First described in 2009 in Japan, the multidrug-resistant human fungal pathogen Candida auris represents a new global public health threat [Citation1,Citation2]. As of February, 2021, C. auris cases have been reported in over 40 countries across six continents, with several outbreaks occurring simultaneously in different hospitals [Citation3–6]. C. auris is often called a “superbug” due to its multidrug-resistant and highly transmissible features. Some clinical isolates are even resistant to all three of the major classes of antifungal drugs (the azoles, the polyenes, and the echinocandins) [Citation2,Citation7]. In 2019, the Centers for Disease Control and Prevention (CDC) placed C. auris on the list of urgent threats (Antibiotic Resistance Threats in the United States, 2019; https://www.cdc.gov).
In 2018, we isolated the first C. auris strain in China from a 76-year-old hospitalized female patient with hypertension and nephritic syndrome in Beijing, China [Citation8]. The strain (BJCA001) was recovered from the bronchoalveolar lavage fluid (BALF) of the patient, identified by matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) mass spectrometry, and verified using internal transcribed spacer (ITS) sequencing analyses. Interestingly, the strain BJCA001 exhibited notably low minimal inhibitory concentrations (MICs) for all tested antifungal drugs. Subsequently, Chen et al. (2018) reported two cases of bloodstream infections caused by fluconazole-resistant C. auris in the Army General Hospital (Beijing, China) [Citation9], and we recently reported a case of a C. auris bloodstream infection in Beijing Hospital (Beijing, China) [Citation10]. All three of these cases were in newborn infants in neonatal intensive care units (NICUs). Additionally, Tian et al. (2018), in a retrospective study, identified 15 cases of C. auris colonization or infection in a general hospital in Shenyang, China [Citation11]. The isolates from these 15 cases had been initially misidentified as Candida haemulonii, which is phylogenetically closely related to C. auris. All the C. auris strains isolated in this hospital were resistant to fluconazole but showed low MICs to other tested antifungal drugs. Recently, Tian et al. (2021) reported additional cases of C. auris in the same hospital in Shenyang [Citation12]. In total, over the course of this 3-year surveillance study from 2016 to 2019, 93 C. auris strains were isolated from 39 patients within intensive care units (ICUs). All 93 of these strains were resistant to fluconazole and a minority of them exhibited high MICs for echinocandins and amphotericin B. Genomic epidemiology analyses indicated that all of the Shenyang strains originated from the same clone, which was first isolated from a respiratory ICU ward. Given the same origin of these strains, their resistance to echinocandins and amphotericin B could be a recent evolutionary adaptation.
We recently reported a case of C. auris bloodstream infection in the First Affiliated Hospital of Xiamen University in Xiamen, China [Citation13]. This case, which was from a 67‐year‐old man with a 10-year-history of gastric ulcers and diabetes, represents the first isolate of C. auris in South Mainland China. With the exception of strain BJCA001, which belongs to the South Asian clade and carries an a-type mating type locus (MTLa), all the other C. auris isolates in mainland China belong to the South African clade and contain an α-type mating type locus (MTLα). Therefore, there are at least two distinct genetic clades of C. auris (South Asian and South African) present in mainland China. Biological analyses demonstrated that strains from both of these clades were capable of undergoing filamentation [Citation14], a critical virulence feature of pathogenic Candida species. However, it has been suggested that isolates of the South Asian clade could be more virulent than those of the South African clade [Citation10,Citation14]. The former clade often contains high copies of the Zorro 3 non-long-terminal repeat (non-LTR) retrotransposon. The role of this retrotransposon in the evolution of the C. auris genome, virulence, and antifungal resistance mechanisms remains to be investigated. Overall, genomic analyses of the C. auris strains of the South African clade isolated from different hospitals in China indicate that these strains are highly associated with each other by phylogeny.
In 2019, the emergence of C. auris in Taiwan was also reported [Citation15]. A strain was isolated from a 55-year-old man with diabetes mellitus and pemphigus vulgaris. The MIC of this strain from Taiwan to fluconazole was relatively low (8 mg/L); however, the MIC of this strain to amphotericin B was relatively high (4–8 mg/L). Recently, Tse et al. (2021) reported the isolation and genomic sequencing of 19 strains of C. auris, obtained from 15 patients from a public hospital in Hong Kong [Citation16]. All 19 of the strains were identified to be within the South Asian clade based on ITS and genomic sequence analyses. Although the authors did not directly assess antifungal susceptibility, DNA sequence analyses indicated that several hotspot gene mutations related to antifungal resistance (e.g. ERG11 mutations) were observed within the strains.
Geographic and phylogenetic information, genetic clades, mating types, and antifungal resistance-associated hotspot mutations for all of the known clinical isolates from China are summarized in .
Figure 1. Distribution, phylogeny, and antifungal information of C. auris clinical isolates in China. (a) A map detailing the distribution of cases of C. auris infection in China (n = 60). Over 100 C. auris strains were isolated in China, from 60 patients. (b) A phylogenetic tree detailing the clade, mating type, and Erg11 hotspot mutations of the C. auris clinical isolates in China. the phylogenetic tree was constructed with RAxML using the GTRGAMMA nucleotide substitution model with ITS sequences and 1000 bootstrap replicates. F126 L is also referred to as VF125AL in publications.
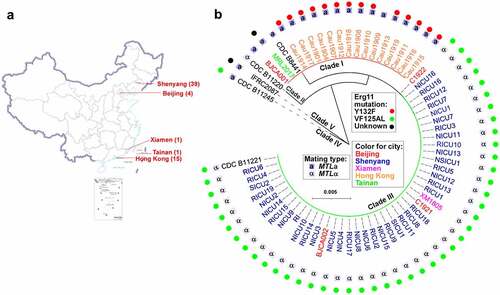
In summary, C. auris infections in China to date have several characteristics. First, most cases have been associated with superficial tissues (e.g. the urinary tract), whereas blood or deep tissue and organ infections have been observed less frequently. Second, most hospitals in China have reported sporadic C. auris cases, rather than clusters of infection, whereas two clusters of C. auris infections occurred in two hospitals (in Shenyang and in Hong Kong). Third, the majority of C. auris clinical isolates reported in China have been resistant to only fluconazole, although several isolates have exhibited high MICs for amphotericin B and echinocandins, perhaps due to a recent evolutionary adaptation in response to antifungal drug use. A F126L (or VF125AL) mutation of the antifungal resistant protein Erg11 has been observed in all fluconazole-resistant isolates in China. Fourth, most C. auris cases in China have occurred in ICU patients aged 55 or older, who often have histories of underlying diseases, such as diabetes and/or hypertension. Finally, it is worth noting that there appears to be a general correlation with long hospital stays and/or treatment with antifungal drugs in predisposing patients to C. auris colonization or infection. In addition, for many of the C. auris clinical isolates obtained in China, other fungal and/or bacterial species were also present in the samples, which could confound identification of C. auris or result in failure to identify and isolate this important emerging pathogen.
Both clinical and basic studies of C. auris in China are limited to date, and many open questions remain to be addressed. For example, what is the original source of C. auris in China? Did C. auris originate from other countries, and if so, how did it get introduced into China? Given the difficulty in accurately identifying this species, can we improve techniques for C. auris isolation and identification in clinical settings? What measures should be taken to prevent clusters of infections and inter- or intra-hospital transmissions, especially during the current coronavirus disease 2019 (COVID-19) pandemic? Is the application of current antifungal drugs in clinical settings associated with the emergence of C. auris? Is C. auris evolving rapidly in terms of its virulence and antifungal drug resistance properties? Given that C. haemulonii is a species closely related to C. auris that is also often resistant to multiple antifungals, and that infection with C. haemulonii has been observed much more frequently in clinical settings in China compared to infection with C. auris, could C. haemulonii be the “superbug” fungal pathogen equivalent of C. auris in China? To address these pressing questions, more efforts are needed to study emerging fungal pathogens, and these efforts would benefit immensely from collaboration between clinical and basic researchers in the field.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article.
Additional information
Funding
References
- Satoh K, Makimura K, Hasumi Y, et al. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009;53:41–44.
- Du H, Bing J, Hu T, et al. Candida auris: epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020;16:e1008921.
- Schelenz S, Hagen F, Rhodes JL, et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control. 2016;5:35.
- Eyre DW, Sheppard AE, Madder H, et al. A Candida auris outbreak and its control in an intensive care setting. N Engl J Med. 2018;379:1322–1331.
- Rhodes J, Abdolrasouli A, Farrer RA, et al. Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg Microbes Infect. 2018;7:43.
- Ruiz-Gaitan A, Moret AM, Tasias-Pitarch M, et al. An outbreak due to Candida auris with prolonged colonisation and candidaemia in a tertiary care European hospital. Mycoses. 2018;61:498–505.
- Lockhart SR, Etienne KA, Vallabhaneni S, et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;64:134–140.
- Wang X, Bing J, Zheng Q, et al. The first isolate of Candida auris in China: clinical and biological aspects. Emerg Microbes Infect. 2018;7:93.
- Chen Y, Zhao J, Han L, et al. Emergency of fungemia cases caused by fluconazole-resistant Candida auris in Beijing, China. J Infect. 2018;77:561–571.
- Fan S, Zhan P, Bing J, et al. A biological and genomic comparison of a drug-resistant and a drug-susceptible strain of Candida auris isolated from Beijing, China. Virulence. 2021;12:1388–1399.
- Tian S, Rong C, Nian H, et al. First cases and risk factors of super yeast Candida auris infection or colonization from Shenyang, China. Emerg Microbes Infect. 2018;7:128.
- Tian S, Bing J, Chu Y, et al. Genomic epidemiology of Candida auris in a general hospital in Shenyang, China: a three-year surveillance study. Emerg Microbes Infect. 2021;10:1088–1096.
- Williams RB, Lorenz MC. Multiple alternative carbon pathways combine to promote Candida albicans stress resistance, immune interactions, and virulence. Mbio. 2020;11:e03070-19.
- Fan S, Yue H, Zheng Q, et al. Filamentous growth is a general feature of Candida auris clinical isolates. Med Mycol. 2021;59:734–740.
- Tang HJ, Lai CC, Lai FJ, et al. Emergence of multidrug-resistant Candida auris in Taiwan. Int J Antimicrob Agents. 2019;53:705–706.
- Tse H, Tsang AKL, Chu YW, et al. Draft genome sequences of 19 clinical isolates of Candida auris from Hong Kong. Microbiol Resour Ann. 2021;10:e00308-20.
