ABSTRACT
Candida albicans is the most prevalent fungal pathogen in humans, particularly in immunocompromised patients. In this study, by screening a C. albicans mutant library, we first identified that the MSS2 gene, an ortholog of Saccharomyces cerevisiae MSS2 required for mitochondrial respiration, mediates chitosan resistance. Upon treatment with 0.2% chitosan, the growth of mss2Δ strains was strikingly impaired, and MSS2 expression was significantly repressed by chitosan. Furthermore, mss2Δ strains exhibited slow growth on medium supplemented with glycerol as the sole carbon source. Similar to the chitosan-treated wild-type strain, the mss2Δ strain exhibited a significantly impaired ATP production ability. These data suggest that an antifungal mechanism of chitosan against C. albicans acts by inhibiting MSS2 gene expression, leading to repression of mitochondrial function. Normal respiratory function is suggested to be required for fungal virulence. Interestingly, the mss2Δ mutant strains exhibited significantly impaired invasive ability in vitro and ex vivo but retained normal hyphal development ability in liquid medium. Furthermore, the MSS2 deletion strains could not form robust biofilms and exhibited significantly reduced virulence. Collectively, these results demonstrated that the antifungal effect of chitosan against C. albicans is mediated via inhibition of mitochondrial biogenesis. These data may provide another strategy for antifungal drug development via inhibition of fungal mitochondria.
Introduction
Mitochondria are specialized organelles found in eukaryotic cells. A primary role of mitochondria is to supply energy through the respiratory chain [Citation1–3]. Moreover, mitochondria perform diverse functions, including roles in controlling the cell cycle, cell growth, signaling, ion homeostasis and metabolite production [Citation2,Citation4]. In humans, mitochondrial dysfunction might be associated with several human diseases [Citation5–7]. In pathogenic fungi, inhibition of the respiratory chain influences the yeast-to-hyphal morphological transition, cell wall biogenesis, drug resistance and virulence [Citation1,Citation3,Citation8–13]. However, the mechanistic link between mitochondria and the physiological roles of fungal pathogens remains largely unclear.
Candida albicans can reside in the human body as a commensal organism [Citation14], but it is also the predominant fungal pathogen that can cause life-threatening systemic infections, particularly in immunocompromised patients [Citation15,Citation16]. The ability of C. albicans to colonize different niches and become pathogenic has been shown to be closely linked to its phenotypic plasticity [Citation17–20]. In fact, several diverse types of C. albicans cells, such as yeast cells, hyphal cells, white cells, opaque cells, gray cells and gastrointestinally induced transition (GUT) cells, [Citation19,Citation21–24], also exhibit the striking ability of Candida cells for morphological transformation in response to different environments and host niches.
Among these phenotypic changes, the reversible morphological transition between yeast and hyphal forms is the central and defining virulence factor in C. albicans [Citation21,Citation25]. The yeast-to-hyphal transition is controlled by Cph1 and Efg1, the downstream targets of Cek1 MAP kinase and cyclic AMP (cAMP) signaling, respectively [Citation26–31]. These signaling cascades and many downstream hypha-specific genes can be induced by different environmental conditions and components, such as high temperature, serum, N-acetylglucosamine (GlcNAc), CO2, and neutral pH [Citation27,Citation30,Citation32–35]. Furthermore, the switch from the yeast to the hyphal form is highly associated with biofilm formation in hosts and on implantable medical devices [Citation36,Citation37], leading to resistance to clinical treatment.
Although mitochondria play major roles in the electron transport chain (ETC), energy production and cellular metabolism [Citation1–3,Citation10,Citation13], information regarding the function of C. albicans mitochondria is currently very limited. In contrast to Saccharomyces cerevisiae, which lacks complex I (NADH:ubiquinone oxidoreductase), C. albicans and most other Candida species have five intact ETC complexes [Citation38–40]. The contribution of ETC complex I to physiological roles is one of the most well studied topics in C. albicans. The NADH:ubiquinone oxidoreductases Nuo1 and Nuo2 are subunits of complex I and are conserved in several fungal pathogens [Citation41]. NUO1 or NUO2 have been shown to be involved in complex I activity, cell wall biogenesis, hyphal formation and biofilm development [Citation9,Citation41]. Additionally, GOA1, originally identified through mutant library screening to be required for oxidant adaptation and present in all members of the CTG clade of pathogenic Candida species except C. glabrata, is involved in mitochondrial complex I activity [Citation42]. Furthermore, the GOA1 deletion mutant showed lower respiratory activity, a reduced ability to form hyphae, high sensitivity to antifungal drugs and cell wall alterations and, moreover, was required for full virulence [Citation41–44]. Further studies showed that the three transcription factors (TFs) Rbf1, Hfl1 and Dpb4, which positively regulate GOA1 directly or indirectly, are involved in mitochondrial biogenesis [Citation12]. Moreover, a recent study identified that several potential mitochondrial proteins involved in the expression of ETC complex I, III or IV are required for respiration, antifungal susceptibility and virulence [Citation13]. However, the functions of most subunits of each ETC complex and mitochondrial proteins have not been studied in C. albicans.
Chitosan, a deacetylated unit of chitin, has numerous commercial applications and biomedical uses [Citation45–50]. Additionally, the positive charge of chitosan endows it with considerable antimicrobial activity against bacteria and fungi if the pH is below 6.5 [Citation51–55]. Although we have identified many transcription factors that potentially mediate chitosan resistance in C. albicans via screening of a TF mutant library [Citation56], the mechanisms underlying the antifungal action of chitosan against C. albicans remain largely unknown. Here, we further screened a library of protein kinase mutant strains of C. albicans to understand the potential cascades required for the response to chitosan. MSS2 was identified from this screen. The MSS2 gene was first identified in S. cerevisiae and is required for ETC activity and mediates membrane insertion of the C-terminus of Cox2p, a subunit of cytochrome c oxidase (complex IV) [Citation57–59]. However, the function of MSS2 in C. albicans is unknown. In this work, we demonstrated that C. albicans MSS2 is necessary for chitosan tolerance and that its expression is inhibited by chitosan. Furthermore, mss2Δ cells exhibited a significantly decreased respiration ability compared with wild-type (WT) cells, similar to that seen in the chitosan-treated wild-type strain. These data suggested that an antifungal mechanism of chitosan against C. albicans acts by inhibiting mitochondrial biogenesis. Interestingly, mss2Δ strains showed significantly impaired invasive growth, biofilm formation and virulence. Hence, inhibition of respiratory activity could be a promising therapeutic strategy to control C. albicans infection.
Materials and Methods
Media and reagents
The media used in these experiments included yeast extract-peptone-dextrose (YPD) medium, YPD medium containing 200 μg/ml of nourseothricin (Werner BioAgents, Jena, Germany), and Spider medium, and they were prepared as previously described [Citation60]. In YPG medium, dextrose was replaced with 3% glycerol. The base RPMI 1640 liquid medium, RPMI 1640 medium supplemented with 0.2% acetic acid (final pH 6.3, buffer control medium), and chitosan medium prepared from RPMI 1640 supplemented with 0.2% chitosan dissolved in 0.2% acetic acid (experimental medium) were prepared as previously described [Citation56,Citation61,Citation62]. The chitosan used in this study was purchased from Shin Era Technology (Taipei, Taiwan). The molecular weight (MW) of chitosan is approximately 20–35 kDa, and its degree of deacetylation is approximately 90% [Citation56]. All chemicals were purchased from Sigma-Aldrich Chemical unless stated otherwise.
Mutant library screening
The C. albicans mutant library strains derived from BWP17 were purchased from different laboratories [Citation63–67]. Each C. albicans kinase mutant strain was grown separately in YPD liquid medium in 96-well microplates at 30°C overnight. The strains were then transferred to 96-well microplates containing RPMI 1640 liquid medium (supplemented with 0.165 M MOPS and 2% glucose), buffer control medium (RPMI 1640 medium supplemented with 0.2% acetic acid) or experimental medium (RPMI 1640 supplemented with 0.2% chitosan dissolved in 0.2% acetic acid). The 96-well microplates were incubated at 30°C for 48 hr, and the optical density of the wells was then measured at 600 nm in a SpectraMax 190 microplate reader (Molecular Devices, CA, USA). The absorbance values of each mutant treated with chitosan or acetic acid were compared to determine the susceptibility of the strains to chitosan, and statistical significance was determined by Student’s t-test.
Plasmid and strain construction
The C. albicans strains and oligonucleotides used in this study are listed in Tables S1 and S2, respectively.
To construct the mss2Δ strains, the 5ʹ flanking and 3ʹ flanking regions of MSS2 were PCR-amplified using primer pairs 1139/1140 and 1141/1142, respectively. The 5ʹ and 3ʹ PCR products were digested with ApaI/XhoI and SacII/SacI, respectively, and cloned into the plasmid pSFS2A [Citation68] to generate the plasmid pSFS2A-MSS2 KO. The plasmid was linearized with ApaI/SacI and transformed into the wild-type strain SC5314 to generate the heterozygous mss2Δ/MSS2 mutant strains. The SAT1 marker was recycled by treatment with 2% maltose. The heterozygous strains were retransformed with the same deletion construct to generate the mss2Δ/mss2Δ strains YL1700/YL1701.
The MSS2 complementation construct was generated by amplification of its endogenous promoter and open reading frame (ORF) using the primer pair 1710/1711. The PCR product was digested with SacII/SacI and cloned into pSFS2A to generate pSFS-MSS2 AB. The plasmid was partially digested with BsmI and transformed into mss2Δ to generate YL1986/YL1987.
Sensitivity assays
Ten-fold serial dilutions of C. albicans cells were generated from suspensions with an OD600 of 1.0. A 5 µl volume of each dilution was spotted onto RPMI 1640 agar, RPMI 1640 agar containing 0.2% acetic acid, or RPMI 1640 agar supplemented with 0.2% chitosan. Images were acquired after the plates were incubated at 30°C for 2–3 days. To test the cell membrane- and cell wall-perturbing agents, final concentrations of 0.04% SDS, 60 μM calcofluor white and 1.6 μg/ml caspofungin were used. Images were acquired after the plates were incubated at 30°C for 2–3 days.
Determination of mitochondrial activities via Seahorse XFe 24 assays
A Seahorse XF analyzer (XFe 24) was used to measure the oxygen consumption rate (OCR) following a previous protocol with slight modifications [Citation69,Citation70]. Overnight cultures of C. albicans cells (4 × 105 cells/well in 50 μl) were washed twice with PBS and distributed into poly-D-lysine-precoated (50 μg/ml) 24-well XFe microplates (Seahorse Bioscience). Four replicates of each strain and chitosan treatment condition were established per experiment. Samples were incubated for 1 hr at 30°C to allow for cell adhesion. The untreated groups were incubated with 50 μl of RPMI 1640 medium, and the treated groups were incubated with 50 μl of 0.2% chitosan for 20 min. Each well was filled with 575 μl of RPMI 1640 medium before analysis. The energy pathway (indicated by the OCR) of each sample was measured using the Seahorse XFe 24 at 9 min intervals. Mitochondrial respiration was first induced by assay medium containing 2 mM L-glutamine. Then, 100 mM triethyltin bromide (TET), 5 μM FCCP and 2 mM antimycin A were sequentially injected into the assay wells during the analyses. Student’s t-test was used for the statistical analyses.
Quantitative reverse transcription-PCR
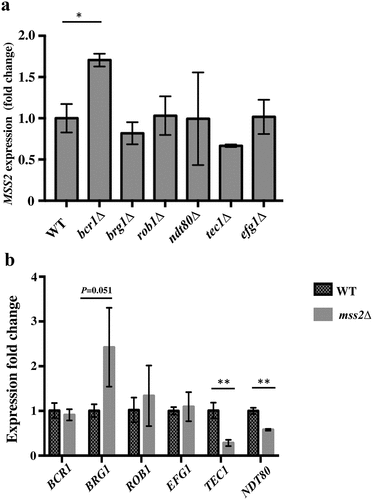
The assay was performed in accordance with the established protocol in our laboratory [Citation71]. Overnight cultures of C. albicans cells (200 μl) were transferred into 10 ml of fresh YPD. Cells were harvested by centrifugation and washed 3 times with sterile water. Cells of each sample were treated with RPMI 1640 medium, RPMI 1640 medium containing 0.2% acetic acid, or RPMI 1640 medium containing 0.2% chitosan at 30°C for 5, 10 or 20 min. Furthermore, to measure the expression of the six core regulatory genes required for biofilm development, cells were collected from biofilms in a silicone model as described in the “Biofilm assays” section of the Materials and Methods. Cells were harvested and washed three times with sterile water. Total RNA was isolated from each sample using a MasterPureTM Yeast RNA Purification Kit (Epicenter, Madison, WI, USA), and DNA was removed with DNase I (Thermo Fisher Scientific, Waltham, MA, USA). RNA was transcribed to cDNA with an iScriptTM cDNA Synthesis Kit (Bio-Rad Laboratories., Inc., CA., USA). Quantitative PCR was performed in a Bio-Rad CFX Manager (Bio-Rad Laboratories, Hercules, CA, USA). Each experiment was independently repeated three times, and the means of the data from the triplicate experiments are shown. The differences between the control group and the experimental group were analyzed with Student’s t-test. The primer pairs 541/542, 1197/1198, 1781/1782, 1783/1784, 1785/1786, 1787/1788, 1791/1792, and 1789/1790 were used to detect ACT1, MSS2, BCR1, BRG1, ROB1, EFG1, TEC1 and NDT80 expression, respectively. Student’s t-test was used for the statistical analyses. The expression of the target genes was normalized to the expression of the ACT1 gene.
Hyphal formation assays
The wild-type, mss2 mutant and complemented strains were grown in RPMI 1640 for 12 hr or in YPD supplemented with or without 50% serum for 4 hr. Hyphal development of the wild-type, mss2 mutant and complemented cells was examined with an Eclipse Ti inverted microscope (Nikon Instruments Inc., Melville, NY, USA).
Agar invasion assays
Overnight cultures of C. albicans cells at an OD600 of 1.0 were subjected to 10-fold serial dilution. A 2 µl volume of each dilution was spotted onto an RPMI 1640 agar plate. The plates were incubated for 4 days at 30°C. Colonies above the agar surface were washed with sterilized deionized water as previously described [Citation72]. Images were acquired before and after the washes.
Cell culture
HeLa (human epithelial cells from a fatal cervical adenocarcinoma) cells were cultured in DMEM (Dulbecco’s Modified Eagle Medium, Thermo Fisher) supplemented with 10% fetal bovine serum (FBS) and antibiotics (10 μg/mL streptomycin and 100 U/mL penicillin) and then incubated at 37°C with 5% CO2.
Cell invasive assays
The experimental procedures of the cell invasive assays followed the established protocol with slight modifications [Citation73,Citation74]. Overnight cultures of HeLa cells on the glass coverslips were washed with PBS and incubated with 5 × 105 C. albicans cells at 37°C with 5% CO2 for 1 hr. Samples were fixed with 4% paraformaldehyde for 30 min. Then, the samples were incubated with 25 μg/mL concanavalin A–fluorescein conjugate (Invitrogen) in PBS for 45 min to stain C. albicans cells localized outside the epithelial cells because concanavalin A-fluorescein cannot penetrate cell membrane of HeLa cells. The membranes of epithelial cells were subsequently permeabilized with 0.1% Triton X-100 detergent for 15 min. Calcofluor white (10 μg/mL, 0.1 M Tris-HCl) was then used to stain C. albicans cells localized outside of and inside epithelial cells for 20 min. Samples on coverslips were washed with PBS three times and then mounted with 100% glycerol. An Eclipse Ti inverted microscope was used to examine the fluorescence of each sample. Three experiments were performed, and 100 C. albicans cells were analyzed in each experiment. Then, the percentage of C. albicans cells that had invaded the epithelial cells was calculated.
Biofilm assays
Biofilm assays were performed following the previously established protocol for measuring biofilm biomass in a silicone model [Citation75]. Silicone squares (Bentec Medical, PR72034-06 N, 1.5 cm × 1.5 cm) placed in a 12-well plastic plate were sterilized and weighed before incubation in bovine serum (Sigma, B-9433). The plates were placed in a shaking incubator at 150 rpm and 37°C overnight. The serum-treated silicone squares were washed with 2 ml of PBS and placed in new 12-well plates supplemented with 2 ml of Spider medium. Notably, 1% mannitol in the Spider medium was replaced with 1% glucose for the C. albicans biofilm analyses because mss2 mutant strains could not utilize mannitol and survive under these culture conditions. Approximately 2 × 107 cells from the overnight YPD cultures of the C. albicans SC5314 wild-type, mss2 mutant and complemented strains were placed on top of the silicone squares. The inoculated samples were incubated at 37°C with shaking at 150 rpm for 90 min to allow for adhesion. The samples were washed with 2 ml of PBS and then incubated in 2 ml of fresh Spider medium for 60 hr at 37°C with shaking at 150 rpm. The supernatant was removed from each sample, and the silicone squares were dried overnight to determine the dry weight of the biofilms.
Virulence assays and fungal burden assessment
Five-week-old male ICR mice purchased from BioLASCO (Taiwan) were used (n = 9). Overnight cultures of C. albicans cells in YPD medium were harvested by centrifugation. Cells from each sample were washed three times with PBS. Mice were inoculated with C. albicans cells of the wild-type, mss2Δ and complemented strains (5 × 105 cells in 200 µl) via tail vein injection. The course of infection was monitored for 25 days to assess survival. Statistical significance was set at a P value of < 0.05 using the log-rank test.
For the fungal burden assessment, mice were infected intravenously with wild-type or mutant C. albicans cells (5 × 105 cells in 200 µl) and sacrificed three days post infection. Kidneys (n = 4 per group) and brains (n = 4 per group) harvested from the mice were dissected and weighed. Each organ was placed in 10 ml of PBS solution and homogenized for 2 min at 30,000 rpm with IKA dispersers (IKA, T10 basic ULTRA-TURRAX® Crushing Disperser, Germany). The tissue homogenates were serially diluted on YPD agar plates supplemented with 100 µg/ml chloramphenicol. The plates were incubated at 30°C for 24 to 48 hr to determine the CFUs per gram of kidney or brain tissue homogenate.
Statistical analyses
For the virulence assays, statistical significance was set at a P value of < 0.05 using the log-rank test. Statistical analyses of data from other experiments were performed using a one-tailed or two-tailed Student’s t-test with a 95% confidence interval.
Results
Determination of the involvement of MSS2 in chitosan resistance by screening a library of protein kinase mutant strains of C. albicans
A total of 166 kinase mutant strains were tested with or without chitosan treatment (Supplementary Table S3 worksheet 1). Twenty-one kinase gene mutants exhibited a significant growth defect upon chitosan treatment ( and Supplementary Table S3 worksheet 1). Screening data showed that several important signaling pathways are involved in the chitosan response. The potential cascades and factors involved in chitosan resistance are summarized in [Citation56]. These pathways included the Hog1 (sln1Δ, pbs2Δ and hog1Δ), Cek1/Cek2 (ste11Δ and cek2Δ), Mkc1 (pkc1Δ, bck1Δ and mkk2Δ) MAP kinases, Ras1-cAMP (tpk2Δ) and calcineurin (cmk1Δ, swe1Δ and hsl1Δ) pathways, as the indicated mutants listed in parentheses were highly susceptible to chitosan ( and Supplementary Table S3 worksheet 2). These findings are not surprising, because it has been proposed that chitosan binds to the fungal surface [Citation52,Citation55,Citation56], and these signaling pathways have been shown to mediate cell wall and cell membrane integrity in C. albicans or budding yeast [Citation76–80]. Additionally, YCK2, FUN31 (PSK1) and CKA1 have been shown to be important for cell surface biogenesis and drug resistance [Citation66,Citation81,Citation82]. The function of three genes, namely, ORF19.4518, PGA43 and PRR2, remains unclear ( and Supplementary Table S3 worksheet 2). Interestingly, several lines of evidence in the screening data showed that the chitosan used in this study targets mitochondria. First, two independent mss2Δ strains exhibited significantly impaired cell growth upon chitosan treatment ( and Supplementary Table S3), although the role of MSS2 in C. albicans has not yet been determined. Second, the Ssn3-regulated gene SEF1 is involved in mitochondrial iron metabolism [Citation83]. Third, both the Ras1-cAMP and Hog1 MAP kinase pathways have been shown to be important for maintaining mitochondrial activity [Citation84,Citation85]. Based on these observations and the finding that its optical density was the lowest observed in the screen ( and Supplementary S3), MSS2 was selected for further investigation.
Figure 1. Screening results of a library of protein kinase mutant strains of C. albicans during chitosan treatment. (a) Each mutant strain was grown separately in 96-well microplates in RPMI 1640 liquid medium (RPMI), RPMI containing 0.2% acetic acid or RPMI containing both 0.2% acetic acid and chitosan. The optical density of the cells after incubation at 37°C for 48 hr was measured at 600 nm. Each mutant strain treated with acetic acid or chitosan was compared to determine the susceptibility of the strains to chitosan. Statistical significance was evaluated by Student’s t-test (unpaired, two-tailed)., P<0.05; P<0.01. (b) Diagrammatic illustration of the potential signaling cascades and factors involved in chitosan resistance in C. albicans. In addition to the electrostatic interaction between chitosan and the C. albicans cell surface, the intracellular response of several potential pathways and factors could be associated with chitosan. These cascades, such as Hog1, Cek1/Cek2, Mkc1, Ras1-cAMP, and calcineruin pathways and other factors [Citation56], are associated with the cell wall or cell membrane biogenesis, stress tolerance, and mitochondrial function, which are required for chitosan resistance. The functional annotation of each gene is listed in the Table S3 (worksheet 2)
![Figure 1. Screening results of a library of protein kinase mutant strains of C. albicans during chitosan treatment. (a) Each mutant strain was grown separately in 96-well microplates in RPMI 1640 liquid medium (RPMI), RPMI containing 0.2% acetic acid or RPMI containing both 0.2% acetic acid and chitosan. The optical density of the cells after incubation at 37°C for 48 hr was measured at 600 nm. Each mutant strain treated with acetic acid or chitosan was compared to determine the susceptibility of the strains to chitosan. Statistical significance was evaluated by Student’s t-test (unpaired, two-tailed)., P<0.05; P<0.01. (b) Diagrammatic illustration of the potential signaling cascades and factors involved in chitosan resistance in C. albicans. In addition to the electrostatic interaction between chitosan and the C. albicans cell surface, the intracellular response of several potential pathways and factors could be associated with chitosan. These cascades, such as Hog1, Cek1/Cek2, Mkc1, Ras1-cAMP, and calcineruin pathways and other factors [Citation56], are associated with the cell wall or cell membrane biogenesis, stress tolerance, and mitochondrial function, which are required for chitosan resistance. The functional annotation of each gene is listed in the Table S3 (worksheet 2)](/cms/asset/3a6315c9-7c35-4f99-b28a-8f98ebf12e7a/kvir_a_1870082_f0001_oc.jpg)
The MSS2 gene is involved in chitosan susceptibility, and its expression is repressed by chitosan in C. albicans SC5314
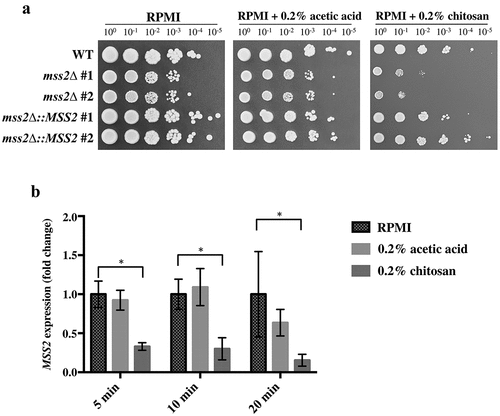
To verify the previous results, MSS2 was knocked out in the C. albicans SC5314 strain, and the chitosan susceptibility of the mutant strains was analyzed. As shown in , the mss2Δ strains in the RPMI 1640 and buffer control (RPMI 1640 + 0.2% acetic acid) groups exhibited a mild growth defect, whereas the mss2Δ strains exhibited more severe growth impairment on medium supplemented with 0.2% chitosan. Chitosan resistance was restored in the complemented strains. These results indicate that C. albicans MSS2 is required for chitosan tolerance. Furthermore, quantitative polymerase chain reaction (qPCR) analysis showed that MSS2 expression was significantly repressed in the wild-type strain after chitosan treatment for 5, 10 and 20 min compared with that in untreated C. albicans cells ().
Figure 2. C. albicans MSS2 mediates chitosan resistance, and its expression is repressed by chitosan. (a) The mss2 null mutant exhibited significant growth defects on medium containing 0.2% chitosan, and complementation restored the resistance to chitosan. A mild growth defect of mss2Δ was observed in the RPMI and buffer control (0.2% acetic acid) groups. (b) Treatment with 0.2% acetic acid for 5, 10 and 20 min did not significantly affect MSS2 expression, whereas SC5314 cells treated with chitosan for each duration exhibited significant reductions in MSS2 expression. The values are presented as the means ± SDs of at least three experimental replicates and were compared with the value for untreated wild-type SC5314 cells. Expression levels were normalized to those of the ACT1 gene. Statistical significance was determined using Student’s t-test, P < 0.05
Respiratory activity is repressed in both mss2Δ cells and chitosan-treated wild-type cells
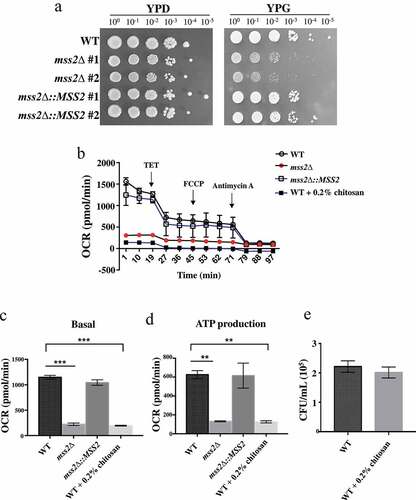
The exact role of MSS2 in C. albicans remains unknown. In the budding yeast S. cerevisiae, deletion of the ScMSS2 gene reduced mitochondrial biogenesis, as evidenced by the failure to integrate the C-terminus of Cox2p, a subunit of complex IV, in the mutant strain [Citation57,Citation59]. We therefore investigated whether the C. albicans MSS2 gene is involved in the ETC. As shown in , mss2Δ strains exhibited more severe growth defects in yeast extract-peptone-glycerol (YPG) medium than in other media, suggesting that C. albicans MSS2 is involved in mitochondrial biogenesis and energy production. To provide support for this hypothesis, a Seahorse analyzer was used to measure mitochondrial respiration. The mss2Δ strain exhibited a significantly decreased OCR under both basal respiration and ATP production conditions (, 3c and 3d). Mitochondrial function was restored in the complemented strain (, 3c and 3d). To determine whether chitosan treatment causes the death of wild-type and/or mss2Δ cells by causing severe mitochondrial deficiency, the viability of the wild-type cells treated with chitosan was assessed. shows that the CFUs of the wild-type and mss2Δ strains were similar after chitosan treatment for 30 min. These data suggest that an antifungal mechanism of chitosan against C. albicans acts by inhibiting mitochondrial biogenesis. Notably, addition of the uncoupler carbonylcyanide p-trifluoromethoxyphenylhydrazone (FCCP) was not sufficient to attain maximal respiration; thus, the maximal respiratory capacity and spare respiratory capacity could not be calculated (, 3c and 3d). This effect might have occurred because FCCP is used mainly to study mammalian cells. As described previously, chitosan repressed MSS2 expression (). These findings imply that chitosan-treated wild-type cells might display mitochondrial activity similar to that seen in the mss2Δ strain. Indeed, compared to the other cells, chitosan-treated wild-type cells exhibited a significantly reduced OCR under both basal respiration and ATP production conditions (, 3c and 3d). These data suggest that chitosan exerts an antifungal effect against C. albicans by inhibiting its respiratory system.
Figure 3. MSS2 deletion mutants cannot efficiently utilize glycerol as the sole carbon source and exhibit strikingly impaired respiratory activity. (a) Compared to the wild-type strain, the mss2Δ strains showed growth defects on YPG medium. (b) Mitochondrial respiratory activity in C. albicans cells was assessed by extracellular flux analysis. Mitochondrial respiration was assessed by measuring the OCR after sequential addition of 100 mM TET, 2 μM CCCP, 5 μM FCCP and 2 mM antimycin A. Quantification of respiratory activity under (c) basal and (d) ATP production conditions, with reference to the mitochondrial respiration data obtained in B. (e) The CFUs were similar between the untreated and chitosan-treated WT cells. The results are shown as the average of the values from three independent experiments and are the mean ± SD values. **, P < 0.01; ***, P < 0.001
Deletion of MSS2 in C. albicans increases susceptibility to sodium dodecyl sulfate (SDS) but confers resistance to calcofluor white
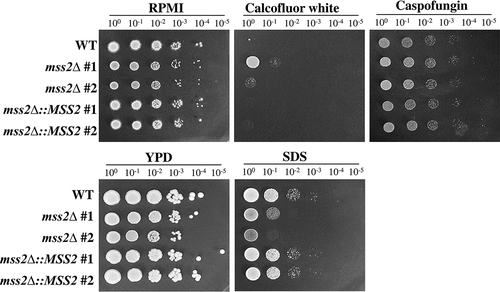
A functional link between the cell wall and mitochondria has been reported in several review articles [Citation1–3,Citation8,Citation10,Citation11]. We therefore sought to determine whether C. albicans MSS2 is involved in cell wall integrity. Calcofluor white has been shown to interfere with cell wall assembly by targeting chitin, leading to an increase in cell wall stress [Citation86,Citation87]. The primary target of the antifungal drug caspofungin in C. albicans is β-1,3-glucan synthase [Citation88]. In contrast, SDS is an amphipathic molecule that has both hydrophilic and hydrophobic regions and damages membranes and lipids [Citation89]. Calcofluor white and caspofungin (cell wall-perturbing agents) and SDS (membrane-perturbing agent) were therefore selected as treatments in cell growth assays. As shown in , the mss2Δ strains exhibited significant growth inhibition on medium supplemented with 0.04% SDS but showed better growth than the other strains on medium supplemented with 60 μM calcofluor white. In addition, the mss2Δ strains showed no difference in growth on medium supplemented with 1.6 μg/ml caspofungin compared to RPMI 1640 medium. These data suggest that MSS2 might be involved in cell wall biogenesis. However, the growth of mss2Δ on medium supplemented with calcofluor white was different from that of the goa1Δ and nuo2Δ strains, which were highly sensitive to calcofluor white [Citation12,Citation41,Citation42,Citation44].
Figure 4. MSS2 deletion enhances resistance to calcofluor white and sensitivity to SDS. The mss2Δ strains did not exhibit changes in caspofungin sensitivity but showed increased resistance to the cell wall-perturbing agent calcofluor white and impaired growth on medium containing SDS
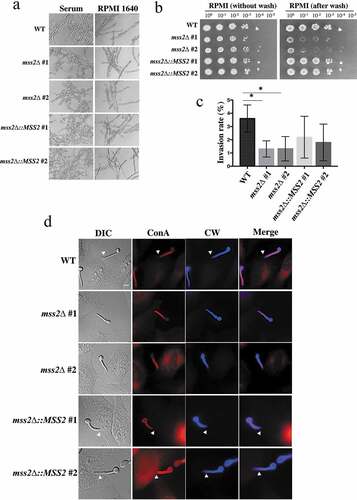
mss2Δ mutant strains exhibit a significantly impaired invasive growth ability on semisolid RPMI 1640 agar and in epithelial cells
Mitochondrial dysfunction can impair hyphal development, as deletion of GOA1, NUO1 and NUO2 results in decreased hyphal formation [Citation41,Citation42]. We sought to determine whether the mss2Δ strains exhibit similar phenotypes. However, the mss2Δ strains exhibited no differences in hyphal formation in YPD + serum and RPMI 1640 liquid media (). Interestingly, that the mss2Δ strains exhibited less filamentous growth than the wild-type strain on RPMI 1640 semisolid agar. The results of agar invasion assays further demonstrated that the mss2Δ strains were more easily washed out than the wild-type and complemented strains (). We further evaluated whether C. albicans MSS2 is involved in the invasion of HeLa epithelial cells. Concanavalin A and calcofluor white were used to differentiate hyphae that contacted the epithelial cell surface and hyphae that had actually invaded the epithelial cells. As shown in and 5d, hyphae of the wild-type strain exhibited stronger invasive ability than those of mss2Δ strains, and the difference was statistically significant. The MSS2-complemented strains had partially recovered invasion rates (), implying that invasion ability might be MSS2 expression-dependent. These data indicate that MSS2 is required for invasive growth on semisolid agar surfaces and in epithelial cells.
Figure 5. Mss2δ mutant strains exhibit a significantly impaired invasive growth ability on solid agar. (a) MSS2 is not involved in hyphal formation in YPD + serum and RPMI 1640 liquid media. (b) MSS2 deletion strains of C. albicans show severe defects in invasive growth on semisolid RPMI 1640 agar. (c) Quantitative analysis of the invasion assays of HeLa cells shows that the presence of MSS2 in C. albicans is able to reinforce the invasion of HeLa epithelial cells. Values are the mean ± SD from three replicates. *, P < 0.05. (d) Representative images of C. albicans hyphae-invaded HeLa cells. Extracellular hyphae were stained with Concanvalin A (red), whereas both extra- and intracellular hyphae were stained with calcofluor white (blue) after the cell membranes of HeLa cells were permeabilized with Triton X-100. The arrows show hyphae invasion. Scale bar: 10 μm
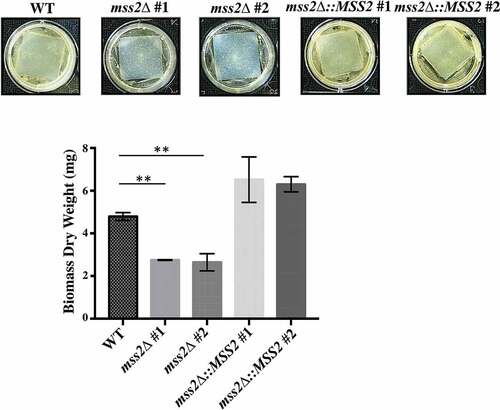
MSS2 deletion strains cannot form robust biofilms
Given that hyphal development on solid surfaces is a key event in successful biofilm formation by C. albicans, biofilm formation was evaluated in the mss2Δ strains. Compared with the wild-type strain (5.62 mg), the mss2Δ strains exhibited a significant reduction in biofilm biomass (2.99 and 2.56 mg) (). Reintroduction of a functional MSS2 gene into each C. albicans mss2Δ mutant restored biofilm formation (7.19 and 7.37 mg) ().
Figure 6. MSS2 deletion strains cannot form robust biofilms. The results of biofilm assays with the C. albicans wild-type, mss2Δ and complemented strains on silicone squares showed that the MSS2 gene is necessary for biofilm development. The presented values are the means ± SDs of the values from three independent experiments with three replicates each. **, P< 0.01
Mss2 regulates the expression of the core biofilm regulatory genes TEC1 and NDT80
Central to the understanding of the mechanisms underlying C. albicans biofilm formation is the understanding that biofilm growth is regulated by a complex regulatory network involving several major transcription factors [16,17,18]. We further investigated whether six core biofilm regulatory factors in C. albicans regulate MSS2 expression and, conversely, whether Mss2 regulates the expression of these six factors. As shown in , deletion of BRG1, ROB1, NDT80, TEC1 or EFG1 did not significantly affect MSS2 expression during the biofilm development stage, whereas bcr1Δ induced MSS2 expression. Furthermore, TEC1 and NDT80 were significantly repressed in the mss2 null mutant (). These data demonstrated the role of MSS2 in biofilm formation.
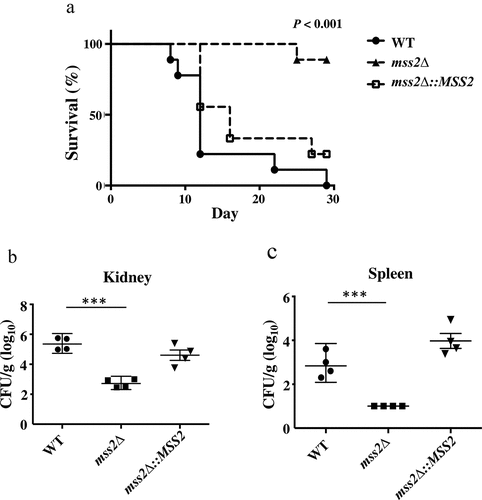
MSS2 gene deletion significantly reduces fungal virulence
The hyphal and biofilm formation abilities are the predominant virulence factors of C. albicans [Citation90,Citation91]. Thus, we further determined whether the C. albicans mss2 null mutant was associated with attenuated virulence in a murine model. As shown in , the virulence of the mss2Δ strain was significantly decreased compared to that of the wild-type strain (P < 0.001, log-rank test). Reintroduction of a functional MSS2 gene into the mss2Δ strain resulted in recovery of virulence. We further analyzed the fungal burden in murine kidneys and spleens. The fungal burdens were significantly decreased in the kidney (P < 0.01) and spleen (P < 0.05) in mice infected with the mss2Δ strain ( and 8c).
Figure 8. MSS2 deletion strains exhibit reduced virulence and result in lower fungal burdens. (a) Survival curves of mice after infection with 5 × 105 C. albicans cells revealed that mss2Δ is involved in pathogenicity. The fungal burden in the kidney (b) and spleen (c) in four mice per strain was measured on day 3 post infection with C. albicans. *, P< 0.05; **, P< 0.01
Discussion
Mitochondria function as a master organelle for energy production. The ability to utilize different carbon sources in glucose-limited environments and host niches is critical for the survival of fungal pathogens. However, mitochondria are also the central hub that coordinates multiple aspects of fungal biology, such as cell wall biogenesis, metabolite production, biofilm development, drug resistance and virulence [Citation1–3,Citation8,Citation9,Citation13,Citation41,Citation42,Citation44,Citation92]. However, the exact mechanisms underlying the ability of mitochondria to manipulate their flexibility to respond to changing environments remain largely unknown. C. albicans is a predominant pathogen affecting humans, particularly immunocompromised patients [Citation15,Citation16]. In this study, we identified 21 genes required for chitosan resistance by screening a library of protein kinase mutant strains. One antifungal mechanism of chitosan acts by targeting the microbial cell surface, leading to an inhibitory effect [Citation52,Citation55,Citation56,Citation93–95]. Indeed, 15 of the 21 genes were found to be involved in cell wall and cell membrane integrity (Supplementary Table 3), as the library mutants exhibited increased sensitivity to chitosan. Few studies have reported that chitosan might target mitochondria in C. albicans and S. cerevisiae [Citation54,Citation94]. Here, the MSS2 gene, an ortholog of S. cerevisiae MSS2, was determined to be involved in chitosan resistance in C. albicans. The functional role of MSS2 in C. albicans has not been studied. In S. cerevisiae, loss of MSS2 causes improper assembly of Cox2 [Citation57–59]. Thus, MSS2 might also control the assembly of the C. albicans cytochrome IV subunit proteins (protein complex) and be required for respiratory activity. To the best of our knowledge, this report is the first to clarify the role of MSS2 in C. albicans and provide direct evidence that the antifungal mechanism of chitosan acts against this fungus by antagonizing mitochondrial activity. However, we cannot rule out the combinatorial effects of several signaling pathways required for the chitosan response, which may thereby contribute to the impacts of chitosan.
Although the links between cell wall biogenesis and mitochondrial function remain largely unknown, mitochondrial dysfunction resulting in fungal cell wall defects has been reported [Citation1,Citation10,Citation12,Citation41,Citation42,Citation44]. Specifically, mutants with loss of GOA1, NOU1, NOU2, NDH51, RBF1 or HFL1 are sensitive to calcofluor white and other cell surface-perturbing agents [Citation12,Citation41,Citation42,Citation44]. Furthermore, GOA1, NUO1, NUO2 and NDH51 are involved in cell wall-related pathways [Citation41,Citation42,Citation44]. Our results showed that the growth of the mss2Δ strains was not affected upon caspofungin treatment, suggesting that the of cell wall β-1,3 glucan content was unaffected in the mutant strains. Interestingly, unlike mutants of the known mitochondrial genes, mss2 null mutants exhibited enhanced resistance to calcofluor white (a cell wall-perturbing agent) but sensitivity to SDS (a cell membrane-perturbing agent). These data suggest that the cell membrane of mss2Δ might be more vulnerable and that the cell wall of mss2Δ might contain more chitin, as calcofluor white binds mainly to chitin [Citation86,Citation87]. These results further indicate that the growth inhibitory effect of chitosan on the mss2Δ strains could be due to damage to the cell membrane and mitochondrial biogenesis.
The ability of C. albicans to undergo the yeast-to-hyphal transition is important in disseminated candidiasis and tissue invasion [Citation21,Citation25]. Unlike the well-known signaling pathways mediating this morphological transition, the precise mitochondrial functions involved in the complex cellular pathways that control hyphal development remain obscure. In general, mitochondrial dysfunction inhibits hyphal formation in C. albicans [Citation1,Citation3,Citation41,Citation42], but the effect of respiratory deficiency on filamentation varies with the inducing environment [Citation9,Citation42]. Interestingly, we found that mss2Δ retained a normal filamentation ability in both YPD + serum and RPMI 1640 liquid medium but exhibited a striking defect in invasive ability in vitro and ex vivo, which may lead to reduced biofilm formation and virulence and have a potential impact on the mucosal infection of this mutant. These data suggest that in addition to the yeast-to-hyphal transition, specific regulatory mechanisms are required for invasive growth on abiotic and biotic surfaces. Indeed, adhesion, turgor-driven force and proteolytic enzyme-mediated mechanisms are directly involved in successful invasion of host cells [Citation96]. Although many genes have been shown to be required for invasive growth on contact surfaces, most genes are also associated with hyphal development in liquid medium; these genes include RAS1, CDC24, RSR1, BUD2, HWP1, MKC1 and many others [Citation97–99]. Interestingly, C. albicans RHR2, encoding a glycerol 3-phosphatase, is required for maintaining intracellular hydrostatic pressure (turgor pressure). RHR2 deletion strains exhibit no defect in hyphal formation [Citation100] but do exhibit adherence defects and reduced biofilm formation [Citation101,Citation102]. Similarly, bcr1Δ exhibits normal hyphal growth in liquid medium but reduced virulence due to its substrate adhesion deficiency and decreased biofilm formation [Citation64,Citation103]. The cell wall protein Dfip is involved in both invasive growth on semisolid agar and virulence, but the null mutant exhibits normal hyphal formation and adhesion ability [Citation104]. These data suggest that MSS2 deletion might repress invasive growth, thereby impacting on biofilm formation and virulence. How MSS2 mediates the invasion process during interactions with abiotic and biotic surfaces and what genes are specifically involved in this process remain open questions.
Biofilms are highly associated with Candida infections. In C. albicans, a core complex regulatory network is critical for biofilm formation [Citation75,Citation105,Citation106]. Here, we showed that the mss2Δ mutant exhibited significantly downregulated expression of TEC1 and NDT80. Previous reports have shown that TEC1 is involved in the interaction with epithelial cells [Citation97], suggesting that the MSS2 deletion strain might exhibit reduced adherence ability in vivo. In addition, we found high levels of MSS2 expression in bcr1Δ. The results imply that the adherence-related transcription factor Bcr1 regulates MSS2 directly or indirectly [Citation88]. Furthermore, the C. albicans ssn3Δ strain displays enhanced biofilm formation and mitochondrial respiration compared with the wild-type strain [Citation107]. Thus, the reduced biofilm formation of the mss2Δ strains may arise from mitochondrial deficiency. Notably, it has been suggested that mitochondrial activity is necessary and beneficial for biofilm formation in the early stage, as the results of some transcriptomic and metabolomic studies showed decreased mitochondrial activity in mature biofilms [Citation75],105[Citation108].
Mitochondria in C. albicans are important contributors to energy production, virulence, drug resistance and cell wall integrity; thus, the respiratory chain is an attractive target for the development of drugs against human fungal pathogens [Citation1,Citation2,Citation10]. Currently, numerous respiratory chain inhibitors have shown considerable antifungal activity [Citation3,Citation11]. Chitosan is a biodegradable and nontoxic natural polysaccharide [Citation50,Citation93], and its broad-spectrum antifungal mechanisms that could target the cell surface and mitochondria and exert several intracellular actions against human pathogens endow it with substantial commercial potential [Citation52,Citation54–56,Citation61,Citation93]. These factors indicate that chitosan is a promising molecule for future antifungal drug development either alone or in combination with current antifungal drugs.
Supplemental Material
Download Zip (44.5 KB)Acknowledgments
We acknowledge the Laboratory Animal Resource Center of National Taiwan University for assistance with the generation of animal study results.
Supplementary material
Supplemental data for this article can be accessed here.
Disclosure statement
The authors have no conflicts of interest to declare.
Additional information
Funding
References
- Calderone R, Li D, Traven A. System-level impact of mitochondria on fungal virulence: to metabolism and beyond. FEMS Yeast Res. 2015;15:fov027.
- McBride HM, Neuspiel M, Mitochondria WS. More than just a powerhouse. Curr Biol. 2006;16:R551–R560.
- Shingu-Vazquez M, Traven A. Mitochondria and fungal pathogenesis: drug tolerance, virulence, and potential for antifungal therapy. Eukaryot Cell. 2011;10:1376–1383.
- Valero T. Mitochondrial biogenesis: Pharmacological approaches. Curr Pharm Des. 2014;20:5507–5509.
- Griffiths KK, Levy RJ. Evidence of mitochondrial dysfunction in autism: Biochemical links, henetic-based associations, and non-energy-related mechanisms. Oxid Med Cell Longev. 2017;2017:4314025.
- Dorn GW 2nd, Vega RB, Kelly DP. Mitochondrial biogenesis and dynamics in the developing and diseased heart. Genes Dev. 2015;29:1981–1991.
- Lesnefsky EJ, Moghaddas S, Tandler B, et al. Mitochondrial dysfunction in cardiac disease: Ischemia–reperfusion, aging, and heart failure. J Mol Cell Cardiol. 2001;33:1065–1089.
- Verma S, Shakya VPS, Idnurm A. Exploring and exploiting the connection between mitochondria and the virulence of human pathogenic fungi. Virulence. 2018;9:426–446.
- Huang XH, Chen XQ, He YM, et al. Mitochondrial complex I bridges a connection between regulation of carbon flexibility and gastrointestinal commensalism in the human fungal pathogen Candida albicans. PLoS Pathog. 2017;13:e1006414.
- Koch B, Traven A. Mitochondrial control of fungal cell walls: models and relevance in fungal pathogens. Curr Top Microbiol Immunol. 2020;425:277-296.
- Duvenage L, Munro CA, Gourlay CW. The potential of respiration inhibition as a new approach to combat human fungal pathogens. Curr Genet. 2019;65:1347–1353.
- Khamooshi K, Sikorski P, Sun N, et al. The Rbf1, Hfl1 and Dbp4 of Candida albicans regulate common as well as transcription factor-specific mitochondrial and other cell activities. BMC Genomics. 2014;15:56.
- Sun N, Parrish RS, Calderone RA, et al. Unique, diverged, and conserved mitochondrial functions influencing Candida albicans respiration. mBio. 2019;10:e00300-00319.
- Neville BA. d’Enfert C, Bougnoux ME. Candida albicans commensalism in the gastrointestinal tract. FEMS Yeast Res. 2015;15:fov081.
- Papon N, Courdavault V, Clastre M, et al. Emerging and emerged pathogenic Candida species: Beyond the Candida albicans paradigm. PLoS Pathog. 2013;9:e1003550.
- Cassone A, Cauda R. Candida and candidiasis in HIV-infected patients: where commensalism, opportunistic behavior and frank pathogenicity lose their borders. AIDS. 2012;26:1457–1472.
- Noble SM, Gianetti BA, Witchley JN. Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat Rev Microbiol. 2017;15:96–108.
- Tao L, Du H, Guan G, et al. Discovery of a “white-gray-opaque” tristable phenotypic switching system in Candida albicans: Roles of non-genetic diversity in host adaptation. PLoS Biol. 2014;12:e1001830. .
- Ene IV, Lohse MB, Vladu AV, et al. Phenotypic profiling reveals that Candida albicans opaque cells represent a metabolically specialized cell state compared to default white cells. mBio. 2016;7:e01269-16.
- Thewes S, Moran GP, Magee BB, et al. Phenotypic screening, transcriptional profiling, and comparative genomic analysis of an invasive and non-invasive strain of Candida albicans. BMC Microbiol. 2008;8:187.
- Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9:737–748.
- Pande K, Chen CB, Noble SM. Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat Genet. 2013;45:1088–1091.
- Sasse C, Hasenberg M, Weyler M, et al. White-opaque switching of Candida albicans allows immune evasion in an environment-dependent fashion. Eukaryot Cell. 2013;12:50–58.
- Slutsky B, Staebell M, Anderson J, et al. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol. 1987;169:189–197.
- Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4:119–128.
- Stoldt VR, Sonneborn A, Leuker CE, et al. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. Embo J. 1997;16:1982–1991.
- Kadosh D, Johnson AD. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: A genome-wide analysis. Mol Biol Cell. 2005;16:2903–2912.
- Lo HJ, Kohler JR, DiDomenico B, et al. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949.
- Leberer E, Harcus D, Broadbent ID, et al. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc Natl Acad Sci U S A. 1996;93:13217–13222.
- Nantel A, Dignard D, Bachewich C, et al. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol Biol Cell. 2002;13:3452–3465.
- Braun BR, Johnson AD. TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics. 2000;155:57–67.
- Taschdjian CL, Burchall JJ, Kozinn PJ. Rapid identification of Candida albicans by filamentation on serum and serum substitutes. AMA J Dis Child. 1960;99:212–215.
- Simonetti N, Strippoli V, Cassone A. Yeast-mycelial conversion induced by N-acetyl-D-glucosamine in Candida albicans. Nature. 1974;250:344–346.
- Mardon D, Balish E, Phillips AW. Control of dimorphism in a biochemical variant of Candida albicans. J Bacteriol. 1969;100:701–707.
- Buffo J, Herman MA, Soll DR. A characterization of pH-regulated dimorphism in Candida albicans. Mycopathologia. 1984;85(1–2):21–30.
- Nobile CJ, Johnson AD. Candida albicans biofilms and human disease. Annu Rev Microbiol. 2015;69:71–92.
- Finkel JS, Mitchell AP. Genetic control of Candida albicans biofilm development. Nat Rev Microbiol. 2011;9:109–118.
- Marcet-Houben M, Marceddu G, Gabaldon T. Phylogenomics of the oxidative phosphorylation in fungi reveals extensive gene duplication followed by functional divergence. BMC Evol Biol. 2009;9:295.
- Gabaldon T, Rainey D, Huynen MA. Tracing the evolution of a large protein complex in the eukaryotes, NADH: Ubiquinone oxidoreductase (Complex I). J Mol Biol. 2005;348:857–870.
- Aguileta G, de Vienne DM, Ross ON, et al. High variability of mitochondrial gene order among fugi. Genome Biol Evol. 2014;6:451–465. .
- She X, Khamooshi K, Gao Y, et al. Fungal-specific subunits of the Candida albicans mitochondrial complex I drive diverse cell functions including cell wall synthesis. Cell Microbiol. 2015;17:1350–1364.
- Bambach A, Fernandes MP, Ghosh A, et al. Goa1p of Candida albicans localizes to the mitochondria during stress and is required for mitochondrial function and virulence. Eukaryot Cell. 2009;8:1706–1720. .
- Sun N, Fonzi W, Chen H, et al. Azole susceptibility and transcriptome profiling in Candida albicans mitochondrial electron transport chain complex I mutants. Antimicrob Agents Chemother. 2013;57:532–542. .
- She X, Calderone R, Kruppa M, et al. Cell wall N-linked mannoprotein biosynthesis requires Goa1p, a putative regulator of mitochondrial complex I in Candida albicans. PLoS One. 2016;11:e0147175.
- Yamada K, Akiba Y, Shibuya T, et al. Water purification through bioconversion of phenol compounds by tyrosinase and chemical adsorption by chitosan beads. Biotechnol Prog. 2005;21:823–829.
- Shahidi F, Arachchi JKV, Jeon YJ. Food applications of chitin and chitosans. Trends Food Sci Technol. 1999;10:37–51.
- Kumar MNVR. A review of chitin and chitosan applications. React Funct Polym. 2000;46:1–27.
- Kim HJ, Chen F, Wang X, et al. Effect of chitosan on the biological properties of sweet basil (Ocimum basilicum L.). J Agric Food Chem. 2005;53:3696–3701.
- Haque T, Chen H, Ouyang W, et al. Superior cell delivery features of poly(ethylene glycol) incorporated alginate, chitosan, and poly-L-lysine microcapsules. Mol Pharm. 2005;2:29–36.
- Cheung RC, Ng TB, Wong JH, et al. Chitosan: an update on potential biomedical and pharmaceutical applications. Mar Drugs. 2015;13:5156–5186.
- Sudarshan NR, Hoover DG, Knorr D. Antibacterial action of chitosan. Food Biotechnol. 1992;6:257–272.
- Rabea EI, Badawy MET, Stevens CV, et al. Chitosan as antimicrobial agent: Applications and mode of action. Biomacromolecules. 2003;4:1457–1465.
- Raafat D, von Bargen K, Haas A, et al. Insights into the mode of action of chitosan as an antibacterial compound. Appl Environ Microbiol. 2008;74:3764–3773.
- Pena A, Sanchez NS, Calahorra M. Effects of chitosan on Candida albicans : Conditions for its antifungal activity. Biomed Res Int. 2013;2013:527549.
- Hosseinnejad M, Jafari SM. Evaluation of different factors affecting antimicrobial properties of chitosan. Int J Biol Macromol. 2016;85:467–475.
- Shih PY, Liao YT, Tseng YK, et al. A potential antifungal effect of chitosan against Candida albicans is mediated via the inhibition of SAGA complex component expression and the subsequent alteration of cell surface integrity. Front Microbiol. 2019;10:602.
- Simon M, Seraphin B, Faye G. The nuclear-encoded MSS2 gene is involved in the expression of the mitochondrial cytochrome-c oxidase subunit 2 (Cox2). Biochim Biophys Acta. 1995;1228:95–98.
- Saracco SA, Fox TD. Cox18p is required for export of the mitochondrially encoded Saccharomyces cerevisiae cox2p C-tail and interacts with Pnt1p and Mss2p in the inner membrane. Mol Biol Cell. 2002;13:1122–1131.
- Broadley SA, Demlow CM, Fox TD. Peripheral mitochondrial inner membrane protein, Mss2p, required for export of the mitochondrially coded Cox2p C tail in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:7663–7672.
- Liang SH, Cheng JH, Deng FS, et al. A novel function for Hog1 stress-activated protein kinase in controlling white-opaque switching and mating in Candida albicans. Eukaryot Cell. 2014;13:1557–1566.
- Alburquenque C, Bucarey SA, Neira-Carrillo A, et al. Antifungal activity of low molecular weight chitosan against clinical isolates of Candida spp. Med Mycol. 2010;48:1018–1023.
- Chien HF, Chen CP, Chen YC, et al. The use of chitosan to enhance photodynamic inactivation against Candida albicans and its drug-resistant clinical isolates. Int J Mol Sci. 2013;14:7445–7456.
- Richard ML, Nobile CJ, Bruno VM, et al. Candida albicans biofilm-defective mutants. Eukaryot Cell. 2005;4:1493–1502.
- Nobile CJ, Mitchell AP. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr Biol. 2005;15:1150–1155.
- Norice CT, Smith FJ Jr., Solis N, et al. Requirement for Candida albicans Sun41 in biofilm formation and virulence. Eukaryot Cell. 2007;6:2046–2055.
- Rauceo JM, Blankenship JR, Fanning S, et al. Regulation of the Candida albicans cell wall damage response by transcription factor Sko1 and PAS kinase Psk1. Mol Biol Cell. 2008;19:2741–2751.
- Davis DA, Bruno VM, Loza L, et al. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics. 2002;162:1573–1581.
- Reuss O, Vik A, Kolter R, et al. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127.
- Zhang P, Li H, Cheng J, et al. Respiratory stress in mitochondrial electron transport chain complex mutants of Candida albicans activates Snf1 kinase response. Fungal Genet Biol. 2018;111:73–84.
- Song Y, Li S, Zhao Y, et al. ADH1 promotes Candida albicans pathogenicity by stimulating oxidative phosphorylation. Int J Med Microbiol. 2019;309:151330.
- Chang WH, Liang SH, Deng FS, et al. The conserved dual phosphorylation sites of the Candida albicans Hog1 protein are crucial for white-opaque switching, mating, and pheromone-stimulated cell adhesion. Med Mycol. 2016;54:628–640.
- Zupan J, Raspor P. Quantitative agar-invasion assay. J Microbiol Methods. 2008;73:100–104.
- Almeida RS, Brunke S, Albrecht A, et al. The hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog. 2008;4:e1000217.
- Maza PK, Bonfim-Melo A, Padovan ACB, et al. Candida albicans: The ability to invade epithelial cells and survive under oxidative stress is unlinked to hyphal length. Front Microbiol. 2017;8:1235.
- Nobile CJ, Fox EP, Nett JE, et al. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148:126–138.
- Cruz MC, Goldstein AL, Blankenship JR, et al. Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J. 2002;21:546–559.
- Eisman B, Alonso-Monge R, Roman E, et al. The Cek1 and Hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryot Cell. 2006;5:347–358.
- Navarro-Garcia F, Sanchez M, Pla J, et al. Functional characterization of the MKC1 gene of Candida albicans, which encodes a mitogen-activated protein kinase homolog related to cell integrity. Mol Cell Biol. 1995;15:2197–2206.
- Roman E, Alonso-Monge R, Miranda A, et al. The Mkk2 MAPKK regulates cell wall biogenesis in cooperation with the Cek1-pathway in Candida albicans. PLoS One. 2015;10:e0133476.
- Huang G, Huang Q, Wei Y, et al. Multiple roles and diverse regulation of the Ras/cAMP/protein kinase A pathway in Candida albicans. Mol Microbiol. 2019;111:6–16.
- Bruno VM, Mitchell AP. Regulation of azole drug susceptibility by Candida albicans protein kinase CK2. Mol Microbiol. 2005;56:559–573.
- Jung SI, Rodriguez N, Irrizary J, et al. Yeast casein kinase 2 governs morphology, biofilm formation, cell wall integrity, and host cell damage of Candida albicans. PLoS One. 2017;12:e0187721.
- Ror S, Panwar SL. Sef1-regulated iron regulon responds to mitochondria-dependent iron-sulfur cluster biosynthesis in Candida albicans. Front Microbiol. 2019;10:1528.
- Grahl N, Demers EG, Lindsay AK, et al. Mitochondrial activity and Cyr1 are key regulators of Ras1 activation of C. albicans virulence pathways. PLoS Pathog. 2015;11:e1005133. .
- Alonso-Monge R, Carvaihlo S, Nombela C, et al. The Hog1 MAP kinase controls respiratory metabolism in the fungal pathogen Candida albicans. Microbiology. 2009;155:413–423.
- Ram AFJ, Wolters A, Hoopen RT, et al. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to calcofluor white. Yeast. 1994;10:1019–1030.
- Roncero C, Durán A. Effect of calcofluor white and congo red on fungal cell wall morphogenesis: In vivo activation of chitin polymerization. J Bacteriol. 1985;163:1180–1185.
- Bachmann SP, VandeWalle K, Ramage G, et al. In Vitro activity of caspofungin against Candida albicans biofilms. Antimicrob Agents Chemother. 2002;46:3591–3596. .
- Fujimoto K. Freeze-fracture replica electron microscopy combined with SDS digestion for cytochemical labeling of integral membrane proteins. Application to the immunogold labeling of intercellular junctional complexes. J Cell Sci. 1995;108:3443–3449.
- Cuellar-Cruz M, Lopez-Romero E, Villagomez-Castro JC, et al. Candida species: new insights into biofilm formation. Future Microbiol. 2012;7:755–771.
- Negri M, Silva S, Henriques M, et al. Insights into Candida tropicalis nosocomial infections and virulence factors. Eur J Clin Microbiol Infect Dis. 2012;31:1399–1412.
- She X, Zhang L, Chen H, et al. Cell surface changes in the Candida albicans mitochondrial mutant goa1Δ are associated with reduced recognition by innate immune cells. Cell Microbiol. 2013;15:1572–1584.
- Kong M, Chen XG, Xing K, et al. Antimicrobial properties of chitosan and mode of action: A state of the art review. Int J Food Microbiol. 2010;144:51–63.
- Lopez-Moya F, Suarez-Fernandez M, Lopez-Llorca LV. Molecular mechanisms of chitosan interactions with fungi and plants. Int J Mol Sci. 2019;20:332.
- Zakrzewska A, Boorsma A, Brul S, et al. Transcriptional response of Saccharomyces cerevisiae to the plasma membrane-perturbing compound chitosan. Eukaryot Cell. 2005;4:703–715.
- Desai JV. Candida albicans hyphae: From growth initiation to invasion. J Fungi (Basel). 2018;4:10.
- Wachtler B, Wilson D, Haedicke K, et al. From attachment to damage: Defined genes of Candida albicans mediate adhesion, invasion and damage during interaction with oral epithelial cells. PLoS One. 2011;6:e17046.
- Kumamoto CA. A contact-activated kinase signals Candida albicans invasive growth and biofilm development. Proc Natl Acad Sci U S A. 2005;102:5576–5581.
- Bassilana M, Blyth J, Arkowitz RA. Cdc24, the GDP-GTP exchange factor for Cdc42, is required for invasive hyphal growth of Candida albicans. Eukaryot Cell. 2003;2:9–18.
- Bonhomme J, Chauvel M, Goyard S, et al. Contribution of the glycolytic flux and hypoxia adaptation to efficient biofilm formation by Candida albicans. Mol Microbiol. 2011;80:995–1013.
- Desai JV, Cheng S, Ying T, et al. Coordination of Candida albicans invasion and infection functions by phosphoglycerol phosphatase Rhr2. Pathogens. 2015;4:573–589. .
- Desai JV, Bruno VM, Ganguly S, et al. Regulatory role of glycerol in Candida albicans biofilm formation. mBio. 2013;4:e00637–00612.
- Nobile CJ, Andes DR, Nett JE, et al. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2006;2:e63.
- Zucchi PC, Davis TR, Kumamoto CA. A Candida albicans cell wall-linked protein promotes invasive filamentation into semi-solid medium. Mol Microbiol. 2010;76:733–748.
- Fox EP, Bui CK, Nett JE, et al. An expanded regulatory network temporally controls Candida albicans biofilm formation. Mol Microbiol. 2015;96:1226–1239.
- Lohse MB, Gulati M, Johnson AD, et al. Development and regulation of single- and multi-species Candida albicans biofilms. Nat Rev Microbiol. 2018;16:19–31.
- Lindsay AK, Morales DK, Liu Z, et al. Analysis of Candida albicans mutants defective in the Cdk8 module of mediator reveal links between metabolism and biofilm formation. PLoS Genet. 2014;10:e1004567.
- Zhu Z, Wang H, Shang Q, et al. Time course analysis of Candida albicans metabolites during biofilm development. J Proteome Res. 2013;12:2375–2385.