ABSTRACT
Feline viral diseases, such as feline panleukopenia, feline infectious peritonitis, and feline coronaviral enteritis, seriously endanger the health of cats, and restrict the development of pet industry. Meanwhile, there is a current lack of effective vaccines to protect against feline viral diseases. Thus, effective therapeutic agents are highly desirable. Interferons (IFNs) are important mediators of the antiviral host defense in animals, particularly type I IFNs. In this study, a novel feline IFN omega (feIFN-ω) gene was extracted from the cat stimulated with feline parvovirus (FPV) combined with poly(I:C), and following codon optimization encoding the feIFN-ω, the desired gene (feIFN-ω’) fragment was inserted into plasmid pPICZαA, and transformed into Pichia pastoris GS115, generating a recombinant P. pastoris GS115 strain expressing the feIFN-ω’. After induction, we found that the expression level of the feIFN-ω’ was two times more than that of feIFN-ω (p < 0.01). Subsequently, the feIFN-ω’ was purified and modified with polyethylene glycol, and its antiviral efficacy was evaluated in vitro and in vivo, using vesicular stomatitis virus (VSV) and FPV as model virus. Our results clearly demonstrated that the feIFN-ω’ had significant antiviral activities on both homologous and heterologous animal cells in vitro. Importantly, the feIFN-ω’ can effectively promote the expression of antiviral proteins IFIT3, ISG15, Mx1, and ISG56, and further enhance host defense to eliminate FPV infection in vivo, suggesting a potential candidate for the development of therapeutic agent against feline viral diseases.
Introduction
Interferon (IFN) is an antiviral protein found from influenza virus-infected chicken embryos in 1957, and subsequent studies have demonstrated that human and animal cells also can produce IFNs, which are considered as the main natural immune barrier for the host against viral infections [Citation1–5]. IFNs are the main choice of antiviral drugs in the current clinical use. According to the structure, functional characteristics, and receptor of IFNs, IFNs can be divided into three subtypes including type I, type II and type III [Citation6]. Among them, type I IFNs play an important role in promoting host defense against virus infections, including IFN-α, IFN-β, IFN-ε, IFN-ω, IFN-κ, IFN-δ, IFN-τ, and IFN-ζ, in particular IFN-α, and IFN-ω, with potent immunomodulatory, antiviral, and antiproliferative properties [Citation7]. Type II IFN, also known as IFN-γ, can induce a series of immune responses and regulate immune system, which acts as an antiviral agent by mainly inhibiting viral activity [Citation8]. Type III IFN, or IFN-λ family, is composed of IFN-λ1, IFN-λ2, IFN-λ3, and IFN-λ4, with antiviral activity [Citation9,Citation10].
Nowadays, it is more and more common to keep companion animals, especially pet cats. However, cat viral diseases, such as feline infectious peritonitis caused by feline infectious peritonitis virus (FIPV), feline panleukopenia caused by feline parvovirus (FPV), and cat enteritis caused by feline enteric coronavirus seriously endanger the health of cats and the development of the pet cat industry [Citation11–14]. In addition, there is a current lack of effective vaccines to protect against cat viral diseases. Therefore, effective therapeutic agents for cat viral diseases are highly desirable. IFN-ω, first reported in 1985 [Citation15], is secreted primarily by leukocytes, which has now been found in humans [Citation16] and some animals including cats [Citation17,Citation18], pigs [Citation19], and horses [Citation20], etc. IFN-ω combines with IFN receptor complex and activates phosphatidylinositol-3-kinase/protein kinase B (P13K/Akt) signal pathway to exert antiviral activity, thereby achieving the effect of inhibiting viruses [Citation21,Citation22]. Compared with IFN-α, IFN-ω can significantly inhibit virus replication [Citation23–25], suggesting a promising antiviral agent.
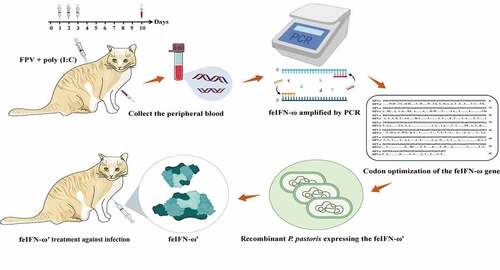
In this study, in order to develop a potent antiviral agent for the treatment of cat viral diseases, a gene encoding a novel feline IFN-ω (feIFN-ω) was obtained from the peripheral blood of the cat stimulated with FPV combined with poly(I:C). Following codon optimization of the gene encoding feIFN-ω, the desired feIFN-ω’ was produced by a recombinant Pichia pastoris strain, and after being modified with polyethylene glycol, the antiviral efficacy of the feIFN-ω’ was evaluated in vitro and in vivo.
Materials and methods
Animal, viruses, bacterial strain, cells, and plasmid
Healthy 4-month-old Dragon Li cats (n = 20) were purchased from a pet market in China. Animal experiments were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, and were approved by the Ethical Committee for Animal Experiments of Zhejiang A&F University (ZAFUAC2021021), China. Vesicular stomatitis virus (VSV) and feline parvovirus (FPV) were kept in our laboratory. Pichia pastoris strain GS115 (P. pastoris GS115) kept in our laboratory was cultured in Yeast-extract Peptone Dextrose (YPD) medium (Solarbio, China) at 30°C. Madin–Darby bovine kidney (MDBK) cells, Madin–Darby canine kidney (MDCK) cells, and feline kidney (F81) cells were cultured in Dulbecco’s Modified Eagle Medium, DMEM (Gibco, USA) supplemented with 10% fetal bovine serum, FBS (Gibco, USA) in a 5% CO2 incubator at 37°C. The plasmid pPICZαA kept in our laboratory was used to construct recombinant P. pastoris expressing feline IFN-ω.
Cloning of gene encoding feIFN-ω
The cat was co-stimulated by 500 µL of FPV (103 TCID50) and 500 µL of 1.0 mg/mL poly(I:C) (Sigma, USA) through subcutaneous injection route for three consecutive days. On day 10 post-stimulation, the peripheral blood of the cat was collected, and the total RNA was extracted by Total RNA Isolation kit (Invitrogen, USA) followed by reverse transcription, generating first-strand cDNA. Next, using the cDNA as template, a gene encoding feIFN-ω was obtained by PCR amplification with the primer pair listed in . After that, the PCR product of interest was purified and subcloned into pMD-19 T plasmid, generating recombinant plasmid pMD-ω, followed by gene sequencing (Kumei, China).
Table 1. Primers used in this study
Gene sequence analysis
The gene encoding feIFN-ω were deposited in GenBank under accession number MT754935. Homology and phylogenetic tree analysis of the feIFN-ω gene were performed using DNASTAR and MEGA7 software.
Codon optimization of the feIFN-ω gene sequences
In this study, we aimed to produce the feIFN-ω by yeast to test its antiviral efficacy in vitro and in vivo, and therefore we optimized the codons of the gene encoding the feIFN-ω according to the codon usage bias of P. pastoris, including predominant codons usage, the adjustment of GC content, and AT-rich repeat region. However, no amino acid sequences of the feIFN-ω were changed. Subsequently, the optimized gene (named feIFN-ω’) was synthesized (Kumei, China), which was harbored in a recombinant plasmid pMD-ω’.
Expression of the feIFN-ω’ by recombinant P. pastoris and modification with polyethylene glycol
The recombinant plasmid pMD-ω containing the feIFN-ω gene (or pMD-ω’ containing the optimized feIFN-ω’ gene) and vector pPICZαA were, respectively, digested by BamH I and Kpn I (NEB, USA), and the gene fragments of interest were purified, followed by ligation with T4 DNA ligase (Takara, China), generating the recombinant plasmid pPICZαA-ω (or pPICZαA-ω’). Next, the recombinant plasmid pPICZαA-ω (or pPICZαA-ω’) was linearized by AVRII enzyme digestion (NEB, USA), and then the linearized pPICZαA-ω (or pPICZαA-ω’) was electroporated into the P. pastoris GS115 competent cells, followed by screening positive clones on YPD plates supplemented with 100 µg/mL of Zeocin (Sigma, USA), generating the recombinant strains named GS115-pPICZαA-ω or GS115-pPICZαA-ω’. In order to produce the feline interferon, the recombinant strain GS115-pPICZαA-ω (or GS115-pPICZαA-ω’) was grown overnight to an OD600 of approximately 1.5 in BMGY medium (Solarbio, China). After centrifugation, the cells pellets were transferred into BMMY medium containing 1.5% YNB, biotin, and 0.05% methanol (Solarbio, China), and cultured for 120 h. Sterile-filtered methanol was supplemented every 24 h to maintain induction condition. The expression level of the feIFN-ω in GS115-pPICZαA-ω (and GS115-pPICZαA-ω’) and its culture supernatants was analyzed by 12% SDS-PAGE. After that, the feIFN-ω’ was purified by Ni2+ affinity chromatography column, and PEGylation was conducted in sodium borate buffer with the feIFN-ω’ and 40 kDa mPEG-2-N-hydroxysuccinimide, and then the polyethylene glycol-modified feIFN-ω’ was purified by a Q-Sepharose column [Citation26].
Determination of antiviral activity of the feIFN-ω’ in vitro
The antiviral activity of the recombinant feIFN-ω’ was determined by microdose cytopathogenic effect inhibition assay (MCIA) in vitro using VSV and FPV as model viruses. The supernatants from induced GS115-pPICZαA-ω’ were serially 10-fold diluted in DMEM supplemented with 5% FBS, and were then added into a 96-well plate (8 replicates per dilution) followed by addition of MDBK cells to a concentration of 5 × 104 cells/well. Then, the MDBK cells were incubated at 37°C in a 5% CO2 incubator for 24 h. After discarding the supernatants, the MDBK cells were infected with VSV (or FPV) at an MOI of 1.0. When the cytopathic effect (CPE) reached 100% in virus control group (untreated with feIFN-ω’), the cells were stained with 0.1% (w/v) of crystal violet, and incubated at 37°C for 30 min. After elution with 50% ethanol-0.1% acetic acid, absorbance was measured at 595 nm. In parallel, the supernatants from GS115-pPICZαA were used as negative control. Subsequently, the antiviral activity of the feIFN-ω’ purified by Ni2+ affinity chromatography column at a concentration of 0.001 ng ~ 10 ng was determined by MCIA, using FPV as model virus and INTERCAT IFN (Toray Industries, Japan) as feline IFN positive control. The test was repeated five times. Furthermore, the species-specific antiviral activity was also determined by the ability of the feIFN-ω’ to inhibit the CPE of VSV on F81 cells, MDCK cells, and MDBK cells. In parallel, the INTERCAT IFN was used as feline IFN control. The assay was repeated three times (8 replicates each sample).
Determination of antiviral activity of the polyethylene glycol-modified feIFN-ω’ in vivo
In order to determine the antiviral efficacy of the polyethylene glycol-modified feIFN-ω’ in vivo, eighteen 4-month-old Dragon Li cats were randomly divided into three groups: FPV infection group (cat was infected with 105 TCID50 FPV via subcutaneous and digestive routes; n = 6), feIFN-ω’ treatment group (cat was infected with FPV followed by treatment with the polyethylene glycol-modified feIFN-ω’ at a dose of 150 μg/0.5 mL, twice, 5 days apart; n = 6), and normal control group (mock group; n = 6). On days 5 and 15 after challenge, three cats were randomly selected from each group, respectively, and the viral loads of the FPV in blood, kidney, liver, spleen, intestine, and feces of the cats were determined using a SYBR Green I-based real-time quantitative RT-PCR (RT-qPCR) assay with the primer pair targeting the FPV VP2 gene (listed in ). Meanwhile, the presence of viral antigen in the intestinal tract of the cats from each group was detected on day 10 after the primary feIFN-ω’ treatment by an immunohistochemistry (IHC) assay using a mouse anti-FPV VP2 polyclonal antibody (prepared in our laboratory; diluted at 1:100) as the primary antibody and HRP-conjugated goat anti-mouse IgG antibody (Abcam, USA; diluted at 1:1000) as the secondary antibody. Moreover, the cats in each group were bled on days 0, 5, 10, and 15 after FPV infection using the blood-diluting pipettes, followed by total white blood cell (WBC) counts with Fuchs-Rosenthal counting chambers. At the same time, using β-actin as internal control, the mRNA transcript levels of antiviral proteins Mx1, ISG15, ISG56, and IFIT3, and cytokines including IL-1β, TNF-α, IL-4, IL-6, IL-10, and IL-12 in the blood samples of the cats from each group were determined on day 10 after feIFN-ω’ treatment by RT-qPCR assay with the primer pairs listed in . In addition, the protein expression levels of the antiviral proteins Mx1, ISG15, ISG56, and IFIT3 were determined by Western blot using rabbit anti-Mx1/ISG15/ISG56/IFIT3 polyclonal antibody (ABclonal, USA; diluted at 1:1000) as the primary antibody and HRP-conjugated goat anti-rabbit IgG antibody (Abcam, USA; diluted at 1:2000) as the secondary antibody, respectively. After that, the immunoblot band was visualized using chemiluminescent substrate reagent (Thermo Fisher Scientific, USA).
Statistical analysis
In this work, data were shown as mean ± standard error (SE) values, and Tukey’s multiple comparison tests and one-way analysis of variance (ANOVA) were used to analyze the differences among groups by GraphPad Prism V8.0 software.
Results
Cloning and phylogenetic analysis of feIFN-ω gene
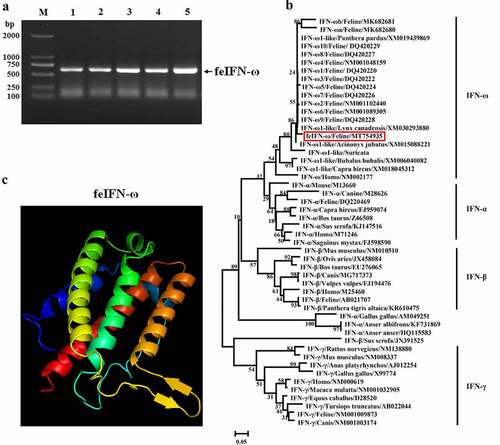
After being stimulated by FPV combined with poly(I:C), a gene of approximately 612 bp encoding feIFN-ω was amplified by RT-PCR from the peripheral blood of the cat (), and phylogenetic analysis of the feIFN-ω gene was performed by the neighbor-joining method (1000 replicates) using MEGA 7 software to explore the evolutionary relationships between the feIFN-ω gene obtained in this study and other IFNs published in GenBank. As shown in , the feIFN-ω gene that was obtained in this study belonged to the type I IFN family, which shared relatively distant genetic relationships with other feIFN-ωs published in GenBank, indicating that a novel feIFN-ω was obtained in this study. We further predicted the three-dimensional (3D) structure of the feIFN-ω by an online tool (http://www.sbg.bio.ic.ac.uk), and result demonstrated that the 3D structure of the feIFN-ω conformed to the 3D structural characteristics of type I IFN ().
Figure 1. A: Amplification of feIFN-ω gene by RT-PCR. M: DNA Marker DL2000; lanes 1–5: the feIFN-ω gene amplified from the peripheral blood of cat. B: Phylogenetic tree analysis of the feIFN-ω gene by neighbor-joining method (1000 replicates) using MEGA 7 software. C: Predicted three-dimensional structure of the feIFN-ω.
Codon optimization of the gene encoding feIFN-ω and its expression by P. pastoris
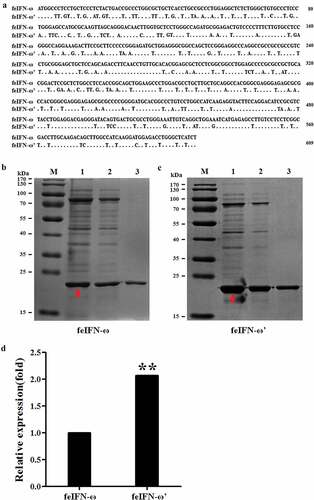
In order to achieve highly efficient expression of the feIFN-ω by P. pastoris, the gene sequences encoding the feIFN-ω were optimized according to the codon usage bias of P. pastoris (). Subsequently, the recombinant strain GS115-pPICZαA-ω expressing feIFN-ω and strain GS115-pPICZαA-ω’ expressing feIFN-ω’ (optimized feIFN-ω) was constructed, respectively. Following induction with methanol, the IFN protein of interest that was expressed by the recombinant strains was detected by SDS-PAGE, and results showed that the feline IFN-ω can be effectively produced by the recombinant strain GS115-pPICZαA-ω () and strain GS115-pPICZαA-ω’ (). By contrast, we found that the expression level of optimized feIFN-ω’ was two times more than that of unoptimized feIFN-ω (p < 0.01) ().
Figure 2. A: Codon optimization of the feIFN-ω according to the codon usage bias of P. pastoris. B: The expression level of feIFN-ω by the GS115-pPICZαA-ω before codon optimization. M: Protein marker; lane 1: feIFN-ω in cells; lane 2: feIFN-ω in supernatants; lane 3: purified feIFN-ω. C: The expression level of feIFN-ω’ by the GS115-pPICZαA-ω’ after codon optimization. M: Protein marker; lane 1: feIFN-ω’ in cells; lane 2: feIFN-ω’ in supernatants; lane 3: purified feIFN-ω’. D: Comparison of feIFN-ω expression levels before and after codon optimization.
Biological activity of the recombinant feIFN-ω’ in vitro
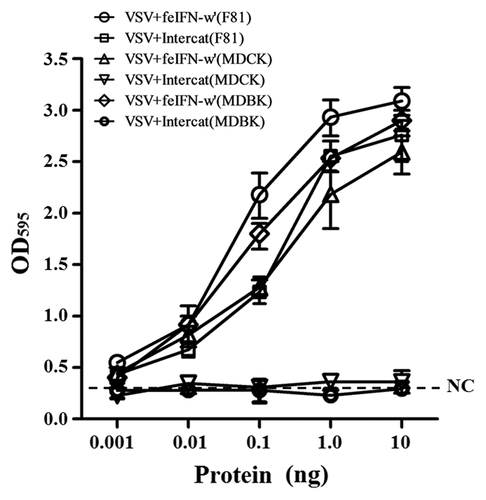
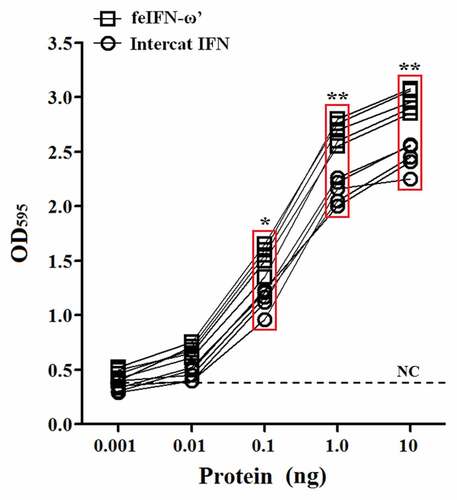
In order to evaluate the biological activity of the recombinant GS115-expressed feIFN-ω’, we first tested the antiviral activity of the supernatants of the recombinant strain GS115-pPICZαA-ω’ induced with methanol, using VSV and FPV as model viruses. As shown in , a significant reduction in the VSV- and FPV-induced CPE on the MDBK cells can be observed, exhibiting an effective antiviral activity with a dose-dependence. Subsequently, we tested the antiviral activity of the purified recombinant feIFN-ω’ expressed by the GS115-pPICZαA-ω’ using INTERCAT IFN as feline interferon-ω positive control, and results showed that the purified feIFN-ω’ displayed significant antiviral activity with a dose-dependence (). By contrast, the antiviral activity of the recombinant feIFN-ω’ was better than that of the INTERCAT IFN control. Furthermore, we tested the species-specific antiviral activity of the purified feIFN-ω’ by determining the ability to inhibit the CPE of VSV on F81 cells, MDCK cells, and MDBK cells, using INTERCAT IFN as a control. As shown in , a significant reduction in the VSV-induced CPE on the F81 cells, MDCK cells, and MDBK cells can be observed with a dose-dependence in the feIFN-ω’ treatment cell groups. However, in INTERCAT IFN treatment cell groups, significant reduction in the VSV-induced CPE on F81 cells can only be observed, but not on MDCK cells, and MDBK cells.
Figure 3. Antiviral activity of the supernatants of the GS115-pPICZαA-ω’ induced by methanol. F81 cells were incubated in the presence of the indicated dilutions of supernatants containing feIFN-ω for 24 h, followed by infection with VSV and FPV, respectively. When CPE in virus control group reached 100%, the cells were stained with crystal violet followed by measuring the absorbance at 595 nm. The connecting curve (red line in the figure) in each group was set up using GraphPad Prism 8.0 software.
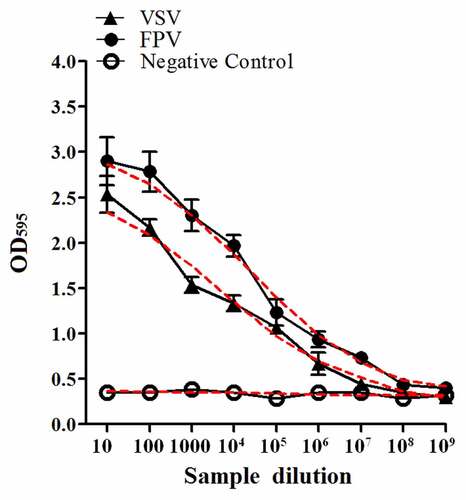
Figure 4. Antiviral activity of the purified feIFN-ω’ using FPV as model virus. F81 cells grown to confluence were treated with the indicated amounts of purified feIFN-ω’ for 24 h, followed by infection with FPV at an MOI of 1.0. When CPE in virus control group reached 100%, the cells were stained with crystal violet followed by measuring the absorbance at 595 nm. In parallel, Intercat IFN was used as positive control. *p < 0.05; **p < 0.01.
Antiviral activity of the recombinant feIFN-ω’ in vivo
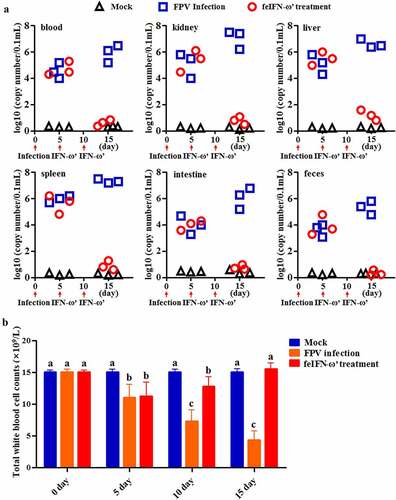
In order to evaluate the antiviral activity of the recombinant feIFN-ω’ in vivo, the cats were infected with FPV followed by the feIFN-ω’ therapy. Our results showed that after the feIFN-ω’ treatment, virus loads in blood, kidney, liver, spleen, intestine, and feces of the cats in the feIFN-ω’ treatment group significantly decreased on days 10 after feIFN-ω’ treatment, indicating effective antiviral activity of the recombinant feIFN-ω’ in vivo (). However, in FPV-infected cat group (without feIFN-ω’ treatment), the virus titers in blood, kidney, liver, spleen, intestine, and feces of the cats significantly increased, and some cats developed severe clinical symptoms and were euthanized after the experiment. During a 15-day monitoring period, we counted the total WBC in blood of the cats in each group, and results showed that compared to the normal control group, the total WBC counts in FPV-infected cat group significantly decreased, while the total WBC counts of the FPV-infected cats were gradually returned to normal after treatment with the feIFN-ω’ ().
Figure 6. A: Changes of viral loads in blood, kidney, liver, spleen, intestine, and feces of the cats in normal control group, FPV infection group, and feIFN-ω treatment group determined by a RT-qPCR assay. B: The total WBC counts of the cats in each group.
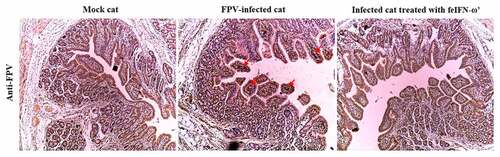
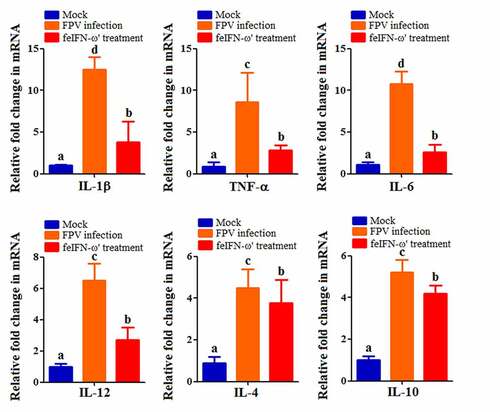
In addition, on day 10 after the feIFN-ω’ treatment, we determined the mRNA transcript levels and protein expression levels of the antiviral proteins Mx1, ISG15, ISG56, and IFIT3 in the blood of the cats from each group using RT-qPCR assay and Western blot assay, respectively. As shown in , both the mRNA transcript levels () and protein expression levels () of these antiviral proteins were significantly higher than those in mock control group (p < 0.01) and FPV infection group (p < 0.05), indicating that the feIFN-ω’ can effectively promote the expression of antiviral proteins. Furthermore, we also determined the mRNA transcript levels of cytokines IL-1β, TNF-α, IL-4, IL-6, IL-10, and IL-12 in the blood samples of the cats from each group by RT-qPCR, and results showed that compared to the FPV infection group, the levels of these cytokines significantly decreased (p < 0.05 or p < 0.01) in the feIFN-ω’ treatment, but still significantly higher (p < 0.05) than those in the mock control group (). Finally, we used the IHC assay to detect the FPV loaded in the intestinal tract of the cat from each group on day 10 after feIFN-ω’ treatment, and found that there was no virus detected in the feIFN-ω’ treatment group, while large amounts of viruses were observed in the FPV infection group ().
Figure 7. The mRNA transcript levels (a) and protein expression levels (b) of antiviral proteins Mx1, ISG15, ISG56, and IFIT3 in the blood of the cats from each group were detected by SYBR Green I RT-qPCR and Western blot, respectively, using β-actin as internal control. The lowercase letters “a versus b, and b versus c” indicate significant difference of p < 0.05; “a versus c” indicates significant difference of p < 0.01.
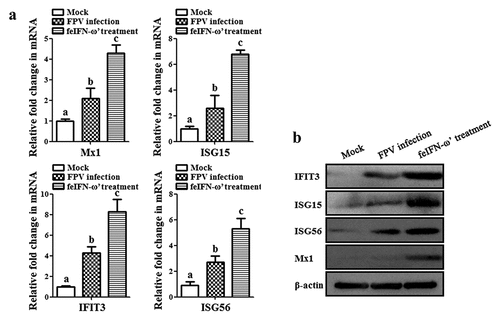
Figure 8. The mRNA transcript levels of cytokines IL-1β, TNF-α, IL-4, IL-6, IL-10, and IL-12 in the blood samples of the cats from each group were determined by RT-qPCR assay on day 10 after feIFN-ω’ treatment. The lowercase letters “a versus b, b versus c, and c versus d” indicate significant difference of p < 0.05; “a versus c, and b versus d” indicates significant difference of p < 0.01; “a versus d” indicates significant difference of p < 0.001.
Discussion
Viral diseases in pet cats such as feline leukemia, feline panleukopenia, and feline infectious peritonitis, with high incidences, are still difficult to prevent due to the lack of effective vaccine products. IFN with good antiviral activity shows positive effects for treating viral diseases in cats [Citation27–30]. In this study, a gene encoding a novel feIFN-ω was cloned from the peripheral blood of cat stimulated with FPV combined with ploy (I:C), and with codon optimization of the feIFN-ω gene, the IFN-ω was produced by a recombinant P. pastoris induced with methanol, and its antiviral activity was evaluated in vitro and in vivo.
Protein expression system includes prokaryotic and eukaryotic systems. Although prokaryotic expression system is most commonly used [Citation18], the activity of expressed proteins is not good. For example, using prokaryotic system to produce interferon, it is necessary to extract the inclusion bodies in cells to obtain the protein of interest, followed by denaturation, refolding, conformational change in vitro, which is time-consuming and laborious, increases the production cost, and also leads to serious protein loss with some biological activity loss [Citation31,Citation32]. Yeast expression system is a kind of eukaryotic expression system, and the expressed target protein can complete the modified folding closest to the natural protein in cells [Citation33], and can be secreted into the supernatants, which is conducive to target protein extraction and purification, achieving the goal of low cost and high return [Citation34]. Therefore, we used a yeast (P. pastoris) expression system to produce the feIFN-ω in this study, and our result showed that the feIFN-ω can be effectively expressed by P. pastoris and secreted into supernatants.
Importantly, the efficient expression of exogenous protein in P. pastoris is related to the conformity of expression system, selection of vectors, expression conditions, and other factors [Citation35,Citation36]. Moreover, rare codons will affect the transcription and translation; the difference between the foreign gene and the host will affect the expression of the target protein; continuously repeated AT bases will end transcription in advance [Citation35]. Therefore, codon optimization is a commonly adopted strategy for improved protein expression [Citation36]. In this study, in order to further improve the expression level of the feIFN-ω in recombinant P. pastoris, the gene sequences encoding the feIFN-ω were subjected to optimization according to the codon usage bias for P. pastoris, such as using high-frequency dominant codon, reducing the GC content, adjusting AT-rich regions, etc. [Citation37–39]. With the codon optimization, the expression level of the feIFN-ω’ (after optimization) by the recombinant GS115-pPICZαA-ω’ was nearly two times higher than that of the feIFN-ω (before optimization) expressed by the recombinant GS115-pPICZαA-ω, indicating that the optimization of codon usage bias for P. pastoris could significantly improve protein expression of the interest.
Subsequently, we systematically assessed the antiviral activity of the feIFN-ω’ in vitro and in vivo. In vitro, we first analyzed the antiviral activity of the supernatants secreted by GS115-pPICZαA-ω’, and results showed that the supernatants displayed effective antiviral activity with a dose-dependence, and significant reduction in CPE on the MDBK cells infected by VSV and FPV was observed, while not observed in negative control group. Then, we purified the recombinant feIFN-ω’ and further tested its antiviral activity, and our data showed that the purified feIFN-ω’ also displayed effective antiviral activity, and significant reduction in the FPV-induced CPE can be observed. By contrast, the antiviral effect of the feIFN-ω’ obtained in this study was better than that of INTERCAT IFN. Utilizing a standard VSV assay, we evaluated the antiviral effect of the feIFN-ω’ on F81 cells (feline), MDBK cells (bovine), and MDCK cells (canine) using the INTERCAT IFN as positive control, and our data showed that the feIFN-ω’ showed highly efficient antiviral effect against VSV on homologous and heterologous animal cells. However, the INTERCAT IFN only showed effective antiviral effect against VSV on homologous animal cells, but not on MDBK and MDCK, which was consistent with previous report [Citation17].
Before performing in vivo experiments to evaluate the antiviral effect of the recombinant feIFN-ω’, in order to enhance the stability of the feIFN-ω’ and extend its half-life in vivo, the feIFN-ω’ was modified with polyethylene glycol. And then, the animal experiment was carried out. The cats were infected with feline parvovirus (FPV), followed by feIFN-ω’ treatment, using FPV-infected cats without interferon treatment as control. We found that the viral loads in blood, kidney, liver, spleen, intestine, and feces of the FPV-infected cats with the feIFN-ω’ treatment (feIFN-ω’ treatment group) significantly decreased, and the virus could be effectively eliminated detected by IHC assay, indicating that the FPV replication was effectively inhibited. However, the viral loads in FPV-infected cats without feIFN-ω’ treatment (FPV infection group) continued to increase. FPV infection can cause panleukopenia in cats. So, we determined the total WBC levels in the cats from mock control group, FPV infection group, and feIFN-ω’ treatment group. Our data showed that the total WBC counts significantly decreased after virus infection, and with the feIFN-ω’ treatment, the total WBC counts in feIFN-ω’ treatment group were gradually returned to normal, but not in FPV infection group, indicating effective therapeutic effect of the feIFN-ω’. To further explore the antiviral effect of the feIFN-ω’, the mRNA levels and protein levels of the antiviral proteins Mx1, ISG15, ISG56, and IFIT3 in the blood of the cats from each group were determined, and from these results, we clearly see that the feIFN-ω’ treatment can significantly promote the expression of antiviral proteins Mx1, ISG15, ISG56, and IFIT3, which would further exert their antiviral functions to inhibit the replication of FPV. Many studies have also demonstrated that interferon ω could stimulate the expression of the antiviral proteins ISGs [Citation40,Citation41]. In addition, we also found that the mRNA transcript levels of pro-inflammatory cytokines IL-1β, TNF-α, IL-6, and IL-12, and anti-inflammatory cytokines IL-4, and IL-10 tended to decrease (p < 0.05 or p < 0.01) in the cats from the feIFN-ω’ treatment group, compared with the FPV infection group. These findings are consistent with an overall reduction of pro-inflammatory pathways in cats treated with IFN [Citation24].
A global overview of the gene clone, codon optimization, expression, and antiviral effect evaluation of a novel feIFN-ω is given in . In conclusion, we cloned a gene encoding a novel feIFN-ω, optimized its codons according to the codon usage bias of P. pastoris, and achieved highly efficient expression of the feIFN-ω from a recombinant P. pastoris. The antiviral efficacy of the feIFN-ω was subsequently evaluated, and our results clearly demonstrated that the feIFN-ω displayed effective antiviral activity against VSV and FPV, suggesting a promising therapeutic agent for viral diseases in cats.
Acknowledgments
This work was supported by the Research and Development Fund of Zhejiang A&F University (grant number 2021FR034), the National Key R&D Program of China (grant number 2016YFD0501003), and Heilongjiang Provincial Natural Science Foundation of China (grant number LH2021C046).
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials. https://www.ncbi.nlm.nih.gov/nuccore/MT754935.1
Additional information
Funding
References
- Sedger LM. MicroRNA control of interferons and interferon induced anti-viral activity. Mol Immunol. 2013;56(4):781–793.
- Russell KD, Sergei VK, Joan ED. Interferon induction and function at the mucosal surface. Immunol Rev. 2013;255(1):25–39.
- Mou C, Pan S, Wu H, et al. Disruption of interferon-β production by the N pro of atypical porcine pestivirus. Virulence. 2021;12(1):654–665.
- Fanunza E, Carletti F, Quartu M, et al. Zika virus NS2A inhibits interferon signaling by degradation of STAT1 and STAT2. Virulence. 2021;12(1):1580–1596.
- Shang J, Zheng Y, Mo J, et al. Sox4 represses host innate immunity to facilitate pathogen infection by hijacking the TLR signaling networks. Virulence. 2021;12(1):704–722.
- Lin JD, Feng N, Sen A, et al. Distinct roles of type I and type III interferons in intestinal immunity to homologous and heterologous rotavirus infections. PloS Pathog. 2016;12(4):e1005600.
- Minayoshi Y, Maeda H, Yanagisawa H, et al. Development of Kupffer cell targeting type-I interferon for the treatment of hepatitis via inducing anti-inflammatory and immunomodulatory actions. Drug Deliv. 2018;25(1):1055–1065.
- Platanias LC. Mechanisms of type I and type II interferon-mediated signaling. Nat Rev Immunol. 2005;5(5):375–386.
- Lopusna K, Rezuchova I, Betakova T, et al. Interferons lambda, new cytokines with antiviral activity. Acta Virol. 2013;57(2):171–179.
- Gao D, Yu X, Zhang B, et al. Role of autophagy in inhibiting the proliferation of A549 cells by type III interferon. Cell Biol Int. 2019;43(6):605–612.
- Stuetzer B, Hartmann K. Feline parvovirus infection and associated diseases. Vet J. 2014;201(2):150–155.
- Cheng N, Zhao Y, Han Q, et al. Development of a reverse genetics system for a feline panleukopenia virus. Virus Genes. 2019;55(1):95–103.
- Cave T, Thompson H, Reid S, et al. Kitten mortality in the United Kingdom: a retrospective analysis of 274 histopathological examinations (1986 to 2000). Vet Rec. 2002;151(17):497–501.
- Domenech A, Miro G, Collado VM, et al. Use of recombinant interferon omega in feline retrovirosis: from theory to practice. Vet Immunol Immunopathol. 2011;143(3–4):301–306.
- Freaney JE, Zhang Q, Yigit E, et al. High density nucleosome occupancy map of human chromosome 9p21-22 reveals chromatin organization of the type I interferon gene cluster. J Interf Cytok Res. 2014;34(9):676–685.
- Xu C, Song X, Fu L, et al. Antiviral potential of exogenous human omega interferon to inhibit pandemic 2009 A (H1N1) influenza virus. Viral Immunol. 2011;24(5):369–374.
- Yang LM, Xue QH, Sun L, et al. Cloning and characterization of A Novel feline IFN-ω. J Interf Cytok Res. 2007;27(2):119–127.
- Wang X, Li F, Han M, et al. Cloning, prokaryotic soluble expression, and analysis of antiviral activity of two novel feline IFN-ω proteins. Viruses. 2020;12(3):335.
- Sang Y, Rowland RR, Hesse RA, et al. Differential expression and activity of the porcine type I interferon family. Physiol Genomics. 2010;42(2):248–258.
- Detournay O, Morrison DA, Wagner B, et al. Genomic analysis and mRNA expression of equine type I interferon genes. J Interf Cytok Res. 2013;33(12):746–759.
- Seo Y, Kim M, Choi M, et al. Possible role of phosphoinositide-3-kinase in Mx1 protein translation and antiviral activity of interferon-omega-stimulated HeLa cells. Pharmacology. 2011;87(3–4):224–231.
- Klarquist J, Cantrell R, Lehn MA, et al. Type I IFN drives experimental systemic lupus erythematosus by distinct mechanisms in CD4 T cells and B cells. Immunohorizons. 2020;4(3):140–152.
- de Mari K, Maynard L, Sanquer A, et al. Therapeutic effects of recombinant feline interferon-omega on feline leukemia virus (FeLV)-infected and FeLV/feline immunodeficiency virus (FIV)-coinfected symptomatic cats. J Vet Intern Med. 2004;18(4):477–482.
- Leal RO, Gil S, Duarte A, et al. Evaluation of viremia, proviral load and cytokine profile in naturally feline immunodeficiency virus infected cats treated with two different protocols of recombinant feline interferon omega. Res Vet Sci. 2015;99:87–95.
- Skorvanova L, Svancarova P, Svetlikova D, et al. Protective efficacy of IFN-ω AND IFN-λs against influenza viruses in induced A549 cells. Acta Virol. 2015;59(4):413–417.
- Du Y, Tian H, Gao XD, et al. Pharmacokinetic properties of a 40 kDa branched polyethylene glycol-modified form of consensus interferon-α (PEG-CIFN) in Rhesus monkeys. Biopharm Drug Dispos. 2008;29(8):481–484.
- Leal RO, Gil S, Sepúlveda N, et al. Monitoring acute phase proteins in retrovirus infected cats undergoing feline interferon-ω therapy. J Small Anim Pract. 2014;55(1):39–45.
- Matsumoto H, Teshima T, Iizuka Y, et al. Evaluation of the efficacy of the subcutaneous low recombinant feline interferon-omega administration protocol for feline chronic gingivitis-stomatitis in feline calicivirus-positive cats. Res Vet Sci. 2018;118:324–330.
- Liu Y, Liu X, Kang H, et al. Identification of feline interferon regulatory factor 1 as an efficient antiviral factor against the replication of feline calicivirus and other feline viruses. Biomed Res Int. 2018;8:1–10.
- Fiorito F, Cantiello A, Granato GE, et al. Clinical improvement in feline herpesvirus 1 infected cats by oral low dose of interleukin-12 plus interferon-gamma. Comp Immunol Microbiol Infect Dis. 2016;48:41–47.
- Vincentelli R, Canaan S, Campanacci V, et al. High-throughput automated refolding screening of inclusion bodies. Protein Sci. 2004;13(10):2782–2792.
- Feng Y, Zhang M, Zhang L, et al. An automatic refolding apparatus for preparative-scale protein production. PLoS ONE. 2012;7(9):e45891.
- Cregg JM, Cereghino JL, Shi J. Recombinant protein expression in Pichia pastoris. Mol Biotechnol. 2000;16(1):23–52.
- Wang QH, Gao LL, Liang HC, et al. [Research advances of the influence factors of high level expression of recombinant protein in Pichia pastoris]. Acta Pharma Sin. 2014;49(12):1644–1649.
- Baghban R, Farajnia S, Ghasemi Y, et al. New developments in Pichia pastoris expression system, review and update. Curr Pharm Biotechnol. 2018;19(6):451–467.
- Xu Y, Liu K, Han Y, et al. Codon usage bias regulates gene expression and protein conformation in yeast expression system P. pastoris. Microb Cell Fact. 2021;20(1):91.
- Kudla G, Lipinski L, Caffin F, et al. High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol. 2006;4(6):e180.
- Kiktev DA, Sheng Z, Lobachev KS, et al. GC content elevates mutation and recombination rates in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci. 2018;115(30):E7109–E7118.
- Scott VL, Shack LA, Eells JB, et al. Immunomodulator expression in trophoblasts from the feline immunodeficiency virus (FIV)-infected cat. Virol J. 2011;8(1):336.
- Shields LE, Jennings J, Liu Q, et al. Cross-species genome-wide analysis reveals molecular and functional diversity of the unconventional interferon-ω subtype. Front Immunol. 2019;10:1431.
- Gao M, Guo Y, Luo X, et al. Design, biological activity and signaling pathway of bovine consensus omega interferon expressed in Pichia pastoris. Mol Immunol. 2019;106:46–52.
- Taglinger K, Van Nguyen N, Helps CR, et al. Quantitative real-time RT-PCR measurement of cytokine mRNA expression in the skin of normal cats and cats with allergic skin disease. Vet Immunol Immunopathol. 2008;122(3–4):216–230.