ABSTRACT
Candida auris is an emerging multidrug-resistant fungal pathogen responsible for nosocomial outbreaks of invasive candidiasis. Although several studies on the pathogenicity of this species have been reported, the knowledge on C. auris virulence is still limited. This study aims to analyze the pathogenicity of C. auris, using one aggregating isolate and eleven non-aggregating isolates from different clinical origins (blood, urine and oropharyngeal specimens) in two alternative host models of candidiasis: Caenorhabditis elegans and Galleria mellonella. Furthermore, possible associations between virulence, aggregation, biofilm-forming capacity, and clinical origin were assessed. The aggregating phenotype isolate was less virulent in both in vivo invertebrate infection models than non-aggregating isolates but showed higher capacity to form biofilms. Blood isolates were significantly more virulent than those isolated from urine and respiratory specimens in the G. mellonella model of candidiasis. We conclude that both models of candidiasis present pros and cons but prove useful to evaluate the virulence of C. auris in vivo. Both models also evidence the heterogeneity in virulence that this species can develop, which may be influenced by the aggregative phenotype and clinical origin.
Introduction
Candida albicans is the predominant etiological agent of invasive candidiasis [Citation1,Citation2]. However, recently Candida auris is emerging as a major cause of invasive nosocomial infections with multidrug-resistance and high morbidity and mortality [Citation3,Citation4]. Since this species was first described in 2009, several clinical presentations, such as colonization, mucosal infection or bloodstream infection have been reported worldwide, mostly associated with outbreaks in surgical and medical intensive care units (ICU) [Citation5,Citation6]. Phenotypic and genotypic characterization studies of C. auris isolates from different countries have revealed the high diversity of this species identifying up to five possible different phylogeographical clades, four clearly distinct and a potential fifth clade, around the world [Citation6,Citation7].
A wide array of virulence mechanisms is encoded in the genome of C. auris [Citation8]. Expression of phospholipase, proteinase, and hemolytic activities, adherence and biofilm formation, antifungal drug and environmental stress resistance are described among the most relevant virulence traits [Citation9–11]. However, the knowledge on C. auris virulence remains scarce and further studies are required to understand its pathogenic behavior, which has been reported to be substantially variable among clades [Citation12]. Currently, about 30 studies have incorporated in vivo models in the analysis of C. auris virulence, murine models being the most widely used [Citation13–18]. However, ethical and economical issues for using these models of candidiasis are encouraging the use of alternative systems such as invertebrate models [Citation19,Citation20]. Several virulence studies of C. auris infections have been reported in invertebrate models using Caenorhabditis elegans, Drosophila melanogaster, Danio rerio or Galleria mellonella [Citation11,Citation21–29]. Moreover, non-mammalian hosts have been successfully implemented for studying host-pathogen interactions and invasive candidiasis caused by other Candida species, and to evaluate the effectiveness of new therapeutic approaches [Citation11,Citation30–33].
Advances in the knowledge of the virulence potential of C. auris will contribute to the control of infections by this emerging pathogen. Recently, two different phenotypes, aggregating and non-aggregating (single-cell phenotype), have been described among C. auris clinical isolates; the latter presenting a higher virulence [Citation21,Citation22,Citation26]. Transcriptional profiles were evidently different between isolates with these two different phenotypes, and differences in the host response, depending on whether there is loss of tissue integrity, were also reported [Citation12]. Although attempts have been made to correlate the site of origin of clinical isolates with their virulence in different Candida species [Citation34–37], no association has been reported between the pathogenicity of C. auris and the origin of clinical specimens.
Therefore, the aim of this study was to analyze the virulence of C. auris isolates including both, aggregating and non-aggregating phenotypes that were retrieved from different clinical specimens. The ability of several C. auris isolates to form biofilms and produce hemolytic and enzymatic activity was assessed in vitro and the virulence traits of these isolates were probed in vivo using the invertebrate model hosts, C. elegans and G. mellonella. Our results demonstrate that both model organisms can be killed by aggregative as well as non-aggregative C. auris isolates. Moreover, this study reaffirmed that the variability among C. auris isolates as well as the site of infection and the different immune response of the host could affect this species virulence, being the C. auris blood isolates the most virulent.
Materials and methods
Candida auris isolates and growth conditions
Twelve clinical C. auris isolates from different patients suffering from candidiasis were analyzed (). These isolates included five from blood, five from urine, and two from oropharynx. All but one of the isolates came from patients of the Hospital Universitario y Politécnico La Fe of Valencia, Spain (Dr. Alba Ruiz Gaitán and Dr. Javier Pemán), three of them being registered in the CBS-KNAW culture collection of Westerdijk Fungal Biodiversity Institute. One blood isolate, C. auris JMRC:NRZ 1101 (Jena Microbial Resource Collection) was recovered from a patient attended at the Institut für Hygiene und Mikrobiologie, Würzburg, Germany (Dr. Oliver Kurzai).
Table 1. Biofilm biomass and metabolic activity levels of each C. auris isolate measured with Crystal violet and XTT reduction assays, respectively
C. auris isolates were stored in vials containing sterile distilled water at room temperature and cultured on Sabouraud dextrose agar (Difco, Becton Dickinson, USA) at 37°C for 24 h before use. The assays in the C. elegans model were performed by culturing C. auris on brain heart infusion (BHI, Panreac, Spain) agar plates supplemented with kanamycin (90 µg/ml) and incubated at 37°C for 24 h. C. auris isolates used for the G. mellonella model assay and for biofilm studies were cultured in yeast extract peptone dextrose broth (YEPD, Panreac) and incubated overnight at 30°C in shaking conditions.
Microscopic visualization
The cellular morphology of the 12 clinical C. auris isolates was observed microscopically. C. auris isolates were grown in YEPD broth for 24 h at 30°C in shaking conditions, washed three times with sterile phosphate-buffered saline solution (PBS, Sigma-Aldrich, USA) and collected by centrifugation at 2500 rpm for 10 min. Cell suspensions of each C. auris isolate were adjusted to a final concentration of 1 × 108 cells/ml with sterile PBS after cell counting by microscopy using a Burker hemocytometer. Microscopic appearance of each sample was visualized using a Nikon Eclipse 80i fluorescence microscope (Melville, NY, USA).
Biofilm development
The ability of C. auris isolates to develop biofilms was assessed with C. albicans strain SC5314 being included as control. Biofilms were produced in sterile, flat-bottomed honeycomb 100-well polystyrene microtiter plates (Labsystems, Finland). Precultured cells were harvested and washed three times with sterile PBS. After cell counting by microscopy, suspensions at a concentration of 1 × 106 cells/ml were prepared in RPMI 1640 medium supplemented with L-glutamine and buffered at pH 7 with 0.165 M 3-(N-morpholine) propanesulfonic acid, MOPS (Sigma-Aldrich). A volume of 100 µl of each cell suspension was dispensed into the wells of the plate. All outer wells from the first and last column of the plate were kept empty to avoid the “edge-effect”. Microplates were incubated at 37°C for 24 or 48 h. Afterward, to carefully eliminate planktonic and poorly adhered cells, spent RPMI was removed and the biofilms were washed three times by adding and removing 100 µl of sterile PBS.
Quantification of biofilm biomass was performed by Crystal violet (CV) staining [Citation38]. After drying the washed biofilms at room temperature for 30 min, 100 µl of 0.4% CV solution (Merck, Germany) was added to each well and incubated for 20 min at room temperature. Each well was then washed twice with 250 µl of sterile distilled water, and finally 150 µl of 33% acetic acid was added to solubilize the CV-stained biomass for spectrophotometric measurement.
Biofilm metabolic activity was evaluated by reduction of 2,3-bis (2-methoxy- 4-nitro-5-sulfophenyl)-5-[(phenylamino)-carbonyl]- 2 H-tetrazolium hydroxide (XTT, Sigma-Aldrich) [Citation39]. A volume of 100 µl of XTT with 1 μM of menadione was added to each washed well, and microplates were incubated in darkness for 2 h at 37°C.
Absorbance measurements were conducted with a BioScreen C MBR microplate reader (Growth Curves Ltd, Finland) at a wavelength of 600 nm for biomass quantification and at 492 nm for metabolic activity determination. Each experiment was performed with biological triplicates on separate days.
Determination of phospholipase, proteinase and hemolytic activities
Phospholipase activity was determined using a precipitation assay as described by Polak (1992) [Citation40] with malt agar plates supplemented with egg yolk [Citation41]. Phospholipase activity was defined as the ratio of the colony diameter compared to the total diameter of colony plus precipitation zone. C. albicans isolate UPV/EHU 04–125 was used as a control with high phospholipase activity.
Aspartyl proteinase activity was assayed as described by Cassone et al. (1987) [Citation42]. Proteinase activity was determined as the diameter of the lytic area around growing colonies. High aspartyl proteinase producer strain Candida dubliniensis UPV/EHU 00–134 was used as a control.
Hemolytic activity was analyzed as described by Luo et al. (2001) [Citation43] by performing the plate assay described by Manns et al. (1994) [Citation44]. C. albicans ATCC 90028 with high hemolytic activity was used as a control.
Enzymatic and hemolytic activities were analyzed at least with biological triplicates on separate days.
Survival assays in Caenorhabditis elegans
C. elegans strain AU37 (glp-4(bn2); sek-1(km4)) was obtained from the Caenorhabditis Genetics Center (University of Minnesota, USA). The double mutation in this strain generates nematodes that are not able to reproduce at 25°C (glp-4) and are more susceptible to infection (sek-1). The nematodes were kept in the laboratory at 15°C in plates with nematode growth medium (NGM, 3 g of NaCl, 17 g of agar, 2.5 g of peptone, 1 ml of 1 M CaCl2, 1 ml of 5 mg/ml cholesterol in ethanol, 1 ml of 1 M MgSO4, 25 ml 1 M KPO4, 975 ml H2O) seeded with the nonpathogenic Escherichia coli strain OP50 as nourishment. Prior to survival experiments, nematodes were synchronized to the same growth stage, as described by Ortega-Riveros et al. (2017) [Citation32]. Nematodes were then placed on BHI agar plates supplemented with kanamycin (90 µg/ml) and seeded with the isolate of C. auris to be assayed to feed the nematodes with the yeast [Citation45]. After incubating the plates at 25°C for 2 h, the nematodes were washed with M9 buffer (3 g of KH2PO4, 6 g of Na2HPO4, 5 g of NaCl, 1 ml of 1 M Mg SO4 and H2O to 1 l) supplemented with kanamycin (90 µg/ml) and placed in plates with NGM agar for 15 min to eliminate, by friction, C. auris cells that might have adhered to the nematode cuticle. Nematodes were placed in groups of 20 individuals in 24-well plates containing M9 buffer supplemented with 10 μg/ml of cholesterol in ethanol and kanamycin (90 µg/ml). Nematode survival was monitored every 24 h until 120 h with a stereomicroscope (Nikon SMZ-745, Japan). A minimum of 60 nematodes were used in each experiment for each C. auris isolate, and groups of uninfected nematodes were included as controls. The experiments were carried out in triplicate on different days.
Survival tests in Galleria mellonella
G. mellonella larvae with a weight between 0.3 and 0.5 g were obtained from Bichosa (Spain). Experiments were started one day after caterpillars arrived, which were placed in groups of 20 individuals in Petri dishes. The last left pro-leg of each larvae was cleaned with 70% ethanol and 10 µl of a C. auris suspension were inoculated with a precision syringe (Agilent, USA). Inoculum of C. auris cells was prepared by washing overnight yeast cultures with PBS supplemented with ampicillin (20 μg/ml) to remove remnants of YEPD and prevent bacterial contamination. Concentrations of 1 × 105, 1 × 106 and 1 × 107 cells/larva were assayed to monitor the virulence of each C. auris isolate. In all trials, two uninfected groups were used as controls; one with untouched larvae and a sham group with larvae inoculated with 10 µl PBS-ampicillin to observe a possible effect of the injection. Survival counts of alive and dead larvae were determined by visual inspection of movement and melanization every 24 h until 120 h after infection. Trials were conducted in triplicate on three different days.
Statistics
The quantitative results obtained in biofilm production were analyzed using the Student’s t test of the statistical program SPSS v24.0 (IBM, Chicago, IL, USA). The analysis of the virulence of C. auris isolates according to the origin of clinical specimens was analyzed by one-way ANOVA using SPSS v24.0. Survival curves were prepared with the Kaplan–Meier method using GraphPad Prism 5 software (GraphPad Software, La Jolla, CA, USA). Differences between the survival rates in both invertebrate models infected with C. auris were analyzed by the log-rank test of SPSS v24.0. For all statistical analyses values of p < 0.05 were considered statistically significant.
Results and discussion
In the present study, we characterized virulence traits of 12 clinical isolates of C. auris in vitro and compared their capability to infect and kill C. elegans and G. mellonella in vivo. These C. auris isolates, that were recovered from patients at two different locations (Spain and Germany), were observed microscopically, and only the blood isolate JMRC:NRZ 1101 from Germany displayed an aggregating phenotype, which was not observed in the other C. auris isolates from Spain (Figure S1).
Biofilm formation, enzymatic activity, and hemolytic activity of Candida auris
The ability of the 12 C. auris clinical isolates to develop biofilms is summarized in . None of the 12 isolates produced a biofilm denser than the C. albicans SC5314 control strain (). The ability to form a biofilm is an important factor in Candida pathogenicity because fungal cells within biofilms are protected from the action of the immune system and antifungal agents. The lower C. auris biofilm production compared to C. albicans SC5314 has been reported previously [Citation9]. Blood isolate JMRC:NRZ 1101 with aggregating phenotype showed the highest biofilm biomass production and metabolic activity (p ≤ 0.0001). The non-aggregating urine isolate CR14 produced the second most dense biofilm with a significantly higher biomass (p ≤ 0.0001) and metabolic activity (p ≤ 0.02, except at 48 h for most of isolates) than the other C. auris isolates. Although C. auris biofilms are thinner than those of C. albicans [Citation46], the capacity of C. auris to form a biofilm has been clearly associated with a lower antifungal susceptibility [Citation16,Citation47]. The ability of C. auris to form a biofilm has been reported for both aggregating and non-aggregating phenotypes [Citation12]. Biomass values obtained in the current study were similar to those reported by Sherry et al. (2017) [Citation22]. However, these authors reported the highest biofilm-forming capacity for non-aggregating isolates. Brown and coworkers [Citation12] also detailed differences in gene expression between both phenotypes: genes related to cellular components (membrane and cell wall constituents) were upregulated in isolates with aggregating phenotype whereas genes related to biological processes and metabolic functions were expressed higher in non-aggregating isolates. Moreover, Short et al. (2019) [Citation48] suggested that the ability of C. auris to form cellular aggregates increases survival of the yeast, which coincided with the upregulation of biofilm-associated genes. In addition, in C. albicans, the ability to produce biofilm has been associated with the expression of secreted aspartyl proteinases [Citation49]. Genes involved in biofilm formation and in the production of phospholipase, proteinase and hemolysin activity have been reported in C. auris, with genes involved in cell adhesion and invasion (ALS and SAP families) showing stronger expression in aggregating than in non-aggregating isolates [Citation8,Citation12,Citation13,Citation50]. However, in the present study, phospholipase and proteinase activities were not detected in any clinical isolate nor any hemolytic activity was observed (Table S1).
Virulence of Candida auris in Caenorhabditis elegans and Galleria mellonella
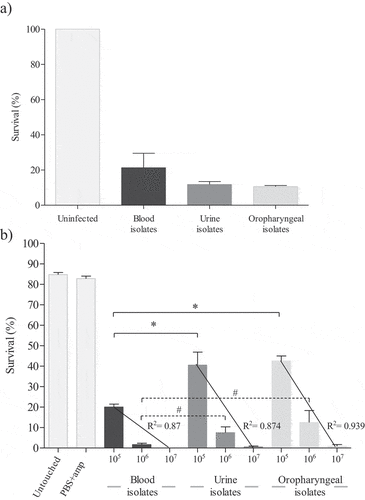
C. auris virulence has been studied in murine [Citation13–15,Citation17,Citation24], fish [Citation28], and fly models [Citation25]. However, there have only been a few studies in the nematode C. elegans [Citation23,Citation29] and the moth larvae G. mellonella [Citation21,Citation22,Citation24,Citation26,Citation27]. In the present study, all C. auris isolates were able to cause the death of at least 47.7% of C. elegans and G. mellonella individuals after 120 h ().
Figure 1. Survival average of C. elegans (a) and G. mellonella (b) at 120 hours post-infection with twelve different C. auris isolates according to the origin of the clinical specimen. Untouched control groups of nematodes and larvae and a group of larvae inoculated with PBS and ampicillin were also included. Larvae of G. mellonella were infected with three inocula of C. auris isolates (1 × 105, 1 × 106 and 1 × 107 cells/larva) and correlation coefficients were calculated. Absent bars indicate 0% survival rate of G. mellonella. Error bars represent standard errors. Solid lines with asterisks denote statistically significant differences of the inoculum 1 × 105 cells/larva between C. auris blood isolates and the other C. auris isolates from urine and oropharyngeal. Dotted lines with hashtag denote statistically significant differences of the inoculum 1 × 106 cells/larva between C. auris blood isolates, without including the aggregative German isolate, and the other C. auris isolates from urine and oropharyngeal. The statistical analysis was performed using the one-way ANOVA test (p < 0.05)
Uninfected C. elegans nematodes used as controls remained viable (100% survival) during the 120 h. A high amount of non-aggregating yeast cells was observed at 120 h post-infection for all C. auris isolates, except for the C. auris JMRC:NRZ 1101 isolate that caused yeast aggregates at 120 h post-infection in C. elegans (Figure S2). This cellular pattern was also observed in infected G. mellonella (data not shown). These results are consistent with those reported by Borman et al. (2016) [Citation21] who first described the formation of aggregates by some C. auris isolates, which later was corroborated by Sherry et al. (2017) [Citation22] and Muñoz et al. (2020) [Citation11]. These studies reported that C. auris did not form hyphae unlike C. albicans in the G. mellonella model of candidiasis, and that non-aggregating C. auris isolates were more virulent, even when compared to some C. albicans isolates [Citation11,Citation21,Citation22]. In a murine model, Ben-Ami et al. (2017) [Citation13] also identified yeast cells during the infection and recovered aggregates from murine tissues. Yue et al. (2018) [Citation15] observed a filamentous morphology of C. auris in cultures on YEPD medium from liver, kidney, brain, lung, and spleen specimens of mice suffering from invasive candidiasis. However, two days of incubation at 30 °C followed by five additional days at 25 °C were needed to observe this morphological switch from yeast to hyphae.
Killing assays in G. mellonella were performed with inocula of 1 × 105, 1 × 106 and 1 × 107 cells/larva for each C. auris isolate. In both control groups, most larvae were alive by 120 h post-infection with survival averages of 85% ± 3.2% in the case of the untouched group control and 82.3% ± 3.6% in larvae inoculated with PBS-ampicillin (). Survival of G. mellonella significantly decreased when a higher inoculum of C. auris was administered, as reported for other species of Candida [Citation31,Citation51,Citation52]. There was a strong correlation (R2 ≥ 0.87) between the injected inocula of C. auris and the survival of G. mellonella. Interestingly, there were statistically significant differences between the virulence of C. auris blood isolates and that of urine and oropharyngeal isolates using the inoculum of 1 × 105 cells/larva (p ≤ 0.009). Differences were also detected with the inoculum of 1 × 106 cells/larva without considering the isolate with aggregating phenotype (p ≤ 0.032) (). The inoculum of 1 × 106 cells/larva appeared the most appropriate for analyzing the virulence of C. auris in the G. mellonella model based on the findings with all the C. auris isolates at the three different inocula and the mortality rates obtained. Therefore, the results obtained with 1 × 106 cells/larva are presented and discussed in detail in the main text while those obtained with other inocula are presented in Figure S3. Inocula of 1 × 107 cells/larva caused the death of more than 60% of the larvae during the first 24 h of infection, and of more than 98.3% at 120 h (Figure S3). On the opposite, with the lowest inoculum of 1 × 105 cells/larva, none of the C. auris isolates had killed more than 60% of G. mellonella larvae at 48 h, with mortality ranging from 43.7% to 83.3% at 120 h (Figure S3). In this in vivo model the highest mortality was observed for blood isolates ().
Virulence of C. auris isolates according to the origin of the clinical specimens
Differences in the expression of virulence factors have been observed between C. auris isolates from different geographical origins, such as the lack of adhesion or few biofilm production associated with clade II isolates (East Asian clade), which frequently cause otitis [Citation12,Citation53,Citation54]. Several studies have associated the expression of specific virulence factors of Candida with the site of infection [Citation34–37]. In the present study, C. auris isolates were not grouped within any specific clade but the eleven isolates from the Spanish outbreak are phylogenetically close to clade III isolates (South African clade) [Citation5,Citation6]. This clade III, as well as clade II, is associated with bloodstream infections and with aggregate formation [Citation55].
We compared the virulence of the C. auris isolates according to their clinical origin in both in vivo models. The applied inoculum size of 1 × 106 cells/larva or lower (2.5–5 × 105 cells/larva) has also previously been used to establish the virulence of C. auris [Citation21,Citation22,Citation24,Citation26,Citation27] and other species of Candida [Citation30,Citation51,Citation52]. Strikingly, the C. auris isolates presented different virulence potential according to their clinical origin. In the G. mellonella model, the following general virulence categorization was observed when comparing the eleven non-aggregating isolates: blood > urine > oropharyngeal isolates with average survival percentages of infected larvae after 120 h of 1.7% for blood, 7.5% for urine, and 12.5% for oropharyngeal isolates. In contrast, mortality of C. elegans caused by infection with C. auris isolates was not clearly associated to their clinical origin. The average survival of nematodes infected with the non-aggregating C. auris isolates was very similar for the three groups of clinical isolates: 13.6% for the infection with blood isolates, 11.8% with urine isolates, and 10.6% with oropharyngeal isolates ().
The virulence of the five individual blood isolates, the five urine isolates and the two oropharyngeal isolates is presented in , and , respectively. The survival rates of G. mellonella infected with blood isolates ranged from 0% to 10% at 120 h, and from 7.6% to 52.3% in the C. elegans model. Moreover, most of the C. auris isolates killed more than 70% of the G. mellonella larvae at 24 h but took more than 96 h in the C. elegans model (). Survival rates for G. mellonella infected with urine isolates were between 1.7% and 18.3% at 120 h (7.8–17.2% in C. elegans). However, all urine isolates killed more than 50% of the G. mellonella larvae at 48 h, while in the case of C. elegans at least 96 h were required (). Finally, for the two oropharyngeal isolates there were no significant differences in virulence between the two non-mammalian models: survival rates of G. mellonella at 120 h ranged from 6.7% to 18.3%, whereas those of C. elegans were between 10% and 11.2%. Once again, in G. mellonella both isolates killed more than 60% of the larvae after 48 h but in C. elegans they took 96 h to eliminate the same percentage of host organisms (). In all cases, in the first 24–48 h post-infection C. auris caused higher mortality in the G. mellonella model than in the C. elegans model (). These results are comparable to those obtained in other studies, which observed that C. auris caused death of G. mellonella within 48 h [Citation21,Citation22] and of C. elegans between 48 and 96 h post-infection [Citation23,Citation29]. Our findings corroborate the idea that both models successfully demonstrate the virulence potential of C. auris, which was similar to or higher than that of other Candida species [Citation11,Citation30,Citation32,Citation33,Citation52]. The observed virulence of C. auris was comparable to C. albicans [Citation21,Citation22,Citation29] and higher than isolates from C. haemulonii complex species [Citation11,Citation29]. However, also within the species C. auris differences in virulence between isolates from different clades have been noted, probably associated with their genomic diversity [Citation12].
Figure 2. Survival curves of G. mellonella (a) and C. elegans (b) infected with C. auris blood isolates. Larvae of G. mellonella were infected with 1 × 106 cells/larva and the control groups used were a group of untouched larvae and larvae inoculated with PBS and ampicillin (PBS+amp) as a puncture (sham) control group. C. elegans worms were infected by C. auris cell ingestion for 2 h. c) Survival percentages at 120 h post-infection of G. mellonella and C. elegans infected with C. auris blood isolates. The C. auris isolates were sorted from highest to lowest survival percentages of C. elegans. Statistically significant differences in pathogenicity of C. auris blood isolates compared to the least virulent isolate JMRC:NRZ 1101 (* C. elegans; ** G. mellonella) and the second least virulent isolate CJ94 (#) calculated using the log-rank test (p < 0.05) are indicated. Absent bars indicate 0% survival rate
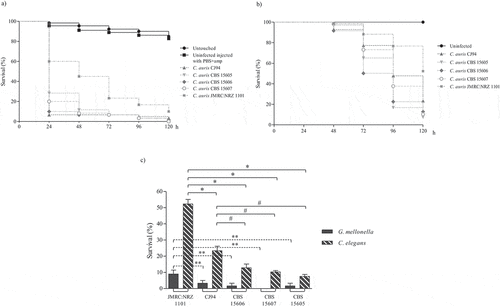
Figure 3. Survival curves of G. mellonella (a) and C. elegans (b) infected with C. auris urine isolates. Larvae of G. mellonella were infected with 1 × 106 cells/larva and the control groups used were a group of untouched larvae and larvae inoculated with PBS and ampicillin (PBS+amp) as a puncture (sham) control group. C. elegans worms were infected by C. auris cell ingestion for 2 h. c) Survival percentages at 120 hours post-infection of G. mellonella and C. elegans infected with C. auris urine isolates. The C. auris isolates were sorted from highest to lowest survival percentages of C. elegans. Statistically significant differences in pathogenicity of C. auris urine isolates compared to the least virulent isolate in G. mellonella, C. auris CR14 (*), and the highest virulent isolate in C. elegans, C. auris CR424 (#), calculated using the log-rank test (p < 0.05) are indicated
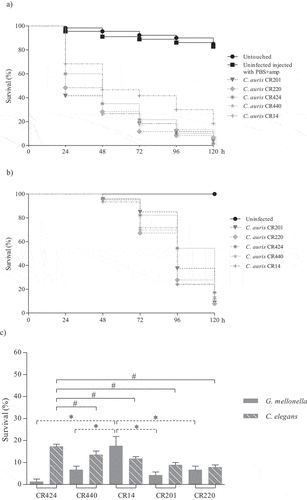
Figure 4. Survival curves of G. mellonella (a) and C. elegans (b) infected with C. auris oropharyngeal isolates. Larvae of G. mellonella were infected with 1 × 106 cells/larva and the control groups used were a group of untouched larvae and larvae inoculated with PBS and ampicillin (PBS+amp) as a puncture (sham) control group. C. elegans worms were infected by C. auris cell ingestion for 2 h. c) Survival percentages at 120 hours post-infection of G. mellonella and C. elegans infected with C. auris oropharyngeal isolates. The C. auris isolates were sorted from highest to lowest survival percentages of C. elegans.

The C. auris JMRC:NRZ 1101 blood isolate, with aggregating phenotype, was significantly less virulent than the other blood isolates in both models (p ≤ 0.0001) (Table S2 and S3). This isolate killed more than 80% of the G. mellonella larvae in 96 h, but it took 120 h to achieve a mortality rate of 52.3% of C. elegans nematodes (). Association of an aggregative phenotype of C. auris with lower pathogenicity compared to non-aggregating counterparts has also already been described earlier [Citation21,Citation22]. Furthermore, the virulence of this aggregating isolate in C. elegans was also significantly lower than that of all non-aggregating isolates from different infection sites (p ≤ 0.0001) ( and ; Table S3). In G. mellonella, the aggregating isolate caused a mortality comparable to that of most isolates from other clinical origins ( and Table S2) and only the mortality caused by this isolate was significantly lower than of urine isolate CR201 (p = 0.016).
Regarding non-aggregating C. auris isolates, blood isolate CBS15607 killed 100% of G. mellonella at 120 h but no significant differences were observed with other non-aggregating blood isolates (). High virulence was observed for four out of five urine isolates (80%), which killed G. mellonella with a mortality rate ranging from 80% to 93.3% between 72 and 120 h without significant differences among them (). The CR312 oropharyngeal isolate also killed 93.3% of larvae at 120 h (); however, its killing kinetics were similar to the two isolates with the lowest virulence, urine isolate CR14 and oropharyngeal isolate CR243, which achieved 81.7% mortality at 120 h. Conversely, there were significant differences between the latter two isolates and the aforementioned four urine isolates (p ≤ 0.006 and p ≤ 0.045, respectively). A higher virulence of Candida isolates from blood specimens has previously been reported [Citation35–37]. C. elegans infection by non-aggregating isolates caused a mortality rate of more than 76.6% at 120 h and C. auris isolates took more than 72 h to exceed a mortality of 50% (). Eldesouky et al. (2018) [Citation23] employed 30 min in the infection of worms and reported similar killing kinetics, whereas Lima et al. (2020) [Citation29] infected C. elegans for four hours and detected a mortality rate of more than 70% at 72 h. Blood isolate CBS15605 and urine isolate CR220 killed the highest percentage of nematodes at 120 h, 92.7% and 92.2%, respectively. However, no significant virulence differences were observed among them and the CBS15606 blood isolate and two urine isolates (CR440 and CR14) (Table S3). Five out of eleven isolates (45.5%) showed the same killing kinetics. Moreover, the oropharyngeal isolates also showed high lethality, causing a mortality rate of 88.8% at 120 h without significant differences with the mortality caused by the CBS15607 blood isolate. Strikingly, the less virulent isolates in the C. elegans model were the CJ94 blood isolate and the CR424 urine isolate with similar killing kinetics, both being significantly different to the rest of the isolates (Table S3). However, both isolates were among the five most virulent against G. mellonella, and their mortality rates in the C. elegans and G. mellonella models differed: 82.8% versus 98.3% for the blood isolate and 76.6% versus 96.7% for the urine isolate, respectively.
It is noteworthy that non-aggregating isolates were more pathogenic than the aggregating isolate and that the aggregating isolate used in this study formed more biofilm than the non-aggregating isolates. Sherry et al. (2017) [Citation22] reported the opposite, a higher biofilm formation capacity in non-aggregating than in aggregating isolates. The aggregating C. auris JMRC:NRZ 1101 blood isolate could be expressing genes implicated in biofilm formation and also genes involved with other virulence factors. The expression of these virulence genes could render a virulence of this isolate similar to that of some non-aggregating isolates, yielding similar killing kinetics during infection of G. mellonella. Transcriptional analysis of C. auris isolates revealed a high number of upregulated genes involved in biofilm formation and other pathogenic traits, which can be associated with the high resistance of this species to antifungal drugs [Citation10,Citation12]. In addition, differences in the infection profile and the host response to infection were observed for aggregating and non-aggregating phenotypes using a three-dimensional skin epithelial model. Although both phenotypes were more virulent in the presence of a wound, an aggregating isolate was more cytotoxic and proinflammatory than a non-aggregating one with more ability to evade the immune system [Citation12]. Trying to explain the differences found between both models of candidiasis, it is important to mention that in G. mellonella C. auris inoculum was injected parenterally, achieving higher concentrations in the tissues. Moreover, a higher lethal capacity of the non-aggregating isolates versus the aggregating isolate was noted in this model, likely due to the differential immune response of the host. Arias et al. (2020) [Citation26] suggested that, due to a higher ability to move and disseminate within G. mellonella tissues, yeast cells might be more virulent than aggregates of cells, with which we concur. In addition, C. elegans infection occurs by yeast ingestion during a defined time. Variations in exposure time consequently might result in a higher or lower fungal burden, leading to variations in the killing kinetics [Citation32]. It may also be possible that C. elegans has greater difficulty in ingesting cellular aggregates than individual yeasts. This fact could lead to a lower intake of fungal burden resulting in a milder infection rather than an aggregating isolate being less virulent. A stronger cytokine response and lower macrophage lysis capacity and neutrophil recruitment have been reported for C. auris compared to C. albicans [Citation18,Citation28]. Interestingly, structurally unique C. auris mannoproteins contribute to a strong innate host defense in all five C. auris clades, although clade-specific differences were observed, clade V being the least immunogenic [Citation28]. Both infection models, C. elegans and G. mellonella, present mechanisms and responses of the innate immune system that are conserved in mammals and may contribute to a better understanding of host-C. auris interactions.
Overall, in both models, isolates of C. auris behaved differently depending on their clinical origin, although the response to C. auris infection of the two models of candidiasis was also different. These differences in the virulence of C. auris according to the site of infection seem consistent with reports about the phenotypic plasticity of this species of Candida in response to environmental conditions, such as the passage through a mammalian body and variations in temperature to colonize a specific niche [Citation15]. However, further studies are required using other C. auris isolates with aggregating phenotype, if possible, from different geographic clades, to assess more accurately their virulence in vivo. There may be a level of heterogeneity in virulence among aggregating C. auris isolates similar to that observed for non-aggregating isolates in this and other studies [Citation12,Citation18].
In conclusion, we demonstrated that both C. elegans and G. mellonella models of candidiasis are simple and clearly appropriate to assess the virulence of C. auris isolates. Likewise, these models are useful to detect variations in the virulence of clinical isolates with different origin and/or capacity to form cell aggregates. The model host G. mellonella, which allows a more precise and direct inoculation of the pathogen into the host tissues, revealed a significantly higher virulence for C. auris isolates from blood specimens. Among the successful strategies of C. auris to cause infection are its ability to evade neutrophil attack and to resist treatments with commonly used antifungal drugs. Therefore, obtaining a more effective and accurate therapy is one of the main targets on which to focus actions against this pathogen. These low-cost and manageable in vivo models are promising tools to analyze host-pathogen interactions or the effectiveness of current and new antifungal drugs against C. auris.
Supplemental Material
Download MS Word (25.1 MB)Acknowledgments
Alba Ruiz Gaitán and Javier Pemán (Hospital Universitario y Politécnico La Fe of Valencia, Spain) and Oliver Kurzai (Institut für Hygiene und Mikrobiologie, Würzburg, Germany) are thanked for kindly providing clinical isolates. This work was supported by grants from the Spanish Ministry of Economy and Competitiveness (MINECO) [SAF2017-86188-P] and from the Consejería de Educación, Universidades e Investigación of Gobierno Vasco-Eusko Jaurlaritza [GIC15/78 IT-990-16]. Ainara Hernando-Ortiz and Aitzol Perez-Rodriguez were funded by Ph.D. grants from the University of the Basque Country (PIF 16/39 and PIF17/167, respectively).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Quindós G, Marcos-Arias C, San-Millán R, et al. The continuous changes in the aetiology and epidemiology of invasive candidiasis: from familiar Candida albicans to multiresistant Candida auris. Int Microbiol. 2018;21(3):107–119.
- Sabino R, Veríssimo C, Áa P, et al. Candida auris, an agent of hospital-associated outbreaks: Which challenging issues do we need to have in mind? Microorganisms. 2020;8(2):181.
- Lockhart SR, Etienne KA, Vallabhaneni S, et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;2017(64):134–140.
- Arikan-Akdagli S, Ghannoum M, Meis JF. Antifungal resistance: specific focus on multidrug resistance in Candida auris and secondary azole resistance in Aspergillus fumigatus. J Fungi. 2018;4(4):129.
- Ruiz-Gaitán A, Moret AM, Tasias-Pitarch M, et al. An outbreak due to Candida auris with prolonged colonisation and candidaemia in a tertiary care European hospital. Mycoses. 2018;61(7):498–505.
- Chow NA, Muñoz JF, Gade L, et al. Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. MBio. 2020;11(2):e03364–19.
- Chow NA, De Groot T, Badali H, et al. Potential Fifth Clade of Candida auris, Iran, 2018. Emerg Infect Dis. 2019;25(9):1780–1781.
- Muñoz JF, Gade L, Chow NA, et al. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat Commun. 2018;9(1):1–13.
- Chybowska AD, Childers DS, Farrer RA. Nine things genomics can tell us about Candida auris. Front Genet. 2020;11(April):1–18.
- Kean R, Delaney C, Sherry L, et al. Transcriptome assembly and profiling of Candida auris reveals novel insights into biofilm-mediated resistance. mSphere. 2018;3(4):e00334–18.
- Muñoz JE, Ramirez LM, Dias LDS, et al. Pathogenicity levels of Colombian strains of Candida auris and Brazilian strains of Candida haemulonii species complex in both murine and Galleria mellonella experimental models. J Fungi (Basel). 2020;6(3):E104.
- Brown JL, Delaney C, Short B, et al. Candida auris phenotypic heterogeneity determines pathogenicity in vitro. mSphere. 2020;5(3):e00371–20.
- Ben-Ami R, Berman J, Novikov A, et al. Multidrug-Resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg Infect Dis. 2017;23(2):195–203.
- Fakhim H, Vaezi A, Dannaoui E, et al. Comparative virulence of Candida auris with Candida haemulonii, Candida glabrata and Candida albicans in a murine model. Mycoses. 2018;61(6):377–382.
- Yue H, Bing J, Zheng Q, et al. Filamentation in Candida auris, an emerging fungal pathogen of humans: passage through the mammalian body induces a heritable phenotypic switch. Emerg Microbes Infect. 2018;7(1):1.
- Kean R, Brown J, Gulmez D, et al. Candida auris: a decade of understanding of an enigmatic pathogenic yeast. J Fungi. 2020;6(1):30.
- Torres SR, Pichowicz A, Torres-Velez F, et al. Impact of Candida auris infection in a neutropenic murine model. Antimicrob Agents Chemother. 2020;64(3):e01625–19.
- Bruno M, Kersten S, Bain JM, et al. Transcriptional and functional insights into the host immune response against the emerging fungal pathogen Candida auris. Nat Microbiol. 2020;5(12):1516–1531.
- Kumar A, Baruah A, Tomioka M, et al. Caenorhabditis elegans: a model to understand host–microbe interactions. Cell Mol Life Sci. 2019. DOI:10.1007/s00018-019-03319-7.
- Jemel S, Guillot J, Kallel K, et al. Galleria mellonella for the evaluation of antifungal efficacy against medically important fungi, a narrative review. Microorganisms. 2020;8(3):390.
- Borman AM, Szekely A, Johnson EM. Comparative pathogenicity of United Kingdom isolates of the emerging pathogen Candida auris and other key pathogenic Candida species. mSphere. 2016;1(4):4–6.
- Sherry L, Ramage G, Kean R, et al. Biofilm-forming capability of highly virulent, multidrug-resistant Candida auris. Emerg Infect Dis. 2017;23(2):328–331.
- Eldesouky HE, Li X, Abutaleb NS, et al. Synergistic interactions of sulfamethoxazole and azole antifungal drugs against emerging multidrug-resistant Candida auris. Int J Antimicrob Agents. 2018;52(6):754–761.
- Wang X, Bing J, Zheng Q, et al. The first isolate of Candida auris in China: Clinical and biological aspects article. Emerg Microbes Infect. 2018;7(1):0–8.
- Wurster S, Bandi A, Beyda ND, et al. Drosophila melanogaster as a model to study virulence and azole treatment of the emerging pathogen Candida auris. J Antimicrob Chemother. 2019;74(7):1904–1910.
- Arias LS, Butcher MC, Short B, et al. Chitosan Ameliorates Candida auris Virulence in a Galleria mellonella infection model. Antimicrob Agents Chemother. 2020;64(8). DOI:10.1128/AAC.00476-20
- Romera D, Aguilera-Correa JJ, García-Coca M, et al. The Galleria mellonella infection model as a system to investigate the virulence of Candida auris strains. Pathog Dis. 2020;78(9):ftaa067.
- Johnson CJ, Davis JM, Huttenlocher A, et al. Emerging fungal pathogen Candida auris evades neutrophil attack. MBio. 2018;9(4):1–9.
- Lima SL, Rossato L, Salles De Azevedo Melo A. Evaluation of the potential virulence of Candida haemulonii species complex and Candida auris isolates in Caenorhabditis elegans as an in vivo model and correlation to their biofilm production capacity. Microb Pathog. 2020;22:104461. 148.
- Gago S, García-Rodas R, Cuesta I, et al. Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis virulence in the nonconventional host Galleria mellonella. Virulence. 2014;5(2):278–285.
- Ames L, Duxbury S, Pawlowska B, et al. Galleria mellonella as a host model to study Candida glabrata virulence and antifungal efficacy. Virulence. 2017;8(8):1909–1917.
- Ortega-Riveros M, De-la-pinta I, Marcos-Arias C, et al. Usefulness of the nonconventional Caenorhabditis elegans model to assess Candida virulence. Mycopathologia. 2017;182(9–10):785–795.
- Hernando-Ortiz A, Mateo E, Ortega-Riveros M, et al. Caenorhabditis elegans as a model system to assess Candida glabrata, Candida nivariensis, and Candida bracarensis Virulence and Antifungal Efficacy. Antimicrob Agents Chemother. 2020;64(10):e00824–20.
- Oksuz S, Sahin I, Yildirim M, et al. Phospholipase and proteinase activities in different Candida species isolated from anatomically distinct sites of healthy adults. Jpn J Infect Dis. 2007;60(5):280–283.
- Ade J, Silva AF, Rosa FC, et al. In vitro differential activity of phospholipases and acid proteinases of clinical isolates of Candida. Rev Soc Bras Med Trop. 2011;44(3):334–338.
- L’Ollivier C, Labruère C, Jebrane A, et al. Using a multi-locus microsatellite typing method improved phylogenetic distribution of Candida albicans isolates but failed to demonstrate association of some genotype with the commensal or clinical origin of the isolates. Infect Genet Evol. 2012;12(8):1949–1957.
- Atalay MA, Koc AN, Demir G, et al. Investigation of possible virulence factors in Candida strains isolated from blood cultures. Niger J Clin Pract. 2015;18(1):52–55.
- Peeters E, Nelis HJ, Coenye T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J Microbiol Methods. 2008;72(2):157–165.
- Ramage G, VandeWalle K, Wickes BL, et al. Characteristics of biofilm formation by Candida albicans. Rev Iberoam Micol. 2001;18(4):163–170.
- Polak A. Virulence of Candida albicans mutants. Mycoses. 1992;35(1–2):9–16.
- Price MF, Wilkinson ID, Gentry LO. Plate method for detection of phospholipase activity in Candida albicans. Sabouraudia. 1982;20(1):7–14.
- Cassone A, De Bernardis F, Mondello F, et al. Evidence for a correlation between proteinase secretion and vulvovaginal candidosis. J Infect Dis. 1987;156(5):777–783.
- Luo G, Samaranayake LP, Yau JY. Candida species exhibit differential in vitro hemolytic activities. J Clin Microbiol. 2001;39(8):2971–2974.
- Manns JM, Mosser DM, Buckley HR. Production of a hemolytic factor by Candida albicans. Infect Immun. 1994;62(11):5154–5156.
- Breger J, Fuchs BB, Aperis G, et al. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog. 2007;3(2):e18.
- Larkin E, Hager C, Chandra J, et al. The emerging pathogen Candida auris: growth phenotype, virulence factors, activity of antifungals, and effect of scy-078, a novel glucan synthesis inhibitor, on growth morphology and biofilm formation. Antimicrob Agents Chemother. 2017;61(5):1–13.
- Romera D, Aguilera-Correa JJ, Gadea I, et al. Candida auris: a comparison between planktonic and biofilm susceptibility to antifungal drugs. J Med Microbiol. 2019;68(9):1353–1358.
- Short B, Brown J, Delaney C, et al. Candida auris exhibits resilient biofilm characteristics in vitro: implications for environmental persistence. J Hosp Infect. 2019;103(1):92–96.
- Tobouti PL, Casaroto AR, De Almeida RS, et al. Expression of secreted aspartyl proteinases in an experimental model of Candida albicans-associat ed denture stomatitis. J Prosthodont. 2016;25(2):127–134.
- Chatterjee S, Alampalli SV, Nageshan RK, et al. Draft genome of a commonly misdiagnosed multidrug resistant pathogen Candida auris. BMC Genomics. 2015;16(1):1–16.
- Mesa-Arango AC, Forastiero A, Bernal-Martínez L, et al. The non-mammalian host Galleria mellonella can be used to study the virulence of the fungal pathogen Candida tropicalis and the efficacy of antifungal drugs during infection by this pathogenic yeast. Med Mycol. 2013;51(5):461–472.
- Scorzoni L, De Lucas MP, Mesa-Arango AC, et al. Antifungal efficacy during Candida krusei infection in nonconventional models correlates with the yeast in vitro susceptibility profile. PLoS One. 2013;8(3):e60047.
- Kwon YJ, Shin JH, Byun SA, et al. Candida auris Clinical Isolates from South Korea: Identification, Antifungal Susceptibility, and Genotyping. J Clin Microbiol. 2019;57(4):e01624–18.
- Jf M, Rm W, Shea T, et al. Chromosomal rearrangements and loss of subtelomeric adhesins linked to clade-specific phenotypes in Candida auris. bioRxiv preprint;2019doi: 10.1101/754143.
- Szekely A, Borman AM, Johnson EM. Candida auris isolates of the Southern Asian and South African lineages exhibit different phenotypic and antifungal susceptibility profiles in vitro. J Clin Microbiol. 2019;57(5):e02055–18.
