quantitative polymerase chain reaction (qRT-PCR)
Bacteria were cultured overnight, and RNA extraction was performed using RNAprep Pure Kit (TianGen). The extracted RNA was reversed to cDNA using the All-in-OneTMFirst-Strand cDNA Synthesis Kit (TaKara). Then, qRT-PCR was performed using the 2*SYBR Green qPCR Master Mix (Low Rox). The CT value was obtained by using the 7500 Fast DX instrument, the ΔCT value was obtained by subtracting from the internal parameter (16S rRNA), and the normalized relative expression level of the target genes was calculated by the comparative cycle threshold (2−ΔΔCT).
Bacteria two-hybrid assay
BacterioMatch® II Two-Hybrid system (Stratagene, La Jolla, CA, USA) was used for bacteria two-hybrid assay. The hcp fragments were cloned into pBT and pTRG to construct pBT-hcp1, pBT-hcp3, pTRG-hcp2 and pTRG-hcp3 plasmids, respectively. Then, pBT and pTRG plasmids were paired into XL1-Blue Reporter Strain. Bacteria strains that were successfully transferred with the two corresponding plasmids were cultured at 37°C overnight. Then they were normalized to OD600 ~ 0.3 and resuspended with M9 media. Finally, 10 μL of the suspensions was spotted on dual selective screening plate and nonselective screening plate, respectively, at 30°C for 24 h.
Statistical analyses
The data obtained were analyzed and plotted with Graphpad prism version 5.0. The comparison between the wild type strain and mutant strains was performed by student’s t test, and the mean ±SD from more than three independent experiments was calculated. Analysis of mouse model’s survival was performed by chi-square test using the SAS 9.0 software.
Authors’ contributions
QHZ and LL conceived and designed research. PW conducted experiments. RQL and JFD contributed new reagents or analytical tools. PW performed the experiments and analyzed the data. QHZ and PW wrote the manuscript. All authors read and approved the manuscript.
Ethics approval and consent to participate
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Supplemental Material
Download Zip (239.8 KB)Disclosure statement
The authors declare that they have no conflict of interest.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Lasica AM, Ksiazek M, Madej M, et al. The type IX secretion system (T9SS): highlights and recent insights into its structure and function. Front Cell Infect Microbiol. 2015;7:215.
- Abby SS, Rocha EPC. Identification of protein secretion systems in bacterial genomes using MacSyFinder. Sci Rep. 2016;6. DOI:https://doi.org/10.1007/978-1-4939-7033-9_1
- Stefan P, Ma AT, Derek S, et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A. 2006. DOI:https://doi.org/10.1073/pnas.0510322103
- Macintyre DL, Miyata ST, Maya K, et al. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci U S A. 2010;107:19520–19524.
- Sana TG, Flaugnatti N, Lugo KA, et al. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci U S A. 2016;113:E5044-E5051.
- Sandra S, Hood RD, Mougous JD. What is type VI secretion doing in all those bugs? Trends Microbiol. 2010. DOI:https://doi.org/10.1016/j.tim.2010.09.001
- Boyer F, Fichant G, Berthod J, et al. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics. 2009;10:104.
- Kirk MD, Pires SM, Black RE, et al. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med. 2015. DOI:https://doi.org/10.1371/journal.pmed.1001921
- Havelaar AH, Kirk MD, Torgerson PR, et al. World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med. 2015;12:e1001923.
- Fields PI, Swanson RV, Haidaris CG, et al. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci U S A. 1986;83:5189–5193.
- Herrero-Fresno A, Olsen JE. Salmonella Typhimurium metabolism affects virulence in the host – a mini-review. Food Microbiol. 2018;71:98.
- Mulder DT, Cooper CA, Coombes BK. Type VI secretion system-associated gene clusters contribute to pathogenesis of Salmonella enterica serovar Typhimurium. Infect Immun. 2012;80:1996–2007.
- Pezoa D, Blondel CJ, Silva CA, et al. Only one of the two type VI secretion systems encoded in the Salmonella enterica serotype Dublin genome is involved in colonization of the avian and murine hosts. Vet Res. 2014;45:2.
- Holden DW. Trafficking of the Salmonella vacuole in macrophages. Traffic. 2010. DOI:https://doi.org/10.1034/j.1600-0854.2002.030301.x
- Riquelme S, Varas M, Valenzuela C, et al. Relevant genes linked to virulence are required for salmonellatyphimurium to survive intracellularly in the social amoeba dictyostelium discoideum. Front Microbiol. 2016;7. DOI:https://doi.org/10.3389/fmicb.2016.01305
- Smriti V, Srikanth CV. Understanding the complexities of Salmonella-host crosstalk as revealed by in vivo model organisms. IUBMB Life. 2015. DOI:https://doi.org/10.1002/iub.1393
- Ji L, Ji-Tao G, Yong-Guo L, et al. The type VI secretion system gene cluster of Salmonella typhimurium: required for full virulence in mice. J Basic Microbiol. 2013. DOI:https://doi.org/10.1002/jobm.201200047
- Pezoa D, Yang HJ, Blondel CJ, et al. The type VI secretion system encoded in SPI-6 plays a role in gastrointestinal colonization and systemic spread of Salmonella enterica serovar Typhimurium in the chicken. PLoS One. 2013;8:e63917.
- Journet L, Cascales E. The type VI secretion system in Escherichia coli and related species. Ecosal Plus. 2016;7. DOI:https://doi.org/10.1128/ecosalplus.ESP-0009-2015
- Osipiuk J, Xu X, Cui H, et al. Crystal structure of secretory protein Hcp3 from Pseudomonas aeruginosa. J Struct Funct Genomics. 2011;12:21–26.
- Brunet YR, Henin J, Celia H, et al. Type VI secretion and bacteriophage tail tubes share a common assembly pathway. EMBO Rep. 2014. DOI:https://doi.org/10.1002/embr.201337936
- Silverman J, Agnello D, Zheng H, et al. Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol Cell. 2013;51:584–593.
- Lin QP, Gao ZQ, Geng Z, et al. Crystal structure of the putative cytoplasmic protein STM0279 (Hcp2) from Salmonella typhimurium. Acta Crystallogr F Struct Biol Commun. 2017;73:463–468.
- Stefan P, Ma AT, Revel AT, et al. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A. 2007. DOI:https://doi.org/10.1073/pnas.0706532104
- Gallique M, Decoin V, Barbey C, et al. Contribution of the pseudomonas fluorescens MFE01 Type VI secretion system to biofilm formation. PLoS One. 2017;12:e0170770.
- Andersson JA, Sha J, Erova TE, et al. Identification of new virulence factors and vaccine candidates for yersinia pestis. Front Cell Infect Microbiol. 2017;7:448.
- Decoin V, Gallique M, Barbey C, et al. Pseudomonas fluorescens type 6 secretion system is related to mucoidy, motility and bacterial competition. BMC Microbiol. 2015;15. DOI:https://doi.org/10.1186/s12866-015-0405-9
- Wang N, Liu J, Pang M, et al. Diverse roles of Hcp family proteins in the environmental fitness and pathogenicity of Aeromonas hydrophila Chinese epidemic strain NJ-35. Appl Microbiol Biotechnol. 2018. DOI:https://doi.org/10.1007/s00253-018-9116-0
- Peng Y, Wang X, Shou J, et al. Roles of Hcp family proteins in the pathogenesis of the porcine extraintestinal pathogenic Escherichia coli type VI secretion system. Sci Rep. 2016. DOI:https://doi.org/10.1038/srep26816
- Wang J, Zhihui Z, Fang H, et al. The role of the type VI secretion system vgrG gene in the virulence and antimicrobial resistance of Acinetobacter baumannii ATCC 19606. PLoS One. 2018. DOI:https://doi.org/10.1371/journal.pone.0192288
- Tamang MD, Gurung M, Nam HM, et al. Antimicrobial susceptibility and virulence characteristics of Salmonella enterica Typhimurium isolates from healthy and diseased pigs in Korea. J Food Prot. 2014;77:1481–1486.
- Schmitt CK, Ikeda JS, Darnell SC, et al. Absence of all components of the flagellar export and synthesis machinery differentially alters virulence of Salmonella enterica serovar Typhimurium in models of typhoid fever, survival in macrophages, tissue culture invasiveness, and calf enterocolitis. Infect Immun. 2001;69:5619–5625.
- Guan J, Xiao X, Xu S, et al. Roles of RpoS in Yersinia pseudotuberculosis stress survival, motility, biofilm formation and type VI secretion system expression. J Microbiol. 2015;53:633–642.
- Storey D, McNally A, Åstrand M, et al. Klebsiella pneumoniae type VI secretion system-mediated microbial competition is PhoPQ controlled and reactive oxygen species dependent. PLoS Pathog. 2020;16:e1007969.
- Weber B, Hasic M, Chen C, et al. Type VI secretion modulates quorum sensing and stress response in Vibrio anguillarum. Environ Microbiol. 2009;11:3018–3028.
- Aubert DF, Flannagan RS, Valvano MA. A novel sensor kinase-response regulator hybrid controls biofilm formation and type VI secretion system activity in Burkholderia cenocepacia. Infect Immun. 2008;76:1979–1991.
- Iyoda S, Kamidoi T, Hirose K, et al. A flagellar gene fliZ regulates the expression of invasion genes and virulence phenotype in Salmonella enterica serovar Typhimurium. Microb Pathog. 2001;30:81–90.
- Fernanda DP, Gerson N, Alline P, et al. The type VI secretion system plays a role in type 1 fimbria expression and pathogenesis of an avian pathogenic Escherichia coli strain. Infect Immun. 2010. DOI:https://doi.org/10.1128/IAI.00531-10
- Brunet YR, Khodr A, Logger L, et al. Silencing of the salmonella pathogenicity island 6-encoded type VI secretion system limits salmonella enterica serovar typhimurium interbacterial killing. Infect Immun. 2015;83:2738–2750.
- Chakib M, Friederike H, Henry H, et al. Conflicting needs for a Salmonella hypervirulence gene in host and non-host environments. Mol Microbiol. 2010. DOI:https://doi.org/10.1046/j.1365-2958.2002.03070.x
- Parsons DA, Heffron F. sciS, an icmF homolog in Salmonella enterica serovar Typhimurium, limits intracellular replication and decreases virulence. Infect Immun. 2005;73:4338–4345.
- Ho TD, Slauch JM. Characterization of grvA, an antivirulence gene on the gifsy-2 phage in Salmonella enterica serovar typhimurium. J Bacteriol. 2001. DOI:https://doi.org/10.1128/JB.183.2.611-620.2001
- Saliba A-E, Lei L, Westermann AJ, et al. Single-cell RNA-seq ties macrophage polarization to growth rate of intracellular Salmonella. Nat Microbiol. 2016. DOI:https://doi.org/10.1038/nmicrobiol.2016.206
- Grant AJ, Morgan FJE, McKinley TJ, et al. Attenuated salmonella typhimurium lacking the pathogenicity island-2 type 3 secretion system grow to high bacterial numbers inside phagocytes in mice. PLoS Pathog. 2012;8(12):e1003070.
- Petra F, Kowal AS, Pascale G, et al. Protocols for growth and development of Dictyostelium discoideum. Nat Protoc. 2007. DOI:https://doi.org/10.1038/nprot.2007.178
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645.
- Zhai YJ, Huang H, Liu J, et al. CpxR overexpression increases the susceptibility of acrB and cpxR double-deleted Salmonella enterica serovar Typhimurium to colistin. J Antimicrob Chemother. 2018;73:3016–3024.
