ABSTRACT
Talaromyces marneffei tends to induce systemic infection in immunocompromised individuals, which is one of the causes of the high mortality. The underlying molecular mechanisms of T.marneffei-induced abnormal liver function are still poorly understood. In this study, we found that T.marneffei-infected patients could develop abnormal liver function, evidenced by reduced albumin and increased levels of aspartate aminotransferase (AST) and AST/alanine aminotransferase (ALT). T. marneffei-infected mice exhibited similar characteristics. In vitro investigations showed that T.marneffei induced the death of AML-12 cells. Furthermore, we determined that T.marneffei infection induced pyroptosis in hepatocytes of C57BL/6J mice and AML-12 cells, demonstrated by the increase of AIM2, caspase-1/-4, Gasdermin D(GSDMD) and pyroptosis-related cytokines in T.marneffei-infected mice/cells. Importantly, cell death was markedly suppressed in the presence of VX765 (an inhibitor of caspase-1/-4). Furthermore, in the presence of VX765, T.marneffei-induced pyroptosis was blocked. Nevertheless, necroptosis and apoptosis were also detected in infected animal model at 14 days post-infection. In conclusion, T.marneffei induces pyroptosis in hepatocytes through activation of the AIM2-caspase-1/-4-GSDMD axis, which may be an important cause of liver damage, and other death pathways including necroptosis and apoptosis may also be involved in the later stage of infection.
Introduction
Talaromyces marneffei (T. marneffei) was included in the list of “critical pathogens” released at ameeting of the World Health Organization expert group on anti-fungal diseases [Citation1]. T.marneffei, previously known as Penicillium marneffei, is the only dimorphic member of the species and is an emerging pathogenic fungus that can cause afatal systemic mycosis known as talaromycosis (formerly called penicilliosis) [Citation2,Citation3]. This disease is endemic in southeast Asia and southern China, and about 50,000 people living with HIV are infected with T.marneffei everyyear in high-risk regions [Citation4,Citation5]. Southern provinces accounted for 99% of talaromycosis cases in China, 43% of which were in Guangxi. Among HIV+ populations, the proportion of T.marneffei infection is 9–18%, and the mortality rate is 25per 100person-months [95% confidence interval: 21.5–26.7] [Citation4,Citation6,Citation7]. Increasing globalization, rising HIV prevalence, and emerging iatrogenic immunodeficiency conditions mean that the disease burden attributed to talaromycosis will continue to rise.
Talaromycosis is the leading cause of death in patients with advanced HIV disease and is associated with amortality of more than 30% if not given timely and appropriate systemic antifungal therapy [Citation8–11]. Based on the natural route of infection, the most commonly involved organ is the lung, but the liver, although frequently overlooked, is also acommonly involved organ [Citation12–14]. Previous studies and clinical observations have shown that T.marneffei infection may cause liver damage. While the liver can show different pathological alterations depending on immune status after T.marneffei infection, aspectrum of pathological damage to the liver has been observed in cases of hepatic talaromycosis in some clinicopathological studies [Citation15,Citation16]. However, the specific pathological mechanism is not clear. Currently, the National Free Antiretroviral Treatment Program requires that the start of antiretroviral treatment should be delayed when liver function abnormalities are greater than level 3, but this increases the risk of opportunistic infections as the restoration of the patient’s immune system will be delayed [Citation17]. Therefore, it is clinically imperative to resolve T.marneffei-mediated liver damage.
Among the numerous factors linked to inflammation-induced liver injury, pyroptosis is gaining increasing attention. Pyroptosis is an acute inflammatory response triggered by either caspase-1 or caspase-4/-11, caspase-5, and pro-inflammatory cytokine maturation [Citation18–20]. Several pathogens have been demonstrated to induce liver damage through the stimulation of pyroptosis. NLRP3 inflammasome is activated by Aspergillus flavus and provoked hepatocyte pyroptosis and oxidative stress, which led to liver damage [Citation21,Citation22]. Aspergillus fumigatus (A.fumigatus) enhanced the expression of pyroptosis-related proteins in macrophages [Citation23]. Pseudomonas aeruginosa (P.aeruginosa) activated the AIM2 receptor through DNA released from host mitochondrial damage, thereby up-regulating the inflammasomes and causing liver damage [Citation24,Citation25]. In addition, the TNF-α signaling pathway and lipid metabolism pathways are also involved in liver injury in some hepatic diseases. Accumulating evidences suggested that innate immune sensors, including NOD-like receptor (NLR) family members (NLRP1B, NLRP3, and NLRC4, etc.), and other non-NLR receptors, such as AIM2, played important roles in the pathophysiology of infectious, inflammatory, and autoimmune diseases. The DNA released during microbial lysis directly binds to AIM2, resulting in oligomerization with ASC and caspase-1 to form afunctional inflammasome complex that promotes caspase-1-dependent pyroptosis by activating the AIM2/caspase-1 pathway [Citation26–29]. In this study, we first analyzed epidemiological data to quantify markers of liver damage in HIV/T.marneffei co-infected patients. Afterward, we explored the mechanism of liver injury caused by T.marneffei in an invitro model using cultured hepatocytes and in an invivo model using immunocompetent mice. Our study explains clinical changes in the liver function of T.marneffei positive patients, in order to help healthcare providers optimize the clinical management of HIV/T.marneffei co-infected patients.
Materials and methods
Ethics statement
Guangxi Medical University Research Ethics Committee approval was obtained (approval number: 2019-SB-065), and all patients gave their informed consent. Animal care and protocols were approved by Animal Welfare Committee of Guangxi Medical University (approval number: 202,006,001).
Fungal strain
The T.marneffei strain used in this experiment was isolated from an HIV/T.marneffei co-infected patient and identified by PCR amplification and 16S-23S rRNA internal transcribed spacer region (ITS) sequences analysis. The strain was incubated on Sabouraud’s dextrose agar (SDA) and Potato dextrose agar (PDA) medium and maintained at 4℃. T.marneffei conidia were cultured on SDA and PDA at 25℃ for 10–14 days and collected by flooding the surface of aculture with phosphate-buffered saline (PBS) and counted with ahemocytometer. Lactophenol cotton blue staining was used to observe T.marneffei conidia and fungal morphology.
Cells culture and treatments
AML-12 (normal mouse hepatic cells) was obtained from the Cell Bank of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences and cultured in Dulbecco’s Modified Eagle’s Medium/F12 (DMEM/F12) (GIBCO, USA) containing 10% fetal bovine serum, 10 µg/ml insulin, 5.5 µg/ml transferrin, 5 ng/ml selenium and 40 ng/ml dexamethasone. Cells in this medium were cultured at 37℃ in ahumidified atmosphere containing 5% CO2 and seeded in 6-well plates at adensity of 2 × 105 cells/well or in 24-well plates at adensity of 5 × 104 cells/well, respectively. For stimulation, hepatic cells were treated with T.marneffei [(Multiplicity of Infection, MOI) = 3)] for 12 h, 24 h and 48 h, respectively.
Animal experiments
Six to eight-week-old male C57BL/6J mice were purchased from Guangxi Medical University Laboratory Animal Center. Mice in the infection group were injected intravenously with 200 microliters of T.marneffei conidia at adose of 105 colony-forming units per microliter, and the control group was injected with the same volume of saline. liver tissues and plasma were collected after 3, 7 and 14 days of infection for following experiments.
Transmission electron microscopy (TEM)
Cells or tissue samples were fixed overnight at 4℃ in 3% glutaraldehyde. The cell pellets or tissues were washed three times with PBS and subsequently stained with osmium solution for 1 h at 4℃. The stained samples were washed three times with PBS, and then dehydrated with 50%, 70%, 90% acetone, respectively for 15 min, finally with 100% acetone three times, 15 min each time. The samples were embedded with Epon-618 resin, with 2 h in amixture of acetone and embedding agent (1 :1), 3 h in amixture of acetone and embedding agent (1 :3), overnight in embedding agent at 37℃, and 60℃ incubation for 48 h. The embedded samples were cut into 60–80 nm sections, and stained with 2% uranium acetate saturated alcohol solution and lead citrate for 15 min respectively. The sections were examined using an H-7650 transmission electron microscope (Hitachi, Japan).
Immunostaining and histology
Liver tissue sections from each group of mice were prepared and fixed in 4% paraformaldehyde at 4℃ overnight and embedded in paraffin. Paraffin sections (3–4 μm) were stained with hematoxylin–eosin (H&E) reagent, periodic acid-Schiff (PAS), and Gomori’s methenamine silver (GMS) separately. After mounting, the sections were observed by an inverted light microscope (Nikon Diaphot Microscope, Japan).
Total RNA extraction, library construction and sequencing
The C57BL/6J mice infected with T.marneffei at 3, 7 and 14 days were compared with control group. Total RNA was extracted from mouse liver tissue using Trizol reagent (Invitrogen, USA) according to the manufacture’s protocol. RNA quality was assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies, USA). Then, mRNA was enriched by Oligo(dT) beads, while prokaryotic mRNA was enriched by removing rRNA by Ribo-ZeroTM magnetic kit (Epicenter, USA). Subsequently, the enriched mRNA was fragmented into short fragments using fragmentation buffer and reverse transcripted into cDNA, which was purified with QiaQuick PCR extraction kit (Qiagen, Netherlands), and ligated to Illumina sequencing adapters. The library was sequenced using Illumina Novaseq6000 System (Guangzhou, China). In this study, atotal of 21 samples at 3 time points were sequenced, with 3 control samples and 4 T.marneffei-infected samples at each time point.
Differential analysis of transcriptome
The reads were obtained from the sequencing machines and were further filtered by fastp software (version 0.18.0) to get high quality clean reads. Paired-end clean reads were mapped to the reference genome using HISAT2. 2.4 to calculate read counts for each unigene. Differentially expressed genes (DEGs) between different experimental conditions were identified using atwo-sided Wald test in DESeq2 package, with statistical significance set as |log2 fold of change| > 1 and adjusted P-value <0.05. Because the DESeq2 package requires input in the form of raw count, which is normalized by the estimateSizeFactors function, we did not calculate reads per kilobase of exon model permillion mapped reads before this. The heatmap was constructed using the pheatmap Rpackage to inflect the expression intensity and direction of the DEGs in different time points. The volcano plots were constructed based on ggplot2 R package to exhibit the DEGs whose |log2 fold of change| were larger than 1.
Function enrichment analysis
The potential function of DEGs among different times infected with T.marneffei, modules enrichment analyses were predicted using Clusterprofiler Rpackage. Genes with |log2 fold of change| > 1 and adjusted P-value <0.05 were selected for Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analysis. Gene set enrichment analysis (GSEA) was performed on RNA expression profiling using the Rpackage ‘clusterProfiler’.
Flow cytometry analysis
Flow cytometry was used to detect cell damage. Briefly, after stimulationwith T.marneffei for 24 h, 48 h and 72 h, the samples were harvested to stain with propidium iodide (PI, BD Biosciences, USA), which was used at afinal concentration of 500 ng/ml. Then samples were run and analyzed using Cyto FLEX2 (Beckman Coulter, USA).
Assessment of lactate dehydrogenase (LDH)
Released LDH in culture supernatants from damaged cells was measured with LDH assay kit (Nanjing Jiancheng Bioengineering Institute, China), and the LDH activity in cell supernatants was calculated according to the instructions. The optical density (OD) value was read at 440 nm with amicroplate reader (BioTek, USA).
Analysis of alanine aminotransferase (ALT) and aspartate aminotransferase (AST)
ALT assay kits and AST assay kits (Nanjing Jiancheng Bioengineering Institute, China) were purchased to examine ALT and AST released by plasma and hepatic cells supernatants.
Primers designing and reverse transcription quantitative PCR (RT-qPCR) analysis
Primers for the amplification of Aim2, caspase-1, caspase-4, Il-1β, Il-18, Tnf-α, Gsdmd, Gapdh and β-actin were designed in PrimerBank website (https://pga.mgh.harvard.edu/primerbank/). Primers were optimized prior to quantification experiments using RT-qPCR. Primer sequences were shown in Table S1. Total RNA was purified using Trizol reagent and chloroform extraction, followed by precipitation in isopropyl alcohol and 75% ethanol. Reverse transcription was performed using areverse transcription kit (TAKARA, Japan). RT-qPCR was performed using SYBR Premix ExTaqTM(TAKARA, Japan) by StepOnePlus Real-time PCR system (Thermo Fisher Scientific, USA).
Protein extraction and western blot analysis
AML-12 cells and liver tissues were harvested and subjected to protein extraction using cell lysis buffer (CST, USA) according to the manufacture’s protocol. After protein extraction, protein concentrations were determined using BCA protein concentration assay kit (Beyotime, China). An equal amount of protein extracted from AML-12 cells or liver tissues were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to polyvinylidene difluoride membrane. After blocking with 5% nonfat milk, membranes were incubated at 4°C overnight with the specific primary antibodies, such as caspase-1, cleaved caspase-1, gasdermin D(GSDMD), cleaved-GSDMD and β-actin, and then incubated for 1 h with the corresponding secondary antibody. Images were developed using the Odyssey CLX two-color infrared laser imaging system (LI-COR, USA).
Lentivirus transfection
The lentivirus vector with caspase-1 siRNA (ID NM_009807) was constructed by Genechem (Shanghai, China). caspase-1 gene expression was interfered using caspase-1-RNAi-lentivirus transfection technology. The sequences were designed and synthesized as follows: RNAi-caspase-1-F:5’-CCGGGGGCAAAGAGGAAGCAATTTACTCGAGTAAATTGCTTCCTCTTTGCCCTTTTTG-3’, RNAi-caspase-1-R:5’-AATTCAAAAAGGGCAAAGAGGAAGCAATTTACTCGAGTAAATTGCTTCCTCTTTGCCC-3’. Non-transfected hepatic cells were used as the blank control, and negative control (NC) was transfected with empty lentivirus vector. The caspase-1-RNAi-lentiviruses were transfected for the knock down (KD) groups.
Data analysis and statistical analysis
For RT–qPCR data, relative gene expression was defined as aratio of target gene expression versus Gapdh and β-actin genes expression. The relative levels of RNA were calculated using the comparative CT (2 −ΔΔ Ct) method.
For western blot data, relative protein levels were calculated as the density ratios of target protein to β-actin, that is, the gray density of the target protein band was divided by that of the internal reference protein of the corresponding sample (target protein/β-actin) to make the corresponding statistical data. Protein quantification was performed using Image Jsoftware.
For statistical analysis, quantitative data with normal distribution were presented as mean ± standard deviations (SD), and nonnormal data were presented as median and the interquartile range. Comparisons were performed by Student’s ttest or one-way ANOVA analysis for normally distributed data, and Wilcoxon rank-sum test for nonnormal data. The results of P-value of <0.05 was considered statistically significant, P < 0.01 indicated astatistically very significant difference, while P < 0.001 indicated astatistically extremely significant difference. SPSS 23, Rstudio and GraphPad Prism 8 was used to analyze. Furthermore, propensity score matching (PSM) analysis was used to reduce selection bias and potential confounding effects caused by CD4+ Tlymphocytes, leukocytes, and platelets, etc. The HIV-1 mono-infected group and HIV/T.marneffei co-infected group were matched in a1 : 1 ratio using the nearest neighbor matching technique without replacement, with acaliper of 0.06.
Results
T.marneffei infection resulted in abnormal liver function in HIV/T. marneffei co-infected patients
In order to investigate the existence of abnormal liver function in T.marneffei-infected patients, we collected samples from 200 HIV-positive patients from Liuzhou People’s Hospital for analysis. Laboratory tests were launched before initiating antifungal therapy to examine the differences in biochemical indicators between the two groups. The PSM was performed to compensate for selection bias. In total, all routine blood variables were similar between the contrast and noncontrast groups via PSM. When the blood routine indexes were comparable between the two groups, serum albumin decreased by 5 g/L in HIV/T.marneffei co-infected patients. In patients with co-infections, AST increased to 59 U/L (interquartile range: 38, 142), exceeding the upper normal limit, and AST/ALT increased from 1.52 to 2.63 (Table S2 and Figure S1). These data indicate impaired liver function in patients with co-infections. Therefore, the results of the epidemiological study showed that T.marneffei infection might contribute to abnormal liver function.
T.marneffei infection resulted in liver damage in C57BL/6J mice
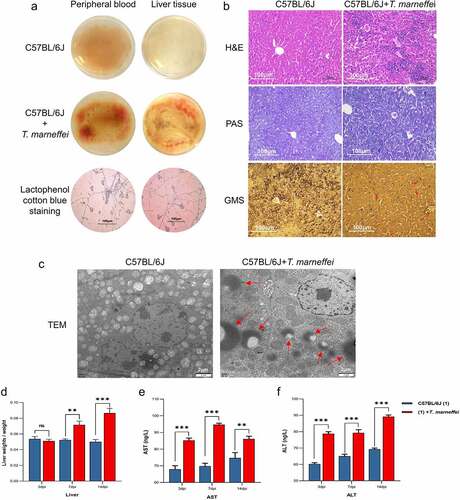
To confirm the epidemiological findings, we constructed aT.marneffei-infected model using 6–8 week old male C57BL/6J mice. At 3, 7 and 14 days post-infection (dpi), whole blood, plasma and liver tissues were collected for analysis. Whole blood and liver tissue homogenates were incubated on PDA plates, and alarge number of conidia were found in the T.marneffei-infected group(). H&E staining of the control group liver tissue showed aregular tissue architecture, alack of fat accumulation in hepatocytes, and the absence of inflammatory cells infiltration. In contrast, the infection group showed inflammatory lesions, mainly manifested as inflammatory cell infiltration (such as monocytes/macrophages, mDC cells, and CD4+ Tcells, etc.), the disordered arrangement of cell cords, and an unclear structure. Using PAS and GMS staining, round to oval sporangia with internal divisions consistent with T.marneffei were visible both free and within liver tissue (). TEM revealed intracellular disturbances: distension of the endoplasmic reticulum; mitochondrial enlargement, swelling and vacuolar degeneration of the mitochondrial cristae; and alighter matrix and fewer cristae. T.marneffei were clearly present in liver tissue (). The liver index (liver weight/body weight) indicated that the liver of the infection group increased significantly by 1.73-fold at 14 dpi (). Plasma levels of AST and ALT were significantly increased (more than 1-fold) in the infection group at 3, 7 and 14 dpi, respectively ().
Figure 1. T.marneffei infection resulted in liver injury in C57BL/6J mice. (a) Whole blood and liver tissue homogenates were incubated at 28℃ for 3d on PDA plates. Large numbers of hyphae and conidia were found on the surface of the medium in the T.marneffei infection group. (b) H&E staining, PAS staining and GMS staining, 200 × . The infection group exhibited obvious inflammatory lesions, inflammatory cell infiltration, disordered arrangement of cell cords, unclear structure and hepatocyte necrosis. In GMS staining, the red arrows point to T.marneffei. (c) TEM, the red arrows pointed to T.marneffei. (d) Liver index (liver weight/body weight) between control group and infection group at 3, 7 and 14 dpi, respectively. (E and F) AST and ALT were determined to estimate hepatic damage. (*P<0.05, **P<0.01, ***P<0.001).
Transcriptomics profiles and the expression levels of cell death-related pathways in T. marneffei-infected liver tissues
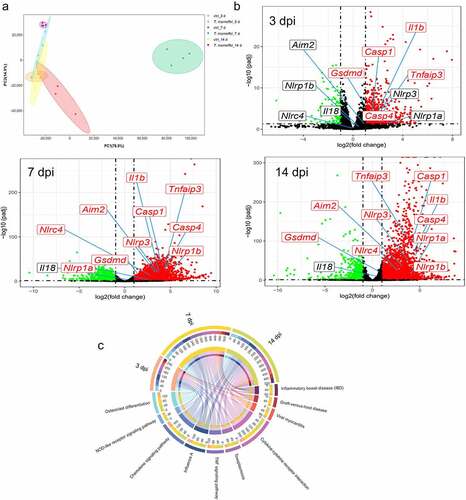
We performed RNA-seq analysis of liver tissue to explore the molecular profile of C57BL/6J livers after T.marneffei infection. Mice were divided into the infection and control groups by injection of T.marneffei conidia and normal saline. The Principal Component Analysis (PCA) was used to describe the differences between different groups of samples (). Atotal of 931 (at 3 dpi), 4749 (at 7 dpi) and 3816 (at 14 dpi) differentially expressed genes were obtained based on the threshold chosen (|log2 Fold of change| > 1 and adjusted P-value <0.05) (). Then, the underlying functions of the DEGs in each time point were predicted via Clusterprofiler Rpackage. The DEGs were significantly involved in immune system process, regulation of immune system process (biological process, BP), cytoplasm, cytoplasmic part, intracellular part (cellular component, CC), protein binding and enzyme binding (molecular function, MF) (Figure S2). Figure S3 showed the KEGG enrichment analysis in metabolism, genetic information procession, cellular processes, etc. Figure S4 displayed the top 10 GO and 15 KEGG pathways according to adjusted P-value, and the most enriched KEGG included pathways in infectious diseases, immune system, and signal transduction. The chord diagram showed the abundance of top 10 KEGG pathways according to adjusted P-value at 3, 7 and 14 dpi (). These DEGs, regulated by T.marneffei infection, were tightly associated with several signaling pathways, including the NOD-like receptor signaling pathway, TNF signaling pathway, and cytokines-cytokines receptor interaction, etc., indicating that T.marneffei infection significantly promoted the inflammatory response of liver tissue.
Figure 2. RNA-Seq analysis of C57BL/6J mice liver tissues at 3, 7 and 14 days post T.marneffei infection. (a) Principal component analysis on RNA-seq data. The data showed clear grouping of replicate samples for same developmental stage and infection groups. PC1 on the X-axis captures the greatest variation and accounts for 77% of the total variance around the PCs, while PC2 on the Y-axis captures thesecond greatest variation and accounts for 15% of the total variance around the PCs. (b) Volcano plots of DEGs at 3, 7 and 14 dpi. The mean expression level for control group vs. infection group was assessed for fold-change (X-axis) vs P-value (Y-axis) by Wald test based on DEseq2 package. Each gene is represented by afilled circle. Red indicates ahigher expression level in infected group compared with control group, while green means the opposite. The genes marked in the figures are: Nlrp1a, Nlrp1b, Nlrp3, Nlrc4, caspase-1, caspase-4, Aim2, Il-1β, Il-18, Tnf-α, Gsdmd. (c) the chord diagram. The abundance of top 10 KEGG pathways according to adjusted P-value in the different time points was exhibited.
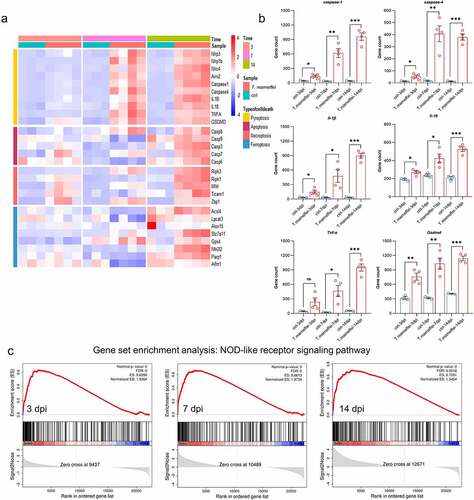
Based on the KEGG pathway analysis, we further analyzed the pathways related to infection, inflammation and signal transduction, as well as cell death-related pathways that may lead to liver damage. After RNA-seq read count data were obtained, the differences in transcription factors were calculated using the DEseq2 package, which was fitted with amodel for anegative binomial distribution, and hypothesis testing was performed using the Wald test. As shown in , the transcriptional levels of pyroptosis-related factors were significantly increased, such as caspase-1 from 2.24-fold at 3 dpi to 22.80-fold at 14 dpi, and Gsdmd from 1.38-fold to 2.21-fold, respectively. The activation time of the apoptosis pathway was later, and there was no difference in the expression in the early stage, caspase-3 was only up-regulated by 0.61-fold at 14 dpi, indicating that apoptosis may just function at later times of infection (Figure S5). The expression of key factors of ferroptosis showed the opposite tendency, that is, the Gpx4 was significantly up-regulated (27% at 14 dpi) when C57BL/6J mice were infected with T.marneffei, indicating that the ferroptosis might not occur (Figure S6). The expression levels of necroptosis associated factors (Ripk1/3, Mlkl, Ticam1, Zbp1) were also upregulated, but the change level was lower than the up-regulated level of pyroptosis-related factors (Figure S7). Therefore, we deduced that pyroptosis may be the dominant form of cell death in the livers of C57BL/6J mice infected with T.marneffei but did not rule out asynergistic role played by necroptosis. The bar plots showed the expression levels of pyroptosis associated factors (), and the different inflammasomes (Nlrp1b, Nlrp3, Nlrc4 and Aim2), upstream stimulatory factors of pyroptosis, were shown in Figure S8. We then employed the GSEA approach, aconventional method for identifying pathways associated with gene expression (). The enrichment score of the NOD-like receptor signaling pathway was higher than 0.6 at three time points, and the core genes in this gene set were highly expressed in the infected group. However, the ES of the apoptosis pathway (mmu04210) was relatively low (0.390), and the difference was not statistically significant (P.adjust = 0.052) between the infection and non-infection groups. Similar to apoptosis pathway, necroptosis pathway (mmu04217) showed alow ES value (0.386), and the difference was not statistically significant (P.adjust = 0.066) between the two groups. In addition, GSEA did not find enriched pathways related to ferroptosis and autophagy.
Figure 3. The expression levels of pyroptosis associated factors. (a) Heatmap of genes involved in different types of cell death. Red represents high expression, and blue represents low expression. (b) the mRNA expression level of different pyroptosis related genes. All of these genes are involved in the NOD-like receptor signaling pathway. (c) Results of GSEA analysis. Pyroptosis related genes were all enriched in the NOD-like receptor signaling pathway in different time points when C57BL/6J mice infected with T.marneffei. Enrichment score (ES) and FDR were shown. Red curve indicates enrichment score (ES), the gray shadow at the bottom represents the Log2 fold of change value of each gene. (*P< 0.05, **P< 0.01, ***P< 0.001).
T.marneffei up-regulated the expression levels of pyroptosis-related genes in C57BL/6J mice
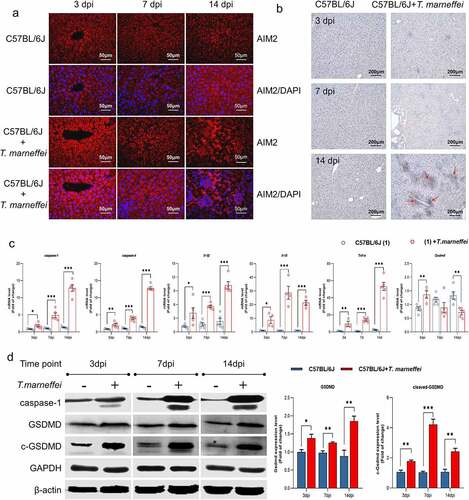
Afterward, we verified that the pyroptosis related factors were induced in the liver of T.marneffei-infected mice. Immunofluorescence imaging revealed the increased expression of AIM2 in T.marneffei-infected liver tissue at 3, 7 and 14 dpi (). To determine the critical factor of pyroptosis, we used immunohistochemistry to detect caspase-1 in mice livers. As shown in , there was increased caspase-1 within the inflammatory infiltrate foci and parenchymal cells in the liver of T.marneffei-infected C57BL/6J mice at 14 dpi. RT-qPCR showed that the mRNA levels of caspase-1/-4, Il-18 and Tnf-α were upregulated at 3 dpi, and continued to increase with ongoing infection. Gsdmd showed asignificantly increased level at 3 dpi, but was significantly decreased at 14 dpi (). The protein expression profile of caspase-1 was consistent with the mRNA profile, while the level of GSDMD and cleaved-GSDMD gradually increased from 3 dpi to 14 dpi (). These results were consistent with the trends identified by RNA-seq analysis, suggesting that proteins central to pyroptosis may play apathological role during acute T.marneffei infection.
Figure 4. The expression level of pyroptosis associated factors in liver tissue of T.marneffei-infected C57BL/6J mice. (a) Immunofluorescence result of the infected group vs. control group in different time points. (b) Representative immunohistochemistry images showing increased staining of caspase-1 protein expression at 14 din samples from T.marneffei-infected mice. The red arrows point to increased caspase-1 protein. (c) the transcriptional level of different pyroptosis related genes. caspase-1/-4, Il-18 and Tnf-α were up-regulated at 3 dpi, and continued to rise with ongoing infection, while Gsdmd exhibited higher level at 3 dpi, and the expression significantly decreased at 14 dpi. (d) the protein expression level of pyroptosis critical proteins by western blot. (*P< 0.05, **P< 0.01, ***P< 0.001).
T.marneffei up-regulated the expression levels of pyroptosis-related genes in AML-12 cells
We established an invitro infection model using the AML-12 cell line to verify the effect of T.marneffei on the pyroptosis-related pathway. Light microscopy indicated that there was asignificant increase in cell death after 48 h of T.marneffei infection (). TEM analysis revealed that T.marneffei were found inside the AML-12 cells. In addition, alarge number of cavities were produced; the cells swelled and ruptured, and the cellular contents were released (). showed that LDH levels were significantly reduced by 24% in culture supernatants when the T.marneffei infected cells were pretreated with VX765 (the inhibitor of pyroptosis), indicating that pyroptosis is involved in the cell damage induced by T.marneffei. This trend was almost similar to necrosulfonamide (NSA, the inhibitor of GSDMD and MLKL) intervention. Flow cytometry was used to detect the damage of T.marneffei-infected AML-12 cells, cell damage was increased significantly after infection with T.marneffei (13% at 24 h, 22% at 48 h and 33% at 72 h) (Figure S9a). Then, total RNA and protein were extracted to determine the expression levels of pyroptosis related genes, and the results showed that the mRNA and protein expression levels of key pyroptosis factors (caspase-1/-4, IL-18, TNF-α, GSDMD) in the infected group were higher than in the control group (). It is worth mentioning that GSDMD protein can induce pyroptosis only after being cleaved and split into cleaved-GSDMD protein by caspase, which may lead to the upregulation of cleaved-GSDMD protein later than the Gsdmd gene level. In this study, cleaved-GSDMD protein showed an upward trend only after 48 h of infection. The detection of inflammatory factors upstream of pyroptosis showed that only Aim2 expression was up-regulated by 26% and 96% after 24 hours and 48 hours of infection, respectively, and the difference was statistically significant(Figure S10).
Figure 5. The expression levels of pyroptosis associated factors in AML-12 cells. (a) Light and transmission electron micrographs of AML-12 cells infected by T.marneffei. There was asignificant increase in cell death after 48 hof T.marneffei infection, and T.marneffei conidia were found inside the AML-12 cells. The red arrows pointed to T.marneffei. (b) the cell supernatant was collected to detect the LDH expression after 48 hof infection. The level of cell damage was decreased when treated with VX765 (the inhibitor of caspase-1/-4) and NSA (the inhibitor of GSDMD and MLKL) (c) the mRNA expression level of different pyroptosis related genes. (d) the protein expression level of pyroptosis critical proteins by western blot. (*P< 0.05, **P< 0.01, ***P< 0.001).
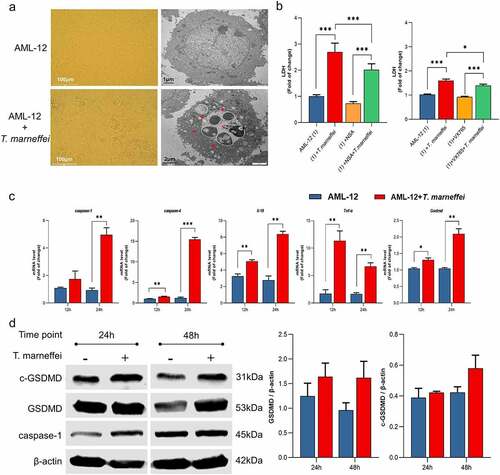
Inhibition of pyroptosis decreased the inflammatory response of AML-12 cells
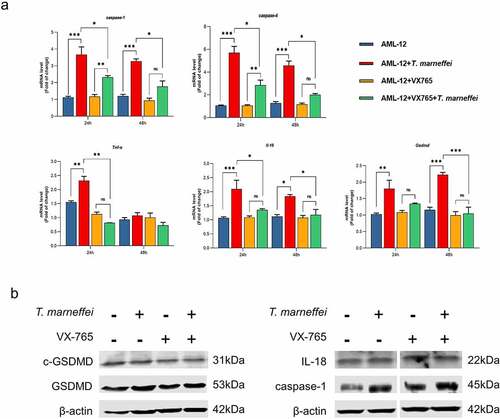
The role of caspase-1-dependent pyroptosis in T.marneffei-induced hepatocytic inflammation was investigated. GFP is afluorescent protein encoded in the lentiviral vector, which is used to determine the transfection efficiency of lentiviral vector. GFP fluorescence in AML-12 cells was observed at 72 h following transfection, which suggested the successful transfection of RNAi-caspase-1 lentivirus (Figure S11). Compared with the cells transfected with empty lentivirus vectors (NC group), the expression level of caspase-1 was reduced by 93% in RNAi-caspase-1 group (Figure S11). However, the mRNA level of downstream inflammatory factors was not suppressed, and caspase-4 showed ahigher mRNA expression level (0.83-fold increase) (), possibly as acompensatory or redundant mechanism. The expression levels of GSDMD and cleaved-GSDMD protein were also elevated, although caspase-1 protein was inhibited (). Therefore, we pre-treated cells with VX765, an inhibitor of both caspase-1/-4. The cell viability result demonstrated that 50 μM was the optimal concentration of VX765 (Figure S9b). Therefore, 50 μM VX765 was selected as the concentration used for subsequent experiments. As shown in , the LDH cytotoxicity assay showed that LDH levels were significantly reduced by 24% in culture supernatants when the T.marneffei infected cells were pre-treated with VX765, indicating that pyroptosis is involved in the cell damage induced by T.marneffei. The mRNA expression levels of caspase-1 and -4 were all suppressed, and the downstream inflammatory factors Il-18, Tnf-α and Gsdmd were reduced by 35%, 65% and 53%, respectively. (). The expression of proteins was consistent with the mRNA results (), indicating that when the pyroptosis is inhibited, the inflammatory injury of liver cells will be alleviated.
Figure 6. The expression level of pyroptosis associated factors after inhibiting caspase-1 in AML-12 cells. (a) After lentiviral transfection, the expression of pyroptosis related factors in T.marneffei-infected cells was detected at 48 hpost infection. Compared with the cells transfected with NC group, the expression level of caspase-1 was decreased in RNAi-caspase-1 group, but the mRNA level of downstream inflammatory factors was not suppressed, and caspase-4 reflected ahigher expression level. (b) the expression levels of pyroptosis related proteins in caspase-/- AML-12 cells at 48 hpost infection. Although the expression of caspase-1 protein was significantly decreased after siRNA transfection, the expression of downstream GSDMD and cleaved-GSDMD proteins were still increased. (*P< 0.05, **P< 0.01, ***P< 0.001).
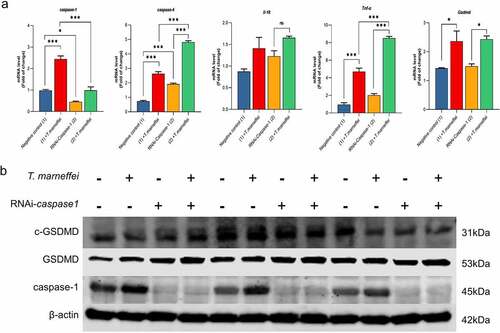
Figure 7. The expression level of pyroptosis associated factors after VX765 pretreatment in AML-12 cells. (a) After VX765 pretreatment, the expression of pyroptosis related factors in T.marneffei-infected cells was detected after 48 hof infection. Caspase-1 and -4 were all suppressed, and downstream inflammatory factors were also decreased. (b) the expression levels of pyroptosis related proteins in VX765-pretreated AML-12 cells at 48 hpost infection. Compared with infection group, VX765 pretreatment group decreased the expression of caspase-1 protein, and the GSDMD and cleaved-GSDMD proteins also decreased. (*P< 0.05, **P< 0.01, ***P< 0.001).
Discussion
T.marneffei is adimorphic fungus which can infect both HIV/AIDS and non-HIV hosts. At present, the pathogenic mechanism of T.marneffei is not well understood. In this study, we found that the infection rate of T.marneffei was 10% in HIV-1 infected patients with CD4+ Tcell counts less than 1500 cells/µl, 17% in those less than 200 cells/µl, and 22% in those less than 100 cells/µl. Overall, 25% of HIV/T.marneffei co-infected patients had hepatosplenomegaly on admission, while 75% showed abnormal AST levels. Then, we established invivo and invitro infection models to explore the molecular mechanism of liver damage. The results showed that T.marneffei could rapidly invade into the liver and hepatocytes. However, it is still unclear how T.marneffei gains entry into hepatocytes. The main function of the liver is to participate in biological metabolism and transformation, and hepatocytes can not only synthesize bile acid, serum albumin, digestive enzymes and various clotting factors, but also participate in the metabolism of lipids, sugars and hormones [Citation30]. In addition, hepatocytes can transform endogenous or exogenous toxins into nontoxic or low-toxic components [Citation31]. At present, the research into pathogen-mediated hepatocyte injury is mainly focused on viral infection [Citation32–34]. Firstly, the innate immunity of hepatocytes provides the first line of defense against pathogens, including hepatitis viruses, and activates both protective direct antiviral and inflammatory responses. For instance, liver damage caused by HBV is due to chronic immune responses by liver-infiltrating immune cells that target infected hepatocytes [Citation35,Citation36]. In the case of fungal infection, many Aspergillus species produce Sterigmatocystin (STC), which is arisk factor of liver cancer [Citation37]. It has been reported that tissue examination in mice showed liver cell injury and necrosis after being fed adiet containing STC. Also, STC is considered to be involved in chronic liver disease of people living in Africa [Citation38]. However, few studies indicated that fungi could actively enter liver parenchymal cells. Interestingly, in this study, fungal spores could be cultured on PDA medium in mouse liver tissue infected with T.marneffei; TEM revealed disordered liver tissues and mitochondria, with T.marneffei found inside the hepatocytes, which were partially degraded. Similar phenomena were observed invitro. Therefore, we can infer that T.marneffei may bind to unidentified receptors on the surface of hepatocytes that enable active invasion into the cells. Other instances of pathogens that are free-living but can invade cells include: P.aeruginosa, which invade epithelial cells (human alveolar epithelial cells and rabbit corneal epithelial cells, etc.) during infection [Citation39]and Burkholderia pseudomallei, which invades and survives intracellularly within A549 human lung epithelial cells [Citation40]. In addition, phagocytosis of A.fumigatus conidia by epithelial cells has also been demonstrated [Citation41,Citation42]. After 4 h of incubation with A.fumigatus showed conidia internalized in amembrane-bound vacuole. The expression levels of factors such as IL-8, TNF-α and GM-CSF were significantly up-regulated [Citation41]. It is worth mentioning that, due to preexisting lung disease, defects in the clearance of internalized pathogens have been shown to increase intracellular survival and further increase the risk of pathogens invading the host and thus predisposing to disease [Citation43–45].
We collected key factors associated with major programmed death modes such as apoptosis, pyroptosis, necroptosis, autophagy and ferroptosis. The results showed that the expression of pyroptosis pathway-related factors was continuously up-regulated and the difference was statistically significant. The activation time of the apoptosis pathway was later and there was no difference in the expression in the early stages. The expression of necroptosis was different, but the change levels were lower than the up-regulated levels of pyroptosis-related factors; also, the expression of key factors of ferroptosis showed the opposite result. On the other hand, we detected factors on different death pathways on AML-12 cells and found that the result of pyroptosis is stable and persistently high expression. Comprehensively, we believe that the acute phase of T.marneffei infection is mainly involves the pyroptosis-related pathway. Pyroptosis has emerged as akey mechanism by which inflammasomes promote the host defense against microbial pathogens, but excessive pyroptosis can aggravate the inflammatory response, as observed during septic shock, atherosclerosis, epilepsy and cardiomyopathy, etc [Citation46,Citation47]. With regard to upstream regulators of pyroptosis, avariety of inflammasomes play an important role. In this study, we found that NLRs such as Nlrp1, Nlrp3 and Nlrc4 were up-regulated by 36.96, 13.98 and 8.97-fold at 7 dpi, respectively, and the expression of Aim2-like receptor was up-regulated by 11.51-fold. However, only Aim2 showed acontinuous upward trend during invitro and invivo validation (Figure S10). According to the records of KEGG pathway database, AIM2 senses the presence of double-stranded DNA (dsDNA) in the cytoplasm, regardless of whether it is of bacterial, viral or host cellular origin. The role of the AIM2 inflammasome in response to fungi is less clear. Aprevious study showed that AIM2 and NLRP3 cooperatively induced the cytoplasmic inflammasome platform in A.fumigatus-infected mouse bone marrow-derived cells. The Legionella pneumophila secretory effector protein SDHA is known to inhibit bacterial DNA release and thus interferes with AIM2 inflammasome recognition and activation [Citation48]. Mice lacking both AIM2 and NLRP3 were highly susceptible to aspergillosis, indicating that AIM2 might be co-activated with NLRP3. In this study, we found that fungi could invade liver cells and cause lysis, suggesting that the activation of AIM2 by dsDNA may induce pyroptosis. In addition to AIM2, caspase-4 is another key factor that plays an important role in activating pyroptosis pathway [Citation49]. Independently of the NLRP3 inflammasome and caspase-1, as areceptor for intracellular LPS, caspase-4 promotes pyroptosis through GSDMD cleavage and activation, and through the release of IL-1, IL-18 and HMGB1. However, we showed that T.marneffei, athermally dimorphic fungus, could also activate caspase-4. Some studies revealed that mitochondrial permeability transition activated caspase-4 by promoting the assembly of protein complexes (named APAF-1 pyroptosome) to promote pyroptosis [Citation50]. In this study, we found the following using TEM evidence of severe mitochondrial dysfunction and damage in the T.marneffei infected group: mitochondrial enlargement, swelling and vacuolar degeneration of the mitochondrial cristae, lighter mitochondrial matrix, and alower overall density of cristae. Therefore, we hypothesized that there might be aconnection between mitochondrial damage and the activation of caspase-4, but the specific mechanism still needs to be further explored.
Numerous factors are responsible for liver injury, such as drugs, alcohol, toxic chemicals, and pathogenic microbes [Citation51]. Many cytokines participate in the development and progression of liver damage, and the interplay between cytokines constitutes acomplex pathogenic process. IL-1β is an important regulator of inflammation, which upregulates the expression of cytokines, including TNF-α and NO, thus amplifying the inflammatory response. IL-17 is acytokine associated with inflammation and autoimmunity and has also been shown to amplify inflammatory responses. However, IL-10 can alleviate inflammatory reactions and inhibit the production and release of many proinflammatory factors. Furthermore, there is evidence that the TGF-β1/STAT3 pathway is an important inflammatory pathway which causes liver fibrosis and cirrhosis [Citation52]. Additionally, various stimuli cause liver damage through activated Tcells and macrophages, which produce ahigh level of inflammatory cytokines, including IL-1β, TNF-α, IFN-γ, and NO, thus ultimately leading to the induction of apoptosis. In this study, we found that in T.marneffei infected patients, plasma AST levels were significantly higher than the upper limit of normal (ULN), while ALT exhibited arelatively normal expression level (P = 0.531). This distinction may be clinically useful for the diagnosis and treatment of patients infected with T.marneffei.
Talaromycosis is asystemic disease with multi-organ involvement. Therefore, anew drug which could inhibit multi-organ injury is urgently required for patients infected with the fungus. VX765, an inhibitor of caspase-1/-4, is an orally absorbed prodrug of the active metabolite and has been demonstrated to be safe for humans as tested in aphase 2b human clinical trial against epilepsy. VX765 reduced disease severity and the expression of inflammatory mediators in models of rheumatoid arthritis and skin inflammation. In this study, we showed that cell damage in T.marneffei-infected AML-12 cells was alleviated by VX765, as indicated by decreased expression levels of inflammatory cytokines. Therefore, VX765 may act as apromising agent against multi-organ damage caused by T.marneffei infection. Some studies using caspase-1-/- mice to explore the role of pyroptosis in traumatic injury or noncognate tuberculosis showed that pyroptosis-related proteins were less prominent in caspase-1-/- mice than those in the WT mice [Citation53,Citation54]. Other studies using intraperitoneal injection of VX765 in an animal pyroptosis model indicated that VX765 at the concentration of 100 or 200 mg/kg could significantly inhibit the expression of IL-1β, IL-18 and GSDMD [Citation55,Citation56]. These invivo studies demonstrated that VX765 or caspase-1−/− inhibited the expression levels of pyroptosis-related proteins in mice induced by external stimuli (such as T.marneffei infection).
However, there are still several limitations that need to be considered further. First, we collected key factors associated with major programmed death modes including apoptosis, pyroptosis, necroptosis, autophagy and ferroptosis. The results showed that the pyroptosis pathway-related factors were continuously up-regulated with adifference that was statistically significant; the activation time of apoptosis pathway was later, and there was no difference in the expression in the early stages; and the expression of necroptosis-related factors was different, but the altered levels were lower than those of pyroptosis-related factors. Therefore, T.marneffei-induced cell death might not just be attributed to pyroptosis. In addition, the ES of the apoptosis pathway was relatively low, and the difference was not statistically significant between infection and non-infection groups. Similar to the apoptosis pathway, the necroptosis pathway showed alow ES value, and the difference was not statistically significant between the two groups. GSEA did not find enriched pathways related to ferroptosis and autophagy. According to the results of GSEA, we believed that pyroptosis may be the main cause of cell death after infection with T.marneffei, but we cannot draw adefinite conclusion. An additional deficiency of the present study is that our invitro data did not fully support the invivo data. Overall, the invitro and invivo data were consistent. At the transcriptional level, the invitro results were consistent with the invivo data. At protein level, although the invitro results were not as significant as those invivo, the similar trends were observed. Nonetheless, we acknowledge that this is one of the limitations. Finally, we would like to state that we do not deny the synergistic effect of various cell death modes, especially necroptosis. As shown in , NSA appeared to inhibit cell damage more than VX765, which may be another indication that other pathways play arole in T.marneffei-mediated liver damage. Cell death is avery complex game in which different core players have the ability to disrupt the delicate balance of the cellular environment from birth to death and from pro-inflammatory to anti-inflammatory signaling. Apoptosis has been the subject of intensive research in the past three decades while pyroptosis has become ahot trend in recent years. Therefore, with the concept of pan-apoptosis (PANoptosis), the research prospect of necroptosis is foreseeable as the last link of the triangle relationship [Citation57,Citation58]. PANoptosis is amultifaceted immune response with important pathophysiological implications for infectious diseases, autoimmunity, and cancer, reflecting the complexity of host responses to pathogens and the interconnectedness of signaling pathways. Subsequent research may be further developed from this perspective.
Conclusion
In conclusion, T.marneffei-infected patients demonstrate abnormal liver function, indicative of liver inflammation and damage. The same phenomenon occurs in the mouse models of T.marneffei infection, in which pyroptosis elicited apathological inflammatory response. T.marneffei infection induces pyroptosis in hepatocytes through activation of the AIM2-caspase-1/-4-GSDMD axis, which may be an important cause of liver damage. Nevertheless, necroptosis and apoptosis were also activated in infected animal model at14 days post-infection, indicating that death pathways other than pyroptosis may also be involved in the later stage of infection. In addition, we found that T.marneffei could invade into the hepatic parenchymal cells, but the entry mechanism needs to be determined in future studies.
Supplemental Material
Download Zip (10.5 MB)Acknowledgements
Gratitude is expressed to all participants who have been dedicated to this study, and to the individuals who volunteered to participate. This study was supported by the National Natural Science Foundation of China (NSFC; 31970167, 81971934, 81803295, 81760602, 81960602), Guangxi Science Fund for Distinguished Young Scholars (2018GXNSFFA281001), Guangxi Youth Science Fund Project (2021GXNSFBA196004, to Wudi Wei), China Postdoctoral Science Foundation (2020M683212, to Wudi Wei), Guangxi Bagui Scholar (to Junjun Jiang), and Guangxi Medical University Training Program for Distinguished Young Scholars (to Junjun Jiang). We are appreciative of Dr.Wu for her critical reading and language polishing in this paper. Moreover, the authors would like to express their gratitude to all those who have dedicated to the prevention and control of infectious diseases.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The datasets generated for this study are available on request to the corresponding author.
Supplementary materials
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21505594.2022.2080904
Additional information
Funding
References
- WHO. First Meeting of the WHO Antifungal Expert Group on Identifying Priority Fungal Pathogens. World Health Org; 2020.
- ChanJF, LauSK, YuenKY, etal. Talaromyces (Penicillium) marneffei infection in non-HIV-infected patients. Emerging Microbes and Infections. 2016;5:e19.
- HoubrakenJ, de VriesRP, SamsonRA. Modern taxonomy of biotechnologically important aspergillus and Penicillium species. Adv Appl Microbiol. 2014;86:199–249.
- Armstrong-JamesD, MeintjesG, BrownGD. Aneglected epidemic: fungal infections in HIV/AIDS. Trends Microbiol. 2014;22:120–127.
- LimperAH, AdenisA, LeT, etal. Fungal infections in HIV/AIDS. Lancet Infect Dis. 2017;e334. DOI:10.1016/S1473-3099(17)30303-1
- JiangJ, MengS, HuangS, etal. Effects of Talaromyces marneffei infection on mortality of HIV/AIDS patients in southern China: aretrospective cohort study. Clin Microbiol Infect. 2019;25:233–241.
- WangYF, XuHF, HanZG, etal. Serological surveillance for Penicillium marneffei infection in HIV-infected patients during 2004-2011 in Guangzhou, China. Clin Microbiol Infect. 2015;21:484–489.
- Vu HaiV, NgoAT, NgoVH, etal. Penicilliosis in Vietnam: aseries of 94 patients. Rev Med Interne. 2010;31:812–818.
- HuY, ZhangJ, LiX, etal. Penicillium marneffei infection: an emerging disease in mainland China. Mycopathologia. 2013;175:57–67.
- SonVT, KhuePM, StrobelM. Penicilliosis and AIDS in Haiphong, Vietnam: Evolution and predictive factors of death. Médecine Et Maladies Infectieuses. 2014;44:495–501.
- LeT, MarcelW, ChiNH, etal. Epidemiology, seasonality, and predictors of outcome of AIDS-associated Penicillium marneffei infection in Ho Chi Minh City, Viet Nam. Clin Infect Dis. 2011;945. DOI:10.1093/cid/cir028.
- QiuY, ZhangJ, LiuG, etal. Acase of Penicillium marneffei infection involving the main tracheal structure. BMC Infect Dis. 2014;14:242.
- MaE, HoP. Disseminated Penicillium marneffei infection. Br JHaematol. 2005;130:2.
- VanittanakomN, CooperCR, FisherMC, etal. Penicillium marneffei infection and recent advances in the Epidemiology and Molecular Biology aspects. Clin Microbiol Rev. 2006;19:95–110.
- YousukhA, JutavijittumP, PisetpongsaP, etal. Clinicopathologic study of hepatic Penicillium marneffei in Northern Thailand. Archives of Pathology & Laboratory Medicine. 2004;128:191–194.
- PanM, QiuY, ZengW, etal. Disseminated Talaromyces marneffei infection presenting as multiple intestinal perforations and diffuse hepatic granulomatous inflammation in an infant with STAT3 mutation: acase report. BMC Infect Dis. 2020;20:394.
- AIDS and Hepatitis CProfessional Group SoID. Prevention CMACCfDca. Chinese guidelines for diagnosis and treatment of HIV/AIDS(2018). Electron JEmerging Infect Dis. 2019;4:65–84. DOI:10.3760/cma.j.issn.0578-1426.2018.12.002
- FranchiL, EigenbrodT, Muñoz-PlanilloR, etal. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol. 2009;10:241–247.
- BergsbakenT, FinkS, CooksonB. Pyroptosis: host cell death and inflammation. Nature Rev Microbiol. 2009;7:99–109.
- ShiJ, ZhaoY, WangK, etal. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665.
- WangY, LiuF, LiuM, etal. Curcumin mitigates aflatoxin B1-induced liver injury via regulating the NLRP3 inflammasome and Nrf2 signaling pathway. Food Chem Toxicol. 2022;161:112823.
- BriardB, FontaineT, SamirP, etal. Galactosaminogalactan activates the inflammasome to provide host protection. Nature. 2020;588:688–692.
- SpinasGA, HansenBS, LindeS, etal. Interleukin 1 dose-dependently affects the biosynthesis of (pro)insulin in isolated rat islets of Langerhans. Diabetologia. 1987;30:474–480.
- PangZ, SunG, JunkinsR, etal. AIM2 inflammasome is dispensable for the host defense against Pseudomonas aeruginosa infection. Cell Mol Biol (Noisy-le-Grand, France). 2015. 61. 63–70. PMID:26107502.
- KarkiR, LeeE, PlaceD, etal. IRF8 regulates transcription of Naips for NLRC4 Inflammasome activation. Cell. 2018;173:920–33.e13.
- ZhaoJ, Miller-LittleW, LiX. Inflammasome-Independent functions of AIM2. JExp Med. 2021;218. DOI:10.1084/jem.20210273
- LugrinJ, MartinonF. The AIM2 inflammasome: sensor of pathogens and cellular perturbations. Immunol Rev. 2018;281:99–114.
- LammertC, FrostE, BellingerC, etal. AIM2 inflammasome surveillance of DNA damage shapes neurodevelopment. Nature. 2020;580:647–652.
- GuoX, ZhongW, ChenY, etal. Benzene metabolites trigger pyroptosis and contribute to haematotoxicity via TET2 directly regulating the Aim2/Casp1 pathway. EBioMedicine. 2019;47:578–589.
- KmieZ. Cooperation of liver cells in health and disease. Adv Anat Embryol Cell Biol. 2001;161(III). DOI:10.1007/978-3-642-56553-3
- ChenY, TangD, WuH, etal. Assessment of long-term functional maintenance of primary human hepatocytes to predict drug-induced hepatoxicity in vitro. Arch Toxicol. 2021. DOI:10.1007/s00204-021-03050-y.
- SatoC, SaitoT, MisawaK, etal. Impaired mitochondrial β-oxidation in patients with chronic hepatitis C: relation with viral load and insulin resistance. BMC Gastroenterol. 2013;13:112.
- HaqueE, KarimM, Salam TeeliA, etal. Molecular mechanisms underlying Hepatocellular Carcinoma Induction by Aberrant NRF2 Activation-mediated transcription networks: Interaction of NRF2-KEAP1 controls the fate of Hepatocarcinogenesis. IntJMol Sci. 2020;21. DOI:10.3390/ijms21155378.
- ChenW, BianH, XieX, etal. Negative feedback loop of ERK/CREB/miR-212-3p inhibits HBeAg-induced macrophage activation. JCell Mol Med. 2020;24:10935–10945.
- FujiwaraK, MatsuuraK, MatsunamiK, etal. Characterization of hepatitis Bvirus with complex structural variations. BMC Microbiol. 2018;18:202.
- HuangQ, CaiD, YanR, etal. Preclinical profile and characterization of the Hepatitis BVirus core protein inhibitor ABI-H0731. Antimicrob Agents Chemother. 2020;64. DOI:10.1128/aac.01463-20.
- GaoW, JiangL, GeL, etal. Sterigmatocystin-Induced oxidative DNA damage in human liver-derived cell line through lysosomal damage. Toxicol invitro. 2015;29:1–7.
- KusunokiM, MisumiJ, ShimadaT, etal. Long-Term administration of the fungus toxin, sterigmatocystin, induces intestinal metaplasia and increases the proliferative activity of PCNA, p53, and MDM2 in the gastric mucosa of aged Mongolian gerbils. Environ Health Prev Med. 2011;16:224–231.
- AngusA, LeeA, AugustinD, etal. Pseudomonas aeruginosa induces membrane blebs in epithelial cells, which are utilized as aniche for intracellular replication and motility. Infect Immun. 2008;76:1992–2001.
- VellasamyK, MariappanV, ShankarE, etal. Burkholderia pseudomallei differentially regulates host Innate Immune response genes for Intracellular survival in Lung Epithelial cells. PLoS Negl Trop Dis. 2016;10:e0004730.
- KhoufacheK, CabaretO, FarrugiaC, etal. Primary invitro culture of porcine tracheal epithelial cells in an air-liquid interface as amodel to study airway epithelium and Aspergillus fumigatus interactions. Med Mycol. 2010;48:1049–1055.
- JiQ, WangL, LiuJ, etal. Aspergillus fumigatus-stimulated human Corneal Epithelial Cells induce Pyroptosis of THP-1 Macrophages by secreting TSLP. Inflammation. 2021;44:682–692.
- FlotoRA, OlivierKN, SaimanL, etal. US Cystic Fibrosis foundation and European Cystic Fibrosis society consensus recommendations for the management of non-tuberculous mycobacteria in individuals with cystic fibrosis: executive summary. Thorax. 2016;71:88–90.
- LiX, HawkinsGA, AmplefordEJ, etal. Genome-Wide association study identifies TH1 pathway genes associated with lung function in asthmatic patients. JAllergy Clin Immunol. 2013;132:313–20.e15.
- CraneMJ, LeeKM, FitzGeraldES, etal. Surviving deadly lung infections: innate host tolerance mechanisms in the pulmonary system. Front Immunol. 2018;9:1421.
- WuP, ChenJ, ChenJ, etal. Trimethylamine N-oxide promotes apoE mice atherosclerosis by inducing vascular endothelial cell pyroptosis via the SDHB/ROS pathway. JCell Physiol. 2020;235:6582–6591.
- YeZ, ZhangL, LiR, etal. Caspase-11 Mediates Pyroptosis of Tubular Epithelial cells and septic acute Kidney injury. Kidney & Blood Pressure Research. 2019;44:465–478.
- SharmaBR, KarkiR, KannegantiTD. Role of AIM2 inflammasome in inflammatory diseases, cancer and infection. Eur JImmunol. 2019;49:1998–2011.
- ChengQ, PanJ, ZhouZ, etal. Caspase-11/4 and gasdermin D-mediated pyroptosis contributes to podocyte injury in mouse diabetic nephropathy. Acta pharmacologica Sinica 2020. DOI:10.1038/s41401-020-00525-z.
- XuW, CheY, ZhangQ, etal. Apaf-1 Pyroptosome senses Mitochondrial permeability transition. Cell Metab. 2021;33:424–36.e10.
- ChenQU, ZhanQ, YingL, etal. Schisandra Lignan extract protects against Carbon Tetrachloride-induced Liver Injury in Mice by inhibiting Oxidative stress and regulating the NF-κB and JNK signaling pathways. Evidence-Based Complementary Altern Med. 2017 Jan 26;2017:5140297. DOI:10.1155/2017/5140297
- Al-RasheedN, FaddaL, AliH, etal. New mechanism in the modulation of carbon tetrachloride hepatotoxicity in rats using different natural antioxidants. Toxicol Mech Methods. 2016;26:243–250.
- LiuW, ChenY, MengJ, etal. Ablation of caspase-1 protects against TBI-induced pyroptosis invitro and in vivo. JNeuroinflammation. 2018;15:48.
- KupzA, ZedlerU, StäberM, etal. ESAT-6-Dependent cytosolic pattern recognition drives noncognate tuberculosis control in vivo. JClin Invest. 2016;126:2109–2122.
- SunZ, NyanzuM, YangS, etal. VX765 attenuates Pyroptosis and HMGB1/TLR4/NF-κB pathways to improve functional outcomes in TBI mice. Oxid Med Cell Longev. 2020;2020:7879629.
- FloresJ, NoëlA, FoveauB, etal. Pre-Symptomatic caspase-1 inhibitor delays cognitive decline in amouse model of Alzheimer disease and aging. Nat Commun. 2020;11:4571.
- JiangW, DengZ, DaiX, etal. Panoptosis: anew insight into oral infectious diseases. Front Immunol. 2021;12:789610.
- YanWT, YangYD, HuXM, etal. Do pyroptosis, apoptosis, and necroptosis (PANoptosis) exist in cerebral ischemia? Evidence from cell and rodent studies. Neural Regen Res. 2022;17:1761–1768.
