ABSTRACT
Shigella is an intracellular pathogen that primarily infects the human colon and causes shigellosis. Shigella virulence relies largely on the type III secretion system (T3SS) and secreted effectors. VirF, the master Shigella virulence regulator, is essential for the expression of T3SS-related genes. In this study, we found that YhjC, a LysR-type transcriptional regulator, is required for Shigella virulence through activating the transcription of virF. Pathogenicity of the yhjC mutant, including colonization in the colons of guinea pigs as well as its ability for host cell adhesion and invasion, was significantly lowered. Expression levels of virF and nearly all VirF-dependent genes were downregulated by yhjC deletion, indicating that YhjC can activate virF transcription. Electrophoretic mobility shift assay analysis demonstrated that YhjC could bind directly to the virF promoter region. Therefore, YhjC is a novel virulence regulator that positively regulates the virF expression and promotes Shigella virulence. Additionally, genome-wide expression analysis identified the presence of other genes in the large virulence plasmid and a genome exhibiting differential expression in response to yhjC deletion, with 169 downregulated and 99 upregulated genes, indicating that YhjC also functioned as a global regulatory factor.
Introduction
Shigella is a Gram-negative enteropathogenic bacterium that causes shigellosis, a common disease that reportedly occurs worldwide, with higher incidence in developing countries. The infection is characterized by the occurrence of diarrhea, often with blood or mucus present in the feces [Citation1]. Shigella gains entry into the human colonic lumen via the fecal-oral route and primarily targets the M cells of the colon, resulting in cell death, acute inflammation, and tissue edema. The released bacteria further enter the colonic enterocytes from the basolateral side [Citation2]. Virulence proteins directly involved in the entry and dissemination of Shigella in host cells are encoded by a large, approximately 200-kb virulence plasmid, containing a 31-kb region (designated “entry region”) encoding the Type III secretion system (T3SS) [Citation1,Citation3]. VirF, a DNA-binding activator belonging to the family of AraC-type transcription regulators, is considered the master regulator that activates the expression of T3SS and secreted virulence proteins by regulating at least two primary Shigella virulence factors, including virB and icsA [Citation4,Citation5]. VirB can directly enable the activation of the transcription of genes encoding the entry region [Citation6,Citation7]. Therefore, VirF is vital for Shigella invasion and colonization of the colon.
Despite the critical invasion processes and important virulence effectors reported in the existing literature, much remains unknown about the regulators and the corresponding regulatory mechanisms underlying Shigella pathogenesis. For instance, only a few regulators, including Fis, CpxA/R, and IHF, which can acrivate virF expression in response to environmental stimuli, have been investigated [Citation8–10]. However, the existence of other factors that act as virF regulators still remains to be investigated. In this study, we found that a new LysR-type transcriptional regulator (LTTR), YhjC, was required for Shigella virulence. The results showed that YhjC could promote Shigella pathogenicity by activating virF transcription, indicating that YhjC functioned as a new virulence regulator. Furthermore, transcriptome mapping results revealed that more than 200 genes were regulated by YhjC, indicating that it also played roles as a global regulator.
Hence, our study not only identified YhjC, a previously unrecognized regulator in the LysR family that plays a role in virulence and global regulation of transcription in Shigella, but also provided novel insights into virF expression and the regulation of Shigella pathogenesis.
Results
The yhjC mutant exhibits limited efficiency in invading guinea pigs colon
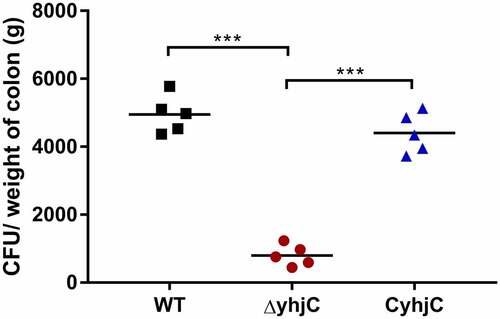
Shigella invades the colonic mucosa, leading to the occurrence of acute inflammatory colitis, dysentery, and intestinal hemorrhage, which destroy the intestinal epithelium [Citation11]. We previously found that yhjC expression was activated during Shigella infection; thus, we investigated whether YhjC was necessary for Shigella pathogenicity. We infected guinea pig colon tissues with the wild type (WT) Shigella flexneri (S. flexneri) 5a strain M90T and the yhjC mutant (ΔyhjC) strain. Colony-forming units (CFU) of the WT bacteria were 6.20 times more than those of the yhjC mutant in the infected colon (), indicating that YhjC was a crucial virulence regulator during the establishment of in vivo Shigella infection. The CFU of complementation was similar to that of WT.
Figure 1. Intrarectal infection of Guinea pigs with the wild type (WT) and ΔyhjC strains. The number of colony-forming units (CFUs) of the WT strains colonizing colon tissues was significantly higher than those of ΔyhjC, and the complementation of yhjC with ΔyhjC restored the ability of mutant Shigella strains to colonize the colon tissues to levels observed with the WT strain. 5 guinea pigs per group were used. Data were obtained from two separate experiments and analyzed using unpaired student’s t-test (*p < 0.05; **p < 0.01; ***p < 0.001)
The yhjC mutant exhibits limited efficiency in adhering to and invading host cells
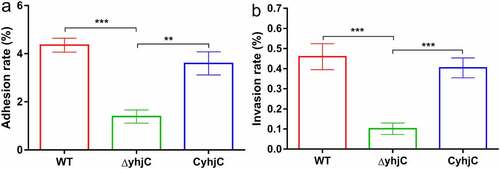
The reduction in the pathogenicity of ΔyhjC may be attributed to the attenuated capacity of the bacteria to infect host cells. To validate this hypothesis, we investigated the ability of the WT and ΔyhjC bacteria to adhere to and invade HeLa cells. The ability of the yhjC mutant to adhere to HeLa cells was approximately 3.14-fold lower than that of the WT strain, and the complement strain (CyhjC) exhibited significantly higher adherence ability compared to the yhjC mutant (). The ability of the yhjC mutant to invade HeLa cells was not as robust as that of the WT, and decreased by 78.01%, although this invasion ability was regained in the CyhjC strain (). These results indicate that yhjC deletion causes pathogen deficiency in terms of host cell adherence and invasion, and YhjC is therefore essential for M90T invasion.
Figure 2. Analysis of the ability of the wild type (WT), ΔyhjC, and complemented Shigella strains to adhere to and invade HeLa cells. Both adhesion and invasion abilities were attenuated in ΔyhjC mutants compared with those observed in the WT, and the complemented strain showed adhesion and invasion abilities similar to the WT. (a) Analysis of adherence ability; (b) analysis of invasion ability. Data were generated from three independent experiments and have been presented as mean ± SD. p-values were determined using unpaired Student’s t-test (*p < 0.05; **p < 0.01; ***p < 0.001)
yhjC deletion decreases the Congo red dye-binding ability of Shigella
Based on the results mentioned above, as T3SS and the secreted virulence proteins contribute to Shigella infection and invasiveness, we speculated that YhjC might affect T3SS activity. The ability of Shigella to bind Congo red dye was positively associated with T3SS activity and the virulence of this pathogen [Citation12,Citation13]. WT bacterial colonies exhibited red coloration with higher intensity than the yhjC mutant colonies (). The OD498/OD600 of WT colonies was 0.290, which was significantly higher than that of yhjC mutant colonies (0.097), indicating that T3SS activity and virulence of WT were higher than those demonstrated by the yhjC mutant. Thus, the results imply that YhjC is essential for T3SS activity, which is activated by VirF.
YhjC promotes Shigella virulence by activating virF
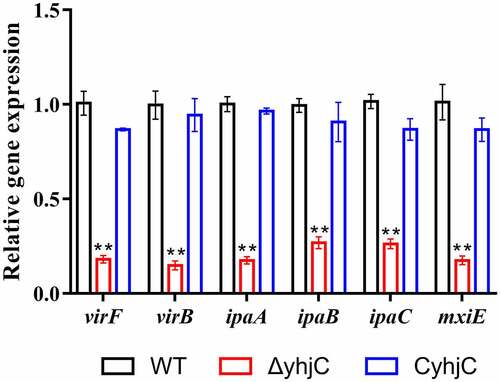
Based on the phenotypic results, we further speculated that YhjC promoted Shigella virulence by controlling the expression of virulence-related genes such as T3SS-related genes, and genes encoding the virulence proteins virB and virF. As the identity of the gene regulated by YhjC was unknown, we selected several invasion-related genes (ipaA, ipaB, and ipaC) and virulence regulators (virF, mxiE, and virB) and compared their expression in the yhjC mutant and the WT strain using quantitative real-time PCR (qRT-PCR). Expression levels of all identified genes were downregulated in response to yhjC deletion (). As the identified genes were regulated by VirF, we concluded that YhjC promote Shigella virulence by activating virF.
Figure 4. yhjC deletion downregulates the expression of the main virulence genes of Shigella. The expression levels of virF, virB, ipaA, ipaB, ipaC, and mxiE were significantly downregulated in ΔyhjC. Data were generated from triplicate experiments and have been presented as mean ± SD. p-values were determined by using unpaired Student’s t-test (*p < 0.05; **p < 0.01; ***p < 0.001)
YhjC binds directly to the promoter region of virF
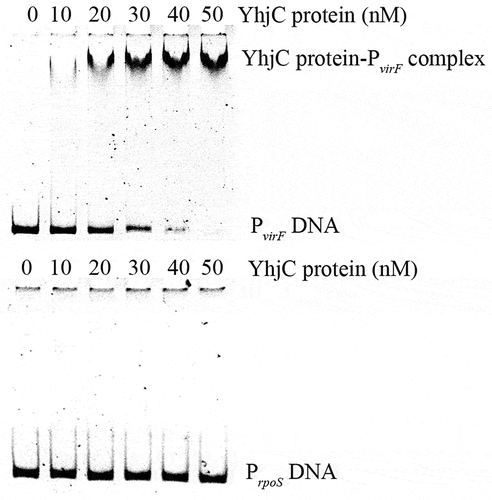
To determine the way of YhjC activating virF expression, we explored the interaction between the purified YhjC-His6 protein and virF promoter DNA using electrophoretic mobility shift assay (EMSA). We observed that a reduced quantity of the virF promoter DNA (PvirF DNA) exhibited migration under increasing concentrations of the YhjC protein, whereas no movement retardation was observed for the rpoS promoter, which was used as the negative control DNA (). Therefore, we confirmed that YhjC bound directly to the virF promoter.
Figure 5. Analysis of the interaction between the purified YhjC-His6 protein and the virF promoter DNA using EMSA. A quantity of 50 ng of DNA and a concentration ranging from 0–50 nM of the purified YhjC-His6 protein were used in each reaction. Less promoter virF fragments exhibited migration with increasing concentrations of the YhjC protein. The DNA of the rpoS promoter was used as a negative control and no binding was observed
RNA sequencing analysis
Transcriptional profile
LTTRs are ubiquitous proteins that regulate the expression of a variety of genes [Citation14]. We further investigated the global regulatory function of YhjC in Shigella by performing sequencing analysis of the RNA samples obtained from the WT and yhjC mutant strains. A total of 15,003,465 and 16,435,097 reads were obtained upon analysis of the WT and yhjC mutants, respectively, with 98.92% and 98.73% of the reads mapping to the reference genome. 268 genes were differentially expressed between the yhjC mutant and the WT strain, including 169 downregulated genes (, Table S2) and 99 upregulated genes (Table S3). Gene ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed. Please check the Supplementary Material (Figure S2 and Figure S3) for the results of these analyses.
Figure 6. RNA sequencing analysis. (a) Transcriptional profiles of T3SS-related genes in the wild type and ΔyhjC Shigella strains. Expression levels of almost all T3SS-related genes were downregulated following yhjC deletion. (b, c) Fold-change in the FPKM of representative differentially expressed genes (DEGs). The fold change in representative DEGs encoding virulence genes is much higher than that observed in non-virulence genes. (d) Verification of the RNA sequencing data through qRT-PCR analysis of non-virulence genes. Changes in the expression of the non-virulence genes are consistent with those of the RNA sequencing data. Data were generated from triplicate experiments. p-values were determined by using multiple t-test and unpaired Student’s t-test (*p < 0.05; **p < 0.01; ***p < 0.001)
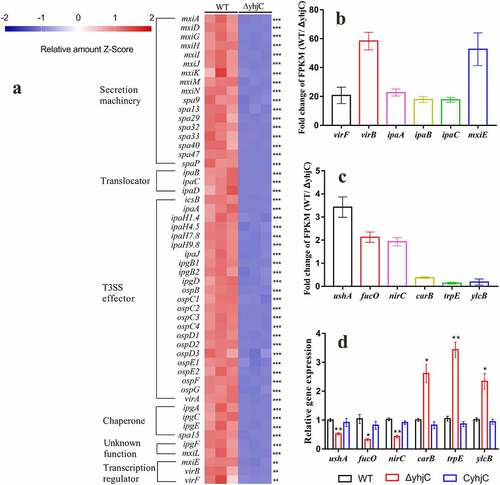
yhjC deletion downregulates the expression of virulence-related genes
RNA sequencing data showed that the expression levels of almost all virulence-related genes were downregulated in the yhjC mutant compared with the WT strain (), and this downregulated expression showed appreciable correlation with the qRT-PCR results (). The expression of virF was downregulated 20.68-fold in the yhjC mutant. Correspondingly, expression levels of virB and icsA, which are directly regulated by VirF, were downregulated by 59.57-fold and 4.90-fold, respectively. VirB can activate the transcription of multiple virulence genes, including genes encoding the T3SS secretion machinery (Mxis and Spas), translocators (Ipas), chaperones (Ipgs), “first-set” effectors (IcsB, OspC2-4, OspD1/2, etc), MxiE, and the outer membrane protease (IcsP) specific for the actin-based motility protein IcsA [Citation15,Citation16]. Consistent with virB downregulation, expression levels of the VirB-regulated genes were also downregulated by 4–71-fold. The transcription of genes encoding the “second-set” T3SS effectors is controlled by the MxiE transcriptional activator, with the IpgC chaperone acting as a coactivator [Citation1,Citation17]. Expression levels of the genes, including virA, ipaHs, ospB, ospC1, ospD3, ospE1/2, ospF, and ospG, were downregulated 2–16-fold.
Validation of RNA sequencing results
To validate the RNA sequencing results, qRT-PCR analysis was performed using six additional randomly selected genes, including ushA, fucO, nirC, carB, trpE, and ylcB. The mRNA levels of these six genes were consistent with the RNA sequencing results (), thus validating the reliability of the RNA sequencing data.
The effect of environmental conditions on yhjC expression
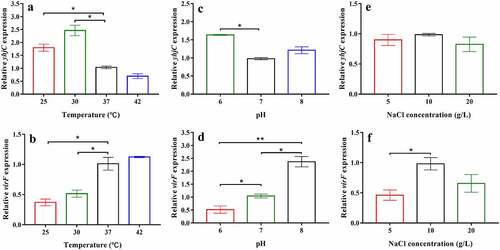
The effects of temperature, pH, and NaCl concentration on the mRNA levels of yhjC and virF were investigated in the current study. The relative gene expression of Shigella at 37 °C, pH 7, or 10 g/L NaCl were set to 1 and used as reference. The mRNA level of yhjC was significantly higher at 30 °C than that observed at 37 °C (), although it continued to exhibit a relatively high expression at 37 °C, based on the Ct values. Consistent with the results reported by a previous study [Citation18], virF mRNA level was significantly lower at 30 °C than that observed at 37 °C and 42 °C (). yhjC transcription at pH 6 was also higher than that at pH 7, whereas virF transcription was suppressed at pH 6 but significantly activated at pH 7 and pH 8 ( and d). The mRNA level of yhjC was not significantly influenced by the NaCl concentrations studied, whereas virF expression was influenced by NaCl concentration ( and f). These results indicate that yhjC activation can occur under permissive environmental stimuli (37 °C, pH 7) as well as under non-permissive environmental conditions. However, the virF transcription was not entirely relevant to the mRNA level of yhjC at different environmental conditions as transcription of virF is controlled by many various factors.
Figure 7. The effects of temperature, pH, and NaCl concentration on the levels of yhjC and virF mRNA. (a, b) Temperature influences the expression of yhjC and virF; (c, d) pH influences the expression of yhjC and virF; (e, f) NaCl concentrations influence the expression of virF but not that of yhjC. The relative gene expression of Shigella at 37 °C, pH 7, or 10 g/L NaCl were set to 1 and used as reference. Data were generated from triplicate experiments and have been presented as mean ± SD. p-values were determined by using unpaired Student’s t-test (*p < 0.05; **p < 0.01; ***p < 0.001)
Discussion
In this study, we investigated the function of the novel transcriptional regulator YhjC in Shigella pathogenicity. Thus far, only one study has reported YhjC, activating csgD transcription during biofilm formation in Escherichia coli K-12 [Citation19]. First, we found that the ability of Shigella to colonize the colon of guinea pigs was significantly weakened by the deletion of yhjC (). Consistent with this observation, the yhjC mutant was less effective at adhering to and invading Hela cells compared with the WT strain (). Thus, we believe that YhjC is a virulence regulator responsible for Shigella infection and invasion, although the underlying mechanism of action is unclear. Most virulence processes associated with Shigella are conferred by the activities of T3SS and its secreted virulence effectors, the activation of which relies on VirF [Citation20]. Further investigation confirmed that YhjC could bind to the promoter region of virF and exercises direct positive transcriptional regulation (). Therefore, we proposed that YhjC contributed to Shigella virulence by activating virF transcription. RNA sequencing results supported this theory, as all the genes controlled by virF including genes encoding T3SS, transcription activators of T3SS, and virulence effectors were obviously downregulated following yhjC deletion ( and b). So the inability of the yhjC mutant to assemble a complete type III secretion apparatus and adequately synthesize virulence effectors ultimately attenuated virulence in vivo. Additionally, the reduced synthesis of translocators, chaperones, and other virulence-associated proteins also considerably attenuated Shigella virulence.
VirF performs transcriptional regulation in response to physical signals mediated by different regulatory factors [Citation4]. The nucleoid-associated protein H-NS represses virF transcription at non-permissive temperatures (below 32 °C) by binding to the promoter region of virF [Citation21,Citation22]. Therefore, YhjC may play an important role in activating virF transcription, especially at temperatures below 32 °C. Perhaps this may explain why yhjC has a robust expression at 30 °C (). However, YhjC also works at the body temperature (around 37 °C), which has been confirmed by the cell and animal experiments in this study. In previous studies, phosphorylated CpxR reportedly induced direct activation of virF transcription by efficiently binding to its target site upstream of the virF gene, while CpxA repressed virF transcription at low pH (6.0) but activated transcription at high pH (7.4) through its action as a CpxR phosphatase [Citation9], consistent with our results. The higher yhjC mRNA level observed at pH 6 might weaken CpxA repression ( and d). Interestingly, NaCl concentration did not affect yhjC expression (). However, virF transcription was markedly suppressed under low osmotic conditions and was activated under physiological osmotic conditions [Citation23], a similar phenomenon was observed in our study (). Considering the results of the RNA sequencing and qRT analysis in addition to these results, YhjC is indispensable to virF transcription, both under permissive and non-permissive environmental conditions (below 37°C, pH 6.0), although it is not the only factor that controls virF expression, as virF is extensively regulated.
Additionally, YhjC seems play much more important role in virF transcriptional regulation than those of Fis, IHF and CpxA/R [Citation8–10], as expression of virF was downregulated by 5.59-fold following yhjC deletion, which was much more significant (). These results strongly suggested that YhjC is vital additional regulator responsible for virF activation during Shigella infection. The sequence of the YhjC target binding site in the virF promoter region will be investigated in a future study. However, it is evident that the interaction between YhjC and the virF promoter is probable, due to the presence of several T-N11-A binding motifs in the promoter (Figure S1), consistent with the characteristics of LTTR-regulated promoters [Citation24].
Gene expression profiling showed that YhjC was a global transcriptional regulator. Gene ontology (GO) enrichment analysis identified the following three functional categories: molecular function, cellular component, and biological process (Figure S2). Under the molecular function category, differentially expressed genes (DEGs) encoding ubiquitin-protein transferase and sequence-specific DNA-binding proteins were likely to be correlated with Shigella pathogenesis. Previous reports have indicated that the IpaH family proteins promote bacterial survival possess E3 ubiquitin ligase activity [Citation25,Citation26]. These DNA-binding proteins probably function as transcription regulators as invasion and cell-to-cell spread of Shigella are largely controlled by several transcription regulators, such as VirF, VirB, and MxiE [Citation4]. GO analysis of cellular components showed that the DEG-encoded proteins were mainly located in the extracellular region, outer cell membrane, host cell cytoplasm, host cell nucleus, host cell plasma membrane, and host cell cytosol. The apparatus for T3SS of Shigella is located in the bacterial membrane and can be inserted into the host cell plasma membrane, while the T3SS effectors are secreted into the host cell cytoplasm. The DEGs associated with biological processes, such as pathogenesis, DNA integration, transposition, DNA recombination, protein secretion, and ion transport, have been identified, further implying that YhjC is a global regulator that can exert influence on Shigella virulence. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was conducted to identify pathways that were enriched in response to yhjC deletion (Figure S3). The most significantly enriched pathways were related to shigellosis, bacterial invasion of epithelial cells, and bacterial secretion system, indicating that YhjC functioned as a global regulator involved in Shigella virulence.
The csgD gene encoding the master regulator of biofilm formation, whose expression is controlled by YhjC, was not identified in the RNA sequencing analysis in this study, and this could be attributable to the absence of biofilm formation during the occurrence of Shigella virulence-induced processes [Citation19,Citation27]. Additional promoters that can establish interactions directly with YhjC should also be investigated. Finally, the effects of differential expression of virF-unrelated genes located on the large virulence plasmid and the genome exerted on bacterial virulence remain unknown and warrant further investigation.
In conclusion, we determined the virulence and global regulatory function of the novel regulator, YhjC, in Shigella pathogenesis. This study suggests that YhjC can activate virF transcription by directly binding to its promoter region. Therefore, YhjC is a critical virulence regulator necessary for promoting Shigella pathogenicity. YhjC expression may also control Shigella growth and survival through certain unidentified mechanisms.
Materials and methods
Bacterial strains and plasmids
The Shigella flexneri 5a strain M90T was used as the wild-type strain in this study. The yhjC mutant was generated using the λ Red recombinase system, supported by the pSim17 plasmid (blastidin-resistant) encoding three proteins (Exo, Beta, and Gam) required for homologous recombination [Citation28]. First, the M90T strain harboring the pSim17 plasmid was established through electrotransformation. Then, DNA fragments composed sequentially (5ʹ→3ʹ) of an upstream 50-bp sequence of the target gene, the chloramphenicol-resistant gene sequence, and the downstream 50-bp reverse complementary sequence of the target gene were subjected to PCR amplification. Chloramphenicol-resistant pKD3 plasmid was used as the template and primers were designed to cover the 50-bp homologous arm sequences. Next, the DNA fragments were introduced into competent M90T cells using electrotransformation. Finally, the cells were recovered at 37°C for 2 h and suspensions of recovered cells were spread on agar plates containing 25 μg/mL chloramphenicol to obtain single mutant colony. To validate the formation of the yhjC mutant strain, the yhjC loci in the chloramphenicol-resistant colony were amplified and preliminarily identified through electrophoresis, and further identification was performed through Sanger sequencing.
The yhjC gene and its upstream 500-bp sequence were cloned into the plasmid pBR322 at a region between NheI and BamHI restriction enzyme digestion sites, and the resulting constructs were electrotransformed into competent yhjC mutants; subsequently, after screening the bacteria for resistance to chloramphenicol and ampicillin on agar plates containing the two antibiotics, the complemented strain (CyhjC) was constructed. To perform expression and purification of the YhjC-His6 protein, the pET-yhjC plasmid was established by cloning the yhjC gene sequence into the pET-28a plasmid between BamHI and XhoI sites downstream of the His-tag. All constructed plasmids and strains were validated using the methods described in the identification of yhjC mutants. Primers used in this study are listed in Table S1.
Growth conditions for bacteria and Hela cells
Bacteria were cultured in either liquid LB broth (1% tryptone, 0.5% yeast extract, and 1% NaCl) or solid LB medium (liquid LB broth supplemented with 1.5% agar) at 37°C. Where necessary, antibiotics were added at the following final concentrations: 25 μg/mL chloramphenicol, 50 μg/mL kanamycin and 100 μg/mL ampicillin. HeLa cells used in this study were cultured in the Dulbecco’s modified Eagle medium (DMEM) with 10% fetal bovine serum at 37°C under 5% CO2. Next, 24 h before infection with bacteria, HeLa cells were dissociated using trypsin, and the cells (1 × 105 cells/well) were seeded into 12-well culture plates to achieve final differentiated cell monolayers.
Intrarectal infection of guinea pigs
Three-week-old female Dunkin-Hartley guinea pigs (< 150 g) were used for conducting the intrarectal infection experiment [Citation29]. To screen for Shigella colonization in the colons, the WT bacterial strain was transformed with the pBR322 plasmid, and the transformation conferred the ability to grow on agar plates containing ampicillin; the yhjC and complemented strains harbor genes for chloramphenicol and ampicillin resistance, respectively. Shigella was first cultured in LB medium at 37°C for approximately 13 h, and then sub-cultured in LB medium at an inoculation ratio of 1:100. When the OD600 of the bacterial culture reached a value of 0.6 (indicating logarithmic phase of the bacteria), the bacterial cells were collected by centrifugation and were suspended in PBS. Guinea pigs were subjected to fasting conditions 15 h prior to intrarectal infection, 5 guinea pigs per group were used. 1010 Shigella CFUs (100 μL) were injected into the rectum of guinea pigs anesthetized with diethyl ether. After an incubation period of 8 h, the guinea pigs were euthanized, and the distal 5-cm specimens of the colon were harvested. Each colon specimen was ground, and bacterial CFUs were counted by conducting surface spread of bacterial suspensions on agar plates containing the corresponding antibiotics.
Adhesion and invasion assays
Adhesion and invasion assays were performed as per methods previously described with slight modifications [Citation5,Citation30]. Briefly, overnight Shigella cultures were sub-cultured into fresh LB medium at an inoculation ratio of 1:100 and grown at 37°C to achieve an OD600 of 0.6. The bacterial cells were then pelleted by centrifugation, and the supernatant was discarded. The retrieved pellets were suspended in fresh DMEM for 20 min. HeLa cells were infected with Shigella cells at a multiplicity of infection (MOI) of 100:1 (ration of the number of bacterial cells to HeLa cells), and the bacteria-HeLa cell mixed suspension was centrifuged at 800 × g for 5 min. Then, HeLa cells and bacterial cells were co-cultured at 37°C to initiate invasion. For adhesion assays, host cells were subjected to washing steps three times using sterile PBS 40 min post-infection to remove bacterial cells that did not adhere to the host cells. Subsequently, the cells were subjected to lysis using 0.1% Triton X-100, and the bacterial cells that could invade HeLa cells were enumerated using the flat colony counting method. The adhesion rate was calculated by dividing the number of recovered bacteria by the total number of bacterial cells used for infection.
For conducting invasion assays, 100 μg/mL gentamicin was added to the cell culture medium 40 min post-infection, and the cells were incubated for 1 h to kill the extracellular bacteria. 100 min post infection, HeLa cells were subjected to washing steps and lysed using 0.1% Triton X-100 to release the intracellular bacterial cells, and the lysates were spread on agar plates for enumeration of bacterial CFUs. The invasion rate was calculated as the ratio of the number of recovered bacteria to the total number of bacterial cells used for infection.
Congo red binding assay
The Congo red binding assay was performed as per protocols previously published [Citation12]. The WT strains and yhjC mutants were cultured overnight in LB broth. The next day, the cultures were serially diluted, and 100 μL of each dilution was added onto trypticase soy broth (TSB) agar plates supplemented with 0.01% (w/v) Congo red (hereafter referred to as Congo red plates). The plates were incubated overnight at 37°C. A density of approximately 6 × 109 cells/mL was used to quantify the relative amount of Congo red bound by the bacterial cells. Ten culture spots for each sample were scraped off the agar plate and suspended in 750 μL 25% ethanol to remove Congo red bound to the cells. The optical density of the cell suspension was measured at 600 nm (OD600) to perform normalization of the samples to the cell number. Cell suspensions were then centrifuged, and the optical density of the supernatant was measured at 498 nm (OD498) to quantify the amount of Congo red released from the bacteria. Relative Congo red binding was determined as: OD498/OD600.
RNA preparation
Shigella cells were cultured overnight in LB at 37°C and sub-cultured at an inoculation ratio of 1:100. When the optical density at 600 nm (OD600) reached a value of 0.6, indicating the logarithmic phase of the bacteria, Shigella cells were collected by centrifugation at 5000 rpm for 5 min and were resuspended in sterile PBS. The bacterial cells were then incubated in 50 µg/mL Congo red solution at 37°C for 20 min to induce T3SS activity in Shigella in vitro [Citation31]. Total RNA extraction was performed using the TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions. The RNA samples were purified using the RNeasy Mini Kit and were treated with DNase I (QIAGEN, Germany) to remove traces of contaminating DNA. The final RNA concentration was determined using the NanoDrop 2000 spectrophotometer (NanoDrop, USA).
qRT-PCR analysis
qRT-PCR was performed using the Applied Biosystems 7500 Real-Time PCR system (Applied Biosystems, USA). The primers used for the qRT-PCR analysis are listed in Table S1. To analyze the expression of virulence genes, RNA samples from the bacteria (collected in the logarithmic phase) were prepared as per methods described in Section 4.6. A total of 1.2 μg RNA from each sample was reverse-transcribed into cDNA using the PrimeScriptTM RT Reagent kit (TaKaRa, Japan). The total qRT-PCR reaction mixture (20 μL) contained 10 μL of the PowerUp™ SYBR™ Green Master Mix (Applied Biosystems, USA), 1 μL of cDNA (approximately 40 ng), 1 μL of forward primer, and 1 μL of reverse primer, each at a final concentration of 0.5 μM. The data were normalized using the 16S rRNA gene as a reference control. Target gene expression levels were calculated using the 2−ΔΔCt method. Each qRT-PCR experiment was performed in triplicate.
Library construction and RNA sequencing
RNA sequencing was performed using the Illumina sequencing platform. The NEBNext Ultra Directional RNA Library Prep Kit (NEB, USA) was used for cDNA library construction following the manufacturer’s instructions. Reads obtained from the sequencing experiment were mapped to the reference genomes of S. flexneri 5a M90T and its large virulence plasmid pWR100. HTSeq (version 0.6.1) was used to count the number of reads. Gene expression levels were calculated using the FPKM method, where FPKM represents fragments per kilobase of exon model per million reads mapped [Citation32]. The DESeq2 (v1.6.3) package was used to compare differences in gene expression between the WT and ΔyhjC strains. Differentially expressed genes were those with |fold-change| ≥ 2 and p < 0.05 [Citation33].
Electrophoretic mobility shift assay
The pET28a-yhjC plasmid was transformed into Escherichia coli BL21 (DE3), and YhjC harboring a C-terminal 6× His tag (YhjC-His6 protein) was expressed after subjection to induction using isopropylthio-β-galactoside (IPTG). The protein was purified from the lysate supernatant of the DE3 using a HiTrap Ni2+-chelating column (GE Healthcare, Germany), as per methods described previously [Citation34]. Protein concentration was determined according to the Bradford method and the samples were stored at −80°C.
EMSA was performed as previously described with some modifications [Citation35]. Briefly, 300-bp DNA fragments of the virF and rpoS promoter regions were amplified by conducting PCR and were subjected to purification steps. A quantity of 50 ng of the promoter fragments and different concentrations (0–50 nM) of the purified YhjC protein were incubated at 26°C for 30 min in a 20-μL solution containing EMSA binding buffer (20 mM Tris HCl pH 7.5, 50 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, and 5% glycerol). Samples were then loaded onto a 6% polyacrylamide gel immersed in 0.5 × Tris-Borate-EDTA for electrophoresis. DNA fragments were stained with Gel Red (Biotium, USA).
Statistical analysis
Data were analyzed using GraphPad Prism (version 7.00; La Jolla, CA, USA). The data presented in each figure or table represent mean ± standard deviation (SD) values obtained from the conduction of three independent experiments. Student’s t-test was used to analyze significant differences between the two groups. Differences were considered significant at p < 0.05 (*p < 0.05; **p < 0.01; ***p < 0.001).
Data availability
The RNA sequencing data acquired in this study are available in the NCBI Sequence Read Archive (SRA, PRJNA721543).
Disclosure of potential conflicts of interest
No potential conflict of interest was reported by the authors.
Supplemental Material
Download Zip (868.5 KB)Supplemental material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Schroeder G, Hilbi H. Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin Microbiol Rev. 2008;21(1):134–156.
- Phalipon A, Sansonetti P. Shigella’s ways of manipulating the host intestinal innate and adaptive immune system: a tool box for survival? Immunol Cell Biol. 2007;85(2):119–129.
- Carayol N, Tran Van Nhieu G. Tips and tricks about Shigella invasion of epithelial cells. Curr Opin Microbiol. 2013;16(1):32–37.
- Di Martino M, Falconi M, Micheli G, et al. The multifaceted activity of the VirF regulatory protein in the Shigella lifestyle. Front mol biosci. 2016;3:61.
- Brotcke Zumsteg A, Goosmann C, Brinkmann V, et al. IcsA is a Shigella flexneri adhesin regulated by the type III secretion system and required for pathogenesis. Cell Host Microbe. 2014;15(4):435–445. .
- Tobe T, Yoshikawa M, Mizuno T, et al. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by virF and repression by H-NS. J Bacteriol. 1993;175(19):6142–6149. .
- Soceaa J, Bowmanb G, Wing H. VirB, a Key Transcriptional Regulator of Virulence Plasmid Genes in Shigella flexneri, Forms DNA-Binding Site-Dependent Foci in the Bacterial Cytoplasm. J Bacteriol. 2021;203(11). DOI:10.1128/JB.00627-20
- Falconi M, Prosseda G, Giangrossi M, et al. Involvement of FIS in the H-NS-mediated regulation of virF gene of Shigella and enteroinvasive Escherichia coli. Mol Microbiol. 2001;42(2):439–452. .
- Nakayama S, Watanabe H. Identification of cpxR as a positive regulator essential for expression of the Shigella sonnei virF gene. J Bacteriol. 1998;180(14):3522–3528.
- Porter M, Dorman C. Positive regulation of Shigella flexneri virulence genes by integration host factor. J Bacteriol. 1997;179(21):6537–6550.
- Ashida H, Kim M, Sasakawa C. Manipulation of the host cell death pathway by Shigella. Cell Microbiol. 2014;16(12):1757–1766.
- Pilla G, McVicker G, Tang C. Genetic plasticity of the Shigella virulence plasmid is mediated by intra- and inter-molecular events between insertion sequences. PLoS Genet. 2017;13(9):e1007014.
- Weatherspoon-Griffin N, Wing H. Characterization of SlyA in Shigella flexneri identifies a novel role in virulence. Infect Immun. 2016;84(4):1073–1082.
- Kim Y, Chhor G, Tsai C, et al. Crystal structure of the ligand-binding domain of a LysR-type transcriptional regulator: transcriptional activation via a rotary switch. Mol Microbiol. 2018;110(4):550–561. .
- McKenna J, Wing H. The antiactivator of type III secretion, OspD1, is transcriptionally regulated by VirB and H-NS from remote sequences in Shigella flexneri. J Bacteriol. 2020;202(10):e00072–20.
- Parsot C. Shigella spp. and enteroinvasive Escherichia coli pathogenicity factors. FEMS Microbiol Lett. 2005;252(1):11–18.
- Gall T, Mavris M, Martino M, et al. Analysis of virulence plasmid gene expression defines three classes of effectors in the type III secretion system of Shigella flexneri. Microbiol (Reading, England).. 2005;151:951–962.
- Falconi M, Colonna B, Prosseda G, et al. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 1998;17(23):7033–7043. .
- Ogasawara H, Ishizuka T, Hotta S, et al. Novel regulators of the csgD gene encoding the master regulator of biofilm formation in Escherichia coli K-12. Microbiol (Reading, England).. 2020;166(9):880–890.
- Di Martino M, Romilly C, Wagner E, et al. One gene and two proteins: a leaderless mRNA supports the translation of a shorter form of the Shigella VirF regulator. mBio. 2016;7(6):e01860–16. .
- Ulissi U, Fabbretti A, Sette M, et al. Time-resolved assembly of a nucleoprotein complex between Shigella flexneri virF promoter and its transcriptional repressor H-NS. Nucleic Acids Res. 2014;42(21):13039–13050. .
- Prosseda G, Falconi M, Giangrossi M, et al. The virF promoter in Shigella: more than just a curved DNA stretch. Mol Microbiol. 2004;51(2):523–537. .
- Mitobe J, Morita-Ishihara T, Ishihama A, et al. Involvement of RNA-binding protein Hfq in the osmotic-response regulation of invE gene expression in Shigella sonnei. BMC Microbiol. 2009;9:110.
- Parsek M, Ye R, Pun P, et al. Critical nucleotides in the interaction of a LysR-type regulator with its target promoter region. catBC promoter activation by CatR. J Biol Chem. 1994;269(15):11279–11284. .
- Ashida H, Sasakawa C. Shigella IpaH family effectors as a versatile model for studying pathogenic bacteria. Front Cell Infect Microbiol. 2015;5:100.
- Ashida H, Mimuro H, Sasakawa C. Shigella manipulates host immune responses by delivering effector proteins with specific roles. Front Immunol. 2015;6:219.
- Kaoukab-Raji A, Biskri L, Allaoui A. Inactivation of the sfgtr4 gene of induces biofilm formation and affects bacterial pathogenicity. Microorganisms. 2020;8(6):841.
- Chan W, Costantino N, Li R, et al. A recombineering based approach for high-throughput conditional knockout targeting vector construction. Nucleic Acids Res. 2007;35(8):e64. .
- Tinevez J, Arena E, Anderson M, et al. Shigella-mediated oxygen depletion is essential for intestinal mucosa colonization. Nat Microbiol. 2019;4(11):2001–2009. .
- Tran Van Nhieu G, Caron E, Hall A, et al. IpaC induces actin polymerization and filopodia formation during Shigella entry into epithelial cells. EMBO J. 1999;18(12):3249–3262. .
- Parsot C, Ageron E, Penno C, et al. A secreted anti-activator, OspD1, and its chaperone, Spa15, are involved in the control of transcription by the type III secretion apparatus activity in Shigella flexneri. Mol Microbiol. 2005;56(6):1627–1635. .
- Mortazavi A, Williams B, McCue K, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. .
- Paudel A, Panthee S, Hamamoto H, et al. YjbH regulates virulence genes expression and oxidative stress resistance in Staphylococcus aureus. Virulence. 2021;12(1):470–480. .
- Breddermann H, Schnetz K. Activation of leuO by LrhA in Escherichia coli. Mol Microbiol. 2017;104(4):664–676.
- Yang B, Feng L, Wang F, et al. Enterohemorrhagic Escherichia coli senses low biotin status in the large intestine for colonization and infection. Nat Commun. 2015;6:6592.

