ABSTRACT
The S-adenosyl-L-homocysteine hydrolase (Sah1) plays a crucial role in methylation and lipid metabolism in yeast and mammals, yet its function remains elusive in filamentous fungi. In this study, we characterized Sah1 in the phytopathogenic fungus F. graminearum by generating knockout and knockout-complemented strains of FgSAH1. We found that the FgSah1-GFP fusion protein was localized to the cytoplasm, and that deletion of FgSAH1 resulted in defects in vegetative growth, asexual and sexual reproduction, stress responses, virulence, lipid metabolism, and tolerance against fungicides. Moreover, the accumulations of S-adenosyl-L-homocysteine (AdoHcy) and S-adenosyl-L-methionine (AdoMet) (the methyl group donor in most methyl transfer reactions) in ΔFgSah1 were seven- and ninefold higher than those in the wild-type strain, respectively. All of these defective phenotypes in ΔFgSah1 mutants were rescued by target gene complementation. Taken together, these results demonstrate that FgSah1 plays essential roles in methylation metabolism, fungal development, full virulence, multiple stress responses, lipid metabolism, and fungicide sensitivity in F. graminearum. To our knowledge, this is the first report on the systematic functional characterization of Sah1 in F. graminearum.
Introduction
The ascomycete fungus Fusarium graminearum (teleomorph: Gibberella zeae) is a predominant causal agent of Fusarium head blight (FHB), which is a disastrous disease on various cereal crops [Citation1,Citation2]. Severe FHB epidemics result in devastating yield loss and the contamination of infected grains with mycotoxins produced by F. graminearum (e.g. deoxynivalenol (DON), zearalenone), which are toxic to humans and livestock [Citation2,Citation3]. To date, no commercially FHB-resistant cultivars are available, and fungicides are heavily relied upon to control infestations worldwide. However, the extensive use of chemical control has negative effects, including the resistance development against fungicide [Citation2,Citation4,Citation5]. Therefore, a better understanding of the mechanisms underlying the procedure of pathogenicity in F. graminearum is essential to develop better target-based control strategies [Citation6].
The methionine cycle is a pathway well documented in yeast and mammals. As summarized in , methionine is first catalyzed into S-adenosyl-L-methionine (AdoMet) by methionine adenosyltransferase, then AdoMet is converted into S-adenosyl-L-homocysteine (AdoHcy) by the removal of a methyl group, subquently AdoHcy is hydrolyzed by Sah1 into homocysteine (Hcy), and methionine is regenerated from Hcy when catalyzed by methionine synthase (Met6) [Citation7,Citation8]. This cycle is tightly linked to various other cellular processes, as AdoMet is the second most widely used enzyme substrate after ATP [Citation9], functioning as the major methyl group donor for AdoMet-dependent methyltransferases reactions, in which acceptor molecules range from nucleic acids to proteins and lipids [Citation8]. At the same time, AdoHcy, which is a by-product of methylation reaction, is a strong competitive inhibitor of all AdoMet-dependent methyltransferases [Citation10]. To relieve AdoHcy inhibition, AdoHcy is reversibly hydrolyzed into adenosine and Hcy by Sah1, which is the singular pathway of AdoHcy catabolism both in yeast and mammals (Sah1 in yeast, AHCY in mammals) [Citation8]. Because the reaction is reversible and there are no other enzymes capable of hydrolyzing AdoHcy, an increase of Hcy levels may induce the formation of AdoHcy and the resulting accumulation may lead to feedback inhibition in the biosynthesis of AdoHcy from AdoMet.
The S-adenosyl-L-homocysteine hydrolase (Sah1) is a protein well conserved from bacteria to mammals with more than 70% amino acid sequence homology between human and yeast orthologs and also among the 90 most highly conserved yeast proteins [Citation11]. Impaired Sah1 activity in most living organisms results in AdoHcy accumulation and severe pathological consequences. For example, mouse embryos cannot survive after deletion of a locus overlapping the AHCY gene [Citation12], and homozygous mutations in the zygotic SAH1 gene were observed to be lethal to Arabidopsis thaliana [Citation13]. Although yeast mutants without Sah1 are viable due to the presence of an alternative pathway for Hcy synthesis via sulfur assimilation, additional mutation in this pathway is lethal for the resulting yeast sah1 mutants [Citation14]. In addition to its integral role in virtually all living organisms, Sah1 is also known to belong to the large family of NAD(P)H/NAD(P)+-binding proteins with a Rossmann-fold [Citation15]. The structures of Sah1/AHCY from various organisms have been characterized [Citation8], of which only the plant Sah1 from Lupinus luteus works as a homodimer [Citation16], while others are tetramers with the NADH/NAD+ cofactor bound at active sites of each subunit [Citation17,Citation18].
Owing to its central role in methionine cycle, Sah1 may be a potential target for fungicide interest. In this study, we identified and characterized Sah1 in F. graminearum (FgSah1). Our results indicate that FgSah1 plays diverse roles in F. graminearum, including methylation metabolism, vegetative growth, asexual and sexual reproduction, stress responses, DON production, full virulence, and lipid metabolism.
Materials and methods
Fungal strains and media
The wild-type (WT) strain of F. graminearum PH-1 and transformants derived from PH-1 were used in this study. All strains were routinely incubated on PDA and CM plates for mycelial growth test [Citation19]. Liquid YEPD (w/v, 1% peptone, 0.3% yeast extract, 2% dextrose) was used to prepare mycelia for the extraction of genomic DNA and total RNA. Mung bean liquid (MBL) broth (30 g mung beans boiled in 1 L water for 20 min, then filtered through cheesecloth) was used to analyze induction of asexual reproduction. Water agar medium was used for conidia germination experiments [Citation20]. Carrot agar (CA) medium was used for self-crossing assays. Potato dextrose broth (PDB) was used for pigmentation production and total RNA extraction. Each experiment was repeated at least three times.
Gene deletion and complementation by protoplast transformation
The gene deletion mutants in the PH-1 background were generated with a homologous recombination strategy (Figure S1A), and the gene replacement cassette (HPH) was constructed with a double-joint PCR approach [Citation21]. Protoplast preparation and transformation were performed following previously described methods [Citation20]. The deletion mutants identified from hygromycin-resistant (100 µg/mL) transformants were verified by diagnostic PCR and further confirmed by Southern blotting (Figure S1B). For complementation assays, the native promoter and open-reading frame of FgSAH1 were amplified from the F. graminearum genome and co-transformed with pYF11 vector (neomycin-resistance marker NEO) harboring green fluorescent protein (GFP) into E. coli using described methods [Citation22]. The resulting pYF11 plasmids were transformed into protoplasts of the ΔFgSah1 mutant and the candidate transformants resistant to G418 (geneticin) at 100 µg/mL were screened with PCR and fluorescence signals. All primers used for these experiments are listed in Supplementary Table S1.
Quantification of AdoMet and AdoHcy production
The mycelia grown in YEPD for 2 d were harvested to measure the content of AdoMet and AdoHcy with HPLC procedures by Poirier et al. [Citation23] with minor modifications. Briefly, mycelial powder (1 g) grinded under liquid nitrogen was added to a 10 mL centrifuge tube. The tube was filled to 5 mL with ultrapure water, then underwent 10 minutes of homogenization and 5 minutes of ultrasonic vibration. The mixture was centrifugated at 13,000 rpm for 20 min, then the supernatant was aspirated, after which the experimental protocol followed the methods of Poirier [Citation23].
Asexual and sexual reproduction assays
For asexual reproduction assays, five mycelial plugs (5 mm in diameter) taken from the edge of a 3-d-old colony of each strain were added to a 50-mL triangular flask containing 30 mL of MBL broth. Each strain was assayed with three flasks and incubated at 25°C for 5 d with shaking (175 rpm). The number of conidia in each flask was calculated using methods by Zheng et al. [Citation24] and the experiment was repeated three times. In conidia germination experiments, the conidia of PH-1, ΔFgSah1 and ΔFgSah1-C were coated evenly on thin layer of water agar plates and incubated at 25°C for 8 h, then examined with an Olympus IX-71 microscope (Tokyo, Japan).
For self-crossing assays, 7-d-old aerial hyphae on carrot agar (CA) cultures were gently removed with a sterile toothpick with the addition of 1 mL autoclaved 2.5% Tween 20 following previously described protocol [Citation25,Citation26]. After additional 2 weeks of incubation at 25°C under a 12 h of light (bright fluorescent lights) −12 h of darkness diurnal cycle, perithecia produced on CA plates were observed and photographed using a stereo microscope (Nikon SMZ25). All the experiments were performed three times with three independent replicates.
Plant infection, penetration, and DON production assays
To test the pathogenicity of ΔFgSah1 on wheat coleoptiles and flowering wheat heads, virulence assays were performed as previously described [Citation27]. Briefly, for coleoptile pathogenicity, 2–3 mm sections from the top of 3-d-old seedlings were removed and the wound was inoculated with a 2.5-μL aliquot conidial suspension (3 × 105 spores/mL) of each strain. For wheat ear pathogenicity, a 10-μL aliquot of conidial suspension (3 × 105 spores/mL) was point-inoculated into a floret of flowering wheat head of Zhenmai 5, and sterile distilled water was used as negative control. More than 20 plants were inoculated with each strain, and disease symptoms were determined at 7 (coleoptiles) or 14 (wheat heads) d post inoculation (dpi). In addition, cellophane penetration assays of ΔFgSah1 mutants were tested using methods described before [Citation28]. Briefly, mycelial plugs (5 mm in diameter) on cellophane membranes placed on CM plates were cultured at 25°C for 3 d in the dark, then fungal colonies from each strain were removed with the cellophane membranes, and the plates were incubated for another 2 d under the same condition to assess the penetration of the mycelia.
DON production was assayed as described previously [Citation28,Citation29]. Briefly, healthy wheat grains soaked in sterile water for 24 h were divided into 250-mL triangular flasks (a 50-g aliquot) and autoclaved for 20 min twice. After being cooled to room temperature, each treatment was inoculated with 1 mL of spore suspension (3 × 105/mL) and shaken at intervals of 12 h to ensure even infection. After the mixture was incubated at 25°C for 20 d in the dark, DON production was determined using a DON ELISA Assay Kit (Wise, Zhenjiang, 465 China) according to methods described before [Citation29,Citation30]. The amount of fungal ergosterol in each sample was estimated and used as the internal control [Citation31]. The experiment was repeated three times with three replicates independently.
Microscopic examinations
Hyphal and conidial morphology were observed with an inverted microscope (Zeiss LSM710/780 Germany). The mycelia of each strain originated from the mycelial plugs which were cultured on cellophane membranes placed on solidified PDA plates for 2 d, and the conidia were harvested from MBL cultures after 5 d of cultivation. To determine spore morphology and septum number, conidia of each strain were stained with 10 µg/mL calcofluor white (CFW) and examined with a confocal laser scanning microscope (Leica TCS SP8, Germany). Lipid droplets (LDs) in mycelia were observed by staining with Nile Red as described before [Citation32].
Sensitivity of mycelial growth to multiple stresses
To test the sensitivity of ΔFgSah1 mutant to multiple stresses, mycelial growth was assayed on PDA plates without or with 1.2 M NaCl or KCl, 0.05% SDS (w/v), 0.05% Congo red (w/v), 8 mM hydrogen peroxide (H2O2). To test the sensitivity of ΔFgSah1 mutant against different fungicides, mycelial plugs (5 mm in diameter) taken from the fringe of a 3-d-old colony of each strain were placed on the center of PDA plates amended with fludioxonil (0, 0.003125, 0.00625, 0.0125, 0.025, 0.05, 0.1, and 0.2 µg/mL) or iprodione (0, 0.5, 1, 2, 4, 8, 16, and 32 µg/mL). The plates were cultivated under 25°C for 3 d in the dark, and all experiments were repeated three times independently.
Investigation of intracellular glycerol content
Two-d-old mycelia of each strain were harvested, and 100 mg samples of mycelia were ground under liquid nitrogen. The glycerol concentrations in mycelia were determined with a Glycerol Assay Kit (Applygen Technologies Inc., Beijing, China). The experiments were repeated three times independently.
Protein manipulation and western blotting
Protein extraction was performed using protocols described previously [Citation33]. The concentrations of the total proteins were measured with a Bradford Protein Assay Kit (Beijing Solarbio Science & Technology Co., Ltd.), and 0.05 mg of each sample was separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were further transferred to Immobilon-P transfer membranes (Millipore, Billerica, USA), then phosphorylation of FgHog1 and total FgHog1 were detected by Phospho-p38 MAPK (Thr180/Tyr182) Antibody #9211 (Cell Signaling Technology Inc., Boston, MA, USA) and anti-Hog1 antibody sc165978 (Santa Cruz, CA, USA), respectively. Protein abundance was quantified with ImageJ software. The experiments were performed three times independently.
Quantitative real-time PCR (qRT‑PCR)
Total RNAs of F. graminearum mycelia harvested from YEPD, PDB, cellophane layers placed on CM plates, and DON induction medium were extracted with the RNAiso reagent (Tiangen Biotech, Beijing, China). First-strand cDNA was synthesized with a HiScript® II 1st Strand cDNA Synthesis Kit* (Vazyme Biotech, Nanjing, China). The actin gene was used as the internal control, and the relative gene expression levels were evaluated by the threshold cycle (2−ΔΔCt) method [Citation34]. The primers used in this study are presented in Table S1.
Statistical analyses
All the experimental data were analyzed with the Date Processing System (DPS, version 7.05), and the means from triplicate samples were analyzed by Fisher’s least significant difference (LSD) test. Each error bar represents the standard deviation (SD) of three independent experiments.
Results
Identification of FgSAH1 in F. graminearum
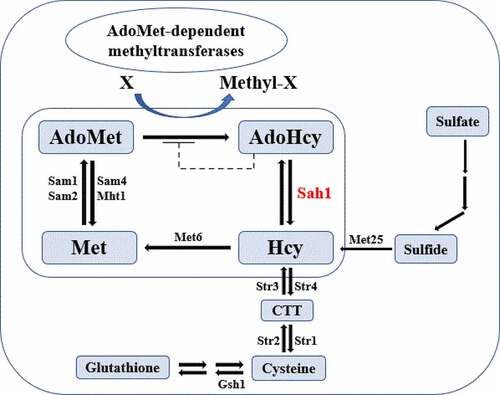
A hydrolase gene (FGSG_05615, designated FgSah1) was retrieved with a BLASTp search of the NCBI database using S. cerevisiae Sah1 (YER043 C) as a query. The FgSAH1 gene is 987 bp and encodes a 328 amino acid protein. Both homologous alignment () and phylogenetic analyses (Figure S2) which were based on the amino acid sequences indicated that the Sah1 protein of F. graminearum is evolutionarily conserved.
Figure 2. Identification of FgSah1 in F. graminearum
The identified Sah1 was primarily detected in the cytosol in yeast and found to localize to the nucleus and cytoplasm in A. thaliana [Citation8]. The signal of green fluorescence protein (GFP), tagged at the C terminus of FgSah1 to visualize its subcellular localization in F. graminearum, was observed to distribute evenly in the hyphae (), suggesting that FgSah1 is localized to the cytoplasm.
AdoMet-dependent methylation metabolism is blocked in ΔFgSah1
As the second most widely used enzyme substrate, AdoMet provides methyl groups for most methylation reactions in all organisms. In this process, it is converted into its demethylated metabolite AdoHcy, whose accumulation can work as the strong competitive inhibitor of all AdoMet-dependent methyltransferases [Citation8,Citation35]. To investigate whether FgSah1 is responsible for catabolizing AdoHcy in F. graminearum, the intracellular content of AdoMet and AdoHcy was determined, and results showed that the FgSAH1 deletion mutants produced sevenfold more AdoHcy and ninefold more AdoMet than those of WT PH-1 and the complemented strain ΔFgSah1-C (). This strongly suggests that the deletion of FgSAH1 leads to the accumulation of AdoHcy, which negatively affects the conversion of AdoMet to AdoHcy and then causes the accumulation of AdoMet as well (). It has also been reported that de novo biosynthesis of phosphatidylcholine (PC) from phosphatidylethanolamine (PE) is based on three sequential AdoMet-dependent methylation steps catalyzed by Cho2 (Phosphatidylethanolamine N-methyltransferase) and Opi3 (bifunctional phosphatidyl-N-methylethanolamine N-methyltransferase/phosphatidyl-N-dimethylethanolamine N-methyltransferase) both in yeast and in mammals [Citation8,Citation36]. In this study, we investigated the expression levels of Cho2 and an Opi3-like gene (FGSG_05066 and FGSG_08613) in ΔFgSah1, finding that the knockout mutants showed dramatically decreased mRNA expression levels of these two AdoMet-dependent methyltransferases (). All results demonstrate that FgSah1 is essential to methylation in F. graminearum.
FgSah1 is involved in vegetative growth and pigmentation in F. graminearum
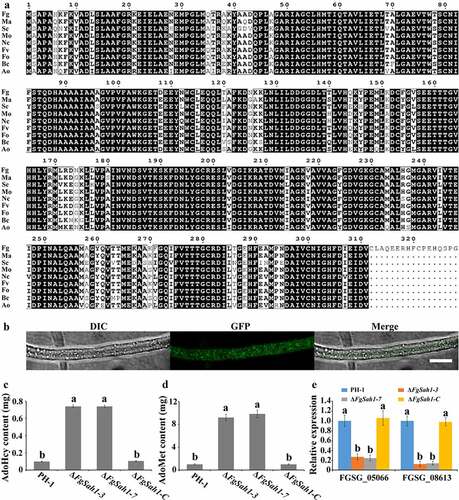
As shown in , the colonial growth rates of the FgSAH1 deletion mutants decreased dramatically compared to PH-1 and ΔFgSah1-C on PDA and CM media, and in contrast to the control strains, which produced abundant aerial hyphae, the ΔFgSah1 mutants produced fewer and shorter aerial hyphae. Moreover, microscopic examination showed that most hyphal branches of the deletion mutants had larger branching angles than those of PH-1 and ΔFgSah1-C (). Furthermore, after incubated in PDB for 5 ds, the ΔFgSah1 mutants produced yellow instead of red pigments (). After 2 weeks, the ΔFgSah1 mutants still failed to produce any red pigments in PDB medium, and the same results were observed on PDA and CM plates (date not shown). To further confirm this phenomenon, 2-d-old mycelia in PDB were harvested for RNA extraction, and relative gene expression levels of PKS12 and AURJ, known to be responsible for the synthesis of the red pigment aurofusarin [Citation37], were determined by qRT-PCR analyses. The results showed that the mRNA expression levels of the two genes were significantly down-regulated in the ΔFgSah1-3 and ΔFgSah1-7 mutants compared to PH-1 and ΔFgSah1-C (), indicating that FgSah1 plays a crucial role in pigment formation in F. graminearum.
Figure 3. Roles of FgSah1 in vegetative growth and pigmentation in F. graminearum
FgSah1 is important for both asexual and sexual reproduction
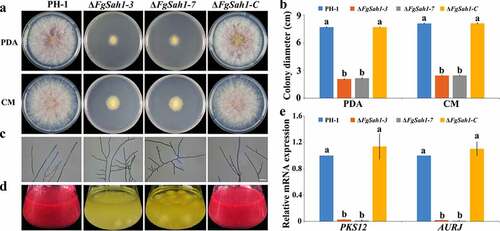
To investigate whether FgSah1 is involved in asexual development, the spore morphology and conidial production of PH-1, ΔFgSah1, and ΔFgSah1-C strains were examined in MBL medium. The number of conidia produced by ΔFgSah1 was significantly lower (about 70% less) compared to that of PH-1 and ΔFgSah1-C strains (), and the conidial germination rates (for 8 h on water agar at 25°C) were also dramatically decreased (). Microscopic observations also revealed that not only was the length of conidia produced by deletion mutants reduced (), but the number of septa of the conidia also declined in ΔFgSah1 in comparison to PH-1 and ΔFgSah1-C (). Apart from asexual reproduction, the sexual reproduction of ΔFgSah1 was assessed on carrot agar plates. After 2 weeks of incubation, the deletion mutants were sterile while the wild-type strain PH-1 and complemented strain ΔFgSah1-C produced abundant perithecia (). The above results suggest that FgSah1 is crucial to both asexual and sexual reproduction, and thus may be a potential fungicide target for controlling the spread of FHB.
Figure 4. Assays for asexual and sexual reproduction in the FgSAH1 deletion mutants
FgSah1 is required for full virulence
To test whether the FgSAH1 gene is involved in the pathogenicity of F. graminearum, infection tests were performed on wheat coleoptiles and flowering wheat heads. The deletion mutants of FgSAH1 exhibited reduced virulence and infections were limited to the wound of coleoptiles or the inoculated spikelet, whereas symptoms on coleoptiles and flowering wheat heads of PH-1 and ΔFgSah1-C were typically severe (). Previous studies reported that loss of virulence may be a result from defective penetration ability of the mutant strains [Citation38,Citation39]. This is affirmed in the present study, where cellophane penetration assays demonstrated that the deletion mutants failed to penetrate cellophane membranes (); even when incubated for an additional 2 weeks, the plates of deletion mutants produced no hyphae or colonies. This result suggests that FgSah1 plays important roles in penetration during infection. Apart from this, the mRNA expression levels of hydrophobins (FGSG_01764 and FGSG_09066), which have been reported to be involved in cellophane penetration and plant infection [Citation40,Citation41], were determined in all tested strains, and results showed that the relative expressions of the two hydrophobins in ΔFgSah1 were greatly down-regulated (). Disease assays on coleoptiles performed by spray inoculations were further confirmed the mutant loses the ability to infect healthy plants, suggesting that FgSah1 affects F. graminearum penetration during plant infection (Figure S3). All these results indicate that FgSah1 is essential to full virulence in F. graminearum.
Figure 5. Pathogenicity and cellophane penetration assays of the FgSAH1 deletion mutants
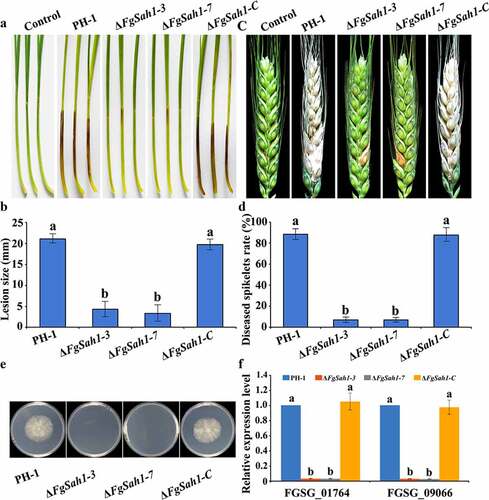
As a key pathogenic factor of F. graminearum [Citation42,Citation43], DON production was assessed in ΔFgSah1 infecting wheat grains. In contrast to tests on pathogenicity, DON production in ΔFgSah1 was significantly increased (). To further confirm this result, the relative expression levels of DON biosynthesis genes TRI5 and TRI6 in minimal synthetic (MS) liquid medium [Citation28] were analyzed by qRT‑PCR, and the results showed that expressions of TRI5 and TRI6 were significantly up-regulated (). All these results indicate that FgSah1 is required for full virulence but also functions redundantly to negatively regulate DON production.
Figure 6. DON production and TRI genes expression assays of the FgSAH1 deletion mutants
Sensitivity of the FgSAH1 deletion mutants to multiple stresses
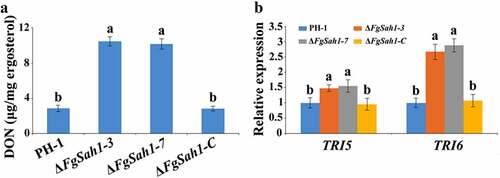
Full virulence of F. graminearum has been reported to be impaired by the depletion of genes involved in various stress responses, including osmotic, cell membrane, cell-wall related, and oxidative stresses [Citation22,Citation44]. Tolerance to environmental stresses is critical for plant infection, and thus the response of ΔFgSah1 to various stressors was examined. The FgSAH1 deletion mutants, wild-type strain PH-1, and the complemented strain ΔFgSah1-C were inoculated on PDA plates with each of 1.2 M NaCl or KCl (osmotic stresses), 0.05% SDS (membrane stress), 0.05% CR (cell wall stress), and 8 mM H2O2 (oxidative stresses). Intriguingly, the mycelia of ΔFgSah1 showed increased sensitivity to NaCl, KCl, SDS, and H2O2, but decreased sensitivity to CR compared to those of PH-1 and ΔFgSah1-C (). These results demonstrate that the hydrolase FgSah1 is also critical in the response to abiotic stresses in F. graminearum.
Figure 7. Sensitivity of the FgSAH1 deletion mutants to osmotic, membrane, cell wall damage, and oxidative stresses
FgSah1 is involved in lipid metabolism
Normally, external hyperosmotic stresses activate the high-osmolarity glycerol (HOG) pathway through which the intracellular glycerol content is regulated by Hog1 phosphorylation [Citation45,Citation46]. In this study, glycerol content and the phosphorylation of FgHog1 were examined. The results demonstrate that both under normal and hyperosmotic conditions, the deletion mutants exhibited decreased glycerol accumulation than PH-1 and ΔFgSah1-C (). In accordance with this result, the phosphorylation levels of FgHog1 in ΔFgSah1 were dramatically decreased compared to that of the wild-type strain PH-1 (). These results indicate that FgSah1 plays a significant role in regulating the activity of FgHog1 and the biosynthesis of glycerol, and in turn maintains osmotic homeostasis in F. graminearum in response to external osmotic stresses.
Figure 8. FgSAH1 is involved in lipid metabolism in F. graminearum
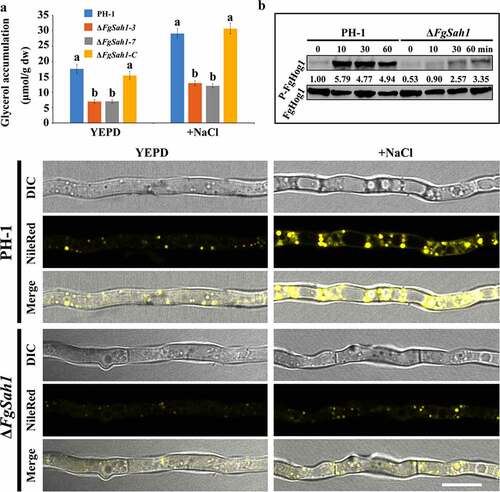
Since glycerol has been reported to be the raw material for LD biosynthesis [Citation47,Citation48], the accumulation of glycerol in ΔFgSah1 can be further confirmed by the formation of LDs. Mycelia of each strain were stained using Nile Red, a selective fluorescent stain for intracellular LDs [Citation49], and observed by fluorescence microscopy. As shown in , the FgSAH1 deletion mutant exhibited fewer LDs than those of PH-1 even under 1.2 M NaCl (2 h). All these results indicate that FgSah1 is required for cellular lipid homeostasis in F. graminearum.
Sensitivity of ΔFgSah1 to different fungicides
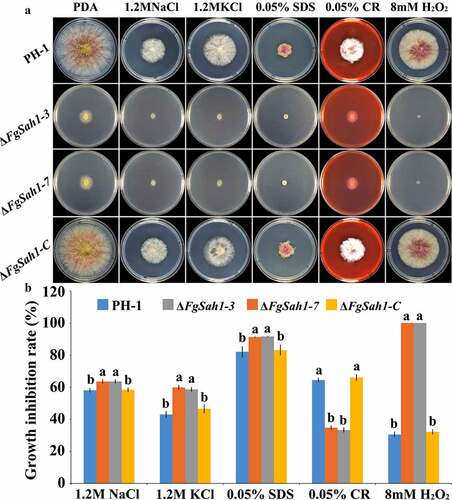
The HOG pathway has also been reported to be related to the sensitivity toward phenylpyrrole and dicarboximide fungicides in some fungal pathogens [Citation50]. Since our results above showed that the decrease of phosphorylation level of FgHog1 affected the accumulation of glycerol in ΔFgSah1, we speculate that FgSah1 is involved in the sensitivity of F. graminearum to fludioxonil and iprodione. To test this hypothesis, the effective concentration for 50% inhibition of mycelial growth (EC50) of fludioxonil and iprodione for each strain was tested, and results showed that the deletion mutants exhibited increased sensitivity to fludioxonil and iprodione than PH-1 and ΔFgSah1-C (), which further confirmed the involvement of FgSah1 in the accumulation of intracellular glycerol in F. graminearum.
Table 1. Sensitivity of ΔFgSah1 to different fungicides
Discussion
Methionine cycle is essential to growth and development of nearly all living organisms, and its dysfunction can cause various diseases and mortality [Citation7,Citation12–14]. In this study, we investigated the function of FgSAH1 which encodes the S-adenosyl-L-homocysteine hydrolase (Sah1). Knowing that the dysfunction of Sah1 is lethal in many organisms including mice and A. thaliana [Citation8], the survival of FgSAH1 deletion mutants indicates that an alternative pathway for Hcy synthesis may be present in F. graminearum, which could be similar to that in yeast (). In the disruption of FgSah1, the contents of AdoMet and AdoHcy in ΔFgSah1 mutants were significantly increased and the expression levels of AdoMet-dependent methyltransferases were remarkably down-regulated, consistent with that in yeast and mammals. The accumulation of AdoHcy functions as a negative regulatory factor to the conversion of AdoMet, to the extent that AdoMet-dependent methylation processes were significantly inhibited. In FgSAH1 deletion mutants, the accumulation of LDs was also decreased drastically, which indicates that FgSah1 is involved in lipid metabolism. All the above results indicate that FgSah1 plays crucial roles in methylation and lipid metabolism.
FgSah1 is essential to methylation metabolism in F. graminearum. Previous studies have demonstrated that only a few AdoMet-dependent methyltransferases are unique to yeast or humans while most are strongly conserved [Citation8]. In this study, we found that the expression levels of the two AdoMet-dependent methyltransferases orthologs (Cho2 and Opi3 in yeast) in F. graminearum were dramatically inhibited (). Consistent with reports in yeast, the lack of FgSah1 resulted in massive accumulation of AdoHcy and AdoMet (), which disrupted the coordinated methylation processes. These results demonstrate that FgSah1 is involved in the regulation of methylation metabolism in F. graminearum.
FgSah1 is also required for vegetative growth and full virulence in F. graminearum. In FgSAH1 deletion mutants, colony growth rate decreased significantly, and abnormal branching angles were observed in mycelia. The mutants also failed to produce the red pigment aurofusarin, an important secondary metabolite [Citation51]. Previous studies reported that the synthesis of aurofusarin is dependent on the PKS12 gene cluster [Citation52–54], whose primary product is nor-rubrofusarin (a new yellow/green compound) [Citation37]. This product is further catalyzed by AurJ to produce rubrofusarin. Interestingly, AurJ is an O-methyltransferase, and its expression may be inhibited by the accumulation of AdoHcy in this study (). The lack of AurJ leads to the accumulation of nor-rubrofusarin and thereby negatively regulates the expression of PKS12; it has also been reported that the ΔFgAurJ mutant lacks aurofusarin [Citation37,Citation55]. Additionally, both asexual and sexual reproduction were defective in ΔFgSah1 mutants; the production and germination rate of spores declined and the deletion mutants even produced no perithecia. Furthermore, the pathogenicity of ΔFgSah1 mutants to coleoptiles and flowering wheat heads were drastically lower than those of PH-1 and ΔFgSah1-C, which may have resulted from the defective penetration capability of the mutant strains. Some of the defects may partly result from the decreased expression of hydrophobins, which have been reported to be involved in many morphogenetic processes such as hyphal growth, attachment, water–air interface penetration, and plant infection [Citation40,Citation41]. However, in contrast to their reduced pathogenicity, production of the key pathogenic factor DON in ΔFgSah1 mutants increased dramatically compared to that in PH-1 and ΔFgSah1-C. Previous studies reported that the accumulation of acetyl-CoA, the basic precursor to DON biosynthesis, will furtherly stimulated DON biosynthesis in F. graminearum [Citation29,Citation30]. Under the catalysis of PKS12, one acetyl-CoA and 6 manonyl-CoA units are condensed and folded into nor-rubrofusarin in F. graminearum [Citation37]. In this study, deficiency in PKS12 may have led to the accumulation of acetyl-CoA, which can explain the increase in DON synthesis. All these results suggest that FgSah1 participates not only in vegetative growth but also in full virulence and DON production.
Tolerance toward environmental pressures is usually a prerequisite for pathogens to carry out normal physiological processes. Our results demonstrate that absence of FgSah1 greatly increased the sensitivity of ΔFgSah1 toward osmotic pressure. Moreover, the ΔFgSah1 mutants had lower tolerance toward membrane stress and oxidative stress, but also showed increased resistance to cell wall stressors compared to PH-1 (). Previous studies have reported that the effects of excessive oxidation stress can be reduced by the accumulation of pigments in filamentous fungi [Citation56]. We inferred that the increased sensitivity of ΔFgSah1 to oxidation stress may have resulted from the decreased ability to remove excessive H2O2. Our results above demonstrate that FgSah1 plays important roles in the response to multiple stresses.
Lastly, FgSah1 is involved in glycerol biosynthesis and lipid metabolism in F. graminearum. The HOG pathway regulates glycerol content in most organisms by MAPK Hog1 phosphorylation to maintain osmotic homeostasis [Citation46]. We determined that the accumulation of glycerol in all the tested strains with or without NaCl, and the results showed decreased glycerol in ΔFgSah1 in comparison to PH-1 and ΔFgSah1-C. Correspondingly, phosphorylation levels of Hog1 were also reduced in mutant strains (). The reduction of glycerol production in ΔFgSah1 also explained its increased sensitivity to fludioxonil and iprodione, which have been reported to be related to osmotic pressure [Citation50]. The lack of glycerol in deletion mutants leads to a sharp decrease in the synthesis of LDs, which has been reported to be crucial to full virulence and proper development [Citation57]. We inferred that FgSah1 may be partly responsible for the impaired infection abilities on the host due to the deficient lipid metabolism in mutant strains.
In summary, this work demonstrates that FgSah1 plays an integral role in the growth and metabolism of the filamentous fungus F. graminearum. We find that it is required for the methionine cycle, methylation metabolism, fungal development, full virulence, multiple stress responses, lipid metabolism, and fungicide tolerance.
Supplemental Material
Download Zip (1,000.1 KB)Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 31672065 and 31801778), the Chinese Key Research & Development Plans (2016YED0201007, 2018YFD0201201 and 2018YFD0201000), the Agricultural Science and Technology Projects of Jiangsu Province, China (BE2018378, BA2018039, PZCZ201715, CX(19)3003, and CX(18)2005), and the Postgraduate Research & Practice Innovation Program of Jiangsu Province, China (KYCX18_0670).We also thank Lian-Shan Li and Zong-Zhe He from Jiangsu Key Laboratory of Pesticide, Nanjing Agriculture University for helping test the content of AdoMet, AdoHcy and ergosterol.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The datasets generated or analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Starkey DE, Ward TJ, Aoki T, et al. Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genet Biol. 2007;44(11):1191–1204. .
- Bai G, Shaner G. Management and resistance in wheat and barley to fusarium head blight. Annu Rev Phytopathol. 2004;42(1):135–161.
- Pestka JJ, Smolinski AT. Deoxynivalenol: toxicology and potential effects on humans. J Toxicol Env Heal B. 2005;8(1):39–69.
- Paul PA, Lipps PE, Hershman DE, et al. Efficacy of triazole-based fungicides for fusarium head blight and deoxynivalenol control in wheat: a multivariate meta-analysis. Phytopathology. 2008;98(9):999–1011. .
- Chen Y, Zhou MG. Characterization of Fusarium graminearum isolates resistant to both carbendazim and a new fungicide JS399-19. Phytopathology. 2009;99(4):441–446.
- Li Y, Bing L, Liu L, et al. FgMon1, a guanine nucleotide exchange factor of FgRab7, is important for vacuole fusion, autophagy and plant infection in Fusarium graminearum. Sci Rep. 2016;5(1):18101. .
- Reed MC, Nijhout HF, Sparks R, et al. A mathematical model of the methionine cycle. J Theor Biol. 2004;226(1):33–43. .
- Tehlivets O, Malanovic N, Visram M, et al. S-adenosyl-L-homocysteine hydrolase and methylation disorders: yeast as a model system. Biochim Biophys Acta. 2013;1832(1):204–215. .
- Cantoni GL. Biological methylation: selected aspects. Annu Rev Biochem. 1975;44(1):435–451.
- Liu S, Wolfe MS, Borchardt RT. Rational approaches to the design of antiviral agents based on S-adenosyl-L-homocysteine hydrolase as a molecular target. Antiviral Res. 1992;19(3):247.
- Mushegian AR, Garey JR, Martin J, et al. Large-scale taxonomic profiling of eukaryotic model organisms: a comparison of orthologous proteins encoded by the human, fly, nematode, and yeast genomes. Genome Res. 1998;8(6):590.
- Miller MW, Duhl DM, Winkes BM, et al. The mouse lethal nonagouti (a(x)) mutation deletes the S-adenosylhomocysteine hydrolase (Ahcy) gene. EMBO J. 1994;13(8):1806–1816.
- Rocha Scf P, Sheikh M, Melchiorre R, et al. The Arabidopsis HOMOLOGY-DEPENDENT GENE SILENCING1 Gene Codes for an S-Adenosyl-L-Homocysteine hydrolase required for DNA Methylation-Dependent gene silencing. Plant Cell. 2005;17(2):404–417. .
- Malanovic N, Streith I, Wolinski H, et al. S-adenosyl-L-homocysteine hydrolase, key enzyme of methylation metabolism, regulates phosphatidylcholine synthesis and triacylglycerol homeostasis in yeast: implications for homocysteine as a risk factor of atherosclerosis. J Biol Chem. 2008;283(35):23989–23999. .
- Rao ST, Rossmann MG. Comparison of super-secondary structures in proteins. J Mol Biol. 1973;76(2):241–256.
- Brzezinski K, Dauter Z, Jaskolski M. High-resolution structures of complexes of plant-adenosyl–homocysteine hydrolase. Acta Cryst. 2012;D68:218–231.
- Guranowski A, Pawelkiewicz J. Adenosylhomocysteinase from yellow lupin seeds: purification and properties. Eur J Biochem. 2010;80(2):517–523.
- Brzezinski K, Bujacz G, Jaskolski M, et al. Purification, crystallization and preliminary crystallographic studies of plant S‐adenosyl‐l‐homocysteine hydrolase (Lupinus luteus). Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64(7):671–673. .
- Chen Y, Zheng SY, Ju ZZ, et al. Contribution of peroxisomal docking machinery to mycotoxin biosynthesis, pathogenicity and pexophagy in the plant pathogenic fungus Fusarium graminearum. Environ Microbiol. 2018;20(9):3224–3245. .
- Zheng ZT, Gao T, Hou YP, et al. Involvement of the anucleate primary sterigmata protein FgApsB in vegetative differentiation, asexual development, nuclear migration, and virulence in Fusarium graminearum. Fems Microbiol Lett. 2013;349(2):88–98.
- Yu JH, Hamari Z, Han KH, et al. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol. 2004;41(11):973–981. .
- Xu L, Wang H, Zhang C, et al. System-wide characterization of subtilases reveals that subtilisin-like protease FgPrb1 of Fusarium graminearum regulates fungal development and virulence. Fungal Genet Biol. 2020;144:103449.
- Poirier LA, Wise CK, Delongchamp RR. et al. Blood determinations of S-adenosylmethionine, S-adenosylhomocysteine, and homocysteine: correlations with diet. Cancer Epidem Biomar. 2001;10(6):649–655.
- Zheng Z, Gao T, Zhang Y, et al. FgFim, a key protein regulating resistance to the fungicide JS399-19, asexual and sexual development, stress responses and virulence in Fusarium graminearum. Mol Plant Pathol. 2014;15(5):488–499. .
- Bowden RL, Leslie JF. Sexual Recombination in Gibberella zeae. Phytopathology. 1999;89(2):182–188.
- Cavinder B, Sikhakolli U, Fellows KM, et al. Sexual development and ascospore discharge in Fusarium graminearum. J Vis Exp. 2012;61:3895.
- Shi D, Ren W, Wang J, et al. The transcription factor FgNsf1 regulates fungal development, virulence and stress responses in Fusarium graminearum. J Integr Agr. 2021;20(8):2156–2169. .
- Yin Y, Wang Z, Cheng D, et al. The ATP-binding protein FgArb1 is essential for penetration, infectious and normal growth of Fusarium graminearum. New Phytol. 2018;219(4):1447–1466. .
- Duan Y, Lu F, Zhou Z, et al. Quinone outside inhibitors affect DON biosynthesis, mitochondrial structure and toxisome formation in Fusarium graminearum. J Hazard Mater. 2020;398:122908.
- Zhou Z, Duan Y, Zhou M. Carbendazim-resistance associated β2-tubulin substitutions increase deoxynivalenol biosynthesis by reducing the interaction between β2-tubulin and IDH3 in Fusarium graminearum. Environ Microbiol. 2020;22(2):598–614.
- Davey BML, Nybakken AL, Kauserud BH, et al. Fungal biomass associated with the phyllosphere of bryophytes and vascular plants. ScienceDirect. 2009;113(11):1254–1260.
- Ren W, Liu N, Sang C, et al. The autophagy gene BcATG8 regulates vegetative differentiation and plant infection of Botrytis cinerea. Appl Environ Microb. 2018;84(11): 2455–17. DOI:10.1128/AEM.02455-17.
- Yu F, Gu Q, Yun Y, et al. The TOR signaling pathway regulates vegetative development and virulence in Fusarium graminearum. New Phytol. 2014;203(1):219–232. .
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–408.
- Lu SC. S-Adenosylmethionine. Int J Biochem Cell B. 2000;32(4):391–395.
- Tehlivets O, Hasslacher M, Kohlwein SD. S-adenosyl-L-homocysteine hydrolase in yeast: key enzyme of methylation metabolism and coordinated regulation with phospholipid synthesis. FEBS Lett. 2004;577(3):501–506.
- Frandsen R, Nielsen NJ, Maolanon N, et al. The biosynthetic pathway for aurofusarin in Fusarium graminearum reveals a close link between the naphthoquinones and naphthopyrones. Mol Microbiol. 2006;61(4):1069–1080. .
- Lv W, Wu J, Xu Z, et al. The putative histone-like transcription factor FgHltf1 is required for vegetative growth, sexual reproduction, and virulence in Fusarium graminearum. Curr Genet. 2019;65(4):981–994. .
- Gu Q, Zhang C, Liu X, et al. A transcription factor FgSte12 is required for pathogenicity in Fusarium graminearum. Mol Plant Pathol. 2015;16(1):1–13.
- Kershaw MJ, Talbot NJ. Hydrophobins and repellents: proteins with fundamental roles in fungal morphogenesis. Fungal Genet Biol. 1998;23(1):18–33.
- Quarantin A, Hadeler B, Kröger C, et al. Different hydrophobins of fusarium graminearum are involved in hyphal growth, attachment, water-air interface penetration and plant infection. Front Microbiol. 2019;10:751.
- Bai GH, Desjardins AE, Plattner RD. Deoxynivalenol-nonproducing fusarium graminearum causes initial infection, but does not cause disease spread in wheat spikes. Mycopathologia. 2002;153(2):91–98.
- Seong KY, Pasquali M, Zhou X, et al. Global gene regulation by Fusarium transcription factors Tri6 and Tri10 reveals adaptations for toxin biosynthesis. Mol Microbiol. 2010;72(2):354–367.
- Yun Y, Liu Z, Zhang J, et al. The MAPKK FgMkk1 of Fusarium graminearum regulates vegetative differentiation, multiple stress response, and virulence via the cell wall integrity and high-osmolarity glycerol signaling pathways. Environ Microbiol. 2014;16(7):2023–2037. .
- Ren W, Liu N, Yang Y, et al. The sensor proteins BcSho1 and BcSln1 Are involved in, though not essential to, vegetative differentiation, pathogenicity and osmotic stress tolerance in botrytis cinerea. Front Microbiol. 2019;10:328.
- Burg MB, Kwon ED, Kültz D. Osmotic regulation of gene expression. FASEB J. 1996;10(14):1598.
- Farese RV, Walther TC. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 2009;139(5):855–860.
- Walther TC, Farese JRV. Lipid droplets and cellular lipid metabolism. Annu Rev Biochem. 2012;81(1):687.
- Greenspan P, Mayer EP, Fowler SD. Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol. 1985;100(3):965–973.
- Kojima K, Takano Y, Yoshimi A, et al. Fungicide activity through activation of a fungal signalling pathway. Mol Microbiol. 2004;53(6):1785–1796. .
- Westphal KR, Wollenberg RD, Herbst FA, et al. Enhancing the production of the fungal pigment aurofusarin in Fusarium graminearum. Toxins (Basel). 2018;10(11):485. .
- Kim JE, Han KH, Jin J, et al. Putative polyketide synthase and laccase genes for biosynthesis of aurofusarin in Gibberella zeae. Appl Environ Microb. 2005;71(4):1701–1708. .
- Malz S, Grell MN, Thrane C, et al. Identification of a gene cluster responsible for the biosynthesis of aurofusarin in the Fusarium graminearum species complex. Fungal Genet Biol. 2005;42(5):420–433. .
- Gaffoor I, Brown DW, Plattner R, et al. Functional analysis of the polyketide synthase genes in the filamentous fungus Gibberella zeae (Anamorph Fusarium graminearum). Eukaryot Cell. 2005;4(11):1926. .
- Liu Y, Liu N, Yin Y, et al. Histone H3K4 methylation regulates hyphal growth, secondary metabolism and multiple stress responses in Fusarium graminearum. Environ Microbiol. 2016;17(11):4615–4630. .
- Rangel DE, Alder-Rangel A, Dadachova E, et al. Fungal stress biology: a preface to the fungal stress responses special edition. Curr Genet. 2015;61(3):231–238. .
- Liu N, Yun Y, Yin Y, et al. Lipid droplet biogenesis regulated by the FgNem1/Spo7-FgPah1 phosphatase cascade plays critical roles in fungal development and virulence in Fusarium graminearum. New Phytol. 2019;223(1):412–429. .