ABSTRACT
Fungi and bacteria often co-exist and physically or chemically interact with each other in their natural niches. This inter-kingdom species interaction is exemplified by the gram-positive bacterial pathogen Streptococcus mutans and opportunistic fungal pathogen Candida albicans, which co-exist in the human mouth. It has been demonstrated that the dynamic interaction between these two species plays a critical role in their virulence and biofilm development. In this study, we discovered that S. mutans represses filamentous development and virulence in C. albicans through secreting a secondary metabolite, mutanocyclin (a tetramic acid). Mutanocyclin functions by regulating the PKA catabolic subunit Tpk2 and its preferential binding target Sfl1. Inactivation of Tpk2 in C. albicans results in an increased sensitivity to mutanocyclin, whereas overexpression of Tpk2 leads to an increased resistance. Dysfunction of SFL1 and its downstream target genes overrides the hyphal growth defect caused by mutanocyclin. Further investigation demonstrates that three glycosylphosphatidylinositol (GPI)-anchored proteins (Spr1, Hyr4, and Iff8), associated with cell wall biogenesis and remodeling, and a set of filamentous regulators also contribute to the mutanocyclin response. We propose that both transcriptional regulation and cell wall composition contribute to mutanocyclin-mediated filamentous inhibition. This repressive effect of mutanocyclin could function as a natural regulator of filamentous development in C. albicans.
Introduction
Interactions between bacteria and fungi are common in the environment and human body. These interactions include various modes ranging from mutualism to antagonism and may result in changes to the pathogenicity of one or both partners [Citation1]. Candida albicans is a major opportunistic fungal pathogen of humans and exists as a commensal in the mouth, gastrointestinal tract (GI) tract, genital tract, and on the skin [Citation2–4]. In these natural niches, many bacterial species co-exist with the fungus and affect its growth and pathogenicity [Citation5,Citation6]. For example, Streptococcus mutans, a major bacterial pathogen of dental caries, and C. albicans are common residents of the human oral cavity [Citation7]. Both species have been thought to be tightly associated with the development of caries, especially in immunocompromised individuals [Citation8–10]. Therefore, the inter-kingdom interaction between S. mutans and C. albicans has been considered to have a synergistic effect on this disease.
The S. mutans-C. albicans association has pleiotropic effects that could be antagonistic or cooperative. For example, the two species can form a dual biofilm that may enhance the virulence of S. mutans and the onset and severity of dental caries [Citation11]. Within oral biofilms, the two species produce biofilm-derived metabolites that modulate gene expression and affect the cell behaviors and colony formation of each other [Citation11,Citation12]. C. albicans has been shown to induce expression of the S. mutans virulence factor glucosyltransferase B (GtfB). In turn, GtfB catalyzes the production of α-glucan and binds to the mannan layer of the C. albicans cell wall [Citation13,Citation14]. The cell wall-associated polypeptide of S. mutans antigen I/II mediates adhesion to C. albicans cells and promotes acid production within the dual-species biofilm [Citation15]. Conversely, the relationship between C. albicans and S. mutans can be antagonistic. S. mutans produces quorum-sensing molecules (for example, CSP) or peptides (for example, SM-F2), which repress hyphal formation and virulence of C. albicans [Citation16,Citation17]. Therefore, the inter-kingdom species interactions among bacteria and fungi are complex. The interactions between C. albicans and S. mutans could be associated with a healthy and balanced community.
The ability of C. albicans to switch between the yeast and filamentous forms is critical to its virulence [Citation4,Citation18,Citation19]. Filamentous cells are more invasive than yeast-form cells and cause extensive damage to host organs. Many host-associated environmental factors and multiple signaling pathways are involved in the regulation of yeast-filamentous transitions in C. albicans [Citation4,Citation18,Citation19]. For example, high environmental temperatures, neutral ambient pH, high levels of CO2, and serum induce the development of filaments, whereas an acidic pH, low temperatures, and rich nutrient conditions are conducive to the yeast form of C. albicans. A recent report has shown that saliva facilitates the growth of C. albicans hyphae under in vitro culture conditions [Citation20]. Among the regulators of morphological changes, the conserved cAMP/protein kinase A (PKA) pathway plays a major role in C. albicans [Citation21,Citation22]. Moreover, the Ste11-Hst7-Cek1/Cek2-mediated MAPK pathway, Rim101-mediated pH-sensing pathway, and Tup1/Nrg1 transcriptional repressors also play critical roles in the filamentous growth of C. albicans [Citation23–27].
In this study, we report that S. mutans inhibits the development of filaments and affects the virulence of C. albicans by secreting a tetramic acid, mutanocyclin. This inhibitory effect of mutanocyclin could directly or indirectly affect the activity of the cAMP/PKA signaling pathway. We also found that a set of transcription factors, such as Efg1, Ahr1, Nrg1, Fcr1, and Sfl1, and several GPI-anchored proteins (Spr1, Hyr4, and Iff8) were involved in this regulation. Animal experiments demonstrated a protective effect of mutanocyclin on infections caused by C. albicans. Our findings uncover a novel link between the two species and provide new insights into the interspecies interactions between bacteria and fungi in the host.
Materials and methods
Strains, libraries, media, and culture conditions
C.albicans cells from stocks stored at −80°C were streaked on YPD agar plates (2% peptone, 1% yeast extract, 2% glucose and 2% agar, wt/vol) and incubated overnight at 30°C. Peptone and yeast extract were purchased from Oxoid Ltd. Company (Hants, UK). Glucose was from Beijing Chemical Works (Beijing, China) and agar was from Sangon Biotech (Shanghai, China). For routine culture, C. albicans cells were plated on YPD agar medium and incubated for 24 hours at 30°C. Cells of single colonies were then collected, dilutedand used for morphological analysis. S. mutans strains were cultured in brain heart infusion medium (BHI; Oxoid, Basingstoke, United Kingdom) plus .5% sucrose [Citation28], referred to herein as BHI-sucrose medium [Citation29]. Sucrose was purchased from Beijing Chemical Works (Beijing, China). BHI-sucrose and BHI + Lee’s glucose (50% Lee’s glucose mixed with 50% BHI broth) media were used for the filamentation assays [Citation29,Citation30]. To obtain supernatant, S. mutans was grown in liquid BHI-sucrose broth at 30°C. After 24 hours of incubation, the bacterial cultures were centrifuged, filtered, and stored at 4°C for further use. A deletion mutant library of 165 transcription factors and a mutant library of cell wall-related genes were used to screen essential elements involved in filamentation during interspecies interactions [Citation31,Citation32]. For library screening, C. albicans strain SN250 served as the WT control. All strains used in this study are listed in Table S1.
Disruption of the mutanocyclin synthesis gene cluster in S. mutans
An in-frame deletion mutant of the S. mutans mutanocyclin gene cluster was constructed by two rounds of homologous recombination. The 2.1-kb counter selection marker IFDC2 cassette was amplified using the primer pair ldhF1-BsaI/ermR1-BsaI with pIFDC2 as a template [Citation33]. A .7-kb and .8-kb fragment flanking the gene cluster were PCR amplified using primers 35C2-upF/35C2-upR-BsaI and 35C2-dnF-BsaI/35C2-dnR using the genomic DNA of S. mutans 35, a clinically isolated S. mutans strain, as a template [Citation34]. The two fragments and IFDC2 cassette were ligated using the Golden Gate cloning assay and transformed into strain S. mutans 35. Colonies with resistance to erythromycin on BHI plates were selected and PCR-verified with primers 35C2-upF1/35C2-dnR1. The IFDC2 cassette was then removed. The .7-kb fragments flanking the IFDC2 cassette were PCR-amplified with primers 35C2-upF/35C2-upR3-BsaI and 35C2-dnF3-BsaI/35C2-dnR, ligated using Golden Gate cloning, and transformed to remove the IFDC2 cassette. Colonies resistant to p-Cl-Phe on BHI plates were selected and PCR-verified.
Spectroscopic analysis
Detection of mutanocyclin in the pure culture of S. mutans and S. mutans Δmuc or in the mixed culture with C. albicans was performed using high-performance liquid chromatography (HPLC) with the chemically synthesized standard as a positive control. Briefly, the cells were cultured in BHI-sucrose medium at 30°C for 72 hours and subjected to centrifugation. The supernatants were extracted three times with an equal volume of ethyl acetate, evaporated under a vacuum, and dissolved in methanol. HPLC detection was carried out on a Shimadzu HPLC system (Shimadzu, Kyoto, Japan) using a C18 column (4.6 × 250 mm, 5 μm, Apollo, Alltech, Lexington, Kentucky, USA) under gradient elution conditions. The detection wavelength was 235 nm. The column was developed with solvent A (H2O with .1% (v/v) formic acid) and acetonitrile at a flow rate of 1.0 mL/min. The percentage of acetonitrile was maintained at 5% over 0–5 min, changed from 5 to 40% over 5–25 min and changed from 40 to 100% over 25–40 min.
Chemical synthesis of mutanocyclin
Mutanocyclin was chemically synthesized from Boc-protected D-Leu as previously described [Citation34]. Briefly, Boc-D-Leu was coupled with Meldrum’s acid in the presence of 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide (EDC) hydrochloride and dimethylaminopyridine (DMAP). Cyclization of the lactam ring was achieved by heating in MeOH to provide N-protected tetramic acid. After removing the Boc group by TFA, C3-acetylation to generate mutanocyclin was performed with acetyl chloride (AcCl) in the presence of boron trifluoride-diethyl ether complex (BF3 OEt2).
Growth curve assays
C. albicans cells of the WT strain SC5314 were initially grown in liquid YPD at 30°C for 24 hours and then collected and washed twice with 1x PBS (10 mM phosphate buffer, 2.7 mM KCl, 137 mM NaCl, pH 7.4). Approximately 1 × 105 cells were inoculated into 3 mL of YPD medium plus DMSO containing 8 µg/mL, 16 µg/mL, or 32 µg/mL of mutanocyclin. The cells were incubated at 30°C with shaking at 200 rpm. Cell densities were detected at different time points by measuring the optical density at 600 nm (OD600).
Filamentation assays
C. albicans cells were plated on YPD agar medium and incubated at 30°C for 24 hours. Approximately 1 × 104 cells were inoculated intofilamentous growth medium containing S. mutans cells, the supernatant, or mutanocyclin. The samples were incubated at 30°C for 24 to 48 hours. The degree of filamentation was defined according to both the length and percentage of filamentous cells. Approximately 200 cells were counted in each sample.
Subcellular localization of mutanocyclin
C. albicans cells (1 × 104 cells/well) were incubated in liquid Lee’s glucose medium containing 32 µg/mL mutanocyclin or an equal volume of vehicle (.32% DMSO) at 30°C for 30 min or 24 hours. Cells were then collected and washed with 1x PBS. Images were obtained using a confocal laser scanning microscope. The excitation wavelength for mutanocyclin was 254 nm.
Animals
All animal experiments were performed according to the guidelines approved by the Animal Care and Use Committee of Fudan University, approval code: SYXK(hu)2020–0032. Four-five weeks old female BALB/c mice weighing 18–20 g were used for animal experiments.
Histopathological assay
Ex vivo tongue infection assays were performed as described previously [Citation35]. The tongues were excised from sacrificed female BALB/c mice, and placed in each well of a 24-well plate. C. albicans cells (1 × 107) in 5 μL of PBS containing DMSO (v/v, .32%) or mutanocyclin (32 μg/mL) were spotted on the tongues (ex vivo infection). After 24 hours of incubation at 37°C tongues were fixed in 10% buffered formalin, washed twice with PBS, dehydrated, and embedded in paraffin wax. Paraffin blocks were sectioned and stained with periodic acid-Schiff (PAS) for microscopy assays [Citation36].
Scanning electron microscopy (SEM) assay
SEM assays were performed as described previously with slight modifications [Citation35]. C. albicans cells (1 × 107) in 5 μL of PBS containing DMSO (v/v, .32%) or mutanocyclin (32 μg/mL) were spotted on the tongues excised from sacrificed female BALB/c mice (ex vivo infection). After 24 hours of incubation at 37°C tongues were fixed in 2.5% glutaraldehyde, and washed three times with PBS. The samples were then dehydrated using gradually increasing concentrations of ethanol, dried, and coated with gold. The prepared samples were imaged using a Phenom Prox Scanning Electron Microscope.
Galleria mellonella infection model
To determine the in vivo effects of mutanocyclin, a G. mellonella survival assay was performed according to a previously described methodology [Citation37]. G. mellonella in the final stage of the larval phase and weighing approximately 300 mg were stored in the dark and used within 7 days of shipment. Ten microliters of C. albicans (SC5314) inoculums (1 x 108 CFU/mL) containing only DMSO (v/v, .32%) or different concentrations of mutanocyclin was injected into larvae through a 10-µL microsyringe. A group of 20 larvae was used per concentration, and the larvae inoculated only with 1x PBS were used to show that death was not due to needle trauma. After injection, the larvae were incubated at 30°C and the number of dead G. mellonella was recorded at 12, 24, 36, 48, 60, and 72 hours. A larva was considered dead when it showed no response to touch.
RNA extraction and quantitative real-time PCR (Q-RT-PCR)
Q-RT-PCR assays were performed according to our previous publications with modifications [Citation30]. C. albicans cells were cultured in liquid Lee’s glucose medium as described in the main text. Cells were harvested, and total RNA was extracted using Thermo Scientific GeneJET RNA Purification kits (Waltham, USA) according to the manufacturer’s instructions. cDNA was synthesized with an oligo(dT) 18 primer (Tm, 32.1; 100 μM) from .6 µg of total RNA using Thermo Scientific RevertAid H Minus Reverse Transcriptase (Waltham, USA). Q-RT-PCR was performed using TOYOBO SYBR green master mix (QPS-201) on a Bio-Rad CFX96 real-time PCR detection system. The signal from each experimental sample was normalized to the expression of the ACT1 gene.
RNA-Seq analysis
C.albicans cells were spread on YPD agar plates and incubated at 30°C for 24 hours, after which they were inoculated into liquid Lee’s glucose medium containing DMSO (v/v, .32%) or mutanocyclin (32 µg/mL) and incubated at 30°C for 24 hours. Two biological repeats were performed for each condition. Cells were harvested, and total RNA was extracted as described above. RNA-Seq analysis was performed by the company Berry Genomics (Beijing, China) as described previously [Citation30]. Briefly, approximately 10 million (M) reads were sequenced in each library of samples. The library products were sequenced using an Illumina NovaSeq platform. The raw reads were filtered by removing adapter and low-quality reads (Phred score ≤10) using the FASTX-Toolkit v.0.14. Clean reads were mapped to the genome of C. albicans SC5314 using HiSat v2.0.5 with default parameters. Mismatches less than two bases were allowed in the alignments. Relative gene expression levels were calculated using the Fragments Per kb per Million reads (FPKM) method, FPKM = 106C/(NL/103), where C is the number of fragments uniquely aligned to gene A, N represents the total number of fragments uniquely aligned to all genes, and L is the number of gene A bases. To be considered significantly differentially expressed, a gene must satisfy three criteria: (1) an FPKM value ≥20 at least in one sample, (2) a fold change value ≥1.5 (except for the functional categories sheet of Data set S1, in which a 2-fold change cutoff was used), and (3) an adjusted P-value (false discovery rate [FDR]) < .05. GO functional enrichment analysis was carried out according to GO terminology determined using the online CGD GO Term Finder tool (http://www.candidagenome.org/cgi-bin/GO/goTermFinder).
Results
S. mutans inhibits filamentous growth of C. albicans
When grown in BHI-sucrose or BHI + Lee’s glucose medium at 30°C C. albicans was able to develop robust filaments ( and S1). These culture conditions also support the growth of S. mutans. To explore the effect of S. mutans on the growth of filaments, we mixed C. albicans and S. mutans cells and cultured them statically in the same medium at 30°C and 37°C As shown in and S1a, the growth of C. albicans filaments was almost blocked in both media in the presence of S. mutans cells, revealing a remarkable inhibitory effect of the bacterial cells on the development of C. albicans filaments.
Figure 1. S. mutans inhibits filamentous growth of C. albicans at 30°C . (a) C. albicans SC5314 cells (1 × 106) were mixed with S. mutans cells (1 × 107) and co-cultured in BHI-sucrose medium or BHI + Lee’s glucose medium at 30°C for 24 hours. (b) the supernatant of S. mutans was mixed with C. albicans SC5314 cells (1 × 106) and incubated in BHI-sucrose medium or BHI + Lee’s glucose medium at 30°C for 24 hours. the “-” sign indicates that no filamentous cells were observed; “±”, “+”, “++”, and “+++”, represent 1–10%, 10–30%, 30–50%, and 50–70% of filamentous cells, respectively. Scale bar, 20 µm.
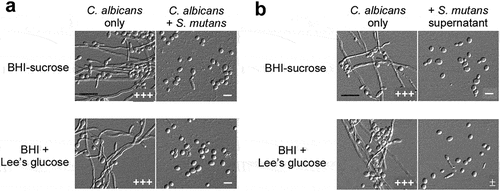
We suspected that S. mutans cells secreted some secondary metabolites into the medium that led to the inhibitory effect on C. albicans filamentation. Therefore, we next examined whether the S. mutans culture supernatant had a similar effect. As expected, the supernatant suppressed the development of filaments of C. albicans in both media ( and S1b). Our findings indicate that S. mutans cells inhibit the growth of C. albicans filaments through secreting some unidentified chemicals.
Mutanocyclin, a tetramic acid, secreted by S. mutans cells inhibits C. albicans filamentation
In a previous study, we identified mutanocyclin as a rich secondary metabolite with anti-infiltration activity in S. mutans [Citation34]. The mutanocyclin-encoding cluster is confined to approximately 15% S. mutans strains (including the S. mutans 35 used in this study) and some closely related species [Citation38]. This chemical is produced under conditions mimicking host niches (for example, anaerobic conditions). We therefore performed high-performance liquid chromatography (HPLC) analysis and examined the production of mutanocyclin in S. mutans and S. mutans-C. albicans mixed cultures. As shown in , a peak with a retention time equivalent to that of chemically synthesized mutanocyclin was observed. High resolution-mass spectrum analysis of this compound from the S. mutans and S. mutans-C. albicans mixed cultures revealed a chemical formula of C10H15NO3 (calculated molecular weight: 198.1125), which was identical to mutanocyclin [Citation34] ().
Figure 2. Identification of a tetramic acid compound, mutanocyclin, produced by S. mutans, as an inhibitor of C. albicans filamentous growth. (a) HPLC analysis of the supernatant extracts of the S. mutans, S. mutans mixed with C. albicans, S. mutans Δmuc mutant, and S. mutans Δmuc mixed with C. albicans cultures. The BHI-sucrose medium served as a negative control. The X-axis represents the retention time in minutes, and the Y-axis is the absorbance unit (mAU) at 235 nm. Chemically synthesized mutanocyclin served as the reference standard (STD). the black arrow indicates the peak of mutanocyclin. (b) Chemical structure of mutanocyclin. MW: molecular weight. (c) Morphologies of C. albicans (SC5314) in the presence of supernatant from S. mutans, S. mutans Δmuc mutant, or E. coli cultures. C. albicans cells (1 × 106) were incubated in BHI + Lee’s glucose medium with or without an equal volume of the bacterial supernatant extracts at 30°C for 48 hours. the number of “+” signs indicates the degree of filamentation. the “-” sign indicates that no filamentous cells were observed; “+” and “++” represent 10–30% and 30–50% of filamentous cells, respectively. Scale bar, 20 µm.
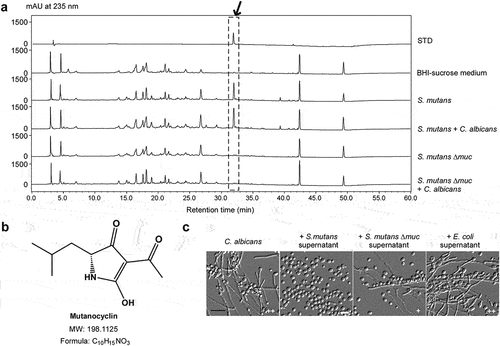
Since the biosynthetic gene cluster (muc) is essential for the production of mutanocyclin in S. mutans, we generated a Δmuc mutant in S. mutans 35 and tested the effect of this mutant on the growth of filaments in C. albicans in a co-culture system. As shown in , the Δmuc mutant of S. mutans was unable to produce mutanocyclin and allowed the growth of filaments in C. albicans. These results suggest that mutanocyclin produced by S. mutans cells has a direct repressive effect on C. albicans filamentation.
Mutanocyclin inhibits C. albicans filamentation in a dose-dependent manner
To verify the inhibitory effect of mutanocyclin on C. albicans filamentation, we examined the dosage effect of chemically synthesized mutanocyclin. As shown in , mutanocyclin at concentrations of 8 µg/mL and 16 µg/mL remarkably reduced filamentation of C. albicans in BHI-sucrose medium after 24 hours or 36 hours of incubation, whereas it completely abolished C. albicans filamentous growth at 32 µg/mL. We then analyzed the expression levels of four filamentous-specific genes, HWP1, ECE1, FLO8, and TEC1 [Citation4]. In the presence of mutanocyclin, the expression levels of these genes were all notably downregulated, consistent with the morphological changes in C. albicans ().
Figure 3. Inhibitory effect of mutanocyclin onC. albicans filamentation. (a) Morphologies of C. albicans cells in the presence of DMSO or 8, 16, and 32 µg/ml of mutanocyclin. C. albicans cells (SC5314, 1 × 104) were incubated in BHI-sucrose medium with or without mutanocyclin at 30°C for 24 to 36 hours. the number of “+” signs indicates the degree of filamentation. the “-” sign indicates that no filamentous cells were observed; “±”, “+”, “++”, and “+++”, represent 1–10%, 10–30%, 30–50%, and 50–70% of filamentous cells, respectively. Scale bar, 20 µm. (b) Relative expression levels of C. albicans filamentous-related genes in DMSO- or mutanocyclin-treated (32 µg/ml) cells. the expression level in the DMSO control was set to 1. Error bars denote the standard deviation (SD). *P < .05 (Student's t test, two tailed). (c) Growth curves of C. albicans cells treated with DMSO (control) containing 8 µg/ml, 16 µg/ml, and 32 µg/ml of mutanocyclin. the X-axis and Y-axis represent the culture time and corresponding cell density (OD600). Bars indicate standard deviations.
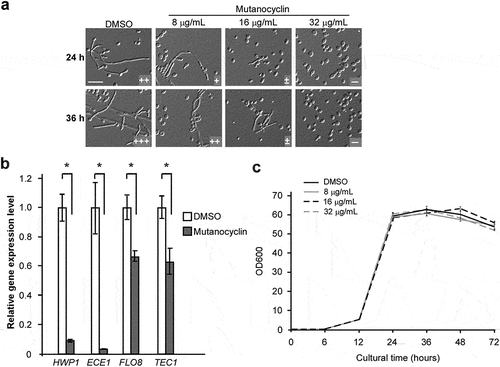
Since the inhibitory activity of mutanocyclin on morphological changes could be due to an indirect effect, we next examined whether this chemical affected the cell growth of C. albicans. As shown in , mutanocyclin had no significant effect on the cell growth rate of C. albicans at different concentrations (from 8 to 32 µg/mL). Moreover, although this chemical belongs to the unacylated tetramic acids, it has an acetyl side chain but does not contain a carboxylic acid terminus (). The pH value of mutanocyclin in water was nearly neutral and addition of this chemical to the media did not obviously affect the pH (data not shown). Taken together, mutanocyclin could function as a specific inhibitor of C. albicans filamentation independent of the changes in the cell growth rate and culture pH.
Global transcriptional profile of C. albicans in response to mutanocyclin
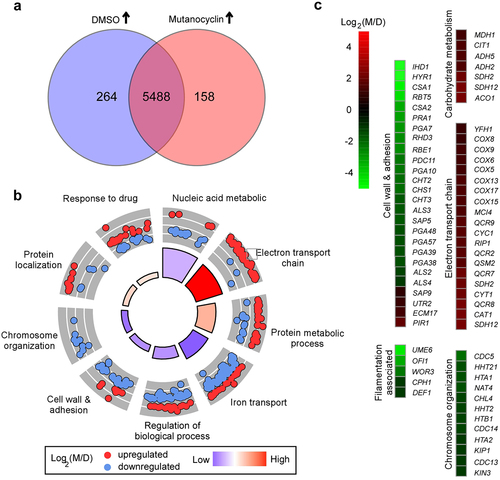
To explore the potential mechanism of mutanocyclin-regulated filamentation in C. albicans, we performed RNA-Seq analysis. Total RNA was extracted from cells grown in liquid Lee’s glucose medium containing DMSO or 32 µg/mL mutanocyclin for 24 hours at 30°C As demonstrated in and Dataset S1, we found 422 differentially expressed genes between the DMSO and mutanocyclin-treated samples (two-fold difference cutoff). Among them, the expression levels of 158 genes were increased and 264 genes decreased in the mutanocyclin-treated cells. The downregulated genes included a large subset of cell wall- and adherence-associated genes, for example, the GPI-anchored protein-encoding genes IHD1, HYR1, RBT5, and RHD3, cell wall surface antigen-encoding gene CSA1, Pry family protein-encoding gene RBE1, ALS family genes (encoding adhesins), and chitin remodeling and synthesis-associated genes CHT2, CHT3, and CHS1. The expression of these genes is affected by Efg1, a downstream target of the cAMP/PKA signaling pathway [Citation39]. Some cell wall biogenesis- and remodeling-related genes, including PIR1 (encoding 1,3-β-glucan-linked cell wall protein), ECM17 (encoding sulfite reductase), and UTR2 (encoding GPI-anchored cell wall glycosidase), were upregulated in response to mutanocyclin treatment. These findings indicate that mutanocyclin treatment has a strong effect on the cell wall of C. albicans. Interestingly, many oxidative metabolism-associated genes, including key members of the tricarboxylic acid (TCA) cycle (CIT1, ACO1, SDH2, SDH12, and MDH1) and electron transport chain (COX5, COX6, COX8, COX9, COX13, COX15, and COX17), were robustly upregulated in response to treatment with mutanocyclin. Numerous transcriptional regulator-encoding genes that are involved in filamentous growth or adherence in C. albicans were markedly downregulated in response to mutanocyclin treatment, for example, UME6, OFI1, WOR3, CPH1, and DEF1. This finding is consistent with the reduced filamentation ability. Moreover, a number of genes involved in chromosome organization or segregation (for example, HHT21, HTA1, HHT2, HTB1, CDC14, HTA2, and KIP1) were also downregulated by mutanocylin. Taken together, these results imply that mutanocylin may have a global impact on the transcriptional profile of C. albicans.
Figure 4. Effect of mutanocyclin on the global gene expression profile of C.albicans. (a) Venn diagram depicting differentially expressed genes. A twofold difference cutoff and false discovery rate (FDRs) <.05 were used to define differentially expressed genes. (b) GO enrichment analysis of differentially expressed genes. Red or blue dots represent upregulated or downregulated genes in response to mutanocyclin treatment, respectively. The inner cycle bars represent the statistical significance. (c) Selected differentially expressed genes indicated by the R package heat map. Log2(M/D), Log2 (read counts of mutanocyclin treatment/read counts of DMSO control).
Role of the GPI-anchored proteins Spr1, Hyr4, and Iff8 in mutanocyclin-regulated filamentation in C. albicans
Since a set of cell wall-associated genes exhibited a differential expression pattern when treated with mutanocyclin, we examined the morphological phenotypes of a subset of cell wall-related gene mutants [Citation40]. We screened three genes (SPR1, HYR4, and IFF8) that could potentially be involved in regulating the mutanocyclin response. SPR1, HYR4, and IFF8 encode GPI-anchored proteins that are required for cell wall biogenesis and remodeling. SPR1 (SPorulation Regulated) encodes a putative glucan 1,3-β-glucosidase that has been identified to participate in cell wall modification [Citation41]. HYR4 (HYphally Regulated) and IFF8 (IPF family file) encode two proteins that belong to the Hyr/Iff cell wall family and play a role in cell wall integrity, cell adhesion, and biofilm formation [Citation42,Citation43]. Unlike the WT control, deletion of SPR1, HYR4, and IFF8 allowed obvious filamentous growth in the presence of mutanocyclin (), suggesting that these cell wall proteins play a critical role in mutanocyclin-regulated filamentation in C. albicans.
Figure 5. Three cell wall-related genes are identified as negative regulators in the presence of mutanocyclin. (a) Cellular morphologies of the hyr4/hyr4, spr1/spr1, and iff8/iff8 mutants in the presence of DMSO or mutanocyclin. C. albicans cells (1 × 104) were grown in Lee’s glucose medium containing DMSO or 32 µg/mL mutanocyclin, and incubated at 30°C for 24 hours. “±”, “++”, and “+++” represent 1–10%, 30–50%, and 50–70% of filamentous cells, respectively. Scale bar, 20 µm. (b) Relative expression levels of HYR4, SPR1, and IFF8 in efg1/efg1mutants treated with DMSO or mutanocyclin (32 µg/mL). The expression level of the WT strain treated with DMSO was set as 1. Error bars denote the standard deviation (SD). *P < .05 (Student's t test, two tailed).
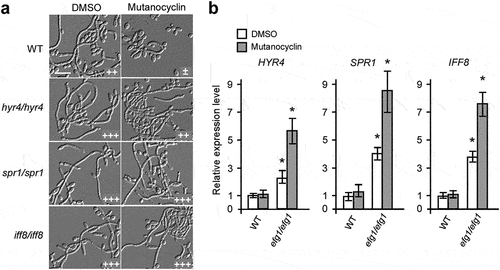
Since the Efg1 transcription factor plays a critical role in the regulation of a number of cell wall-associated genes [Citation44], we detected the expression of SPR1, HYR4, and IFF8 in the efg1/efg1 mutant. As shown in , the expression levels of all three genes were increased in the efg1/efg1 mutant compared with the WT strain, especially when treated with mutanocyclin.
Role of the Ras1-cAMP/PKA signaling pathway of C. albicans in the mutanocyclin-induced response
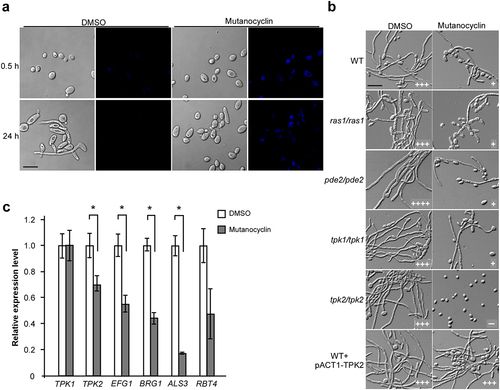
Mutanocyclin is a tetramic acid compound that can be directly observed in fungal cells using a confocal laser scanning microscope (under UV light at 254 nm). As demonstrated in , mutanocyclin accumulation was observed in the cytoplasm of C. albicans cells, whereas no fluorescent signal was detected in the untreated control. Since the conserved Ras1-cAMP/PKA signaling pathway plays a central role in the regulation of filamentation in C. albicans [Citation21,Citation22], we tested whether mutanocyclin targeted this intracellular pathway. We found that deletion of RAS1, TPK1, and PDE2 had no significant effect on the growth of filaments in C. albicans, whereas deletion of TPK2 completely abolished filamentous development in the presence of mutanocyclin (). To confirm the importance of Tpk2 in the mutanocyclin-induced response, we detected overexpressed TPK2 (strain WT + pACT1-TPK2) in C. albicans. As expected, the TPK2-overexpressing strain overrode the inhibitory effect of mutanocyclin on C. albicans filamentation. Consistently, q-RT-PCR analysis revealed that the expression levels of TPK2 and its downstream targets EFG1 and BRG1, ALS3, and RBT4 were downregulated following mutanocyclin treatment. However, no obvious changes were detected in the expression of TPK1 (). Moreover, the expression levels of the cell wall-associated genes HYR4, SPR1, and IFF8 were significantly increased in the efg1/efg1 mutant, which is a target of Tpk2 [Citation45]. Therefore, the PKA catalytic subunit Tpk2, but not Tpk1, plays a crucial role in mutanocyclin-repressed filamentation in C. albicans.
Figure 6. Role of the Ras1-cAMP/PKA signaling pathway in mutanocyclin-regulated filamentation in C.albicans. (a) Confocal laser scanning microscopic images of C. albicans (SC5314) cells treated with mutanocyclin (blue) for 0.5 or 24 hours. Scale bar, 10 µm. (b) Cellular morphologies of the Ras1-cAMP/PKA signaling mutants and TPK2-overexpressing strain in the presence of DMSO or mutanocyclin. Fungal cells were grown in liquid Lee’s glucose medium containing mutanocyclin (32 µg/mL) or DMSO, and incubated at 30°C for 24 hours. The “-” sign indicates that no filamentous cells were observed; “+”, “+++”, and “++++”, represent 10-30%, 50-70%, and >70% of filamentous cells, respectively. Scale bar, 20 µm. (c) Relative expression levels of TPK1, TPK2, and four transcription factor-encoding genes in C. albicans cells treated with DMSO or mutanocyclin (32 µg/mL). The expression level in DMSO was set as 1. Error bars denote the standard deviation (SD). *P < .05 (Student's t test, two tailed).
Roles of transcriptional and cell wall regulators in mutanocyclin-repressed filamentation in C. albicans
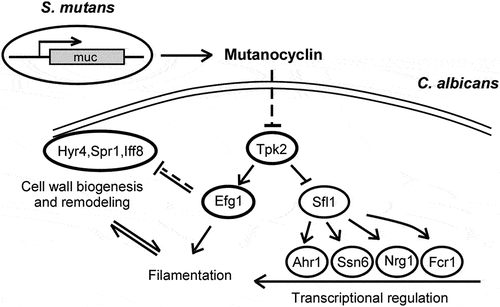
To further explore the molecular mechanism of mutanocyclin, we screened a deletion mutant library of 165 transcription factors [Citation32]. The filamentation assays were performed in Lee’s glucose medium containing 32 µg/mL mutanocyclin or DMSO (control). Five mutants (sfl1/sfl1, ahr1/ahr1, ssn6/ssn66, nrg1/nrg1, and fcr1/fcr1) were able to undergo filamentation even in the presence of mutanocyclin ( a), suggesting that these genes played a negative role in filamentous growth under these culture conditions. It has been reported that Sfl1 (suppressor gene for flocculation 1) is a specific target of Tpk2 and acts as a negative regulator of filamentation in C. albicans [Citation46]. This result is consistent with findings obtained for the tpk2/pk2 mutant and TPK2-overexpressing strain ( b). Ahr1, Nrg1, and Ssn6 are also well-characterized regulators of filamentation, which have been demonstrated to be regulated by Sfl1 in C. albicans [Citation46] ( b).
Figure 7. A library screen identifies a set of transcriptional regulators involved in the response to mutanocyclin treatment. (a) Cellular morphologies of the sfl1/sfl1, ahr1/ahr1, nrg1/nrg1, ssn6/ssn6, fcr1/fcr1 mutants in the presence of DMSO or mutanocyclin. C. albicans cells (1 × 104) were grown inLee’s glucose medium containing DMSO or 32 µg/mL mutanocyclin, and incubated at 30°C for 24 hours. “±”, “++”, “+++”, and “++++” represent 1–10%, 30–50%, 50–70%, and >70% of filamentous cells, respectively. Scale bar, 20 µm. (b) Model of Sfl1-mediated regulation of filamentation in C. albicans. (c) Relative expression levels of FCR1 in the WT (SC5314), tpk2/tpk2, and sfl1/sfl1 strains treated with DMSO or mutanocyclin (32 µg/mL). The expression level of the WT strain treated with DMSO was set as 1. Error bars denote the standard deviation (SD). *P < .05 (Student's t test, two tailed).
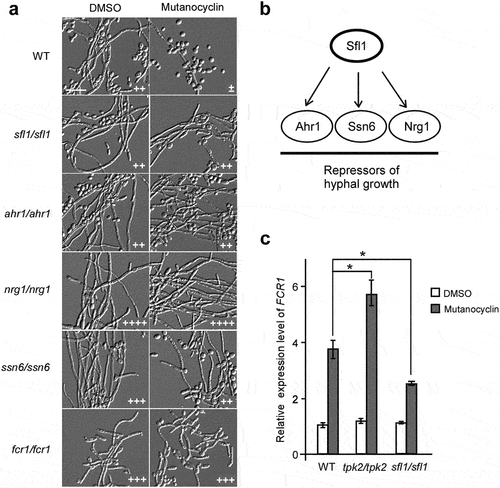
Fcr1 is a predicted zinc cluster transcription factor that functions as a negative regulator of fluconazole and ketoconazole resistance, as well as in the regulation of filament formation [Citation47]. We found that deletion of FCR1 allowed obvious filamentous growth in the presence of mutanocyclin. Consistently, expression of the FCR1 gene was notably enhanced by mutanocyclin treatment (), suggesting that Fcr1 is a key regulator in the response to mutanocyclin. Znaidi et al. reported that Fcr1 is a transcriptional target of Sfl1 in various biological processes [Citation46]. Q-RT-PCR assays demonstrated that compared with the WT strain, the expression level of FCR1 was increased in the tpk2/tpk2 mutant and decreased in the sfl1/sfl1 mutant following treatment with mutanocyclin (). This expression pattern is consistent with the morphological phenotypes of these mutants in the presence of mutanocyclin.
Efficacy of mutanocyclin in ex vivo mouse tongue and G. mellonella infection models of C. albicans
Filamentous growth is important for the virulence of C. albicans. Given the potent inhibitory effect of mutanocyclin on filamentous growth of C. albicans, we predicted that treatment with this chemical would reduce fungal virulence. First, we performed ex vivo mouse tongue infection assays. As shown in , C. albicans cells in the DMSO-treated control formed robust filaments and invaded the tongue tissue. However, C. albicans cells in the mutanocyclin-treated sample maintained the yeast-form. These results imply that mutanocyclin can function as a filamentation repressor during infection and has potential as a therapeutic agent.
Figure 8. Effects of mutanocyclin on the virulence of C.albicans in mouse and G. mellonella infection models. (a) Histopathological assays. C. albicans SC5314 cells (1 × 107) in 5 μL of PBS containing DMSO or mutanocyclin (32 μg/mL) were spotted on the removed tongues of 4 − 5-week-old mice. After 24 hours of infection at 37°C, the infected tongues were stained with PAS and then used for microscopy assays. Scale bar, 20 µm. (b) Scanning electron microscope (SEM) images of the infected tongue tissues. Scale bar, 10 µm. (c) Survival rates of C. albicans-infected G. mellonella larvae treated with DMSO or mutanocyclin. C. albicans SC5314 cells (1 × 108 CFU/mL) and DMSO, or 6.4, 12.8, or 25.6 µg mutanocyclin, were injected individually into each G. mellonella larva (20 larvae in each treatment group). *P < .05 (compared with the DMSO control, Student's t test, two tailed).
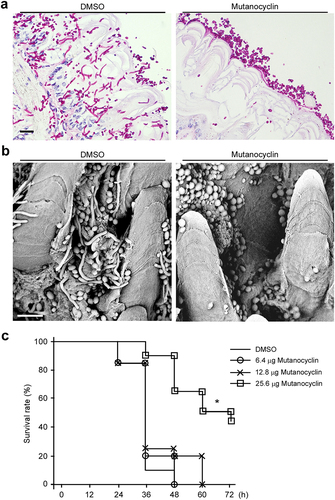
We next performed in vivo virulence assays using the G. mellonella infection model to evaluate the efficacy of mutanocyclin. G. mellonella larvae were infected with C. albicans, and the survival rates were analyzed. As shown in , the groups infected by C. albicans and treated with DMSO or 6.4 µg mutanocyclin showed 100% of death within 48 hours postinfection. However, the survival rate was increased by 20% and 60% following treatment with 12.8 µg and 25.6 µg of mutanocyclin, respectively. These results suggest that mutanocyclin notably attenuates the virulence of C. albicans in the G. mellonella infection model.
Discussion
Cross-kingdom interactions between bacteria and fungi are common and important for infection outcomes. The symbiotic relationship between S. mutans and C. albicans has been well investigated [Citation11,Citation13,Citation14,Citation48]. For example, C. albicans secretes the quorum-sensing molecule, farnesol, to stimulate microcolony and biofilm development of S. mutans [Citation11]. Animals coinfected with the two species display higher levels of microbial carriage and lesion severity [Citation13]. In this study, we report a novel antagonistic relationship between the two species: S. mutans suppresses the filamentous growth and virulence of C. albicans through secreting a secondary metabolite, mutanocyclin. This chemical has a tetramic acid reutericyclin core in its structure [Citation34]. We demonstrated that mutanocyclin regulated morphological transitions in C. albicans through multiple mechanisms including alterations of the Ras1-cAMP/PKA pathway, transcriptional regulation, and cell wall biogenesis/remodeling. These changes finally resulted in its therapeutic effect on C. albicans infections in mouse skin and in G. mellonella infection models.
The Ras1-cAMP/PKA signaling pathway plays a critical role in filamentous growth of C. albicans [Citation21,Citation22]. Here, we found that the catalytic subunit of PKA, Tpk2, functioned as a major regulator in mutanocyclin-repressed filamentous growth. Inactivation of Tpk2 led to complete abrogation of filamentous growth in the presence of mutanocyclin, whereas overexpression of TPK2 was able to override the suppressive effect of this chemical (). Consistently, expression of the downstream target gene EFG1 and cell wall-associated genes ALS3 and RBT4 were significantly decreased following treatment with mutanocyclin. Moreover, dysfunction of HYR4, SPR1, and IFF8, which encode three GPI-anchored proteins associated with cell wall biogenesis and remodeling, allowed obvious filamentous growth in the presence of mutanocyclin. Deletion of EFG1 led to increased expression of HYR4, SPR1, and IFF8 in C. albicans, especially when treated with mutanocyclin (). These results suggest that both transcriptional regulation and cell wall biogenesis are involved in the regulation of mutanocyclin-inhibited filamentous growth of C. albicans.
The Sfl1 transcription factor is a negative regulator of filamentous growth that physically interacts with Tpk2 [Citation46]. In Saccharomyces cerevisiae, Tpk2 acts upstream of Sfl1 in the regulation of pseudohyphal growth through phosphorylation of Sfl1. This phosphorylation relieves the repression of genes (for example, FLO11) regulated by Sfl1 [Citation49]. In C. albicans, Tpk2 also bound preferentially to Sfl1, and deletion of SFL1 allowed the development of filaments even in the presence of mutanocyclin (), suggesting the presence of a conserved regulatory mechanism in the two yeast species. The ahr1/ahr1, ssn6/ssn6, and nrg1/nrg1 mutants were able to undergo filamentation in the presence of mutanocyclin (). These regulators and Sfl1 comprise a network in which Sfl1 acts as a central “switch on/off” regulator that plays a crucial role in filamentous growth of C. albicans in response to mutanocyclin ().
Transcription of the zinc cluster regulator Fcr1 was enhanced in C. albicans cells following treatment with mutanocyclin (). It has been reported that Fcr1 contains a PKA phosphorylation site and an ATP/GTP binding motif, suggesting that Fcr1 is a potential target of Tpk2 [Citation47,Citation50]. Disruption of TPK2 causes a significant increase in FCR1 expression. Moreover, Sfl1 has also been demonstrated to bind to the promoter of FCR1 [Citation47], and deletion of SFL1 leads to reduced expression of FCR1 following treatment with mutanocyclin. These results suggest that Tpk2 and Sfl1 play a major role in the response to mutanocyclin treatment and that FCR1 is a downstream factor of the two regulators.
RNA-Seq analysis demonstrated that mutanocyclin treatment had a global effect on the transcriptional profile in C. albicans (, Dataset S1). A large set of genes involved in metabolism, cell wall remodeling, and chromosome organization were differentially expressed in response to treatment with this chemical. These changes could directly or indirectly affect filamentous or polarized growth in C. albicans. The Ras1-cAMP/PKA signaling pathway and Efg1 have also been demonstrated to function in the regulation of cell wall biogenesis [Citation51–53]. Therefore, the transcriptional profile reflects the outcome of the regulation of corresponding signaling pathways following treatment with mutanocyclin. This chemical could modulate cell wall components predominantly through the Ras1-cAMP/PKA signaling pathway and Efg1, as well as a subset of filamentous regulators through Sfl1. We propose that this transcriptional regulation and cell wall alterations coordinately regulate the response to mutanocyclin in C. albicans ().
Figure 9. Model for the mutanocyclin-mediated interspecies interaction between C. albicans and S. mutans. S. mutans secretes the secondary metabolite, mutanocyclin, which regulates filamentation of C. albicans through the Ras1-cAMP/PKA signaling pathway and a set of transcription factors and cell wall-associated proteins.
Mutanocyclin did not affect the cell growth rate of C. albicans even at a high concentration, but it specifically targeted filamentous growth. The development of filaments is critical for C. albicans to cause infections. It has been demonstrated that C. albicans filaments are produced coordinately with virulence factors such as adhesins and hydrolytic enzymes andgeneration of hydrostatic pressure [Citation54,Citation55]. C. albicans filaments can actively penetrate and rupture the macrophages, thereby conferring the pathogen cells greater resistance to the host immune system [Citation56]. Thus, targeting the regulatory process of filamentous growth could be a promising strategy for anti-C. albicans infection treatments. Furthermore, our virulence assays in two animal models revealed a notable therapeutic effect of mutanocyclin. Fcr1, a potential target of mutanocyclin, has been described as a suppressor of fluconazole and ketoconazole resistance [Citation47], implying a synergistic effect between mutanocyclin and azole drugs. Taken together, we report a novel cross-kingdom relationship between S. mutans and C. albicans. Since both species are commensal microorganisms of the mouth niche, the inhibitory effect of S. mutans on filamentous growth of C. albicans could be critical for the biofilm dispersal process. Additionally, because the increasing prevalence of drug-resistant pathogenic fungi threatens the effectiveness of existing antifungal agents, we propose that mutanocyclin, or similar compounds, may provide a novel therapeutic potential for future applications.
Accession number
The RNA-seq dataset was deposited in the NCBI Gene Expression Omnibus (GEO) portal with accession number GSE188886.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All primary data that support the findings of this study are available from the corresponding author upon reasonable request.
Additional information
Funding
References
- Frey-Klett P, Burlinson P, Deveau A, et al. Bacterial-Fungal interactions: hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol Mol Biol Rev. 2011;75:583–609.
- Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9:737–748.
- Berman J. Candida albicans. Curr Biol. 2012;22:R620–2.
- Huang G. Regulation of phenotypic transitions in the fungal pathogen Candida albicans. Virulence. 2012;3:251–261.
- Hibbing ME, Fuqua C, Parsek MR, et al. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25.
- Morales DK, Hogan DA. Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog. 2010;6:e1000886.
- Simon-Soro A, Tomas I, Cabrera-Rubio R, et al. Microbial geography of the oral cavity. J Dent Res. 2013;92:616–621.
- Becker MR, Paster BJ, Leys EJ, et al. Molecular analysis of bacterial species associated with childhood caries. J Clin Microbiol. 2002;40:1001–1009.
- Xiao J, Huang X, Alkhers N, et al. Candida albicans and early childhood caries: a systematic review and meta-analysis. Caries Res. 2018;52:102–112.
- Vila T, Sultan AS, Montelongo-Jauregui D, et al. Oral candidiasis: a disease of opportunity. J Fungi. 2020;6:15.
- Kim D, Sengupta A, Niepa TH, et al. Candida albicans stimulates Streptococcus mutans microcolony development via cross-kingdom biofilm-derived metabolites. Sci Rep. 2017;7:41332.
- Jakubovics NS, Kolenbrander PE. The road to ruin: the formation of disease-associated oral biofilms. Oral Dis. 2010;16:729–739.
- Falsetta ML, Klein MI, Colonne PM, et al. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect Immun. 2014;82:1968–1981.
- Hwang G, Liu Y, Kim D, et al. Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross-kingdom biofilm development in vivo. PLoS Pathog. 2017;13:e1006407.
- Yang C, Scoffield J, Wu R, et al. Antigen I/II mediates interactions between Streptococcus mutans and Candida albicans. Mol Oral Microbiol. 2018;33:283–291.
- Pestova EV, Havarstein LS, Morrison DA. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol. 1996;21:853–862.
- Jarosz LM, Deng DM, van der Mei HC, et al. Streptococcus mutans competence-stimulating peptide inhibits Candida albicans hypha formation. Eukaryot Cell. 2009;8:1658–1664.
- Biswas S, Van Dijck P, Datta A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol Mol Biol Rev. 2007;71:348–376.
- Whiteway M, Bachewich C. Morphogenesis in Candida albicans. Annu Rev Microbiol. 2007;61:529–553.
- Kim HE, Liu Y, Dhall A, et al. Synergism of Streptococcus mutans and Candida albicans reinforces biofilm maturation and acidogenicity in saliva: an in vitro study. Front Cell Infect Microbiol. 2020;10:623980.
- Hogan DA, Sundstrom P. The Ras/cAMP/PKA signaling pathway and virulence in Candida albicans. Future Microbiol. 2009;4:1263–1270.
- Huang G, Huang Q, Wei Y, et al. Multiple roles and diverse regulation of the Ras/cAMP/protein kinase A pathway in Candida albicans. Mol Microbiol. 2019;111:6–16.
- Kohler JR, Fink GR. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc Natl Acad Sci U S A. 1996;93:13223–13228.
- Braun BR, Johnson AD. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science. 1997;277:105–109.
- Davis D, Wilson RB, Mitchell AP. RIM101-Dependent and-independent pathways govern pH responses in Candida albicans. Mol Cell Biol. 2000;20:971–978.
- Murad AM, Leng P, and Straffon M, et al. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 2001;20:4742–4752.
- Du H, Huang G. Environmental pH adaption and morphological transitions in Candida albicans. Curr Genet. 2016;62:283–286.
- Guan C, Che F, Zhou H, et al. Effect of rubusoside, a natural sucrose substitute, on Streptococcus mutans biofilm cariogenic potential and virulence gene expression in vitro. Appl Environ Microbiol. 2020;86(16). DOI:10.1128/AEM.01012-20
- Bowen WH, Burne RA, Wu H, et al. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 2018;26:229–242.
- Tao L, Zhang Y, Fan S, et al. Integration of the tricarboxylic acid (TCA) cycle with cAMP signaling and Sfl2 pathways in the regulation of CO2 sensing and hyphal development in Candida albicans. PLoS Genet. 2017;13:e1006949.
- Pande K, Chen C, Noble SM. Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat Genet. 2013;45:1088–1091.
- Homann OR, Dea J, Noble SM, et al. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 2009;5:e1000783.
- Xie ZJ, Okinaga T, Qi FX, et al. Cloning-Independent and counterselectable markerless mutagenesis system in Streptococcus mutans. Appl Environ Microb. 2011;77:8025–8033.
- Hao T, Xie Z, Wang M, et al. An anaerobic bacterium host system for heterologous expression of natural product biosynthetic gene clusters. Nat Commun. 2019;10:3665.
- Tao L, Du H, Guan G, et al. Discovery of a “white-gray-opaque” tristable phenotypic switching system in Candida albicans: roles of non-genetic diversity in host adaptation. PLoS Biol. 2014;12:e1001830.
- Jarvensivu A, Hietanen J, Rautemaa R, et al. Candida yeasts in chronic periodontitis tissues and subgingival microbial biofilms in vivo. Oral Dis. 2004;10:106–112.
- Wang X, Bing J, Zheng Q, et al. The first isolate of Candida auris in China: clinical and biological aspects. Emerg Microbes Infect. 2018;7:93.
- Liu L, Hao T, Xie Z, et al. Genome mining unveils widespread natural product biosynthetic capacity in human oral microbe Streptococcus mutans. Sci Rep. 2016;6:37479.
- Sohn K, Urban C, Brunner H, et al. EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol Microbiol. 2003;47:89–102.
- Noble SM, French S, Kohn LA, et al. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 2010;42:590–598.
- Satala D, Karkowska-Kuleta J, Zelazna A, et al. Moonlighting proteins at the candidal cell surface. Microorganisms. 2020;8:1046.
- Richard ML, Plaine A. Comprehensive analysis of glycosylphosphatidylinositol-anchored proteins in Candida albicans. Eukaryot Cell. 2007;6:119–133.
- Boisrame A, Cornu A, Da Costa G, et al. Unexpected role for a serine/threonine-rich domain in the Candida albicans Iff protein family. Eukaryot Cell. 2011;10:1317–1330.
- Braun BR, Johnson AD. TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics. 2000;155:57–67.
- Bockmuhl DP, Ernst JF. A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics. 2001;157:1523–1530.
- Znaidi S, Nesseir A, Chauvel M, et al. A comprehensive functional portrait of two heat shock factor-type transcriptional regulators involved in Candida albicans morphogenesis and virulence. PLoS Pathog. 2013;9:e1003519.
- Talibi D, Raymond M. Isolation of a putative Candida albicans transcriptional regulator involved in pleiotropic drug resistance by functional complementation of a pdr1 pdr3 mutation in Saccharomyces cerevisiae. J Bacteriol. 1999;181:231–240.
- Sztajer H, Szafranski SP, and Tomasch J, et al. Cross-Feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. ISME J. 2014;8:2256–2271.
- Robertson LS, Fink GR. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc Natl Acad Sci, USA. 1998;95:13783–13787.
- Walker JE, Saraste M, and Runswick MJ, et al. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951.
- Jung WH, Warn P, Ragni E, et al. Deletion of PDE2, the gene encoding the high-affinity cAMP phosphodiesterase, results in changes of the cell wall and membrane in Candida albicans. Yeast. 2005;22:285–294.
- Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:262–291.
- Fanning S, Xu W, Beaurepaire C, et al. Functional control of the Candida albicans cell wall by catalytic protein kinase A subunit Tpk1. Mol Microbiol. 2012;86:284–302.
- Liu H. Co-Regulation of pathogenesis with dimorphism and phenotypic switching in Candida albicans, a commensal and a pathogen. Int J Med Microbiol. 2002;292:299–311.
- Kumamoto CA, Vinces MD. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell Microbiol. 2005;7:1546–1554.
- Vylkova S, Lorenz MC. Modulation of phagosomal pH by Candida albicans promotes hyphal morphogenesis and requires Stp2p, a regulator of amino acid transport. PLoS Pathog. 2014;10:e1003995.
