ABSTRACT
Streptococcus suis serotype 2 (SS2), an emerging zoonotic pathogen, causes swine diseases and human cases of streptococcal toxic shock syndrome. RNA-binding proteins (RBPs) can modulate gene expression through post-transcriptional regulation. In this study, we identified an RBP harbouring an S1 domain, named RbpA, which facilitated SS2 adhesion to host epithelial cells and contributed to bacterial pathogenicity. Comparative proteomic analysis identified 145 proteins that were expressed differentially between ΔrbpA strain and wild-type strain, including several virulence-associated factors, such as the extracellular protein factor (EF), SrtF pilus, IgA1 protease, SBP2 pilus, and peptidoglycan-binding LysM’ proteins. The mechanisms underlying the regulatory effects of RbpA on their encoding genes were explored, and it was found that RbpA regulates gene expression through diverse mechanisms, including post-transcriptional regulation, and thus acts as a global regulator. These results partly reveal the pathogenic mechanism mediated by RbpA, improving our understanding of the regulatory systems of S. suis and providing new insights into bacterial pathogenicity.
Introduction
For survival in complex external environments, gene expression in bacteria must be regulated to alter protein production and further satisfy the needs of the bacteria. Such regulation can be achieved through both transcriptional and post-transcriptional mechanisms. Transcriptional regulation is modulated predominantly at the transcription initiation level by two-component signalling systems (TCSs) or transcription factors (TFs) that harbour DNA-binding domains and have the ability to bind specific gene promoters [Citation1,Citation2]. Although RNA synthesis is carried out by RNA polymerase (RNAP), the ability of RNAP to access the gene promoter is determined by TCSs or TFs, and thus transcription initiation is tightly regulated. Regarding post-transcriptional regulation, RNA-binding proteins (RBPs) and small RNAs (sRNAs) play important roles by modulating RNA translation, processing and stability after transcription initiation [Citation3,Citation4]. sRNAs are noncoding RNA molecules and usually participate in base pairing with target mRNAs in the vicinity of the ribosome-binding site (RBS) to compete with the ribosome, which further represses mRNA translation and stimulates mRNA decay [Citation3,Citation4]. RBPs have more diverse mechanisms to interact with their RNA targets than sRNAs; for example, RBPs adjust the target RNAs’ susceptibility to RNases, modulate the accessibility of target RBSs for ribosome binding, act as sRNA chaperones, and modulate the formation of transcription terminator/anti-terminator structures [Citation5–7].
Protein S1 is an important RBP in Gram-negative bacteria that could facilitate ribosomal interactions with mRNA. Among all RBPs, protein S1 possesses the broadest variety of cellular functions, which involve its six homologous repeat S1 domains [Citation8]. The S1 domain adopts an evolutionarily conserved fold that exists in many RNA associated proteins in organisms ranging from bacteria to humans [Citation6,Citation9,Citation10], such as RNases (RNaseE, RNaseII, and PNPase), which are involved in mRNA decay in Escherichia coli. Therefore, protein S1 can directly shield RNase recognition sites to carry out post-transcriptional regulation [Citation8]. Although protein S1 has an affinity for the AU-rich mRNA sites, it also has the ability to bind various heterogenous RNAs, including sRNAs, which suggests that protein S1 is able to regulate the stability of sRNA [Citation11,Citation12]. The S1 protein is truncated in low-G+C Gram-positive bacteria (four S1 domains instead of the six found in E. coli) and loses the ability to bind the ribosome [Citation13]. However, several proteins containing S1 or S1-like domains that facilitate bacterial resistance to various external stresses and are associated with important biological functions were identified in Gram-positive bacteria. For example, GSP13 of Bacillus subtilis is involved in the response to ethanol stress, heat shock, oxidative stress, salt stress, ammonium starvation, and glucose starvation [Citation14,Citation15]. Ygs affects the general stress response and biofilm formation of Staphylococcus epidermidis [Citation16]. The cold shock protein CspR participates in the pathogenicity and the long-term survival of Enterococcus faecalis [Citation17], while another cold shock protein, CspB, affects the antimicrobial susceptibility, pigmentation, and growth of Staphylococcus aureus [Citation18]. These studies suggest that the S1 domain also possesses cellular functions in Gram-positive bacteria, which may depend on its post-transcriptional regulation.
Streptococcus suis serotype 2 (SS2) is an important Gram-positive, zoonotic pathogen responsible for severe swine and human diseases [Citation19,Citation20]. The main entry routes of S. suis into the host are skin injuries, the mouth and the upper respiratory tract [Citation21]. After breaching the mucosal barriers, S. suis disseminates to different tissues and organs through the bloodstream. A number of virulence factors were proposed to contribute to the pathogenicity of S. suis during each infection stage [Citation22], for example, SrtF and SBP2 pilus is used for S. suis adhesion, capsular polysaccharide protects S. suis from neutrophil and macrophage-mediated phagocytosis and killing [Citation22]. Meanwhile, many regulatory systems, including TCSs, TFs, and sRNAs, were shown to coordinate gene expression in response to external stimuli [Citation23–26]. Although these studies have improved the understanding of the mechanism of S. suis infection, the pathogenic mechanisms remain incompletely understood. As important post-transcriptional regulators, RBPs can modulate gene expression to satisfy the needs of bacteria, thereby facilitating bacterial survival in constantly changing environments [Citation5]. However, RBPs have not received enough attention, and there have been few reports on RBPs in S. suis.
In the present study, we carried out whole-genome screening of the ZY05719 strain and identified an RBP harbouring an S1 domain, designated RbpA, which was found to contribute to SS2 adhesion and virulence. Comparative proteomic analysis revealed that RbpA regulates several virulence-associated factors of SS2 and acts as a global regulator, and further work demonstrated that RbpA regulates gene expression through diverse mechanisms, including post-transcriptional regulation. These findings improve the understanding of SS2 regulatory systems and provide valuable insights into bacterial pathogenicity.
Materials and methods
Bacterial strains and eukaryotic cells
The SS2 strain ZY05719 was isolated from an infected pig in Sichuan Province, China. The S. suis was cultured at 37°C in 5% CO2 atmosphere by Todd-Hewitt broth (THB; Becton-Dickinson, USA) or THB agar (THA). The E. coli was cultured at 37°C in Luria – Bertani media (LB, Becton-Dickinson). These antibiotics were used when required: for E. coli, 50 μg/mL kanamycin (Kan) and 50 μg/mL spectinomycin (Spc); for SS2, 50 μg/mL Kan and 100 μg/mL Spc. The bacterial strains and plasmids used in this study are listed in Table S1. The human laryngeal carcinoma cell lines (HEp-2) were cultivated at 37°C containing 5% CO2 in DMEM (Gibco, Thermo) with 10% FBS (foetal bovine serum, Gibco).
Recombinant DNA techniques
Candidate gene deleted strain was constructed via natural DNA transformation as previously described [Citation27]. Briefly, the forward and reverse DNA sequences flanking the candidate gene locus were amplified from ZY05719 chromosomal DNA, which were fused to the sacB-spc cassette using overlap extension PCR by specific primers. SS2 strain was transformed with the fusion fragment by natural transformation and applied to the spectinomycin THA medium. To construct markerless mutants, the proto-positive mutant was transformed with the fusion homologous fragment without the sacB-spc cassette. The positive clone was screened on the THA with 10% (w/v) sucrose.
The genes with the SPA-tag (3 × Flag) sequence in their native locus in both wild-type (WT) and ΔrbpA strains were constructed as above. Briefly, the forward and reverse DNA sequences flanking the candidate gene locus were amplified from ZY05719 chromosomal DNA, which were then fused to the spa-spc cassette. The SS2 strain was transformed with the fusion fragment and applied to the spectinomycin THA medium.
The complemented strains were constructed with the pSET2 vector. Briefly, the DNA fragments containing the ORF of the target gene and its promoter region were amplified from ZY05719 chromosomal DNA, respectively. Thereinto, the putative promoter sequence was predicted by the BProm program (SoftBerry). These two fragments were fused by overlap extension PCR, which was further inserted into the pSET2 vector using ClonExpress Ultra One Step Cloning Kit (Vazyme) [Citation28]. The recombinant pSET2 plasmid was verified by PCR and sequencing, which was further transformed into mutant strain by the above method. All primers are shown in Table S2.
Bioinformatics analysis
The 9 putative RBPs were retrieved from the National Center for Biotechnology Information database (NCBI, Reference Sequence: NZ_CP007497.1), and the S1 domain in RBP was further identified by the Conserved Domain Database of NCBI. Three-dimensional structures and domains of RbpA were analysed by the SWISS-MODEL (https://www.swissmodel.expasy.org/) [Citation29]. The MEGA7 software was applied to construct a phylogenetic tree.
Adhesion assays
The adhesion experiment of SS2 to the HEp-2 cell was performed as before [Citation30], but with some modifications. The SS2 strains at mid-exponential growth phase were washed thrice with PBS, and then suspended in DMEM to an appropriate density. The HEp-2 cells were cultured until 90% confluence in 24-well cell plates and then infected with a bacterial suspension at a ratio of 1:10. The plates then were centrifuged, and then incubated with 5% CO2 at 37°C for 1.5 h. After washed six times with 1 × PBS and trypsinized for ten min, the lysate was plated to THA plates. The adhesion mediated by SS2 strains with pKSM411 plasmid was performed as the above method. After the intracellular nucleoid was stained with DAPI (Solarbio, China), the processed samples were examined by fluorescence microscopy (Carl Zeiss LSM710, Germany). Each assay was performed three times.
Animal test
This test was performed at Laboratory Animal Center of Nanjing Agricultural University and approved by Jiangsu Administrative Committee for Laboratory Animals. The licence number was SYXK(SU) 2010–0005. SPF (Specific pathogen-free) BALB/c mice (six weeks old, female) were purchased from Yangzhou University.
Ten mice in each group were intraperitoneally injected with strains of interest at a dose of 5 × 108 CFU/mouse. Survival rates and clinical signs were monitored until seven days after challenge. To examine the pathological changes produced by SS2 strain, the mice were given intraperitoneal injections with strain of interest (1 × 108 CFU/mouse), and then euthanized at three days after challenge. The lung, liver, and brain were collected and fixed in 4% paraformaldehyde. Thin section was stained by haematoxylin and eosin for further observation. All samples were scored based on pathological evaluations.
Comparative proteomic analysis
The bacteria were collected in mid-exponential growth phase, washed thrice with PBS, and suspended in 8 M urea lysis buffer. After sonication for 50 cycles and incubated at 56°C for 60 min, the bacterial supernatant was further alkylated with iodoacetamide (Sigma-Aldrich) and precipitated in prechilled acetone. The proteins were harvested and digested overnight with trypsin (Promega), which was further desalted on a Strata X C18 SPE column (Phenomenex). After the digested peptide was vacuum-dried, we applied the TMT Sixplex™ Isobaric Label Reagent (Thermo) to process the peptides with equal amounts. The comparisons in the proteomic data with estimated fold changes ≥1.2 and P values <0.05 were considered significant. The identified proteins were analysed by GO categories, KEGG enrichment, and clustering analyses.
Western blotting
The whole bacterial proteins from WT and ΔrbpA strains containing the SPA-tag (3 × Flag) sequence were extracted, and the concentration of the protein was measured by a BCA Protein Assay Kit (Pierce). The protein was diluted to the same concentration and boiled for 10 min, and equal amount of the protein was separated using SDS-PAGE and electro-blotted onto polyvinylidene fluoride membranes (Bio-Rad) in a transblotting chamber. The membrane was firstly incubated for 2 h at 25°C with the primary antibody against FLAG (Beyotime, China), and then incubated with the secondary antibody (HRP-conjugated). The band was visualized with ECL Western Blotting Substrate (Vazyme), and the grey value of these bands was calculated by the Image J software. A total of three independent experiments were performed.
RNA isolation qRT-PCR analysis
The strain was collected in mid-exponential growth phase, which were further washed thrice with PBS and resuspended into 1 mL TRIzol (TaKaRa). After transferred into tubes containing glass beads (MP Biomedicals), the resuspended cells were vortexed by FastPrep-24TM5 G (MP Biomedicals) for 5 min to lyse the cells. Total RNAs were isolated according to the manufacturer’s instructions. The HiScript II 1st strand cDNA synthesis kit with gDNA wiper (Vazyme) was used to synthesis cDNA. The ChamQ Universal SYBR qPCR mix (Vazyme) and the QuantStudio 6 Flex real-time PCR system (Thermo) were further used to validate the genes transcript concentration. The parC served as the reference gene. The relative fold changes in gene expression were analysed by the 2−ΔΔCT method. All primers are listed in Table S2. A total of three independent experiments were performed.
β-galactosidase assays
As the gene native promoters may be regulated by transcription factors of SS2 and thus affect the β-galactosidase activity, the rpoB promoter from Staphylococcus aureus was used in this assay. The leader regions of srtF, sbp2’, and lysM’ were amplified from the ZY05719 chromosomal DNA by using specific primers. A promoter-less lacZ vector pLUG220 [Citation31] was digested with SmaI and ligated with PCR products using ClonExpress Ultra One Step Cloning Kit (Vazyme, China), which were then verified by sequencing and transformed respectively into the WT, ΔrbpA, and CΔrbpA strains. β-galactosidase activity was measured using oNPG as the substrate. The value was normalized to the OD420 and further expressed in Miller units [Citation32]. Each sample procedure was repeated three times.
Data analysis
The data was analysed by GraphPad Prism 8 (GraphPad Prism). The unpaired two-tailed Student’s t test was applied to compare the means between two groups. The log-rank test was applied to analyse the survival rates. P value <0.05 indicated statistically significant difference.
Results
Identification of RBPs harbouring an S1 domain that contribute to SS2 virulence
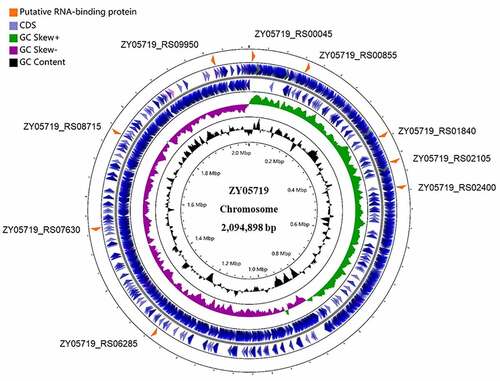
Analysis of the ZY05719 genome identified 9 putative RBPs (), among which only ZY05719_RS02105, ZY05719_RS06285, and ZY05719_RS07630 were predicted to harbour an S1 domain (Table S3). Bioinformatics analysis showed that an S1 domain was located in the N-terminus of ZY05719_RS02105 and ZY05719_RS06285 and in the C-terminus of ZY05719_RS07630. ZY05719_RS07630 was predicted to be a transcriptional accessory protein, while the C-terminal S1 domain in a transcription accessory protein is called Tex [Citation33]; thus, ZY05719_RS07630 may regulate gene expression at the transcriptional level. To analyse the potential post-transcriptional roles of RBPs harbouring an S1 domain in the pathogenesis of SS2, we created ZY05719_RS02105 and ZY05719_RS06285 deletion mutants respectively. Although ZY05719_RS06285 exhibited 33% amino acid sequence identity with the virulence regulatory protein CvfB of Staphylococcus aureus [Citation34], mouse survival assays showed that this gene is not essential for SS2 virulence in this model of bacteraemia (Figure S1). However, the ZY05719_RS02105 mutant of SS2 exhibited significantly attenuated pathogenicity (Figure S1) and thus deserves further investigation.
Figure 1. Distribution of putative RBPs in the genome of S. suis strain ZY05719. The nine putative RBPs screened from the NCBI database were shown in the circular diagram. The blue circles (outermost circles) represent the CDS (coding sequence) on the negative or positive chains. The purple and green circles (middle circles) represent the negative and positive GC skew, respectively. The black circles (innermost circles) represent the GC content. Map was drawn by the CGView ServerBETA (http://cgview.Ca/).
RbpA is conserved among different serotypes of S. suis
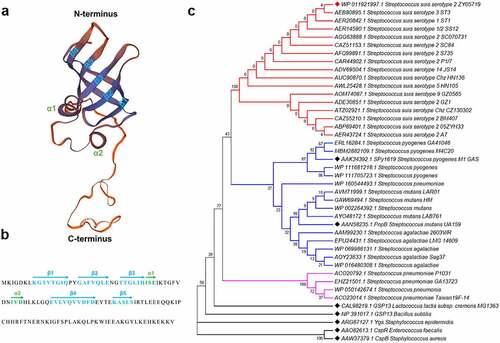
ZY05719_RS02105 comprises 123 amino acid residues and was predicted to contain an S1 domain (residues 8–71) along with a flexible C-terminal tail () by the SWISS-MODEL online server. The N-terminal S1 domain contains two small α-helices and a five-stranded antiparallel β-sheet (). Homology analysis of ZY05719_RS02105 revealed that it shares 49% amino acid sequence identity with GSP13 of B. subtilis and 39% amino acid sequence identity with Ygs of S. epidermidis. As both GSP13 and Ygs were identified as general stress response proteins that help bacteria cope with different stressors [Citation14–16], such as pH, temperature, ethanol, and oxidation, we performed a series of assays to assess if ZY05719_RS02105 plays similar roles in S. suis. However, the ZY05719_RS02105 mutant did not exhibit a significant difference in growth compared to the WT strain when exposed to these stressors (Figure S2), indicating that ZY05719_RS02105 has unknown functions. According to the phylogenetic tree based on protein amino acid sequences, this protein belongs to the same branch in different serotypes of S. suis, while its homolog in other Streptococcus species, as well as GSP13, Ygs, CspR, CspB, are in different branches (). These results indicated that ZY05719_RS02105 is conserved among S. suis strains; thus, it was redesignated RbpA (RNA-binding protein A).
Figure 2. Structural prediction and phylogenetic tree of RbpA. (a) Predicted three-dimensional structure of RbpA by the SWISS-MODEL online server. The two small α-helices and five-stranded antiparallel β-sheet are indicated at the corresponding positions. (b) the secondary structural elements are shown in the RbpA sequences by arrows. (c) Phylogenetic analysis of RbpA from Streptococcus species and several reported RBPs from other bacterial species. The tree was constructed with MEGA7 software using the neighbor-joining method. Corresponding serotypes and strain names are shown after the accession numbers of the proteins. All amino acid sequences used in this analysis were downloaded from the NCBI database.
RbpA facilitates SS2 adhesion and pathogenicity
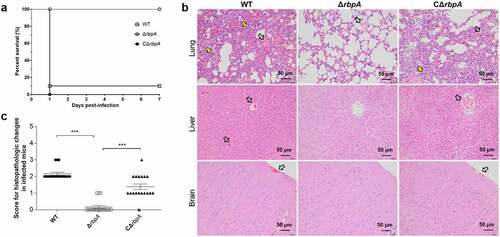
Adhesion to the surface of host cell is important for initiating SS2 dissemination and infection [Citation35]. Our data showed that the adhesion of the ΔrbpA strain to HEp-2 cells was significantly decreased compared to that of the WT and complementary strains (). In addition, mice infected with the ΔrbpA strain showed a significantly higher survival rate and less severe clinical signs than those challenged with the WT and complementary strains, which showed acute clinical signs including rough hair coat, lethargy, and shivering (Figure S1 and 4(c)). To analyse the roles of RbpA during systemic infection, lung, liver, and brain samples from infected mice were examined histologically. As shown in , the mice challenged with the WT and complementary strains exhibited more obvious pathological changes, such as haemorrhage and inflammatory cell infiltration, than the mice challenged with the ΔrbpA strain. These results suggested that RbpA plays a critical role in SS2 pathogenicity.
Figure 3. RbpA facilitates SS2 adhesion to HEp-2 cells. (a) the rbpA deletion affects the SS2 adhesion to HEp-2 cells. The adherent bacterial numbers were evaluated by determining the number of CFU in each well. The statistical significance was calculated by two-tailed unpaired Student’s t tests (*, P < 0.05; **, P < 0.01). (b) Fluorescence microcopy of SS2 adhesion to HEp-2 cells. The WT and ΔrbpA strains were transformed with the pKSM411 plasmid and detected directly with FITC (in green), while the nucleoids of HEp-2 cells were stained with DAPI (in blue). The WT strain without the pKSM411 plasmid was set as the control group.
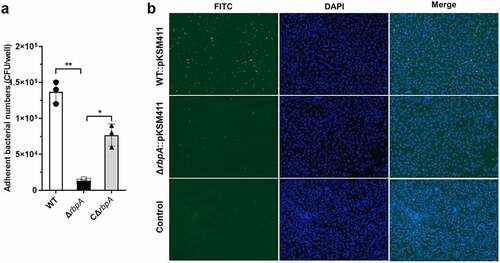
Figure 4. RbpA contributed to the pathogenicity of SS2. (a) Survival curves of mice infected with the WT, ΔrbpA, and CΔrbpA strains. (b) Histopathology of infected tissues from mice at 3 days postinfection with the WT, ΔrbpA, and CΔrbpA strains. The infected tissues were stained by hematoxylin and eosin and examined using light microscopy. Hollow arrows show hemorrhage and yellow arrows show inflammatory cell infiltration. (c) Score for histopathologic changes of infected tissues from mice at 3 days postinfection with the WT, ΔrbpA, and CΔrbpA strains. Histopathologic sections were randomly selected from each infected tissue and scored based on severity and inflammation. The statistical significance was calculated by two-tailed unpaired Student’s t tests (***, P < 0.001).
Identification of the RbpA-regulated proteins by comparative proteomic analysis
As RBPs regulate gene expression in a post-transcriptional manner, proteomic analysis was applied here to identify the mechanism whereby RbpA contributes to SS2 pathogenicity. The results showed that 84 proteins were highly upregulated and 61 proteins were significantly downregulated when the rbpA was deleted in the SS2 ( and Table S4). To further confirm the changes in the proteome, the expression of five proteins with the SPA-tag (3×Flag) (with GroEL used as an internal reference) was detected by western-blotting using the anti-Flag antibody. We found that the expression levels of GlnQ, LysM,’ and IgA1 in the ΔrbpA strain were lower than those in WT strain, while ZY05719_RS01400 was upregulated in the ΔrbpA strain (). In addition, the GroEL as a control showed the similar expression levels in ΔrbpA and WT strains. These results support the expression pattern observed in the proteomic analysis.
Figure 5. Comparative proteomic analysis of the differentially expressed proteins between WT and ΔrbpA strains. (a) Volcano plot of differentially expressed proteins between WT strain and ΔrbpA strain determined based on the proteomic data. The ratio of the protein expression level between the WT strain and ΔrbpA strain was standardized with a P-value < 0.05 and a ratio ≥ 1.2 to define the differentially expressed proteins. (b,c) Western blotting analysis of the proteomics data. ZY05719_RS01400, GlnQ, LysM,’ and IgA1 were chosen to verify the protein expression levels, and GroEL was used as a control protein. Equal amounts of the whole bacterial proteins from the WT and ΔrbpA strains containing the SPA-tag sequence were subjected to SDS-PAGE and then immunoblotting with an anti-FLAG antibody. The grayscale of the reactive protein bands was measured by using a software program. Fold change = grayscale of target protein in ΔrbpA/grayscale of target protein in WT. (d) GO analysis of the RbpA-regulated proteins. These proteins are grouped into three terms: biological process, cellular component, and molecular function. (e) KEGG pathway enrichment of the RbpA-regulated proteins. The x-axis represents the enriched pathways, and the y-axis represents the proteins number of the differentially expressed proteins involved in each pathway.
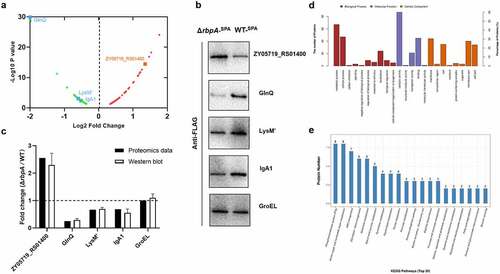
We then employed GO categories to analyse the functional categories of the differentially abundant proteins, and these proteins were found to be enriched in the following terms: catalytic activity and binding (molecular function), metabolic and cellular process (biological process), and membrane part (cellular component) (). KEGG analysis showed that the differentially expressed proteins were enriched in the phosphotransferase system (PTS), amino sugar and nucleotide sugar metabolism, and ABC transporter pathways (). These results revealed that the RbpA-regulated proteins are involved in various physiological processes in SS2, and RbpA works as a global regulator.
RbpA regulates the expression of virulence-associated factors in SS2
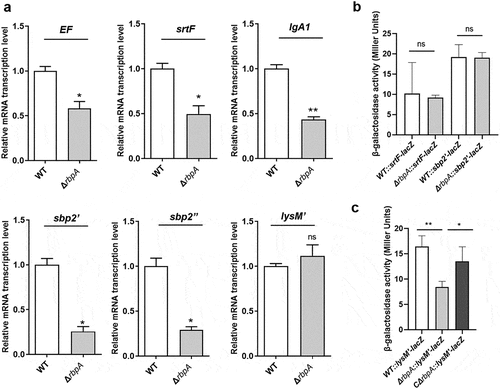
Many virulence-associated factors have been proposed since the first report on S. suis [Citation22]. Here, we observed that the expression of several virulence-associated factors was significantly downregulated according to the proteomic analysis, including the extracellular protein factor (EF), SrtF pilus, IgA1 protease, SBP2 (including SBP2’ and SBP2’’) pilus, and peptidoglycan-binding LysM’ proteins (Table S4). Gene expression can be regulated by RBPs through diverse mechanisms [Citation5], for example, modulation of mRNA stability and adjustment of mRNA translation through interaction with RBSs. We therefore performed qRT-PCR to measure these genes mRNA levels. Our results showed that the mRNA levels of EF, srtF, IgA1, and sbp2 but not lysM’ were significantly decreased in ΔrbpA strain compared to WT strain (), and the expression of these virulence-associated factors was restored in the CΔrbpA strain (Figure S3). In addition, β-galactosidase assays were performed to further determine the underlying mechanisms by which RbpA carries out post-transcriptional regulation, and for this, the 5’ untranslated region (5”UTR) of the putative target gene, which contains the RBS, was ligated to the lacZ vector. The results showed that β-galactosidase synthesis was identical in ΔrbpA and WT strains when the lacZ vector was fused to the leader regions of srtF and sbp2”, while the β-galactosidase level decreased significantly in the ΔrbpA strain carrying the lysM’-lacZ plasmid (). Furthermore, qRT-PCR showed that the RNA levels of these lacZ reporters in the ΔrbpA strain were not significantly different from those in WT strain (Figure S4). These data, coupled with the qRT-PCR results, indicated that RbpA regulates the production of EF, SrtF, IgA1, and SBP2 through modulation of mRNA synthesis, whereas it may interact with the 5”UTR of lysM” and facilitate its translation.
Figure 6. RbpA regulates the expression of virulence-associated factors. (a) Transcriptional analysis of the virulence-associated factors. qRT-PCR expression values are presented as the means plus standard deviations from three independent experiments. (b, c) β-Galactosidase activity (Miller Units) detected for different gene fusions in the WT, ΔrbpA, and CΔrbpA strains. The results represent the means of three independent experiments. The statistical significance was calculated by two-tailed unpaired Student’s t tests (ns, P > 0.05; *, P < 0.05; **, P < 0.01).
Discussion
Recently, a number of bacterial genes have been identified to be regulated post-transcriptionally largely due to the discovery of post-transcriptional regulators, such as RBPs and sRNAs [Citation4,Citation36]. In all organisms, RBPs play important roles in modulating gene expression by regulating the transcription, stability, and translation of cellular RNAs [Citation5,Citation6]. It is well known that Hfq is a regulatory RBP that acts as an RNA chaperone to facilitate the pairing of sRNAs with their target mRNAs, thereby affecting a variety of physiological functions in bacteria [Citation37,Citation38]. However, many Gram-positive bacteria, including Streptococcus species, do not contain any obvious Hfq homolog-encoding genes [Citation39]. Studies on post-transcriptional regulation in S. suis have mainly focused on sRNAs [Citation23,Citation25], while there have been few reports on RBPs. RBPs harbouring an S1 domain were identified to modulate gene expression by post-transcriptional regulation and participate in many cellular processes [Citation5,Citation8]. Although several proteins harbouring an S1 or S1-like domain were identified to facilitate resistance to various external stressors in Gram-positive bacteria [Citation15–18], they have not been described as RBPs, and their functions were not connected to the S1 domain. In the present study, we demonstrated that RbpA, an RBP containing an S1 domain, contributes to SS2 adhesion and virulence through post-transcriptional regulation as a global regulator. To our knowledge, this is the first study to connect the S1 domain with post-transcriptional regulation and biological functions in Streptococcus species, which improves our understanding of bacterial regulatory systems and provides valuable insights into bacterial pathogenicity.
Our results showed that the ΔrbpA deletion mutant strain exhibited lower adhesion and attenuated virulence compared to the WT and complementary strains. Further comparative proteomic analysis identified 145 genes that were differentially expressed between WT and ΔrbpA strains, which included several virulence-associated factors, such as, the EF, SrtF pilus, IgA1 protease, SBP2 (including SBP2’ and SBP2’’) pilus, and peptidoglycan-binding LysM’ proteins. EF has been used historically as a marker of S. suis virulence [Citation40], while the protease IgA1 shows good cleavage activity towards IgA1 and contributes to SS2 virulence [Citation41]. Pili are located on the bacterial surface and contribute to adherence to the surface of host cells, and they also participate in the virulence of many different Gram-positive pathogenic species [Citation42]. In S. suis, SrtF and SBP2 are both pilus-associated proteins [Citation43,Citation44]. Although SBP2 is truncated into SBP2’ and SBP2’’ in some serotypes of S. suis, these proteins were identified to facilitate the adhesion process and pathogenesis of SS2 [Citation44,Citation45]. The LysM domain is one of the most common domains of cell surface proteins of bacteria [Citation46], which was originally reported to have cell wall lyase activity and to be involved in bacterial virulence [Citation47]. LysM’ harbours a LysM domain and was recently demonstrated to have anti-phagocytosis functions in SS2 [Citation48], which may further contribute to SS2 infection. The expression of these virulence-associated factors was significantly downregulated in the RbpA proteomic data, which can partly explain the pathogenesis mechanism mediated by RbpA.
In addition to these virulence-associated factors, many fitness genes and metabolic genes are also affected by the rbpA deletion. The KEGG analysis indicated that the RbpA-regulated proteins are involved in the PTS, amino sugar and nucleotide sugar metabolism, and ABC transporter pathways. PTS function as a phosphorelay system and could couple the transport of carbohydrates with their simultaneous phosphorylation, which is associated with bacterial resistance to oxidative stress [Citation49]. As S. suis encounters oxidative stress during the process of infection, PTS may play an important role in the RbpA-mediated adaptation of SS2 to the host environment. It is well known that capsular polysaccharide is a key virulence factor in S. suis [Citation50]. Previous study showed that capsular polysaccharide biosynthesis of S. suis is affected by the availability of glucose or other carbohydrates [Citation51]. As the RbpA-regulated proteins are involved in amino sugar metabolism and ABC transporter pathways, which may affect the capsular polysaccharide biosynthesis, it is possible these proteins also contribute to the RbpA mediated pathogenicity of SS2.
The mechanisms used by the S1 protein to post-transcriptionally regulate genes mainly depend on binding to mRNAs during the initiation of protein synthesis as a ribosomal component, modulating the formation of transcription anti-terminator structures, and shielding RNase recognition sites [Citation5,Citation8,Citation13]. Although many small proteins (70 residues) are constituted of only a single S1 domain, such as bacterial translation initiation factor 1, these small proteins are still able to bind to RNA [Citation52]. As RbpA contains an S1 domain, we explored its regulatory mechanism by comparing it with the mechanisms of the S1 protein. We found that the RNA levels of EF, srtF, IgA1, and sbp2 were significantly downregulated in ΔrbpA strain compared to WT strain, while the RNA level of lysM’ was not obviously changed. RbpA may regulate the mRNA synthesis of EF, srtF, IgA1, and sbp2 by modulating mRNA stability, for example, by shielding RNase recognition sites to protect mRNAs against cleavage. In addition, given that TCSs and TFs can regulate gene expression at the transcription initiation level, these gene transcriptional changes may be mediated by other potential TCSs and TFs, and may thus be indirectly regulated by RbpA. The β-galactosidase assays indicated that RbpA may interact with the 5”UTR of lysM”, while sequence analysis showed that AU-rich sites are present in the vicinity of the RBS of lysM’ (Figure S5). As the S1 domain showed a higher affinity for AU-rich mRNA sites [Citation8], these results indicated that RbpA may interact with the RBS of lysM’ to conduct post-transcriptionally regulation and further affect this gene translation efficiency.
In summary, RbpA, an RBP containing an S1 domain, was identified to act as a global regulator via diverse regulatory pathways in SS2. We demonstrated that RbpA contributed to SS2 adhesion and virulence by regulating several virulence-associated factors. However, the underlying mechanisms by which RbpA performs post-transcriptional regulation require further investigation.
Supplemental Material
Download Zip (2.1 MB)Acknowledgements
We are grateful to Dr. Pascale Romby from Architecture et Réactivité de l’ARN (UPR 9002), CNRS, for providing pLUG220 plasmid.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All data generated or analysed during this study are included in this article and its supplementary information files. The proteomics raw data of this study have been deposited under the accession number IPX0003548002 at iProX.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21505594.2022.2103233.
Additional information
Funding
References
- Browning DF, Butala M, Busby SJW. Bacterial transcription factors: regulation by pick “N mix”. J Mol Biol. 2019;431:4067–4077.
- Gao R, Bouillet S, Stock AM. Structural basis of response regulator function. Annu Rev Microbiol. 2019;73:175–197.
- Desnoyers G, Bouchard MP, Masse E. New insights into small RNA-dependent translational regulation in prokaryotes. Trends Genet. 2013;29:92–98.
- Nitzan M, Rehani R, Margalit H. Integration of bacterial small RNAs in regulatory networks. Annu Rev Biophys. 2017;46:131–148.
- Van Assche E, Van Puyvelde S, Vanderleyden J, et al. RNA-binding proteins involved in post-transcriptional regulation in bacteria. Front Microbiol. 2015;6:141.
- Holmqvist E, Vogel J. RNA-binding proteins in bacteria. Nat Rev Microbiol. 2018;16:601–615.
- Babitzke P, Lai YJ, Renda AJ, et al. Posttranscription initiation control of gene expression mediated by bacterial RNA-binding proteins. Annu Rev Microbiol. 2019;73:43–67.
- Hajnsdorf E, Boni IV. Multiple activities of RNA-binding proteins S1 and Hfq. Biochimie. 2012;94:1544–1553.
- Bycroft M, Hubbard TJ, Proctor M, et al. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell. 1997;88:235–242.
- Matsumoto Y, Xu Q, Miyazaki S, et al. Structure of a virulence regulatory factor CvfB reveals a novel winged helix RNA binding module. Structure. 2010;18:537–547.
- Koleva RI, Austin CA, Kowaleski JM, et al. Interactions of ribosomal protein S1 with DsrA and rpoS mRNA. Biochem Biophys Res Commun. 2006;348:662–668.
- Windbichler N, von Pelchrzim F, Mayer O, et al. Isolation of small RNA-binding proteins from E. coli: evidence for frequent interaction of RNAs with RNA polymerase. RNA Biol. 2008;5:30–40.
- Salah P, Bisaglia M, Aliprandi P, et al. Probing the relationship between Gram-negative and Gram-positive S1 proteins by sequence analysis. Nucleic Acids Res. 2009;37:5578–5588.
- Bernhardt J, Volker U, Volker A, et al. Specific and general stress proteins in Bacillus subtilis–a two-dimensional protein electrophoresis study. Microbiology. 1997;143:999–1017.
- Kaan T, Homuth G, Mader U, et al. Genome-Wide transcriptional profiling of the Bacillus subtilis cold-shock response. Microbiology. 2002;148:3441–3455.
- Wang X, Niu C, Sun G, et al. Ygs is a novel gene that influences biofilm formation and the general stress response of Staphylococcus epidermidis. Infect Immun. 2011;79:1007–1015.
- Michaux C, Martini C, Shioya K, et al. CspR, a cold shock RNA-binding protein involved in the long-term survival and the virulence of Enterococcus faecalis. J Bacteriol. 2012;194:6900–6908.
- Duval BD, Mathew A, Satola SW, et al. Altered growth, pigmentation, and antimicrobial susceptibility properties of Staphylococcus aureus due to loss of the major cold shock gene cspB. Antimicrob Agents Chemother. 2010;54:2283–2290.
- Bonifait L, Veillette M, Letourneau V, et al. Detection of Streptococcus suis in bioaerosols of swine confinement buildings. Appl Environ Microbiol. 2014;80:3296–3304.
- Goyette-Desjardins G, Auger JP, Xu J, et al. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg Microbes Infect. 2014;3:e45.
- Segura M, Calzas C, Grenier D, et al. Initial steps of the pathogenesis of the infection caused by Streptococcus suis: fighting against nonspecific defenses. FEBS Lett. 2016;590:3772–3799.
- Fittipaldi N, Segura M, Grenier D, et al. Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol. 2012;7:259–279.
- Wu Z, Wu C, Shao J, et al. The Streptococcus suis transcriptional landscape reveals adaptation mechanisms in pig blood and cerebrospinal fluid. Rna. 2014;20:882–898.
- Feng L, Zhu J, Chang H, et al. The CodY regulator is essential for virulence in Streptococcus suis serotype 2. Sci Rep. 2016;6:21241.
- Xiao G, Tang H, Zhang S, et al. Streptococcus suis small RNA rss04 contributes to the induction of meningitis by regulating capsule synthesis and by inducing biofilm formation in a mouse infection model. Vet Microbiol. 2017;199:111–119.
- Zheng C, Li L, Ge H, et al. Role of two-component regulatory systems in the virulence of Streptococcus suis. Microbiol Res. 2018;214:123–128.
- Zhu Y, Dong W, Ma J, et al. Utilization of the ComRS system for the rapid markerless deletion of chromosomal genes in Streptococcus suis. Future Microbiol. 2019;14:207–222.
- Takamatsu D, Osaki M, Sekizaki T. Construction and characterization of Streptococcus suis-Escherichia coli shuttle cloning vectors. Plasmid. 2001;45:101–113.
- Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–w303.
- Zhang Y, Zhong X, Lu P, et al. A novel autolysin AtlASS mediates bacterial cell separation during cell division and contributes to full virulence in Streptococcus suis. Vet Microbiol. 2019;234:92–100.
- Huntzinger E, Boisset S, Saveanu C, et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. Embo J. 2005;24:824–835.
- Aviv G, Gal-Mor O. lacZ reporter system as a tool to study virulence gene regulation in bacterial pathogens. Methods Mol Biol. 2018;1734:39–45.
- He X, Thornton J, Carmicle-Davis S, et al. Tex, a putative transcriptional accessory factor, is involved in pathogen fitness in Streptococcus pneumoniae. Microb Pathog. 2006;41:199–206.
- Matsumoto Y, Kaito C, Morishita D, et al. Regulation of exoprotein gene expression by the Staphylococcus aureus cvfB gene. Infect Immun. 2007;75:1964–1972.
- Segura M, Fittipaldi N, Calzas C, et al. Critical Streptococcus suis virulence factors: are they all really critical? Trends Microbiol. 2017;25:585–599.
- Nogueira T, Springer M. Post-transcriptional control by global regulators of gene expression in bacteria. Curr Opin Microbiol. 2000;3:154–158.
- Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr Opin Microbiol. 2010;13:24–33.
- Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–589.
- Sun X, Zhulin I, Wartell RM. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 2002;30:3662–3671.
- Silva LM, Baums CG, Rehm T, et al. Virulence-associated gene profiling of Streptococcus suis isolates by PCR. Vet Microbiol. 2006;115:117–127.
- Zhang A, Mu X, Chen B, et al. IgA1 protease contributes to the virulence of Streptococcus suis. Vet Microbiol. 2011;148:436–439.
- Telford JL, Barocchi MA, Margarit I, et al. Pili in gram-positive pathogens. Nat Rev Microbiol. 2006;4:509–519.
- Fittipaldi N, Takamatsu D, de la Cruz Dominguez-Punaro M, et al. Mutations in the gene encoding the ancillary pilin subunit of the Streptococcus suis srtF cluster result in pili formed by the major subunit only. PLoS One. 2010;5:e8426.
- Yu Y, Qian Y, Du D, et al. SBP2 plays an important role in the virulence changes of different artificial mutants of Streptococcus suis. Mol Biosyst. 2016;12:1948–1962.
- Shao J, Zhang W, Wu Z, et al. The truncated major pilin subunit Sbp2 of the srtBCD pilus cluster still contributes to Streptococcus suis pathogenesis in the absence of pilus shaft. Curr Microbiol. 2014;69:703–707.
- Eckert C, Lecerf M, Dubost L, et al. Functional analysis of AtlA, the major N-acetylglucosaminidase of Enterococcus faecalis. J Bacteriol. 2006;188:8513–8519.
- Bateman A, Bycroft M. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J Mol Biol. 2000;299:1113–1119.
- Xu B, Zhang P, Li W, et al. hsdS, belonging to the type I restriction-modification system, contributes to the Streptococcus suis serotype 2 survival ability in phagocytes. Front Microbiol. 2017;8:1524.
- Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol. 2008;6:613–624.
- Zhong X, Zhang Y, Zhu Y, et al. Identification of an autorepressing two-component signaling system that modulates virulence in Streptococcus suis serotype 2. Infect Immun. 2019;87. DOI:10.1128/IAI.00377-19
- Willenborg J, Fulde M, de Greeff A, et al. Role of glucose and CcpA in capsule expression and virulence of Streptococcus suis. Microbiology (Reading). 2011;157:1823–1833.
- Yu W, Hu J, Yu B, et al. Solution structure of GSP13 from Bacillus subtilis exhibits an S1 domain related to cold shock proteins. J Biomol NMR. 2009;43:255–259.
