SUMMARY
Using DNA affinity chromatography we demonstrate that the S. aureus regulatory proteins MgrA, Rot, SarA, and SarS bind DNA baits derived from the promoter regions associated with the genes encoding aureolysin, ScpAB, SspABC, and SplA-F. Three of four baits also bound SarR and SarZ, the exception in both cases being the ScpAB-associated bait. Using the USA300, methicillin-resistant strain LAC and the USA200, methicillin-sensitive strain UAMS-1, we generated mutations in the genes encoding each of these proteins alone and in combination with sarA and examined the impact on protease production, the accumulation of high molecular weight proteins, and biofilm formation. These studies confirmed that multiple regulatory loci are involved in limiting protease production to a degree that impacts all of these phenotypes, but also demonstrate that sarA plays a predominant role in this regard. Using sarA mutants unable to produce individual proteases alone and in combination with each other, we also demonstrate that the increased production of aureolysin and ScpA is particularly important in defining the biofilm-deficient phenotype of LAC and UAMS-1 sarA mutants, while aureolysin alone plays a key role in defining the reduced accumulation of alpha toxin and overall cytotoxicity as assessed using both osteoblasts and osteoclasts.
Introduction
Staphylococcus aureus has capacity to cause a wide variety of diseases including acute toxemias, skin and soft tissue infections, sepsis, and deep secondary infections of essentially any tissue including bone [Citation1]. This can be attributed to the arsenal of virulence factors S. aureus has at its disposal coupled with a complex and highly interactive regulatory circuit that allows for precise tailoring of this arsenal to meet the demands of different microenvironments within the host. Included among its many virulence factors are at least 15 extracellular proteases [Citation2]. Five of these are variants of the exfoliative toxins (ETA, ETB, ETC, ETD, and ETE), which are serine proteases that exhibit exquisite specificity for desmoglein 1, a desmosomal cadherin responsible for maintaining the integrity of cell-to-cell adhesive structures in the skin [Citation3]. Although the exfoliative toxins reportedly exhibit superantigen activity, this remains controversial, and it is clear that this highly specific proteolytic activity is the primary factor that defines the association between these proteases and the skin infections scalded skin syndrome and bullous impetigo [Citation4,Citation5]. The exfoliative toxins exhibit further specificity with respect to host species, and collectively they are only produced by approximately 5% of S. aureus strains [Citation3,Citation5].
Of the remaining 10 extracellular proteases, six are “serine protease-like” proteins encoded within the spl operon [Citation6].These proteases (SplA, SplB, SplC, SplD, SplE, and SplF) also exhibit relatively limited substrate specificity and are not produced by all S. aureus strains [Citation7–10]. For instance, a PCR-based survey of 167 S. aureus strains isolated from various forms of infection found that the spl operon is absent in approximately 16% of strains, and the entire operon is present in only 31% [Citation10]. Among other strains, the presence of individual spl genes was found to range from 45% (splD) to 85% (splF). Nevertheless, the spl-encoded proteases have been implicated in the pathogenesis of pneumonia [Citation8], and the highest total abundance of the six spl genes was observed in isolates obtained from pneumonia patients [Citation10]. A recent report also found that mutations in splD and splF were associated with reduced fitness in a murine osteomyelitis model [Citation11].
The other four extracellular proteases are the zinc metalloprotease aureolysin, the cysteine proteases ScpA and SspB, and the serine protease SspA. The genes encoding these proteases (aur, scpA, sspB, and sspA, respectively) are present in at least 98% of all S. aureus strains [Citation10] and are arranged in three transcriptional units, with one containing only aur, the second containing scpA and the gene for its inhibitor staphostatin B (scpB), and the third containing sspA, sspB, and sspC, the latter encoding the inhibitor staphostatin C (sspC) [Citation12–14]. With the exception of ScpA, fully active forms of these proteases are produced as part of a cascade, with aureolysin being required for full activation of SspA, and SspA being required in turn for full activation of SspB [Citation14]. However, this activation cascade is not absolute in that proteolytically active forms of SspA and SspB are produced in an aureolysin mutant. By comparison to the exfoliative toxins and SplA-F, these proteases exhibit broad substrate specificity, and it is these proteases that have been studied most extensively as virulence factors [Citation2,Citation14]. These studies confirmed that these proteases degrade a variety of host proteins and serve important roles in pathogenesis by promoting tissue invasion, nutrient acquisition, and subversion of host defenses [Citation15–25].
Based on such observations, it would be anticipated that eliminating the ability of S. aureus to produce these proteases would result in decreased virulence, but we and others have demonstrated that doing so in the USA300 strain LAC increases rather than decrease virulence in diverse forms of infection including sepsis and osteomyelitis [Citation26–28]. Similarly, it would be anticipated that increased production of these proteases would increase virulence, but we have also demonstrated that mutation of the staphylococcal accessory regulator (sarA) results in the increased production of all of these proteases but reduced virulence in these same animal models [Citation27–29]. These paradoxical observations can be explained by the fact that eliminating protease production results in the increased abundance of S. aureus surface-associated and extracellular virulence factors [Citation26], while the increased production of proteases in sarA mutants results in the decreased accumulation of these virulence factors [Citation27–29].
This suggests that, while it is important that S. aureus have the capacity to produce extracellular proteases, it is equally important that it has the capacity to limit the production of these proteases such that they serve their intended purposes on behalf of the bacterium without compromising the availability of its own virulence factors. S. aureus employs a complex and highly interactive regulatory circuit to ensure the availability of its virulence factors in amounts appropriate for different microenvironments within the host, and this includes extracellular proteases [Citation30,Citation31]. Two of the central elements in this regulatory circuit are the accessory gene regulator (agr), which enhances the production of many extracellular proteins including proteases, and the staphylococcal accessory regulator (sarA), which functions as a repressor of protease production [Citation14,Citation32]. The regulatory functions of these two loci appear to be universal with respect to the genes/operons encoding individual proteases, including the exfoliative toxins [Citation33], with agr activating and sarA repressing their production. However, there is some contradictory evidence in this regard. For instance, several reports concluded that mutation of sarA has no impact on transcription of the spl operon [Citation6,Citation14,Citation34] but we have demonstrated that all six Spl proteases are present in increased amounts in conditioned media (CM) from stationary phase cultures of a LAC sarA mutant by comparison to CM from LAC itself [Citation27–29]. Irrespective of this discrepancy, it is clear that mutation of agr results in a decrease in overall protease activity to a degree that can be correlated with an increased capacity to form a biofilm, while mutation of sarA results in an increase in overall protease activity to a degree that can be correlated with a decreased capacity to form a biofilm [Citation32]. Although sarA enhances transcription of agr [Citation35] this demonstrates that the regulatory impact of sarA on protease production and biofilm formation is independent of its interaction with agr. The phenotype of an agr/sarA double mutant also mimics that of an isogenic sarA mutant with respect to both of these phenotypes, demonstrating that the impact of mutating sarA is epistatic to that of mutating agr [Citation32].
It has also become increasingly evident that the regulatory circuit modulating protease production extends far beyond these two loci. Indeed, mutations in argR2, arlRS, atlR, codY, hisR, mgrA, mntR, msaABCR, nsaR, rbf, rex, rot, rpiRB, saeRS, sigB, sarR, sarS, sarT, sarU, sarV, sarX, sarZ, xdrA, yjbh, and a number of uncharacterized putative regulatory loci have all been shown to impact the production of extracellular proteases [Citation14,Citation34,Citation36–54]. A recent report directly compared the impact of mutating these loci on transcription of each of the four transcriptional units (aur, scpAB, sspAB, and splA-F) that encode aureolysin, ScpA, SspA, SspB, and SplA-F and concluded that the regulatory proteins encoded by seven of these loci (CodY, MgrA, Rot, SaeR, SarR, SarS, and SarA) constitute a primary network while those encoded by an additional seven loci (ArgR1, AtlR, MntR, Rbf, Rex, SarU, and XdrA) constitute a secondary network that functions primarily through its impact on regulatory elements within this primary network [Citation34]. However, the impact of each of these loci with respect to individual protease genes/operons was found to vary widely. For instance, mutation of codY resulted in increased expression of the genes/operons encoding all of these proteases. This was also true of a sarA mutant with the exception of the spl operon, with expression levels of this operon being essentially unchanged in a sarA mutant [Citation34]. As noted above, this is consistent with other reports examining the impact of sarA on spl transcription [Citation6,Citation14], but contradicted by our proteomics studies demonstrating that these proteases are present in increased amounts in CM from a LAC sarA mutant [Citation27–29]. In contrast, mutation of mgrA was associated with decreased expression of all protease genes/operons other than that encoding scpA, which was largely unaffected in an mgrA mutant [Citation34]. Mutation of the other four genes in the putative primary network resulted in increased expression of some protease genes/operons and decreased expression of others.
These observations emphasize the complexity of S. aureus regulatory circuits that modulate the production of extracellular proteases, and this implies that doing so is important in allowing S. aureus to employ post-translational tailoring of its virulence factor repertoire in a fashion that best meets its needs in different host microenvironments. Thus, it is important to better understand critical elements within the regulatory network that modulates protease production and the manner by which they do so. To this end, we employed an unbiased DNA-based protein capture approach to identify S. aureus regulatory proteins that bind to each of the promoter regions associated with aur, scpAB, sspABC, and the splA-F operon. Focusing on those regulatory loci that encode proteins that were captured by at least three of these DNA baits, we then generated mutations in the corresponding genes both alone and in combination with sarA and examined the impact on transcription of each gene/operon, overall protease activity, accumulation of alpha toxin and extracellular protein A (eSpa), and biofilm formation. We also investigated the role of individual proteases alone and in combination with each other on the accumulation of alpha toxin, biofilm formation, and cytotoxicity for osteoblasts and osteoclasts. To account for potential strain-dependent differences, all of these studies were done using the USA300, CC8, methicillin-resistant strain LAC and the USA200, CC30, methicillin-sensitive strain UAMS-1.
Results
Identification of S. aureus regulatory proteins that bind protease-associated promoter regions
We used an unbiased DNA-based protein capture approach to identify S. aureus regulatory proteins that bind to the promoter regions of each of the four genes/operons that encode aureolysin, ScpA, SspA and SspB, and the SplA-F proteases. Specifically, the DNA regions extending ~400 bp upstream of the translational start site of each of these genes/operons was amplified by PCR using 5ʹ biotinylated primers. The amplification products were mixed with whole cell lysates obtained from LAC or its isogenic sarA mutant. The mixtures were then incubated with streptavidin and loaded onto a magnetic column to isolate each DNA bait and its bound proteins. Bound proteins were dissociated from each bait and visualized by SDS-PAGE. All four DNA baits captured a 15 kDa protein (). This protein was eluted at salt concentrations of ≥0.5 M, suggesting equivalent and relatively high-affinity binding to all baits. SarA is a 15 kDa protein, and this protein was not captured with any of the four DNA baits using lysates prepared from a LAC sarA mutant. These observations suggest this protein is likely to be SarA, which was confirmed by Western blot analysis using an anti-SarA antibody ().
Figure 1. Examination of captured proteins by SDS-PAGE and Western blot. The region extending ~400 bp upstream of the genes and/or operons encoding extracellular proteases indicated on the left were used as DNA baits to capture proteins from whole cell lysates prepared from the USA300 strain LAC or its isogenic sarA mutant (ΔsarA). Proteins were eluted with increasing concentrations of salt and examined by SDS-PAGE (left side of each panel) and Western blot using an anti-SarA antibody (right side of each panel). Control samples included experiments done with a LAC lysate and no DNA ait (top)
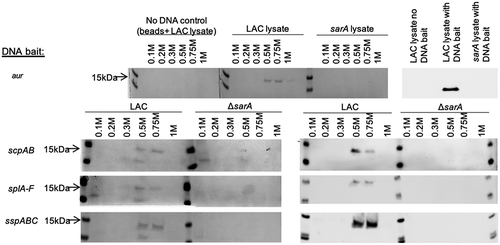
SarA has been shown to bind to the aur and sspABC promoter regions [Citation49,Citation52] but to our knowledge this is the first report demonstrating it also binds the scpA and splA-F promoters. The fact that SarA was captured using the spl bait is particularly significant in light of the discrepancy between reports suggesting that mutation of sarA does not impact expression of the spl operon [Citation34] and our demonstration that these proteases, as well as aureolysin, ScpA, SspA, and SspB are present in increased amounts in conditioned medium (CM) from stationary phase cultures of a LAC sarA mutant by comparison to CM from LAC itself [Citation27–29]. In addition, we previously used pCM11 [Citation55] to generate transcriptional reporters in which DNA fragments upstream of each protease-encoding gene/operon were used to drive expression of the superfolder green fluorescent protein (sGFP), and when these were introduced into the parent strains and their isogenic sarA mutants, fluorescence was significantly higher in the sarA mutants with all four reporters including the spl::gfp reporter, and this was true in both LAC and UAMS-1 [Citation56]. This confirms that SarA represses expression of all four of the transcriptional units encoding aureolysin, ScpA, SspA/SspB, and SplA-F, and the results of our binding assays suggest that it does so by directly binding to cis elements associated with all four of the relevant promoter regions.
Two other proteins, both of which were smaller than SarA, were also visible in some SDS-PAGE gels (). One of these was eluted at low salt concentration (0.1 M) suggesting low affinity and potentially nonspecific binding. The other was a 12–13 kDa protein that was eluted at a salt concentration comparable to that required to elute SarA (0.5 M). However, these studies were done in triplicate, and unlike SarA this protein was not consistently captured with any protease-associated bait in an amount sufficient to be visible by SDS-PAGE. Additionally, it was sometimes captured from lysates prepared from the LAC sarA mutant and in experiments done with LAC lysates using the SAU300_1445 control bait (data not shown). Nevertheless, in an attempt to identify this protein, as well as any additional proteins that may have been captured in amounts that were not visible by SDS-PAGE, we subjected eluates to LC-MS/MS analysis. Given the number of baits, the number of individual fractions obtained with each bait, and the need to carry out these experiments in triplicate to allow for robust statistical analysis, these studies were done using a 1:1 mixture of the 0.2 and 0.3 M NaCl eluates, the 0.5 M eluate alone, and a 1:1 mixture of the 0.75 and 1 M eluates from each triplicate sample. These experiments included a control without any DNA bait, thus allowing us to account for nonspecific binding to the streptavidin beads, and a control in which the DNA bait was derived from the ~400 bp region upstream of the gene encoding the segregation and condensation protein A (SAU300_1445), thus allowing us to distinguish proteins that bind to any functional promoter (e.g. those that constitute the basal transcription machinery) from those that bind specifically to each protease-associated promoter region. For statistical analysis, spectral counts obtained for each protein in each sample were summed and the resulting counts obtained with each bait compared to the spectral counts observed for the same protein in the control bait using a two-tailed t-test. Proteins with a p-value ≥ 0.05 were filtered out and the fold change (FC) was calculated for the remaining proteins. Proteins with an FC ≤ 2 were filtered out along with proteins with less than 8 spectral counts in at least 2 of 3 replicates; this cutoff was chosen based on counts obtained with the no DNA control samples.
Based on this analysis we concluded that the smaller protein eluted only at relatively high NaCl concentrations was most likely to be the nucleoid-associated protein HU. HU is an architectural protein involved in chromatin remodeling, but it is known to impact gene transcription [Citation57]. Interestingly, it has also been suggested that SarA may serve a similar architectural role [Citation58]. Thus, we cannot rule out the possibility that this protein impacts transcription of S. aureus genes including at least some that encode extracellular proteases, although it has never been shown to do so in any studies we are aware of. In any event, this protein did not meet our inclusion criteria for more detailed study, and we chose to focus on those proteins that met these criteria. To identify these, all proteins captured with each protease-related bait and the LC-MS/MS datasets obtained for each were compared with each other using Venny 2.1. The results confirmed that SarA was captured by all four baits as well as MgrA, Rot, and SarS (Fig. S1). With three of four baits, the protein with the highest number of spectral counts was SarA followed by MgrA, the exception being the sspABC bait in which the number of spectral counts observed with SarA and MgrA were comparable (Table S1). MgrA was previously shown to bind DNA baits associated with the aur and sspABC promoters [Citation49], and a recent report demonstrated that mutation of mgrA limits transcription of aur, sspABC, and the spl operon but had little impact on transcription of scpAB [Citation34]. This report was based on qRT-PCR analysis and did not examine MgrA binding specificity, but our results suggest that the regulatory impact of MgrA on the production of all extracellular proteases is likely to occur via a direct mechanism.
The other two proteins that were bound by all four baits are SarS and Rot. The number of spectral counts observed for SarS was generally greater than those observed for Rot with all four protease-associated baits, but even those observed for SarS were consistently lower than those observed for SarA (Table S1). Nevertheless, these findings are consistent with a previous study that found that Rot binds to all four protease promoters as assessed by electrophoretic mobility shift assays (EMSA) done with purified Rot and that mutation of rot could be correlated with increased overall protease activity and a reduced capacity to form a biofilm [Citation59]. In contrast, previous reports that were limited to the sspABC and aur promoters found that SarS binds to the sspABC promoter but not the aur promoter [Citation49,Citation52]. Thus, to our knowledge this is the first report showing that SarS also binds the aur, scpA, and spl promoters. This suggests that, while both SarS and Rot are known to have global regulatory effects on the production of multiple proteins, they are likely to modulate the production of extracellular proteases directly rather than through an indirect mechanism involving other regulatory loci.
SarR and SarZ were bound by the promoter baits associated with aur, sspABC, and the spl operon, but based on our inclusion criteria neither was judged to specifically bind the bait associated with scpAB (Fig. S1). As with the impact of sarA on the spl operon, there are conflicting reports in the literature with respect to the impact of SarR in the protease regulatory circuit. For instance, there are reports concluding that SarR binds the aur and sspABC promoters and enhances transcription of both of these genes/operons [Citation49,Citation52], while another report concluded that mutation of sarR has no significant effect on transcription of the gene encoding aureolysin but does result in decreased transcription of the genes/operons encoding all other extracellular proteases [Citation34]. SarR was first identified as a repressor of sarA transcription, and a sarR mutant was shown to produce increased levels of SarA [Citation60]. Thus, it would be anticipated that mutation of sarA and sarR would have opposite effects on protease production, and transcriptional analysis by qRT-PCR indicate that this is generally the case [Citation34]. However, to our knowledge, direct interactions between SarR and promoter regions associated with scpA and splA-F have not been previously reported. To the extent that the scpA promoter was the one exception with respect to SarR binding, these results also suggest that the impact of SarR on transcription of aur, sspABC, and the spl operon may occur via a direct mechanism, while the impact on scpAB transcription is likely to occur indirectly.
Similarly, SarZ has been implicated as a direct mediator of transcription of sspABC [Citation37,Citation51] but it has not been implicated in the direct transcriptional regulation of any of the other genes/operons encoding extracellular proteases. At the same time, mutation of sarZ has been correlated with altered transcription of a number of other genes known to impact protease production including agr, sarA, sarS, and mgrA [Citation37]. In the recent report that examined the global network impacting protease production in S. aureus, mutation of sarZ was associated with increased protease activity, but it was not considered among the regulatory loci in the primary or secondary regulatory networks defined by this study, and its impact on transcription of individual genes/operons encoding extracellular proteases was not examined [Citation34].
Relative impact of S. aureus regulatory proteins on protease production
These studies suggest that MgrA, Rot, SarA, SarR, SarS, and SarZ are likely to modulate the production of most if not all extracellular proteases directly, but they do not address the phenotypic impact of this binding. To address this, we mutated mgrA, rot, sarA, sarR, sarS, and sarZ in LAC and UAMS-1 and examined the impact on overall protease activity using a gelatin-based FRET assay. In LAC, mutation of rot, sarA, and sarS resulted in a statistically significant increase in protease activity, while mutation of sarR resulted in a slight but significant decrease (). Overall, these results are consistent with a previous report demonstrating that mutation of sarA, rot, and sarS in the USA300 strain JE2 increased overall protease activity as assessed by gelatin zymography [Citation34]
Figure 2. Relative impact of regulatory mutations on protease activity. Protease activity in conditioned medium (CM) from stationary phase cultures from LAC (top), UAMS-1(bottom), and each of the indicated isogenic regulatory mutants was assessed using a gelatin-based FRET assay. Results are reported as the average ± standard error of the mean from two biological replicates, each of which included three experimental replicates. Asterisk indicates statistical significance by comparison to the results observed with the isogenic parent strain. Doubles asterisks indicate statistical significance by comparison to the isogenic sarA mutant
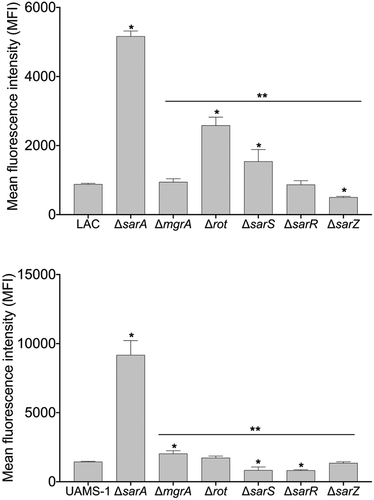
However, the results observed with UAMS-1 regulatory mutants differed by comparison to those observed with the corresponding LAC mutants. Specifically, mutation of sarS and sarZ resulted in a significant decrease in overall protease activity in UAMS-1, while mutation of mgrA resulted in a significant increase (). In fact, only mutation of sarA resulted in a significant increase in protease activity in both LAC and UAMS-1. The increase observed in a UAMS-1 mgrA mutant is noteworthy in that qRT-PCR analysis found that mutation of mgrA in the USA300 strain Houston resulted in decreased expression of several genes/operons encoding extracellular proteases, most notably the spl operon [Citation34].
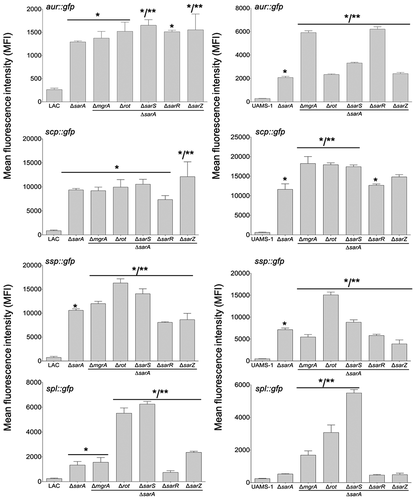
To investigate this further, we employed pCM11-based transcriptional reporters constructed such that the same promoter regions used as DNA baits were used to drive expression of the gfp gene. These reporters were introduced into LAC, UAMS-1, and derivatives of each carrying mutations in each of the regulatory genes encoding proteins captured in our DNA bait assays. Fluorescence levels were significantly increased with all four reporters in sarA mutants generated in both LAC and UAMS-1 (). In LAC, fluorescence was also significantly increased in rot and sarS mutants, but only with the spl::gfp and ssp::gfp reporters, and as with protease activity itself the increased fluorescence levels observed in LAC rot and sarS mutants with both of these reporters were well below those observed in the isogenic sarA mutants (). These results indicate that in LAC the regulatory proteins Rot, SarA and SarS repress the production of at least a subset of extracellular proteases directly at a transcriptional level, but that SarA is the only regulatory protein that does so with all 10 extracellular proteases.
Figure 3. Relative impact of regulatory loci on expression of individual protease genes and operons in LAC and UAMS-1. The indicated pCM11 gfp reporter plasmids were introduced into LAC, UAMS-1 and each of the isogenic regulatory mutants. Results are reported as the average mean fluorescence intensity ± standard error of the mean from two biological replicates, each of which included three experimental replicates. Asterisk indicates statistical significance by comparison to the results observed with the isogenic parent strain. Doubles asterisks indicate statistical significance by comparison to the isogenic sarA mutant
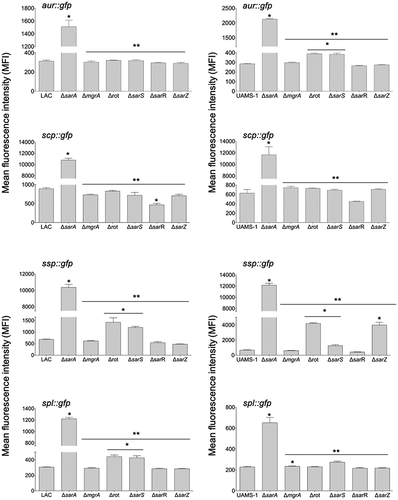
With the exception of the UAMS-1 sarA mutant, the results of our reporter assays with UAMS-1 regulatory mutants differed by comparison to the corresponding LAC mutants. The results were also less consistent with the results of assays assessing overall protease activity. Specifically, mutation of rot and sarS resulted in increased fluorescence with our pCM11 reporters with the aur::gfp and ssp::gfp reporters, but only mutation of sarS resulted in a significant increase in fluorescence with the spl::gfp reporter (). Unlike the corresponding LAC mutant, a UAMS-1 sarZ mutant also exhibited increased fluorescence with the ssp::gfp reporter. However, in UAMS-1 only mutation of mgrA resulted in a significant increase in overall protease activity (). The gelatin-based FRET assay we employed to assess overall protease activity does not reflect the production of all extracellular proteases, but it does readily detect aureolysin, SspA, and SspB, and in this respect, it is somewhat surprising that these changes were not reflected in our protease assays. However, as with the LAC mutants, all of the changes observed in our UAMS-1 reporter assays were minimal by comparison to the UAMS-1 sarA mutant (). Thus, while the results of these reporter assays support the conclusion that multiple regulatory loci that bind protease-associated promoters impact transcription to a statistically significant extent, they also support the conclusion that SarA plays a predominant role in this regard. Indeed, as with protease activity itself (), only mutation of sarA resulted in a significant increase in fluorescence with all reporters in both LAC and UAMS-1, and in both strains the increase observed in sarA mutants was significantly greater than that observed with the isogenic mutations generated in any of these other regulatory loci.
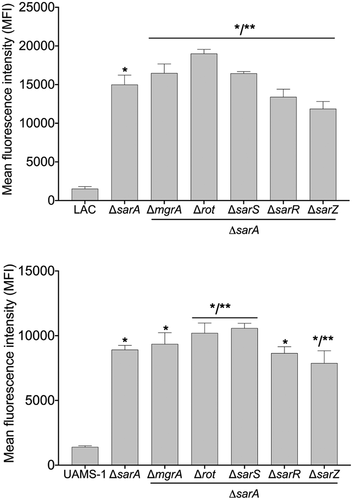
Impact of S. aureus regulatory proteins on the abundance of extracellular proteins
The results discussed above do not preclude the possibility that mutation of regulatory loci other than sarA also impact S. aureus phenotypes of potential clinical relevance. More directly, the increased protease production observed in sarA mutants has been shown to play an important role in defining the reduced accumulation of many S. aureus proteins including alpha toxin and protein A, the reduced capacity to form a biofilm, and reduced virulence of sarA mutants in murine models of sepsis and osteomyelitis [Citation27,Citation28,Citation56,Citation61]. Thus, the question becomes whether the relatively limited impact of other regulatory mutations on protease production also significantly impacts these phenotypes.
To begin to address this, we first examined SDS-PAGE exoprotein profiles in the LAC and UAMS-1 regulatory mutants. Mutation of essentially all six loci had an impact on overall exoprotein profiles, which is not surprising given the global impact of all of these regulatory loci on protein production [Citation30]. Differences in overall exoprotein patterns were apparent between LAC and UAMS-1 mutants, which is not surprising given known differences between these two strains. However, the overall impact of each mutation in both strains was virtually indistinguishable. Specifically, mutation of sarA resulted in a decrease in the abundance of full-length proteins in both strains that were visibly apparent by comparison to all of the other mutants (). Mutation of rot and sarS also limited the accumulation of these proteins, albeit to a lesser extent by comparison to the isogenic sarA mutant. Notably, as assessed by SDS-PAGE, mutation of mgrA had little impact on the abundance of high molecular weight proteins in either strain despite the fact that it resulted in a slight but statistically significant increase in overall protease activity in UAMS-1 ().
Figure 4. Relative impact of regulatory loci on protein abundance. Top: SDS-PAGE of conditioned medium (CM) from LAC (top), UAMS-1 (bottom) and isogenic regulatory mutants generated in each parent strain. Bottom: Western blots of CM from the same strains using antibodies for alpha toxin (Hla, top), protein A (Spa, left), and Nuc1 (right). Purified alpha toxin (Hla) was included as a control for alpha toxin blots (UAMS-1 does not produce alpha toxin), while protein A and nuc1 mutants (Δspa and Δnuc1, respectively) were included as controls for the protein A and Nuc1 blots
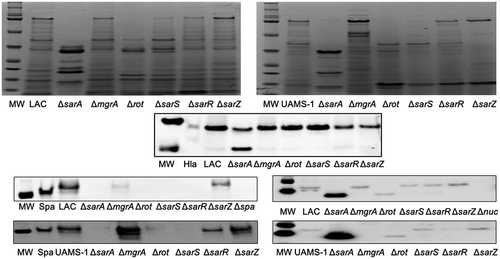
We also assessed the abundance of alpha toxin, protein A, and the Nuc1 extracellular nuclease by Western blot. These proteins were chosen based on our previous demonstration that all of these are impacted in LAC sarA mutants owing largely to protease-mediated degradation [Citation27,Citation28,Citation62]. The abundance of alpha toxin was decreased to a greater extent in a LAC sarA mutant than any other regulatory mutant examined, and a smaller protein reactive with the anti-alpha toxin antibody was evident only in the LAC sarA mutant (). Neither of these was the case with any other regulatory mutant, although mutation of sarR and sarZ in LAC did result in an apparent decrease in the abundance of full-length alpha toxin. In contrast, extracellular protein A was essentially absent in every mutant examined except the LAC mgrA and sarZ mutants (). It was not possible to assess the abundance of alpha toxin in UAMS-1 because this strain does not produce alpha toxin. However, protein A was also absent in CM from every UAMS-1 mutant with the exception of the mgrA, sarR, and sarZ mutants. Indeed, the abundance of protein A was significantly increased in the UAMS-1 mgrA mutant by comparison to the isogenic parent strain despite the fact that mutation of mgrA was correlated with a slight increase in overall protease activity in UAMS-1 (). Interestingly, the abundance of protein A was not increased in a LAC mgrA mutant despite the fact that no increase in protease activity was observed in this mutant.
The impact on Nuc1 varied both with respect to abundance and form. Specifically, Nuc1 is produced as a larger NucA form that is proteolytically cleaved to form a smaller NucB form. In both LAC and UAMS-1, only the NucB form was present in sarA and rot mutants (), suggesting enhanced proteolytic cleavage. Additionally, the overall abundance of NucA was increased in LAC and UAMS-1 sarA mutants, and this was not the case with any other regulatory mutant examined.
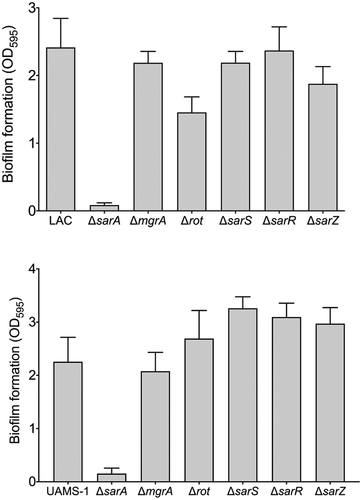
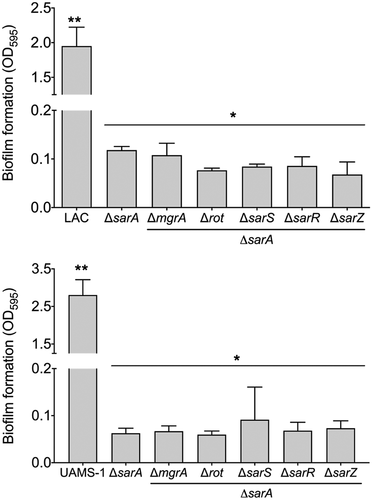
Relative impact of S. aureus regulatory proteins on biofilm formation
Increased protease production has been shown to be a primary contributing factor to the reduced capacity of S. aureus regulatory mutants to form a biofilm [Citation47,Citation59,Citation63–65]. Based on this we also examined the impact of mutations in the genes encoding each of the targeted proteins on biofilm formation. Just as mutation of sarA resulted in a greater increase in protease activity than any other mutation in both LAC and UAMS-1 (), it also limited biofilm formation in both strains to a greater extent than any other regulatory mutation (). In LAC, mutation of rot and sarR limited biofilm formation to a modest but statistically significant degree, while in UAMS-1 no other regulatory mutation was found to significantly limit biofilm formation. Indeed, mutation of sarS, sarR, and sarZ were all associated with a slight but statistically significant increase in biofilm formation in UAMS-1 (). In contrast, mutation of sarS and sarZ had no significant impact on biofilm formation in LAC. Surprisingly, mutation of sarR was found to limit biofilm formation in LAC to a modest but statistically significant degree () despite the fact that it was also correlated with a decrease in overall protease activity ().
Figure 5. Relative impact of regulatory loci on biofilm formation. Biofilm formation was assessed in LAC (top), UAMS-1 (bottom) and their isogenic regulatory mutants. Results are reported as the average ± standard error of the mean from two biological replicates, each of which included six experimental replicates. Asterisk indicates statistical significance by comparison to the results observed with the isogenic parent strain. Doubles asterisks indicate statistical significance by comparison to the isogenic sarA mutant
Impact of the functional status of other regulatory loci on sarA-associated phenotypes
All of the results cited above support the conclusion that multiple regulatory loci bind protease-associated promoter regions and modulate protease production and protease-associated phenotypes. However, they also support the conclusion that none do so more than SarA in terms of their overall impact on these phenotypes or their impact in diverse clinical isolates of S. aureus. To investigate this further, we mutated each of the genes encoding regulatory proteins captured in our primary experiments in combination with sarA and examined these same phenotypes. As expected, protease production was significantly increased in both LAC and UAMS-1 sarA mutants, and mutation of each of the other regulatory loci had relatively little impact on protease production by comparison to the isogenic sarA mutant (). That said, from a statistical perspective some differences were observed. Specifically, by comparison to the isogenic sarA mutant, protease production was increased in LAC mgrA/sarA, rot/sarA, and sarS/sarA mutants, while it was decreased in sarR/sarA and sarZ/sarA mutants. Protease production was also increased in UAMS-1 rot/sarA and sarS/sarA mutants by comparison to the sarA mutant, and slightly decreased in a UAMS-1 sarZ/sarA mutant ().
Figure 6. Impact of the functional status of regulatory mutations on protease production in isogenic sarA mutants. Protease activity in conditioned medium (CM) from stationary phase cultures from LAC, UAMS-1, and each of the indicated isogenic regulatory mutants was assessed using a gelatin-based FRET assay. Results are reported as the average ± standard error of the mean from two biological replicates, each of which included three experimental replicates. Asterisk indicates statistical significance by comparison to the results observed with the isogenic parent strain. Doubles asterisks indicate statistical significance by comparison to the isogenic sarA mutant
Transcriptionally, at least as assessed using our pCM11 reporter assay, the results were clear, particularly in UAMS-1. For instance, in LAC fluorescence levels observed with the aur::gfp, scp::gfp and ssp::gfp reporters were comparable in sarA mutants and sarA mutants with concomitant mutations in other regulatory loci (), and what differences were observed were generally consistent with the results of protease assays using the same strains (). This was particularly true with rot/sarA and sarS/sarA mutants, both of which exhibited increased fluorescence with at least one of the reporters (). This was also true with the spl::gfp reporter, and in fact the increase observed in these double mutants with this reporter was greater even by comparison to the isogenic sarA mutant. However, the spl-encoded proteases are not readily detectable in gelatin-based protease assays, so it is unlikely this contributed to the increased protease activity we observed in these mutants. In UAMS-1, fluorescence levels were increased relative to the isogenic sarA mutant in rot/sarA and sarS/sarA mutants with all four reporters (). Interestingly, this was also true with the UAMS-1 mgrA/sarA mutant, while in LAC this was true only with the ssp::gfp reporter. Nevertheless, while we did observe some differences between strains, these results suggest that in both LAC and UAMS-1 maximal repression of protease production is dependent on multiple proteins, most notably SarA, Rot, and SarS.
Figure 7. Impact of the functional status of regulatory mutations on expression of protease genes/operons in LAC sarA mutants and UAMS-1 sarA mutants. pCM11 gfp reporter plasmids were introduced into LAC or UAMS-1 and the indicated regulatory mutants. Results are reported as the average mean fluorescence intensity ± standard error of the mean from two biological replicates, each of which included three experimental replicates. Asterisk indicates statistical significance by comparison to the results observed with the isogenic parent strain. Doubles asterisks indicate statistical significance by comparison to the isogenic sarA mutant
Such results illustrate the intricacy of S. aureus regulatory circuits that modulate protease production, but once again they do not address the phenotypic impact of the differences we observed in these mutants. Thus, we repeated our exoprotein analysis both by SDS-PAGE and by Western blot. In both LAC and UAMS-1, SDS-PAGE analysis confirmed that all of the double mutants examined exhibited a protein profile directly comparable to that observed in the isogenic sarA mutants (). This was reflected in the decreased accumulation of full-length alpha toxin and/or increased accumulation of its degradation product in all LAC mutants other than the sarR/sarA mutant. It was also reflected in the absence of protein A in all mutants in both LAC and UAMS-1 (). Additionally, only the smaller NucA form of Nuc1 was present in all sarA mutants irrespective of the functional status of any other regulatory gene (). Biofilm formation was also reduced to an extent comparable to the isogenic sarA mutant in all other regulatory mutants examined ().
Figure 8. Relative impact of regulatory loci on protein abundance. Top: SDS-PAGE of conditioned medium (CM) from LAC (left), UAMS-1 (right), their isogenic sarA mutants, and isogenic sarA mutants with additional mutations in the genes encoding each of the other regulatory proteins examined in this study. Bottom: Western blots of CM from the same strains using antibodies for alpha toxin (Hla, top), protein A (Spa, left), and Nuc1 (right). Purified alpha toxin (Hla) was included as a control for alpha toxin blots (UAMS-1 does not produce alpha toxin), while protein A and nuc1 mutants (Δspa and Δnuc1, respectively) were included as controls for the protein A and Nuc1 blots
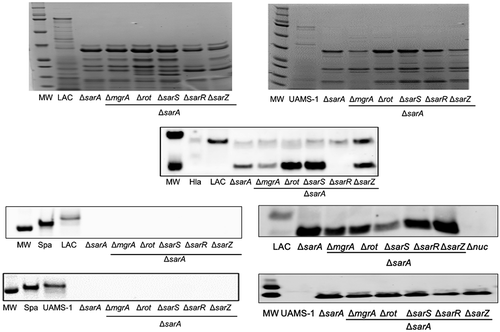
Figure 9. Relative impact of regulatory loci on biofilm formation. Biofilm formation was assessed in LAC (top), UAMS-1 (bottom), their isogenic sarA mutants, and isogenic sarA mutants with additional mutations in the genes encoding each of the other regulatory proteins examined in this study. Results are reported as the average ± standard error of the mean from two biological replicates, each of which included six experimental replicates. Asterisk indicates statistical significance by comparison to the results observed with the isogenic parent strain. Doubles asterisks indicate statistical significance by comparison to the isogenic sarA mutant
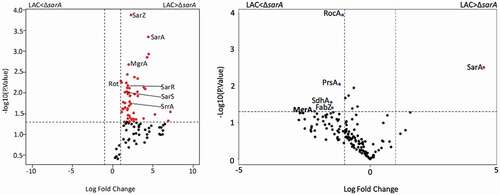
Impact of SarA on the binding of other regulatory proteins
The results cited above demonstrate that multiple regulatory loci impact protease production and protease-associated phenotypes but that SarA plays a predominant role in this regard. They also suggest that all of these proteins impact protease production directly as evidenced by their identification using DNA capture assays. However, this does not preclude the possibility that their impact is mediated through an interaction with SarA. To begin to address this, we repeated our DNA bait assays using whole cell lysates prepared from a LAC sarA mutant. All five of the other proteins prioritized based on experiments done with LAC lysates were identified in these experiments as well, but their overall binding patterns were altered. For instance, none of the other five proteins bound all four baits (Fig. S2). Rot was captured with the aur, splA-F, and sspAB-associated baits, and SarZ bound the baits associated with aur, scpAB, and sspABC, while MgrA, SarR and SarS only bound the baits associated with aur and sspABC. Thus, the only two baits that bound the same proteins in experiments done with both LAC lysates and lysates prepared from its isogenic sarA mutant were those associated with aur and sspABC (Fig. S2). Irrespective of the bait used, the number of spectral counts observed with all proteins was also higher in experiments done with lysates prepared from LAC (Table S1) by comparison to those prepared from its isogenic sarA mutant (Table S2). This was most evident with the spl-associated bait in which the number of spectral counts observed with LAC lysates was higher than those observed with LAC ΔsarA lysates with all five proteins (). Additional proteins were also identified in this comparison that bound to the spl-associated bait at higher levels in the presence of SarA, one example being the regulatory protein SrrA (). One exception was MgrA and the aureolysin-associated bait. Specifically, the number of spectral counts observed in this context were higher with lysates prepared from the sarA mutant than those prepared from LAC itself (Table S2 and ). These results support the hypothesis that the impact of at least some regulatory loci on binding protease-associated promoters, and presumably protease production, may be dependent, at least in part, on the presence or absence of SarA.
Figure 10. Differential protein binding to the spl (left) and the aureolysin (right) promoters in LAC versus the isogenic ΔsarA mutant. Volcano plots were generated based on fold-change of protein levels using the averaged spectral counts from biological triplicates. The x-axis indicates a log2 fold-change, and the y-axis indicates −log10 p-value based on the Student’s t-test. The horizontal line indicates a p-value <0.05, and the vertical lines represent a fold-change >2. Proteins in which the abundance was reduced to a statistically significant degree in ΔsarA lysates compared to wild-type lysates are shown as red dots in the upper right quadrant, whereas those that were present in increased amounts are shown as blue circles in the upper left quadrant. Black dots indicate proteins for which differences in abundance were not statistically significant
Impact of individual proteases on sarA-associated phenotypes
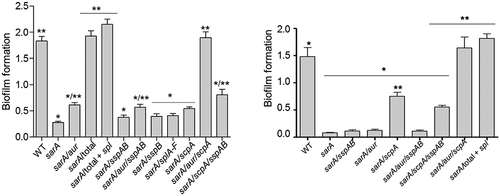
To investigate the role of individual proteases in defining important phenotypes of sarA mutants, we generated mutations in the genes encoding individual proteases alone and in combination with each other in both LAC and UAMS-1 sarA mutants. The accumulation of full-length alpha toxin was essentially abolished in CM from a LAC sarA mutant, and it was restored to levels even beyond those observed in the LAC parent strain by eliminating the production of aureolysin (). Since aureolysin sits at the top of an activation cascade that includes SspA and SspB, it is possible that this is due to the impact of eliminating aureolysin production on the activity of these proteases, but if that were the case it would be expected that the accumulation of alpha toxin would be restored by eliminating production of SspA and/or SspB, and this was not the case. Neither mutation of scpA nor the functional status of the spl-encoded proteases had any impact on the abundance of alpha toxin, thus suggesting that it is aureolysin itself that defines its reduced abundance in a LAC sarA mutant. Similarly, mutation of the gene encoding aureolysin was sufficient to restore the cytotoxicity of CM from a LAC sarA mutant for both osteoblasts and osteoclasts (). In contrast, mutation of the gene encoding aureolysin alone was not sufficient to restore biofilm formation in LAC or UAMS-1 sarA mutants. Rather, in both strains this required concomitant mutation of the genes encoding both aureolysin and ScpA ().
Figure 11. Impact of individual proteases on accumulation of alpha toxin (top) and cytotoxicity (bottom). Top: Conditioned medium (CM) from the indicated strains was resolved by SDS-PAGE prior to Western blotting with an anti-alpha toxin antibody. The control consists of purified alpha toxin. As previously described, this commercially available antibody is cross-reactive with gamma toxin.13,60 Bottom: Cytotoxicity was assessed using conditioned medium (CM) from stationary phase cultures of LAC (WT), its isogenic sarA mutant, and derivatives of the sarA mutant unable to produce the indicted proteases. Assays were done with RAW (left) or MC3T3 cells (right) as surrogates for primary osteoclasts and osteoblasts, respectively. Results are reported as relative viability. Single asterisk indicates a statistically significant increase in viability by comparison to CM from LAC (WT)
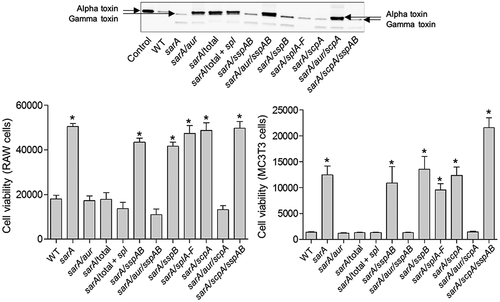
Figure 12. Impact of individual specific proteases on biofilm formation. Biofilm formation was assessed using a microtiter plate assay as previously described.16 WT refers to LAC (left) or UAMS-1 (right). “Total” refers to a sarA mutant unable to produce any extracellular protease. “Total + spl” refers to a sarA mutant unable to produce any extracellular protease other than those encoded by the spl operon. All other designations indicate the genes encoding specific proteases that were mutated in the LAC or UAMS-1 sarA mutants. Single asterisk indicates statistical significance relative to the isogenic parent strain. Double asterisks indicate statistical significance relative to the isogenic sarA mutant
Cis elements involved in SarA-mediated repression of the expression of individual proteases
We chose to focus on SarA in these experiments because it was the most abundant protein identified in our DNA bait assays and had a greater impact on protease production and protease-associated phenotypes than mutation of any of the other regulatory genes examined. Additionally, mutation of sarA had the most consistent impact on all of these phenotypes in both LAC and UAMS-1, and sarA mutants were the only mutants that exhibited increased expression of all four reporters in both strains. Thus, to gain insight into the molecular interaction between SarA and each protease-associated bait, we cloned sequentially truncated versions of the region upstream of each protease gene/operon starting with the same fragment used in our DNA bait assays and decreasing by 20–50 bp each time. These were cloned into our pCM11 gfp reporter [Citation66] and introduced into LAC and its isogenic sarA mutant. Relative fluorescence levels were then assessed to determine whether it was possible to identify a specific region likely to contain one or more SarA binding sites. The rationale behind this approach was that it should be possible to replicate the phenotype of a sarA mutant even in the SarA-positive LAC parent strain in the absence of the relevant SarA binding site(s).
As expected, increased fluorescence was observed in the LAC sarA mutant with all four full-length reporter constructs (). The magnitude of the change did vary in a reporter-dependent manner, with the scp and ssp reporters exhibiting the greatest increase in fluorescence, which was perceptible even to the eye, followed by the spl reporter and lastly by the aur reporter. With the exception of the spl::gfp reporter (see below), this difference was apparent with every truncated construct generated with every reporter until the promoter region was reduced to a region containing 100 bp upstream of the translational start site of each protease-associated gene and/or operon (). With the aur constructs, fluorescence was increased in LAC to a level comparable to that observed in the isogenic sarA mutant, and comparable to each other, with constructs containing 70 and 100 bp. This suggests the presence of one or more SarA binding sites between 100 and 150 bp upstream of the translational start site of the aur gene. Included within this region is a DNA site (ATTTTTA) comparable to the SarA binding site (ATTTTAT) defined by our previous SELEX experiments [Citation67]. This putative SarA binding site is centered 69 bp upstream of the proposed −35 region [Citation14] and 138 bp upstream of the translational start site. This is consistent with the results of our reporter assays and suggests that SarA-mediated repression of aur expression is unlikely to be due to competition for the RNA polymerase binding site.
Figure 13. Characterization of regions containing cis active elements that contribute to SarA-mediated repression of extracellular proteases. Constructs containing the indicated number of base pairs upstream of each protease gene/operon translational start site were used to generate truncated versions of the pCM11 gfp reporter plasmid indicated in each panel. These were then introduced into LAC and its sarA mutant and fluorescence assessed after ON growth. Results are reported as the average mean fluorescence intensity ± standard error of the mean from two biological replicates, each of which included three experimental replicates. The sequence below each panel illustrates the region upstream of each protease gene/operon with the region containing putative SarA binding sites (bold italics), putative −35, −10, and ribosome-binding sites (bold underlined), and transcriptional and translational start sites (underlined)
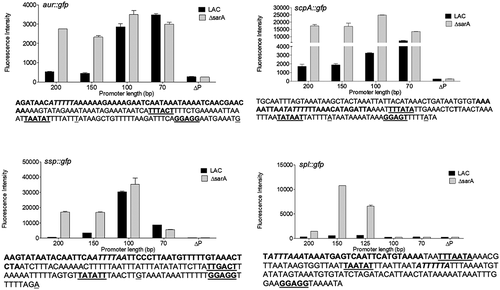
The results observed with the ssp::gfp reporter were similar to those observed with the aur::gfp reporter in that fluorescence was increased to a comparable level in LAC and its sarA mutant with the construct extending 100 bp upstream of the sspABC translational start codon (). This region also contains potential SarA binding sites (ATTTTAA or TTTTATT) as defined based on our previous SELEX analysis [Citation67]. These overlapping sites are centered 68 or 69 bp upstream of the putative −35 box and 128 or 129 bp upstream of the translational start codon. By comparison to the 100 bp construct, fluorescence was significantly decreased in both LAC and its sarA mutant with the reporter construct containing 70 bp. There are several possible explanations, one being the loss of a binding site for an activator of protease production. Alternatively, while the location of the putative SarA binding site associated with the sspABC operon is located the same distance upstream of the proposed −35 site, it is possible the sspABC promoter itself may be compromised to some degree by deletion of the region between 70 and 100 bp. In this respect it is important to note that the key promoter elements have been proposed for the sspABC operon [Citation14], but to our knowledge they have not been experimentally validated.
With the scp::gfp reporter, fluorescence was increased in LAC with the 100 bp construct, but to a lesser degree than was observed with the 70 bp construct (). Additionally, with the 100 bp reporter construct, fluorescence was increased in both LAC and its isogenic sarA mutant. This suggests the existence of a binding site for a protein other than SarA in the region between 100 and 150 bp upstream of the translational start codon. Based on the results of our binding data indicating that SarZ binds the scpAB promoter region to a greater extent in the absence of SarA (Table S1 vs Table S2 and Fig. S1 vs Fig. S2), as well as reporter assays with double mutants (), one candidate in this regard is SarZ. These results also suggest the absence of a SarA binding site in the 70 bp construct and, by extension, the presence of a SarA binding site in the region between 70 and 100 bp. This region upstream contains potential overlapping SarA binding sites (ATATTTT or ATTTTTT) centered 22 and 20 bp upstream of the putative −35 box and 85 or 83 bp upstream of the translational start codon [Citation14].
The results observed with the spl::gfp reporter were different by comparison to all other reporters. Specifically, we observed essentially no fluorescence with a construct extending as far as 100 bp upstream of the translational start codon. To our knowledge, no promoter elements have been proposed for the spl operon, but based on those proposed for the other protease genes/operons the most likely candidates we could find were located further upstream (), in which case this result would presumably reflect the absence of a functional promoter. Interestingly, there is a potential SarA binding site located between the putative ribosome-binding site and −10 box. Our assays do not allow us to assess the possible functionality of this site, but if it is a functional SarA binding site then it suggests that SarA binding may interfere with the binding of RNA polymerase to the splA-F promoter, thus suggesting a different regulatory mechanism by comparison to the other protease genes/operons examined. However, fluorescence was increased in a sarA mutant by comparison to LAC with constructs containing 125 and 150 bp of upstream DNA. This suggests the presence of a SarA binding site. Although no potential binding sites were identified in this region that matched the binding site defined by our SELEX experiments as well as the downstream site discussed above or any of the putative binding sites associated with each of the other promoter regions, there is a similar AT-rich region present in this location (). Interestingly, fluorescence was decreased in both LAC and its sarA mutant with a 200 bp reporter construct by comparison to the constructs containing 150 and 125 bp, although it remained higher in the sarA mutant than in LAC. This suggests the presence of a binding site for an alternative repressor of spl expression in the region between 150 and 200 bp. Based on the results of our reporter assays using full-length reporter constructs (), Rot and SarS would be potential candidates in this regard. This is consistent with the observation that expression of the full-length spl::gfp reporter was increased in rot/sarA and sarS/sarA mutants even by comparison to the isogenic sarA mutant ().
All of these reporter assays were done with constructs generated with the promoter regions from LAC, but DNA sequence comparisons confirm that all of the putative regulatory elements, including the putative SarA binding site, are present and in the same location upstream of the UAMS-1 promoter regions associated with the genes/operons encoding aureolysin, SspA/SspB, and ScpA (data not shown). On the other hand, we did find differences in the promoter sequence upstream of the spl operon in LAC and UAMS-1. This was not surprising given that UAMS-1, as with EMRSA16, MRSA252, MN8, and several other S. aureus strains [Citation10], does not encode SplA or SplB.
Discussion
Changes in the production of extracellular proteases have been correlated with clinically relevant phenotypes of S. aureus including the accumulation of alpha toxin and protein A, biofilm formation, and virulence in diverse animal models [Citation26–28,Citation32,Citation53,Citation56,Citation59,Citation65,Citation68,Citation69]. Indeed, eliminating the production of aureolysin, SspA, SspB, ScpA, and the SplA-F proteases, or increasing the production of these same proteases, has been shown to have a dramatic impact on virulence and the overall S. aureus virulence factor repertoire [Citation26–29]. Such observations underscore the importance of understanding the regulatory mechanisms that modulate protease production as a post-translational means of controlling the abundance of S. aureus virulence factors.
A growing number of regulatory loci have been implicated in the production of extracellular proteases. Indeed, one report found that mutation of >50 of 108 regulatory loci examined resulted in increased or decreased transcription of one or more of the genes encoding these proteases [Citation34]. Similarly, a screen of the 1,920 mutants in the Nebraska Transposon Mutant Library (NTML) identified 62 mutants that exhibited differences in protease activity [Citation70]. The experimental approach taken in the studies we report was based on two considerations. The first is that regulatory proteins that directly impact protease production would likely have a greater phenotypic impact than regulatory loci that impact protease production indirectly. The second is that proteins that regulate the production of all or most extracellular proteases are likely to have a greater impact on these phenotypes than regulatory proteins that have a less global impact.
Based on this, we used an unbiased chromatography approach using DNA baits derived from each of the four transcriptional units encoding the 10 primary S. aureus extracellular proteases (aureolysin, ScpA, SspA/SspB, and SplA-F). Based on SDS-PAGE, Western blot, and LC-MS/MS, we identified six S. aureus proteins (SarA, MgrA, SarS, Rot, SarR, and SarZ) that were bound by at least three of these four DNA baits. All of these have been previously implicated in protease production, in some cases to a degree that can be correlated with altered phenotypes of potential clinical relevance, most notably biofilm formation [Citation27,Citation28,Citation34,Citation59,Citation71]. These studies confirmed that SarA is the most abundant protein captured with all four protease-associated baits followed by MgrA, SarS, and Rot in approximately that order. SarR and SarZ were captured by three of four baits, the exception in both cases being the ScpA-associated bait. However, it is important to note that SarR and SarZ were identified in LC-MS/MS analysis of proteins captured with the ScpA bait, but not to an extent that meet all of our inclusion criteria.
With few exceptions (e.g. Rot), it remains to be determined whether the capture of these proteins reflects a direct interaction with specific DNA targets or perhaps interactions among the proteins themselves. Nevertheless, subsequent studies analyzing derivatives of LAC and UAMS-1 with mutations in the six genes encoding each of these proteins confirmed that mutation of some of the genes encoding other regulatory proteins also resulted in increased protease production, decreased abundance of high molecular weight exoproteins, and decreased biofilm formation. The three that had the largest and most consistent impact were Rot, SarA and SarS, however, mutation of sarA had a significantly greater impact on all of these phenotypes by comparison to all other mutants including rot and sarS mutants. Moreover, sarA mutants were the only mutants that exhibited increased expression of all four protease genes/operons and did so in both LAC and UAMS-1. Additionally, this was also evident in the phenotype of derivatives of LAC and UAMS-1 with concomitant mutations in sarA and each of the other genes encoding regulatory proteins identified and prioritized based on the results of our binding assays. Specifically, mutation of sarA resulted in a greater increase in protease production, a greater decrease in the accumulation of high molecular weight proteins, and a more significant reduction in biofilm formation irrespective of the functional status of any of these other regulatory loci. Thus, we believe these collective results support the hypothesis that SarA plays a predominant role in limiting the production of S. aureus extracellular proteases.
However, this does not preclude an important role for other regulatory proteins in fine-tuning protease production. Indeed, in both LAC and UAMS-1 maximum expression of certain protease genes/operons was only observed when sarA was mutated along with other regulatory loci, the most notable among these once again being rot and sarS. This suggests that the functional status of sarA may influence the role of other regulatory proteins in modulating protease production. To examine this, we repeated our DNA bait assays using lysates prepared from a LAC sarA mutant. Interestingly, in the absence of SarA, all of the same regulatory proteins identified in our primary binding assays were found to bind protease-associated baits, but none were found to bind all four baits as defined by our statistical criteria. This reflects the fact that spectral counts observed with lysates from sarA mutants were generally reduced with all baits by comparison to those observed with lysates prepared from LAC. This was most apparent with the spl-associated bait in which MgrA, Rot, SarS, SarR and SarZ all bound to a significantly greater extent from lysates prepared from LAC than those prepared from its isogenic sarA mutant. However, there were also exceptions. Specifically, MgrA was bound by the aur-associated bait in greater abundance from the LAC sarA lysate than the lysate prepared from LAC itself. Mechanistically, one possible explanation for these findings is that MgrA competes with SarA for a similar or overlapping binding site, in which case the absence of SarA would presumably allow for increased binding. Alternatively, SarA could repress the production of MgrA, thus making it more abundant in a sarA mutant. Addressing these possibilities will require additional experiments that are beyond the scope of this report, but they nevertheless suggest that the functional status of sarA does impact the interaction between other regulatory proteins and specific protease promoter regions either directly at the protein-DNA level or indirectly through protein-protein interactions.
This impact of the functional status of sarA was also reflected in the results of our reporter assays in that we observed significant reporter-dependent differences with the double mutants. This made it important to determine whether specific proteases play a more important role than others in defining these potentially relevant in vitro phenotypes. We addressed this with an experimental focus on sarA for the reasons discussed above. Specifically, we generated sarA mutants that were unable to produce individual proteases alone and in combination with each other and examined the impact on the abundance of alpha toxin, cytotoxicity for osteoblasts and osteoclasts, and biofilm formation. The results confirmed that mutation of the gene encoding aureolysin alone was sufficient to restore the accumulation of alpha toxin in LAC as well as overall cytotoxicity for osteoblasts and osteoclasts. Similarly, mutation of the genes encoding aureolysin and ScpA was sufficient to fully restore biofilm formation in both LAC and UAMS-1 sarA mutants. This suggests that specific targets of these proteases may be particularly important. However, while mutation of sarA has been shown to attenuate virulence in murine models of sepsis and osteomyelitis owing to the increased production of extracellular proteases, these studies utilized sarA mutants unable to produce any extracellular protease [Citation27,Citation28,Citation61], and it remains to be determined whether eliminating the production of these proteases alone or in combination with each other is sufficient to restore the virulence of sarA mutants in these same animal models. Additionally, using our reporter assays we did identify double mutants that exhibited increased expression of the aur::gfp and/or scp::gfp reporters even by comparison to the isogenic sarA mutant in both LAC and UAMS-1, and this raises the possibility that mutation of these regulatory loci may also attenuate virulence.
These results also raise the possibility that regulatory proteins that only bind specific protease-associated promoters are important. Indeed, we identified 11 proteins that bound only to the aureolysin promoter region (Table S3). Included among these was the redox-sensing transcriptional repressor (Rex), which has also been previously associated with protease production. Specifically, mutation of rex was shown to result in decreased transcription of aur as well as sspABC [Citation34]. This suggests that Rex would not impact protease production in a manner that would limit the abundance of S. aureus virulence factors. Similarly, we identified six proteins that bound only to the scpAB promoter. Among these was a transcriptional regulator belonging to Xre family (SAUSA300_1969) that to our knowledge has not been functionally characterized. Interestingly, we identified 20 proteins that were captured only by the splA-F bait. Among these were the staphylococcal respiratory response protein A (SrrA), SarX, SarV, and a transcriptional regulator belonging to the MarR family encoded by SAUSA300_2452. Mutation of sarV has been shown to result in decreased transcription of the genes encoding aureolysin, ScpA and SplA [Citation46], suggesting that SarV may function as a transcriptional activator. There is also at least one report indicating that sarX represses transcription of ssp [Citation37]. Unlike the aur, scpA, and spl-associated baits, we did not identify any proteins that bound only the ssp-associated bait.
Finally, given the predominant role of SarA in repressing the production of all extracellular proteases, we carried out experiments using our reporter constructs that were aimed at identifying common DNA elements associated with SarA-mediated repression. Based on our earlier experiments that employed an unbiased SELEX (systematic evolution of ligands by exponential enrichment) approach to identify a functional SarA binding site [Citation67], these studies allowed us to identify putative SarA binding sites upstream of the genes encoding aureolysin, ScpA, and SspA/SspB, but we were unable to do so in the region upstream of the splA-F operon. This is consistent with the observation that the results observed in all of our experiments focusing on the spl operon were generally distinct by comparison to those focusing on other protease genes/operons, and suggest that the mechanistic basis for the regulatory impact of SarA on expression of the spl operon may differ by comparison to its impact on expression of the genes encoding other proteases. Indeed, the spl-associated bait bound the highest number of proteins that did not bind any other protease-associated bait. Nevertheless, this does not change the observation that mutation of sarA results in increased expression of the splA-F operon and the increased accumulation of all six Spl proteases as previously reported [Citation27,Citation29]. Thus, while the results we report support the conclusion that multiple regulatory loci contribute to the control of S. aureus protease production, they also support the primary conclusion that sarA serves the predominant role in this regard. It is well known that sarA serves global regulatory roles, and we are not suggesting that other factors are not important, but we are confident that the results we report demonstrate that the impact of SarA on the production of extracellular proteases plays a key role in defining the phenotype of S. aureus sarA mutants.
Materials and methods
Bacterial strains and growth conditions
All bacterial strains used in this study are listed in . Those generated specifically for this study were generated by ɸ11-mediated transduction from previously described mutants in our culture collection [Citation14,Citation28,Citation34,Citation71–73] or available in the Nebraska Transposon Mutant Library (NTML). Primarily to generate the strains needed for our reporter assays, it was necessary to switch the resistance marker with the mutants in the NTML using previously described methods [Citation74]. The exception was the UAMS-1 sarS mutant, which was generated using the pKOR1 mutagenesis system as previously described [Citation75]. Strains were maintained at −80°C in tryptic soy broth (TSB) containing 25% (v/v) glycerol. For each experiment, strains were retrieved from cold storage by plating on tryptic soy agar (TSA) with appropriate antibiotic selection. Antibiotics were used at the following concentrations: chloramphenicol (Cm), 10 μg/mL; kanamycin (K), 50 μg/mL; neomycin (N), 50 μg/mL; erythromycin (Erm), 10 μg/mL; spectinomycin (Spec), 1 mg/mL; and tetracycline (Tet), 5 μg/mL. For phenotypic assays, 5 mL liquid cultures were grown overnight (ON) in TSB at 37°C with constant shaking without antibiotic selection.
Table 1. Bacterial strains used in this study
DNA-affinity chromatography pull-down for isolation of proteins that bind protease-associated promoter regions
The DNA baits corresponding to each protease-associated promoter region were amplified by PCR. The oligonucleotides used for each amplification are listed in . In each case one of the oligonucleotides was biotinylated at the 5ʹ end. PCR fragments were purified using the Wizard® SV Gel and PCR Clean-Up System (Promega). Approximately 15 μg (calculated to saturate 50 μL of streptavidin beads (Miltenyi Biotec)) of DNA were incubated with 250 μL of whole cell lysates from S. aureus LAC or the isogenic ΔsarA mutant for 30 minutes at room temperature with constant shaking. The whole cell lysates were prepared using ON cultures standardized to and OD560 = 10. The cultures were centrifuged, the pellets were resuspended in BS/THES buffer [Citation78] and transferred to Lysing Matrix B tubes (MP Biomedicals) the cells were lysed on FastPrep-24™ Classic Instrument. Streptavidin beads were added, and the mixture was immobilized in a μ Column (Miltenyi Biotec) in the magnetic field of the μMACS Separator (Miltenyi Biotec). Columns were previously equilibrated using the protein applications buffer (Miltenyi Biotec) and rinsed twice with 100 μL of BS/THES. After running the beads-DNA-lysates mixture through the column the unbound material was removed by washing 5 times with BS/THES. Bound proteins were eluted using buffers with increasing NaCl concentrations (0.1 M NaCl, 0.2 M NaCl, 0.3 M NaCl, 0.5 M NaCl, 0.75 M NaCl, and 1 M NaCl).
Table 2. List of oligonucleotides used to amplify the protease promoter elements
Identification of bound proteins
Each sample obtained with each protease-associated bait was visualized by SDS-PAGE and Western blot analysis with an α-SarA antibody, as previously described [Citation28,Citation29,Citation71]. Additional analysis was done using liquid chromatography tandem mass spectrometry (LC-MS/MS). Specifically, samples were reduced, alkylated, and digested using filter-aided sample preparation [Citation79] with sequencing grade-modified porcine trypsin (Promega). Tryptic peptides were then separated by reverse phase XSelect CSH C18 2.5 um resin (Waters) on an in-line 150 × 0.075 mm column using an UltiMate 3000 RSLCnano system (Thermo). Peptides were eluted using a 60 min gradient from 98:2 to 65:35 buffer A:B ratio (Buffer A = 0.1% formic acid, 0.5% acetonitrile; Buffer B = 0.1% formic acid, 99.9% acetonitrile). Eluted peptides were ionized by electrospray (2.2 kV) followed by mass spectrometric analysis on an Orbitrap Fusion Lumos mass spectrometer (Thermo). MS data were acquired using the FTMS analyzer in profile mode at a resolution of 240,000 over a range of 375 to 1500 m/z. Following HCD activation, MS/MS data were acquired using the ion trap analyzer in centroid mode and normal mass range with precursor mass-dependent normalized collision energy between 28.0 and 31.0. The analysis was done in triplicate for each DNA bait using a 1:1 mixture of the 0.2 M and 0.3 M NaCl eluates, the 0.5 M eluate, and a 1:1 mixture of the 0.75 M and 1 M NaCl eluates.
Data analysis
Proteins were identified by database search using Mascot (Matrix Science, version 2.5.1) against the USA300 S. aureus database (2653 entries, Genebank accession JTJK01000002). A decoy database (based on the reverse of the protein sequences) was used in the search to calculate the FDR for the search algorithm. The search parameters include fixed modification of carbamidomethyl on cysteine, variable modifications of acetylation on the protein N-terminus, variable modification of oxidation on methionine, a maximum of two missed cleavages with trypsin, parent ion tolerance of 3 ppm, and fragment ion tolerance of 0.5 Da. Scaffold (Proteome Software) was used to verify MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established with less than 1.0% false discovery by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established with less than 1.0% false discovery and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm [Citation80]. Total spectral counts for each replicate were exported from Scaffold into Microsoft Excel for further analysis.
The spectral counts obtained for each identified protein from each of the three eluates obtained with each sample were summed and the resulting counts obtained with each protease promoter compared to the spectral counts found for the same protein present in the samples obtained using the promoter element of the control gene SAUSA300_1445 using two-tailed t-test. Proteins with a p-value ≥ 0.05 were filtered out and the fold change was calculated for the remaining proteins. Proteins with an FC ≤ 2 were filtered out along with proteins with less than 8 spectral counts in at least two out of the three replicates (this number was chosen based on counts obtained from the no DNA control samples). The lists of proteins meeting these criteria were compared and visualized with a Venn diagram created using Venny (version 2.1). The Venn diagrams represent the number of shared and unique proteins identified binding to each promoter element. Volcano plots, generated using R studio, were used to visualize the significant changes in proteins binding to each bait depending on the presence/absence of SarA. The y-axis consists of −log10 p-values based on Student’s t test analysis, and the x-axis consists of the log2 fold change.
Extracellular protease activity
These assays were done as previously described [Citation27]. Briefly, 5 mL overnight cultures grown without antibiotic selection. To account for slight variations in optical density, all cultures were standardized relative to each other based on an optical density (OD560) of 10.0) and filter sterilized to obtain CM. Protease activity was assessed using the EnzChek Gelatinase/Collagenase Assay Kit (Thermo) according to the manufacturer’s instructions. The final gelatin fluorescein conjugate substrate concentration used in each assay was 25 μg/mL. Fluorescence intensity was read after a 23 h incubation period using the FLUOstar Omega Multi-Mode Microplate Reader (BMG LABTECH) (excitation filter = 485 nm emission filter = 520 nm.
Transcriptional reporter assays
For each protease gene/operon amplicons ranging from 150 to 500 bp immediately upstream of the translational start site were cloned using Gibson Assembly Cloning (SGI-DNA) into pCM11 between the HindIII and KpnI sites. Additional constructs containing shorter promoter elements (50–125) were constructed by amplifying the regions of interest by PCR using primers including the HindIII and KpnI restriction sites and ligating into pCM11. The promoterless control plasmid (ΔP) was constructed using the QuikChange II XL Site-Directed Mutagenesis Kit to delete the region between the HindIII and KpnI sites on pCM11. All primers used to generate the inserts for these constructs are listed in . These plasmids were constructed and cloned in E. coli One Shot TOP10 (Thermo) and subsequently transformed into S. aureus strain RN4220 by electroporation [Citation81] before transduction into appropriate S. aureus strains using phage ɸ11. Fluorescence was assessed as previously described [Citation56]. Briefly, ON cultures were standardized by optical density. For single regulatory mutants all assays were done after standardizing conditioned medium (CM) to an OD560 of 10. For mutants containing more than one regulatory mutation, assays were done with CM standardized to an OD560 of seven because some of these mutants did not reach an OD560 of 10 given the high levels of GFP being expressed. 200 µL of the standardized cultures were plated into black clear-bottomed 96-well plates and the fluorescence intensity was measured with a FLUOstar Omega microplate reader (excitation, 485 nm; emission, 520 nm) (BMG Labtech).
Table 3. List of plasmids and oligonucleotides used in this study
Exoprotein profiles and Western blotting
CM collected as described above was resolved by SDS-PAGE using 4% to 12% gradient Novex Bis-Tris Plus gels (Life Technologies). Proteins were visualized by staining with SimplyBlue SafeStain (Life Technologies). Images were obtained using a Bio-Rad ChemiDoc MP imaging system (Bio-Rad Laboratories). The accumulation of alpha-toxin (Hla), protein A (Spa), and the NucA and NucB forms of Nuc1 were assessed in these samples by Western blot as previously described [Citation28,Citation29,Citation71].
Biofilm formation
Biofilms were assessed as previously described [Citation82]. Briefly, 5 mL ON cultures grown in biofilm media (TSB supplemented with dextrose and NaCl) were standardized by optical density OD560 of 0.05 and inoculated into non-tissue culture treated 96-well plates previously coated with 20% human plasma and incubated at 37°C. Biofilms were stained with crystal violet before eluting with ethanol and measuring the absorbance at 595 nm with a FLUOstar Omega microplate reader.
Cytotoxicity
Cell viability was assessed on confluent monolayers of MC3T3-E1 and RAW 264.7 cells (10,000 and 50,000 cells/well, respectively) after 24 hour incubation with a 1:1 mixture of CM from standardized S. aureus stationary cultures and mammalian cell growth media, as previously described [Citation61]. Cytotoxicity was determined using the LIVE/DEAD Viability/Cytotoxicity Kit for mammalian cells (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Statistical analysis
All assays were done in triplicate with at least two independent biological replicates. Statistical analyses were performed using GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA). Statistical significance was determined by one-way ANOVA with Dunnett’s correction. Separate comparisons were made with all strains relative to LAC or to its ΔsarA mutant. A p-value ≤0.05 was considered statistically significant.
Author contributions
Conceived and designed the experiments: AR, KB, MS. Performed the experiments: AR, KB, SB, BG. Analyzed the data: AR, KB, SB, AT, LS, MS. Wrote the paper: AR, KB, MS.
Supplemental Material
Download PDF (425.5 KB)Acknowledgments
This work was supported by NIH grant R01-AI119380 to MSS. Additional support was provided by a generous gift from the Texas Hip and Knee Center and research core facilities supported by the Center for Microbial Pathogenesis and Host Inflammatory Responses (P20-GM103450), the Translational Research Institute (UL1TR000039), the United States Army Congressionally Directed Medical Research Programs (W81X1H-14-PRORP-EA), and NIH grant R01-AI124458 to LNS. AJT acknowledges support from the National Institutes of Health grants P20GM121293, UL1TR000039, S10OD018445, and P20GM103429.
Disclosure statement
The authors report no conflict of interest.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532.
- Tam K, Torres VJ. Staphylococcus aureus secreted toxins and extracellular enzymes. Gram-Positive Pathogens. 2019:640–648.
- Bukowski M, Wladyka B, Dubin G. Exfoliative toxins of Staphylococcus aureus. Toxins (Basel). 2010;2:1148–1165.
- Imanishi I, Nicolas A, Caetano AB, et al. Exfoliative toxin E, a new Staphylococcus aureus virulence factor with host-specific activity. Sci Rep. 2019;9:16336.
- Oliveira D, Borges A, Simoes M. Staphylococcus aureus toxins and their molecular activity in infectious diseases. Toxins (Basel). 2018;10(6):252.
- Reed SB, Wesson CA, Liou LE, et al. Molecular characterization of a novel Staphylococcus aureus serine protease operon. Infect Immun. 2001;69:1521–1527.
- Dubin G, Stec-niemczyk J, Kisielewska M, et al. Enzymatic activity of the Staphylococcus aureus SplB serine protease is induced by substrates containing the sequence Trp-Glu-Leu-Gln. J Mol Biol. 2008;379:343–356.
- Paharik AE, Salgado-pabon W, Meyerholz DK, et al. The Spl serine proteases modulate Staphylococcus aureus protein production and virulence in a rabbit model of pneumonia. mSphere. 2016;1.
- Stach N, Kalinska M, Zdzalik M, et al. Unique substrate specificity of SplE serine protease from Staphylococcus aureus. Structure. 2018;26:572–579 e4.
- Zdzalik M, Karim AY, Wolski K, et al. Prevalence of genes encoding extracellular proteases in Staphylococcus aureus - important targets triggering immune response in vivo. FEMS Immunol Med Microbiol. 2012;66:220–229.
- Wilde AD, Snyder DJ, Putnam NE, et al. Bacterial hypoxic responses revealed as critical determinants of the host-pathogen outcome by TnSeq analysis of Staphylococcus aureus invasive infection. PLoS Pathog. 2015;11:e1005341.
- Kantyka T, Shaw LN, Potempa J. Papain-like proteases of Staphylococcus aureus. Adv Exp Med Biol. 2011;712:1–14.
- Sabat A, Kosowska K, Poulsen K, et al. Two allelic forms of the aureolysin gene (aur) within Staphylococcus aureus. Infect Immun. 2000;68:973–976.
- Shaw L. The role and regulation of the extracellular proteases of Staphylococcus aureus. Microbiology. 2004;150:217–228.
- Elmwall J, Kwiecinski J, NA M, et al. Galectin-3 is a target for proteases involved in the virulence of Staphylococcus aureus. Infect Immun. 2017;85:IAI.00177–17.
- Guerra FE, Borgogna TR, Patel DM, et al. Epic immune battles of history: neutrophils vs. Staphylococcus aureus. Front Cell Infect Microbiol. 2017;7:286.
- Jusko M, Potempa J, Kantyka T, et al. Staphylococcal proteases aid in evasion of the human complement system. J Innate Immun. 2014;6:31–46.
- Laarman AJ, Ruyken M, Malone CL, et al. Staphylococcus aureus metalloprotease aureolysin cleaves complement C3 to mediate immune evasion. J Immunol. 2011;186:6445–6453.
- Lehman MK, Nuxoll AS, Yamada KJ, et al. Protease-mediated growth of Staphylococcus aureus on host proteins is opp3 dependent. mBio. 2019;10.
- Murphy J, Ramezanpour M, Stach N, et al. Staphylococcus aureus V8 protease disrupts the integrity of the airway epithelial barrier and impairs IL-6 production in vitro. Laryngoscope. 2018;128:E8–E15.
- Pietrocola G, Nobile G, Rindi S, et al. Staphylococcus aureus manipulates innate immunity through own and host-expressed proteases. Front Cell Infect Microbiol. 2017;7:166.
- Prokesova L, Potuznikova B, Potempa J, et al. Cleavage of human immunoglobulins by proteinase from Staphylococcus aureus. Adv Exp Med Biol. 1995;371A:613–616.
- Smagur J, Guzik K, Bzowska M, et al. Staphylococcal cysteine protease staphopain B (SspB) induces rapid engulfment of human neutrophils and monocytes by macrophages. Biol Chem. 2009a;390:361–371.
- Smagur J, Guzik K, Magiera L, et al. A new pathway of staphylococcal pathogenesis: apoptosis-like death induced by Staphopain B in human neutrophils and monocytes. J Innate Immun. 2009b;1:98–108.
- Sonesson A, Przybyszewska K, Eriksson S, et al. Identification of bacterial biofilm and the Staphylococcus aureus derived protease, staphopain, on the skin surface of patients with atopic dermatitis. Sci Rep. 2017;7:8689.
- Kolar SL, Ibarra JA, Rivera FE, et al. Extracellular proteases are key mediators of Staphylococcus aureus virulence via the global modulation of virulence-determinant stability. Microbiologyopen. 2013;2:18–34.
- Ramirez AM, Byrum SD, Beenken KE, et al. exploiting correlations between protein abundance and the functional status of saers and sara to identify virulence factors of potential importance in the pathogenesis of Staphylococcus aureus osteomyelitis. ACS Infect Dis. 2020;6:237–249.
- Zielinska AK, Beenken KE, Mrak LN, et al. sarA-mediated repression of protease production plays a key role in the pathogenesis of Staphylococcus aureus USA300 isolates. Mol Microbiol. 2012;86:1183–1196.
- Byrum SD, Loughran AJ, BEENKEN KE, et al. Label-free proteomic approach to characterize protease-dependent and -independent effects of sarA inactivation on the Staphylococcus aureus exoproteome. J Proteome Res. 2018a;17(10):3384–3395.
- Jenul C, Horswill AR. Regulation of Staphylococcus aureus virulence. Microbiol Spectr. 2018;6.
- Priest NK, Rudkin JK, FEIL EJ, et al. From genotype to phenotype: can systems biology be used to predict Staphylococcus aureus virulence? Nat Rev Microbiol. 2012;10:791–797.
- Beenken KE, Mrak LN, GRIFFIN LM, et al. Epistatic relationships between sarA and agr in Staphylococcus aureus biofilm formation. PLoS One. 2010;5:e10790.
- Kato F, Kadomoto N, Iwamoto Y, et al. Regulatory mechanism for exfoliative toxin production in Staphylococcus aureus. Infect Immun. 2011;79:1660–1670.
- Gimza BD, Larias MI, Budny BG, et al. Mapping the global network of extracellular protease regulation in Staphylococcus aureus. mSphere. 2019;4. DOI:https://doi.org/10.1128/mSphere.00676-19
- Chien Y, Manna AC, Cheung AL. SarA level is a determinant of agr activation in Staphylococcus aureus. Mol Microbiol. 1998;30:991–1001.
- Austin CM, Garabaglu S, Krute CN, et al. Contribution of YjbIH to virulence factor expression and host colonization in Staphylococcus aureus. Infect Immun. 2019;87.
- Ballal A, Ray B, Manna AC. sarZ, a sarA family gene, is transcriptionally activated by MgrA and is involved in the regulation of genes encoding exoproteins in Staphylococcus aureus. J Bacteriol. 2009;191:1656–1665.
- Crosby HA, Schlievert PM, Merriman JA, et al. The Staphylococcus aureus global regulator MgrA modulates clumping and virulence by controlling surface protein expression. PLoS Pathog. 2016;12:e1005604.
- Fournier B, Klier A, Rapoport G. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol Microbiol. 2001;41:247–261.
- Gustafsson E, Oscarsson J. Maximal transcription of aur (aureolysin) and sspA (serine protease) in Staphylococcus aureus requires staphylococcal accessory regulator R (sarR) activity. FEMS Microbiol Lett. 2008;284:158–164.
- Ingavale S, Van wamel W, Luong TT, et al. Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infect Immun. 2005;73:1423–1431.
- Ingavale SS, Van wamel W, Cheung AL. Characterization of RAT, an autolysis regulator in Staphylococcus aureus. Mol Microbiol. 2003;48:1451–1466.
- Lucas AL, Manna AC. Phenotypic characterization of sarR mutant in Staphylococcus aureus. Microb Pathog. 2013;57:52–61.
- Luong TT, Dunman PM, Murphy E, et al. Transcription profiling of the mgrA regulon in Staphylococcus aureus. J Bacteriol. 2006;188:1899–1910.
- Majerczyk CD, Dunman PM, Luong TT, et al. Direct targets of CodY in Staphylococcus aureus. J Bacteriol. 2010;192:2861–2877.
- Manna AC, Ingavale SS, Maloney M, et al. Identification of sarV (SA2062), a new transcriptional regulator, is repressed by SarA and MgrA (SA0641) and involved in the regulation of autolysis in Staphylococcus aureus. J Bacteriol. 2004;186:5267–5280.
- Mrak LN, Zielinska AK, Beenken KE, et al. saeRS and sarA act synergistically to repress protease production and promote biofilm formation in Staphylococcus aureus. PLoS One. 2012;7:e38453.
- Nygaard TK, Pallister KB, Ruzevich P, et al. SaeR binds a consensus sequence within virulence gene promoters to advance USA300 pathogenesis. J Infect Dis. 2010;201:241–254.
- Oscarsson J, Tegmark-wisell K, Arvidson S. Coordinated and differential control of aureolysin (aur) and serine protease (sspA) transcription in Staphylococcus aureus by sarA, rot and agr (RNAIII). Int J Med Microbiol. 2006;296:365–380.
- Said-salim B, Dunman PM, Mcaleese FM, et al. Global regulation of Staphylococcus aureus genes by Rot. J Bacteriol. 2003;185:610–619.
- Tamber S, Cheung AL. SarZ promotes the expression of virulence factors and represses biofilm formation by modulating SarA and agr in Staphylococcus aureus. Infect Immun. 2009;77:419–428.
- Tegmark K, Karlsson A, Arvidson S. Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol Microbiol. 2000;37:398–409.
- Trotonda MP, Tamber S, Memmi G, et al. MgrA represses biofilm formation in Staphylococcus aureus. Infect Immun. 2008;76:5645–5654.
- Ziebandt AK, Weber H, Rudolph J, et al. Extracellular proteins of Staphylococcus aureus and the role of SarA and sigma B. Proteomics. 2001;1:480–493.
- Lauderdale KJ, Boles BR, Cheung AL, et al. Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect Immun. 2009;77:1623–1635.
- Rom JS, Ramirez AM, Beenken KE, et al. The impacts of msaABCR on sarA-associated phenotypes are different in divergent clinical isolates of Staphylococcus aureus. Infect Immun. 2020;88.
- Shen BA, Landick R. Transcription of bacterial chromatin. J Mol Biol. 2019;431:4040–4066.
- Fujimoto DF, Higginbotham RH, Sterba KM, et al. Staphylococcus aureus SarA is a regulatory protein responsive to redox and pH that can support bacteriophage lambda integrase-mediated excision/recombination. Mol Microbiol. 2009;74:1445–1458.
- Mootz JM, Benson MA, Heim CE, et al. Rot is a key regulator of Staphylococcus aureus biofilm formation. Mol Microbiol. 2015;96:388–404.
- Manna A, Cheung AL. Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect Immun. 2001;69:885–896.
- Loughran AJ, Gaddy D, Beenken KE, et al. Impact of sarA and phenol-soluble modulins on the pathogenesis of osteomyelitis in diverse clinical isolates of Staphylococcus aureus. Infect Immun. 2016;84:2586–2594.
- Byrum SD, Loughran AJ, Beenken KE, et al. Label-free proteomic approach to characterize protease-dependent and -independent effects of sarA inactivation on the Staphylococcus aureus Exoproteome. J Proteome Res. 2018b;17:3384–3395.
- Atwood DN, Beenken KE, Loughran AJ, et al. XerC contributes to diverse forms of Staphylococcus aureus infection via agr-dependent and agr-independent pathways. Infect Immun. 2016;84:1214–1225.
- Atwood DN, Loughran AJ, Courtney AP, et al. Comparative impact of diverse regulatory loci on Staphylococcus aureus biofilm formation. Microbiologyopen. 2015;4:436–451.
- Loughran AJ, Atwood DN, Anthony AC, et al. Impact of individual extracellular proteases on Staphylococcus aureus biofilm formation in diverse clinical isolates and their isogenic sarA mutants. Microbiologyopen. 2014;3:897–909.
- Lauderdale KJ, Malone CL, Boles BR, et al. Biofilm dispersal of community-associated methicillin-resistant Staphylococcus aureus on orthopedic implant material. J Orthop Res. 2010;28:55–61.
- Sterba KM, Mackintosh SG, Blevins JS, et al. Characterization of Staphylococcus aureus SarA binding sites. J Bacteriol. 2003;185:4410–4417.
- Rom JS, Atwood DN, Beenken KE, et al. Impact of Staphylococcus aureus regulatory mutations that modulate biofilm formation in the USA300 strain LAC on virulence in a murine bacteremia model. Virulence. 2017;8:1776–1790.
- Tsang LH, Cassat JE, Shaw LN, et al. Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS One. 2008;3:e3361.
- Fey PD, Endres JL, Yajjala VK, et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio. 2013;4:e00537–12.
- Beenken KE, Mrak LN, Zielinska AK, et al. Impact of the functional status of saeRS on in vivo phenotypes of Staphylococcus aureus sarA mutants. Mol Microbiol. 2014;92:1299–1312.
- Blevins JS, Beenken KE, Elasri MO, et al. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect Immun. 2002;70:470–480.
- Wormann ME, Reichmann NT, Malone CL, et al. Proteolytic cleavage inactivates the Staphylococcus aureus lipoteichoic acid synthase. J Bacteriol. 2011;193:5279–5291.
- Bose JL, Fey PD, Bayles KW. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl Environ Microbiol. 2013;79:2218–2224.
- Bae T, Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2006;55:58–63.
- Luong TT, Newell SW, Lee CY. Mgr, a novel global regulator in Staphylococcus aureus. J Bacteriol. 2003;185:3703–3710.
- Gillaspy AF, Hickmon SG, Skinner RA, et al. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect Immun. 1995;63:3373–3380.
- Jutras BL, Verma A, Stevenson B. Identification of novel DNA-binding proteins using DNA-affinity chromatography/pull down. Curr Protoc Microbiol. 2012;24:1F–1.
- Wisniewski JR, Zougman A, Nagaraj N, et al. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362.
- Nesvizhskii AI, Keller A, Kolker E, et al. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658.
- Schenk S, Laddaga RA. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett. 1992;73:133–138.
- BEENKEN KE, BLEVINS JS, SMELTZER MS. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect Immun. 2003;71:4206–4211.
