ABSTRACT
Porcine rotavirus (PoRV) is an important pathogen, leading to the occurrence of viral diarrhoea . As the infection displays obvious enterotropism, intestinal mucosal immunity is the significant line of defence against pathogen invasion. Moreover, as lactic acid bacteria (LAB) show acid resistance, bile salt resistance and immune regulation, it is of great significance to develop an oral vaccine. Most traditional plasmid delivery vectors use antibiotic genes as selective markers, easily leading to antibiotic accumulation. Therefore, to select a food-grade marker in genetically engineering food-grade microorganisms is vital. Based on the CRISPR-Cas9D10A system, we constructed a stable auxotrophic Lactobacillus paracasei HLJ-27 (Lactobacillus △Alr HLJ-27) strain. In addition, as many plasmids replicate in the host bacteria, resulting in internal gene deletions. In this study,we used a temperature-sensitive gene editing plasmidto insert the VP4 gene into the genome, yielding the insertion mutant strains VP4/△Alr HLJ-27, VP4/△Alr W56, and VP4/W56. This recombinant bacterium efficiently induced secretory immunoglobulin A (SIgA)-based mucosal and immunoglobulin G (IgG)-based humoral immune responses. These oral mucosal vaccines have the potential to act as an alternative to the application of antibiotics in the future and induce efficient immune responses against PEDV infection.
GRAPHIC ABSTRACT
Introduction
Rotavirus is one of the primary diarrhoea-causing viral pathogens found in animals worldwide. Its effects on livestock production often lead to substantial economic loss. There is currently no effective means to control and treat the diseases caused by this virus, and vaccination is still the only way to prevent infection [Citation1]. Porcine rotavirus (PoRV) proliferates primarily in the villi of the small intestinal epithelium; this is, therefore, a key potential therapeutic target for oral vaccines. Nevertheless, it is challenging to deliver sufficient amounts of antigen to the mucosal surface by oral vaccination due to the stomach acidity [Citation2]. Efforts are currently underway to develop a lactic acid bacteria (LAB) vaccine as an antigen delivery vehicle to overcome the limitations posed by the internal environment and adequately elicit a mucosal immune response [Citation3]. LAB are acid and bile tolerant, which effectively prevents their degradation in the digestive tract and during immune tolerance reactions [Citation4]. Furthermore, they are considered to be safe with beneficial effects on the health of humans and animals [Citation5]. Their impacts range from maintaining intestinal flora balance, neutralizing food mutagens, and alleviating allergic reactions, to lowering serum cholesterol levels [Citation6,Citation7]. These microorganisms are ubiquitous in the intestine and widely utilized in food fermentation, vaccine production, health supplements, and feed additives [Citation8–10]. There have been some studies regarding their effects on the immune cells of the gut. These probiotic bacteria can interact with gut-associated lymphoid tissue, Peyer’s patches, and the intestinal lamina propria [Citation11]. Cells involved in the innate immune response have been reported to be the main targets of LAB, inducing immune stimulation in the gut [Citation12]. Lactobacillus paracasei (L. paracasei) is an example of one LAB strain with the advantage of being susceptible to heterologous delivery using a vector. It persists in intestinal and vaginal tissues, regulating the balance of flora and reducing the levels of Escherichia coli and Shigella spp. in the human gut microflora [Citation13]. The researchers tested the probiotic effects of L. paracasei in mice, and the results showed that the strain could regulate the balance of intestinal flora and enhance the specific immune response in the body similar to antigens [Citation14]. These studies indicate that L. paracasei strains show a powerful potential for application in vaccine delivery.
To date, most of the expression systems of LAB used plasmids as antigen delivery vehicles. However, there are several disadvantages to traditional plasmid expression as the recombinant bacteria generally require the selection pressure of an antibiotic [Citation15,Citation16]. The application of antibiotics is generally not accepted in genetically modified organisms, as it might cause the transmission of antibiotic resistance from one organism to another. Therefore, food-grade markers have been selected to replace antibiotic resistance markers in genetically engineering food-grade microorganisms, especially LAB. So far, there are many studies concentrated on complementary selection markers for LAB, the premise of which was to construct auxotrophic LAB [Citation17,Citation18]. The auxotrophic type of bacteria with certain genes deletion, such as housekeeping genes, which code for synthesizing substances to catalyse the essential metabolic reactions of bacteria. Mutation or deletion of these genes results in the absence of the corresponding products. Thus, bacterial strains in the external environment or basic media cannot grow unless the corresponding substrate is supplemented. Alanine racemase (Alr), a key enzyme that participates in bacterial cell wall biosynthesis and catalyses the conversion of L-alanine (L-Ala) to D-alanine (D-Ala), is an important element for bacterial cell wall biosynthesis of peptidoglycan [Citation19]. D-Ala does not exist naturally, so LAB lacking the Alr gene can only grow normally in MRS basal medium supplemented with D-Ala [Citation18]. Recent studies have shown that deletion of the Alr gene affects the growth of strains such as Lactobacillus plantarum, E. coli, and Bacillus subtilis [Citation20–22]. Therefore, the construction of Alr-deficient strains would provide new candidates for selecting food-grade screening markers.
In addition, many plasmids are replicated through the rolling circle mechanism in host bacteria, generating unstable single-stranded intermediates, resulting in internal deletions [Citation4,Citation23]. Therefore, as a way to remove the resistance selection pressure and guarantee stable gene expression, the integration of heterologous genes into the genome for expression has been proposed [Citation4]. Currently, homologous recombination based on plasmids is utilized in LAB for genome editing. The disadvantages of this system include its time-consuming and complicated nature [Citation24]. Single-stranded oligonucleotide-mediated recombineering has been applied to Lactobacillus to generate site-directed mutations [Citation25], though the mutagenesis efficiency was lower [Citation26]. Therefore, developing a faster and more accurate method to improve the editing efficiency of LAB is vital. The clustered regularly interspaced short palindromic repeat (CRISPR) system, an adaptive bacterial immunity system for prokaryotes to resist foreign gene invasion, has been successfully developed as a powerful tool for genome editing [Citation27,Citation28]. In CRISPR-Cas9 systems, the Cas9 nuclease is guided by CRISPR RNA (crRNA) and transactivating crRNA (tracrRNA) to distinguish and break the target DNA based on Watson – Crick base pairing between the single-guide RNA (sgRNA) and protospacer of target DNA, with the feature of an adjacent protospacer adjacent motif (PAM) [Citation29]. Chimeric sgRNA, an artificially designed, optimized form of the crRNA-tracrRNA complex, can simplify the design and operation process [Citation30]. The CRISPR-Cas9-mediated genome editing system has been developed and widely applied in various organisms; yet there are currently few studies to apply it to gene editing of Lactobacillus [Citation31]. The Cas9 protein causes DNA double-strand breaks (DSBs), which are repaired by nonhomologous end joining in eukaryotes; however, most bacterial strains lack this repair mechanism. The occurrence of DSB could thus lead to the death of bacteria [Citation32]. Song et al. utilized a Cas9D10A nickase (Cas9D10A) mutant that only triggered DNA single-strand breaks, as a replacement for the wild-type Cas9 protein, by optimizing the promoters of sgRNA and the Cas9D10A protein to achieve fragment knockout and site-specific insertion in the Lactobacillus genome, proving that the CRISPR-Cas9D10A system was an effective tool to overcome the lethality induced by DSB [Citation33].
VP4, the dimeric spike protein of the PoRV, could induce the production of neutralizing antibodies [Citation34]. Studies have shown that newborn mice born from female mice immunized with this protein can gain passive immunity [Citation35]. This study aimed to use the CRISPR-Cas9D10A system to integrate the coding sequence of the neutralizing epitopes of a fragment of PoRV VP4 and the promoter P776 of the pyruvate hydratase gene [Citation4], a constitutively highly expressed gene, into Lactobacillus W56, auxotrophic Lactobacillus W56 (Lactobacillus △Alr W56), and auxotrophic Lactobacillus paracasei HLJ-27 (Lactobacillus △Alr HLJ-27) isolated from pig intestines, in order to observe the expression of the heterologous protein VP4. The immunogenicity of these strains were then analysed in mice through oral administration with the strains VP4/△Alr HLJ-27, VP4/△Alr W56, and VP4/W56. The immune response induced by oral immunization with recombinant strains in mice could provide a reference for the immunization of piglets. The immunity assessment of piglets is in progress.
Materials and methods
Bacteria, viruses, and plasmids
In the laboratory, L. paracasei HLJ-27 isolated from pig intestines [Citation36] and wild-type Lactobacillus W56 were grown in de Man, Rogosa, and Sharpe broth (MRS; Sigma-Aldrich, St Louis, MO, USA). Lactobacillus △Alr W56, stored in our laboratory, and △Alr HLJ-27 were grown in MRS medium containing 200 ng/mL D-alanine (Sigma-Aldrich, St Louis, MO, USA) at 37°C without shaking. The PoRV JL94 strain was isolated in our laboratory and stored at −140°C. Erythromycin (EMC; Sigma-Aldrich, St Louis, MO, USA) was applied at a concentration of 10 ng/mL to strains containing the pLCNICK-Alr/VP4 plasmid. The gene-editing backbone plasmid pLCNICK was gifted by Yang Sheng from the Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. All E. coli strains containing the gene-editing plasmids were grown anaerobically at 30°C in Luria – Bertani (LB) broth containing 50 mg/L kanamycin.
Construction of the gene-editing plasmids
The gene-editing plasmids were constructed as shown in . shows the deletion plasmid, and lists the primers used. Cloning was performed following the procedure described below. The Alr gene was designed as the auxotrophic site. The backbone pLCNICK, containing the Cas9 gene, LAB replicon, EMC, two homologous arms (Has), and sgRNA, is a temperature-sensitive plasmid only able to replicate at 30°C. The upper and lower homology arms (Alr-up and Alr-down, respectively) were amplified from the genome of Lactobacillus W56 and extracted with the bacterial Genome Extraction Kit (Novizan, Nanjing, China). sgRNA was acquired from the plasmid pUC57-sgRNA, designed using predecessors targeting the Alr site and synthesized by Kumei Biotechnology Co., Ltd. (Jilin, China). All polymerase chain reaction (PCR) fragments were gel purified and fused to one fragment with the homologous sequence, by fusion PCR in the order of Alr-up, Alr-down, and sgRNA sequences. The Alr-up-Alr-down-sgRNA fragment was inserted into the vector pLCNICK at the Apa I and Xba I restriction sites to generate the deletion gene-editing plasmid pLCNICK-Alr-up-Alr-down-sgRNA (pLCNICK-Alr). The plasmid pLCNICK-Alr was identified using PCR, enzyme digestion, and sequencing (data not shown). The plasmid pLCNICK-VP4 (sequence as shown in ) was constructed to exchange the heterologous genes into the genome (). The thymidylate synthase gene (thyA) was selected as the insertion site, and the expression cassette was composed of the P776 promoter, RBS, and VP4-flag sequences. The plasmid was successfully constructed and subjected to enzyme digestion and sequence identification.
Figure 1. Schematic drawing of the construction of gene editing pLCNICK series plasmids (a,b) and the plasmids-mediated Alr knockout and VP4 expression cassette insertion (replacement) in Lactobacillus chromosome (c,d). The black boxes shown in the figure are the replacement parts in different plasmids. a: the plasmid pLCNICK-Alr construction process. The PCR products of upstream (Alr-up) and downstream homologous arms (Alr-down) were acquired from the genome of Lactobacillus W56 stored in our laboratory. The sgRNA sequence was designed by predecessors targeting the Alr site, synthesized by Kumei Biotechnology Co., Ltd (Jilin, China). The fragments were fused into a sequence with the repetitive sequence in the primers and inserted into the vector pLCNICK at Apa I and Xba I restriction sites, generating the gene editing plasmid pLCNICK-Alr. b: the plasmid pLCNICK-VP4 construction process. The genes coding the promoter P776 and the fragments RBS-VP4 were amplified with the plasmids pPG-P776-EGFP and pMD19T-RBS-VP4, respectively, constructed in the laboratory. The upstream (thyA-up) and downstream homology arms (thyA-down) and sgRNA sequence are obtained as described earlier, yet the targeted site was thyA. The fragments were amplified by fusion PCR in the order thyA-up, P776, RBS-VP4, thyA-down, and sgRNA with the complementary sequence in the primers and inserted into the vector pLCNICK at Apa I and Xba I restriction sites, yielding the gene editing plasmid pLCNICK-VP4.
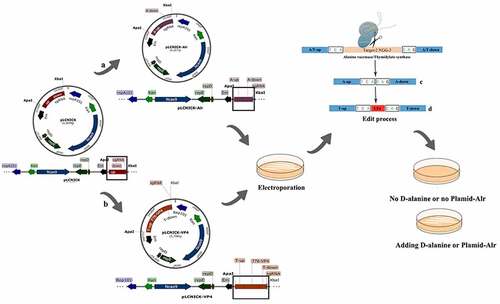
Table 1. The sequence of primers.
Table 2. The sequence of VP4.
Electroporation
Many plasmids of pLCNICK-Alr and pLCNICK-VP4 were extracted and then electrotransformed into competent cells, as previously described with some modifications [Citation37,Citation38]. In brief, bacterial cells were inoculated into MRS medium with (special MRS medium) or without (basic MRS medium) D-Ala at a ratio of 1% and cultured at 37°C until the OD600 value reached 0.4–0.8. The cells were placed in an ice bath for 30 min and then centrifuged at 3500 rpm for 5 min to acquire the bacterial pellet. They were then washed with ice-cold EPWB and EPB buffer twice and resuspended in 1 mL EPB buffer (200 μL per tube) for electrotransformation. The electroporation process was performed on ice at 2.2 kV with approximately 1 μg of gene-editing plasmid DNA added to 200 μL of competent cells in a 0.4-cm cuvette. Transformed cells were cultured statically at 30°C for 4 h. Subsequently, they were plated on 5 μg/mL EMC special MRS medium or basic MRS medium.
Construction of deletion strain Lactobacillus △alr HLJ-27 and insertion mutation strains using the CRISPR-Cas9D10Asystem
A schematic diagram of the gene-editing process of the plasmid in the host strain is shown in (c: deletion; d: insertion). The recombinant strains with the gene-editing plasmids were cultivated for 48 h at 30°C following electroporation. Single clones were picked and cultured in the corresponding MRS medium (special MRS medium or basic MRS medium), and the chromosomes were extracted for identification by PCR and sequencing with a pair of primers that targets the genome. Agarose gel electrophoresis identification results were used to confirm the results of deletions and substitution, and the hybrid mutant strains were acquired and named △Alr HLJ-27, VP4/△Alr W56, VP4/△Alr HLJ-27, and VP4/W56.
In order to obtain mutant strains, a purification process was conducted as follows: the hybrid strains were streaked on the corresponding MRS without antibiotics. Then, the purified strains were obtained, and the editing plasmids were eliminated. The hereditary stability of the recombinant strains was evaluated by continuous inoculation (1%, 20 times) and culturing (at 37°C in corresponding MRS medium), with every two generations’ genome extracted for identification by PCR. These mutant strains were then streaked onto MRS agar plates to verify the degree of bacterial demand.
Protein expression
In an effort to analyse the expression of VP4 protein in mutant bacterial strains, VP4/△Alr W56, VP4/△Alr HLJ-27, and VP4/W56 were inoculated in a corresponding MRS medium at 37°C without shaking, and the bacterial precipitation was acquired by centrifugation at 12,000 rpm for 2 min. After lysis with lysozyme, ultrasonic breaking, and centrifugation, the proteins in the supernatants were detected by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) in a 15% gel, followed by a western blot assay. The proteins were further electrotransferred onto polyvinylidene fluoride membranes (Millipore, Milford, MA, USA). Next, the proteins were incubated with mouse anti-flag monoclonal antibody (Sigma, St. Louis, MO, USA) at a dilution of 1:1500 and then with horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody (Sigma, St. Louis, MO, USA) as a secondary antibody with a dilution of 1:8000. The colour development of immunoblots was carried out using an ECL reagent (Thermo Scientific, Durham, NC, USA).
Immunization of mice
Five-week-old female pathogen-free BALB/c mice were purchased from Harbin Veterinary Research Institute, China, and raised under specific pathogen-free conditions with free access to a standard chow diet and water. Prior to oral administration, the mutant insertion strains were inoculated into an MRS medium. The bacterial precipitate was collected by centrifugation at 4500 rpm for 5 min and resuspended in PBS to adjust the bacterial count to 2 × 1010 CFU mL−1. A total of 240 mice were randomly divided into eight groups with 30 mice in each group: PBS, ΔAlr W56, ΔAlr HLJ-27, W56, VP4/ΔAlr W56, VP4/ΔAlr HLJ-27, and VP4/W56. The immunization doses and procedures are presented in . All groups of mice were immunized for three consecutive days, with the first immunization on days 0, 1, and 2, booster immunization on days 14, 15, and 16, and second booster immunization on days 28, 29, and 30.
Figure 2. Immunization procedures of mice and the process of samples collection. Mice were immunized once a day for 3 consecutive days (days 0, 1, and 2) and boosted twice at 2-week intervals (days 14, 15, and 16 and days 28, 29, and 30). Feces, sera, vaginal fluid, and intestinal mucus were collected (black check marks) at different time points. The red circle means the time point to detect the level of cytokines and CD4/CD8 T-cell and lymphocyte proliferation. Sampling time points were calculated post the primary immunization.
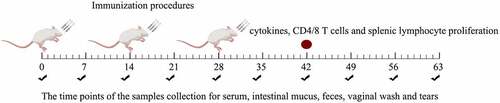
Serum, faecal, intestinal mucus, and vaginal lavage samples were collected from seven days post-immunization for 63 d and stored at −40°C until analysis. The preparation of samples prior to detection was slightly modified from the previously described method [Citation39]. In brief, 0.1 g of faecal pellet was mixed with 500 μL faecal lysate in PBS containing 0.05 mmol/L EDTANa2 and incubated overnight at 4°C. Intestinal mucus was acquired with a glass slide by scraping the gut mucosa, and the mouse vagina was repeatedly rinsed with PBS to collect vaginal lavage, which was then directly used for detection.
Detection of the IgG and SIgA antibody levels
The detection of the sera IgG level and the SIgA level in the vaginal lavage, intestinal tract, and faecal pellets, was performed by indirect ELISA. PoRV propagated on MA104 cells were utilized as an antigen with which to coat polystyrene microtiter plates, which were then incubated overnight at 4°C. Cell cultures were used as negative controls for the antigen. After discarding the waste solution, the well plates were washed three times with PBS containing 0.05% Tween 20 (PBST) for 5 min and blocked with 5% skim milk in PBS at 37°C for 2 h. After the well plates were washed three times with PBST, the collected samples were incubated for 2 h at 37℃, and each sample repeated three times. The serum was diluted 10 times with 5% skim milk, and the vaginal lavage, intestinal tract, and faecal lysate were centrifuged to collect supernatant for use. Subsequently, an HRP-conjugated goat anti-mouse IgG or IgA antibody (Invitrogen, USA) was added to wells with a dilution of 1:5000 and incubated for 1 h at 37°C. The colour development was carried out under dark conditions with a TMB solution at a ratio of 1:1, and the OD450 value was determined.
Neutralizing activity of the sera
In order to detect the neutralizing activity of the sera IgG in mice with the mutant strains VP4/△Alr W56, VP4/△Alr HLJ-27, and VP4/W56, on day 42 post-immunization, the serum was collected through eyeball blood and then diluted 2× serially. It was mixed with an equal volume of 200 × 50% tissue culture infectious dose (TCID50) of PoRV propagated on MA104 cells and incubated at 37°C for 1 h. Then, 100 μL of the mixture was added to MA104 monolayer cells grown to 80% confluence, with each dilution having five replicates. PoRV-specific cytopathies were observed after incubation for 60 h in a 37°C, 5% CO2 incubator. Data statistics were calculated by Reed – Muench.
Lymphocyte proliferation
Three mice in each group were sacrificed by cervical dislocation on day 42 after the primary immunization. Splenic lymphocytes were obtained under sterile conditions for the proliferation assay as previously described with minor modifications [Citation40]. Briefly, splenocytes at a density of 5 × 106 cells/mL were added to 96-well plates with eight replicates and incubated with RPMI-1640 plus containing 20% (v/v) of foetal calf serum at 37°C in a 5% CO2 incubator. The cells were stimulated for 66 h with 5 μg/mL concanavalin A (ConA) as a positive control, RPMI-1640 medium as a negative control, and 1, 5, or 25 μg/mL purified VP4 protein, prepared and stored in our laboratory, as the specific antigen stimulation [Citation41]. By measuring the absorbance at 490 nm, lymphocyte proliferation was assessed using the CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay (Promega, Madison, WI, USA).
T cell analysis
Flow cytometry was used to analyse the proportion of CD4+ and CD8+ in mouse spleen T lymphocytes by adjusting the density to 106 cells/mL, as previously described [Citation42]. Three mice in each group were sacrificed under aseptic conditions by cervical dislocation 42 d after primary immunization to acquire splenic lymphocytes. T cells were suspended in complete RPMI 1640, and 0.5 μL P-phycoerythrin (PE)-conjugated mouse anti-CD4 antibody was added. Allophycocyanin (APC)-labelled mouse anti-CD8-allophycocyanin antibodies were used as primary antibodies and incubated with the cells under dark conditions at 37°C for 30 min. Cells were then washed and suspended in PBS (pH 7.2) for analysis using a fluorescence-activated cell sorting (FACS) calibre cytometer. Cells were gated based on side scatter/forward scatter (SSC/FSC) parameters and CD4+/CD8+ T cells were gated based on APC/PE-labelled fluorescence parameters.
Cytokine detection
For cytokine detection, mouse serum was collected 40 d after primary immunization. Interferon (IFN)-γ, interleukin (IL)-2, IL-4, IL-10, IL-12, and IL-17 levels were tested using OptEIA Set Mouse series ELISA Kits (Biosource International, USA). The cytokine content was calculated using a linear regression equation and by drawing a standard curve of the absorbance values of the samples [Citation43].
Statistical analysis
Data are represented as the mean ± standard error of triplicates and were analysed with GraphPad Prism 7. A one-way analysis of variance and Tukey’s multiple-comparison test was used to determine the significance between treatment and control groups. A single asterisk represents P <0.05, and double asterisks represent P <0.01.
Results
Construction of the deletion and VP4 insertion mutant strains
The CRISPR-Cas9D10A series plasmid pLCNICK-Alr or pLCNICK-VP4 was electroporated into Lactobacillus competent cells HLJ-27 or △Alr W56, △Alr HLJ-27, and W56, respectively, yielding the mutant strains △Alr HLJ-27, VP4/△Alr W56, VP4/△Alr HLJ-27, and VP4/W56. The results of the agarose gel electrophoresis are shown in . In the deletion strains, the lengths of amplified wild-type and mutant-type genome using the primers Alr-F/R were 2700 bp and 1500 bp (), while in the insertion strains, the lengths were 2583 bp and 1893 bp (primers thyA-F/R; , respectively. These results were in line with the anticipated results. The precise gene insertion and deletion were confirmed by sequencing, and the deletion and insertion mutant strains △Alr HLJ-27, VP4/△Alr W56, VP4/△Alr HLJ-27, and VP4/W56 were successfully constructed.
Figure 3. Identification and analysis of the stability Lactobacillus strains △Alr HLJ-27 and VP4/△Alr W56, VP4/△Alr HLJ-27 and VP4/W56. a: PCR amplification fragments of 1500 bp and 2700 bp correspond to the deletion mutant and wild type. b–d: PCR amplification fragments of 1893 bp and 2583 bp correspond to the insertion mutant and wild type. The stability was detected for every two generations using the primers Alr-F/R (a) and the A-F/R to identify PCR products of mutant strains genome DNA. f–i: Western blot analysis the protein stability of insertion mutant strain VP4/△Alr W56, VP4/△Alr HLJ-27 and VP4/W56. Expression of VP4 protein by the strains were detected with anti-flag tag monoclonal antibody every four generations for VP4/W56, VP4/△Alr W56 and VP4/△Alr HLJ-27, respectively (j).
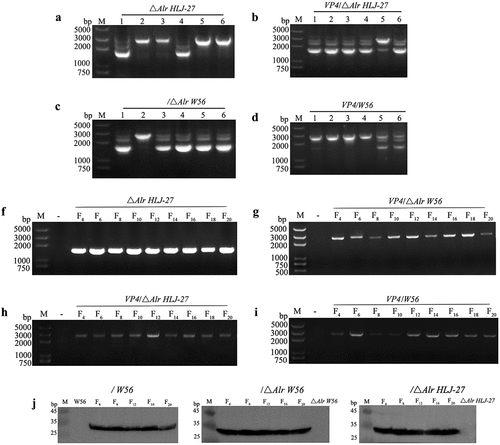
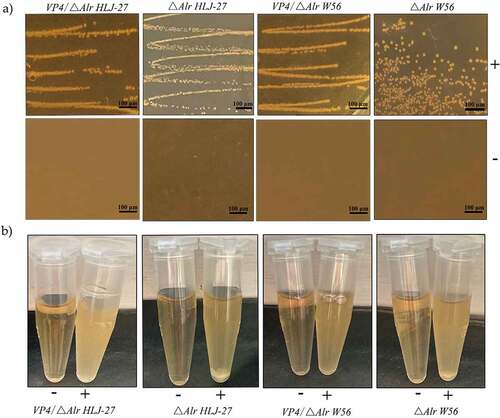
The stability of the deletion and insertion expression cassette was assessed by inoculating the strains △Alr HLJ-27, VP4/△Alr W56, VP4/△Alr HLJ-27, and VP4/W56, continuously for 20 generations at a ratio of 1% in the corresponding MRS medium and PCR amplifying every two generations. The results revealed that these strains were stably inherited over 20 generations, with each passage containing the same DNA sequences (). In order to verify the requirement for D-Ala by the deletion and insertion strains △Alr HLJ-27, VP4/△Alr W56, and VP4/△Alr HLJ-27, the strains were inoculated in the corresponding liquid MRS medium on MRS agar plates, and the results are shown in . The mutant strains grew only in the presence of D-Ala medium and not in the normal MRS medium.
Expression of VP4 by insertion mutant strains
To detect the expression of VP4 protein, labelled with a flag tag in the mutant strains, the Lactobacillus was processed with lysozyme and sonication to prepare protein samples. The mouse anti-flag-labelled monoclonal antibody was applied as the primary antibody for Western blot analysis. As shown in , the predicted immunoblot bands of VP4 protein were approximately 35 kDa in VP4/△Alr W56, VP4/△Alr HLJ-27, and VP4/W56, but no bands were observed in the control strains △Alr W56, △Alr HLJ-27, W56, and HLJ-27. Our data indicated that the insertion mutant strains could efficiently express VP4 protein.
Detection of IgG and SIgA antibody levels
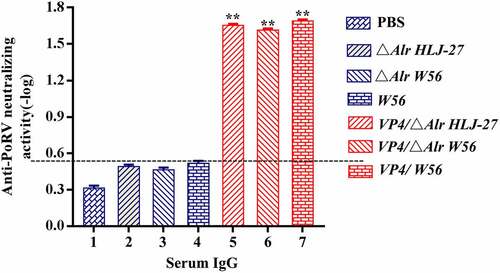
The immunogenicity of insertion mutants VP4/ΔAlr W56, VP4/ΔAlr HLJ-27, and VP4/W56 was evaluated by measuring the levels of anti-PoRV IgG and SIgA antibodies. These antibodies were detected by indirect ELISA after oral immunization of mice as the standard to assess systemic and mucosal immune responses. Serum, faecal, intestinal mucus, and vaginal lavage samples to detect the anti-PoRV IgG and SIgA were collected on days 7, 14, 21, 28, 35, 42, 49, 56 and 63 post primary immunization. As shown in , there were no data differences observed between immune (VP4/△Alr W56, VP4/△Alr HLJ-27, and VP4/W56) and control groups (△Alr W56, △Alr HLJ-27, W56, HLJ-27, and PBS; P>0.05). A significant rise was observed in anti-PoRV-specific IgG antibodies in VP4/△Alr W56, VP4/△Alr HLJ-27, and VP4/W56 at 7 d post primary immunization, and their levels significantly increased after the booster immunization compared to that of the control groups (△Alr W56, △Alr HLJ-27, W56, HLJ-27, and PBS; P <0.01). The levels peaked at 42 d post-immunization. The specific IgG level of VP4/W56 was the highest among the three groups, and the levels in the group VP4/△Alr HLJ-27 was higher than that in VP4/△Alr W56. There were no significant changes among the vaccine groups VP4/△Alr W56, VP4/△Alr HLJ-27, and VP4/W56. Moreover, at 42 d post-immunization, the PoRV-neutralizing activity of the mouse sera was detected using a virus neutralization assay, as shown in . The results indicated that the sera IgG antibody activity in mice activated by VP4/△Alr W56, VP4/△Alr HLJ-27, and VP4/W56 exhibited anti-PoRV-neutralizing activity significantly different from that obtained from the control groups (△Alr W56, △Alr HLJ-27, W56, HLJ-27, and PBS; P <0.01). The antibody activity in the group VP4/W56 was slightly higher than that in the VP4/△Alr HLJ-27 and VP4/△Alr W56 groups. However, there were no statistical differences among the three groups. As expected, the mutant strains groups VP4/△Alr W56, VP4/△Alr HLJ-27, and VP4/W56 could elicit an anti-PEDV immune response.
Figure 5. Anti-PoRV-Specific IgG antibody levels in the sera (a) and mucosal SIgA antibody in genital tract, intestinal mucus, and feces (b) by ELISA using PoRV as the coating antigen. Bars represent the mean SE in each group (**P < 0.01 compared to the control groups: △Alr HLJ-27, △Alr W56, W56 and PBS; Vaccine group strains: VP4/△Alr HLJ-27, VP4/△Alr W56 and VP4/W56).
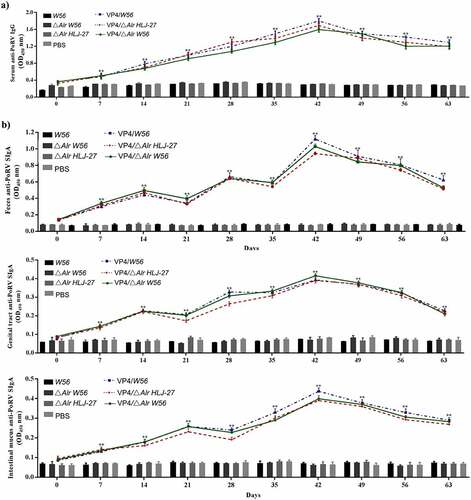
Figure 6. Levels of the anti-PoRV-specific neutralizing antibody titer in mice postimmunization serum. The results were determined by measuring the anti-PEDV IgG antibody and the anti-PEDV-neutralizing antibody titer with serial dilution in sera from immunized mice, ** P < 0.01 compared to the control groups.
Moreover, to determine the production of anti-PoRV-specific SIgA in faeces, vaginal lavage fluid, and intestinal mucus of immunized mice, an ELISA analysis was performed. As shown in , anti-PoRV-specific mucosal SIgA was detected on day 7 post primary immunization. The SIgA antibody levels were significantly increased in the faeces, vaginal lavage, and intestinal mucus of mice orally immunized with VP4/△Alr W56, VP4/△Alr HLJ-27, and VP4/W56 compared to that in the control groups (P <0.01). The order of the levels of SIgA antibodies was VP4/W56> VP4/△Alr HLJ-27 > VP4/△Alr W56, and there were no significant differences among the three groups. After booster immunization on day 14, the levels of SIgA antibody increased quickly and reached the highest point on day 42. Our results revealed that the application of the insertion mutant strains could significantly induce systemic and mucosal antibody responses.
Lymphocyte proliferation and T-cell analysis
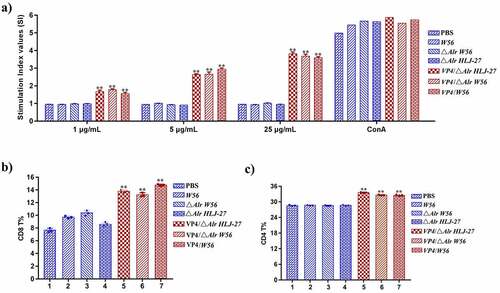
To analyse the proliferation of splenic lymphocytes by the MTT assay, mice were sacrificed to obtain the cells and stimulated with the VP4 protein. As shown in , splenocyte stimulation index markedly increased in mice orally immunized with the insertion mutant strains VP4/△Alr W56, VP4/△Alr HLJ-27, and VP4/W56, but not in mice immunized with △Alr W56, △Alr HLJ-27, W56, HLJ-27, or PBS (P <0.01). A significant change was noted in the insertion mutant strain groups stimulated with increasing concentrations of purified VP4. These results indicated that the test bacterial strains could induce cellular immunity in mice. The effect of the cellular immune response of insertion mutant strains VP4/△Alr W56, VP4/△Alr HLJ-27, and VP4/W56 post immunization was determined by analysing the levels of CD4+ and CD8+ cells using a FACS calibre cytometer. The results indicated significant changes in the percentage of CD4+ and CD8+ in mice immunized with mutant strains VP4/△Alr W56, VP4/△Alr HLJ-27, and VP4/W56 than those in the control groups (P <0.01), with no statistical difference among the mutant strains (). These results revealed that strains VP4/△Alr W56, VP4/△Alr HLJ-27, and VP4/W56 could induce cellular immunity in mice.
Figure 7. Determination of the level of lymphocyte proliferation (a) and specific T cell, CD8+% (b) and CD4+% (c) in splenic lymphocytes in immunized mice. The lymphocyte proliferation in immunized mice responding to PoRV VP4 antigen restimulation was determined by an MTT assay with ConA as a positive control. **P < 0.01 compared to the control groups: △Alr HLJ-27, △Alr W56, W56 and PBS.
Analysis of cytokine responses
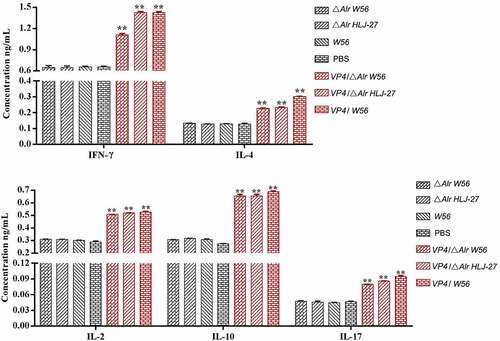
The cellular immune responses were evaluated by measuring the levels of IFN-γ, IL-2, IL-4, IL-10, IL-12, and IL-17 induced by the administered mutant strains VP4/△Alr W56, VP4/△Alr HLJ-27, and VP4/W56 using mouse series ELISA kits. As shown in , a significant increase was noted in the levels of IFN-γ, IL-2, IL-4, IL-10, IL-12, and IL-17 at 42 d post-immunization in the serum of mice immunized with VP4/△Alr W56, VP4/△AlrHLJ-27, and VP4/W56, compared to those in the control groups (△Alr W56, △Alr HLJ-27, W56, HLJ-27, and PBS; P <0.01). There were no significant changes in levels between the strains VP4/△Alr W56, VP4/△Alr HLJ-27, and VP4/W56.
Discussion
Lactobacillus, a food-grade microorganism with adjuvant properties and weak immunogenicity, is widely used as a mucosal live vector vaccine. However, traditional expression systems are mostly plasmid-based with antibiotics as selection markers; these systems typically exhibit unstable expression of heterologous proteins [Citation4]. To solve these problems, an efficient genome editing tool was developed for chromosomal deletion and insertion with a site of interest. The rapid development of the CRISPR-Cas9 system for genetic editing has made modifying various organisms increasingly convenient, particularly for use in cases that are lacking in effective genetic tools for gene manipulation. The applications of CRISPR are mainly dependent on DSB induction by Cas9. However, DSBs have been observed to induce a high mortality rate in certain bacteria [Citation44,Citation45]. In this study, Cas9D10A, a Cas9 protein mutant, was utilized to cause only DNA single-strand breaks to solve this problem. Song et al. revealed that the CPISPR-Ca9D10A system could rapidly and efficiently manipulate chromosomal deletions and site-specific insertions [Citation33].
In this study, Alr was selected as the auxotrophic site for deletion with CPISPR-Ca9D10A. Alr participates in converting L-Ala to D-Ala, and D-Ala, a peptidoglycan component of the bacterial cell wall, is essential for bacterial growth. Studies have shown that only the double mutant of the dadX gene is responsible for Alr activity, and the Alr gene in E. coli is auxotrophic to D-Ala [Citation46,Citation47]. LAB, such as Lactococcus lactis and Lactobacillus plantarum, encode Alr to catalyse the conversion of D-Ala and L-Ala [Citation18,Citation48]. Due to the stable expression of the Alr gene and the absence of D-Ala in most industrial fermentation media, the Alr gene of LAB is an important candidate marker for food-grade LAB that helps assess the biosafety of these microbes and prevent various side effects caused by antibiotic resistance in animals. Bron et al. utilized the gene, encoding Alr as a promoter-screening tool to clone a random fragment of the genome of Lactobacillus WCFS1 upstream of the Alr gene without the promoter. The library was set up by introducing the plasmid into the Lactobacillus ΔAlr strain, and the effectiveness of the Alr complementary screening method in identifying the condition or compositional activity of the promoter was confirmed [Citation48]. Nguyen et al. used the Alr gene from Lactobacillus to replace erythromycin in the pSIP expression vector and successfully overexpressed the β-galactosidase gene of Lactobacillus reuteri L103. This new selection marker provides a reference for and suggests the potential for its use as a replacement for antibiotics [Citation49].
In this study, the CRIPSR-Cas9D10A gene-editing system was utilized to acquire an auxotrophic mutant, △Alr HLJ-27, by cutting a single strand of the genome of Lactobacillus HLJ-27, isolated from piglet intestines. After the plasmid pLCNICK-Alr was electrotransformed into L. paracasei HLJ-27 to obtain the deficient strain, agarose gel analysis revealed the appearance of two bands, indicating that the cultured bacteria were a mixture of the mutants and wild type, consistent with the results of Song et al. [Citation33]. The D-Ala demand experiment is an essential screening indicator for constructing Alr auxotrophic Lactobacillus. The demand experiment was performed with the deletion strain successfully constructed. The results showed that D-Ala played a vital role in the screening methods by replacing antibiotic screening of defective strains, making it possible to meet the requirements of safety standards for genetically modified biological products. Furthermore, the results revealed that the VP4 gene could exist stably in the genome.
PoRV is a common virus that causes diarrhoea in pigs worldwide. It primarily infects the mucosal surfaces, especially the intestinal mucosal epithelium. VP4, an outer capsid protein of PoRV, could effectively stimulate the production of neutralizing antibodies, to provide protective immunity for animals [Citation45,Citation50]. For expressing the PoRV VP4 protein, Lactobacillus was chosen as an oral vaccination vector, as it can elicit specific humoral and mucosal immunity safely [Citation51]. Qiao et al. exploited the heat-labile B subunit of E. coli (LTB) and VP4 protein for fusion expression using SDS-PAGE and western blotting analysis in L. casei. The mice were immunized orally with recombinant strains, and they observed higher levels of IgG and SIgA production [Citation41]. Yin et al. expressed the VP4 gene at the pyruvate hydratase site in the genome of Lactobacillus, and the results indicated that the heterologous gene could be inherited stably and induce an effective immune response [Citation4]. In this study, in the laboratory-preserved Lactobacillus W56, the auxotrophic strains △Alr W56 and △Alr HLJ-27, were chosen as the host strains. The heterologous protein gene VP4 was combined with the promoter P776 of the constitutively expressed gene pyruvate hydratase from LAB to constitute the expression cassette for genome expression. The gene-editing plasmid was successfully constructed and then electrotransformed into the three competent cells. Two bands were present in the genome PCR amplification, in line with the principle of CRIPSR-Cas9D10A to cut one chain. The demand experiment results indicated that the insertion mutant strains VP4/△Alr W56 and VP4/△Alr HLJ-27 could only grow with exogenously added D-Ala. Genetic stability results also showed that these strains could be stably inherited. The insertion mutant strains VP4/W56, VP4/△Alr W56 and VP4/△Alr HLJ-27 could effectively induce the production of mucosal and humoral immune responses to fight against PoRV, as evidenced by significantly higher levels of serum IgG and mucosal SIgA. Moreover, genetically-engineered vaccines are safe, and the mode of administration is practical and convenient.
SIgA play a key role in protecting piglets from pathogens invading the gastrointestinal tract, and the production of SIgA reflects the capability of the body to fight viral invasion and the degree of a vaccine’s protection [Citation52]. In the present study, levels of anti-PoRV SIgA antibodies were observed in the faecal pellets, intestinal mucus, and genital lavage of the mice immunized with these insertion mutant strains. We observed that levels of SIgA antibodies increased and reached their peak at 42 d post primary immunization. The local production of SIgA could prevent pathogens from binding to small intestinal cells and deter the virus from crossing the mucosal barrier [Citation53].Although SIgA antibody levels reflect the primary mechanism of mucosal surface defence, serum-derived IgG also plays a vital role in immune defence. It impairs the ability of the virus to pass through the intestinal mucosa and blocks the spread of pathogens throughout the body. It also plays a synergistic protective role with SIgA [Citation54]. In this study, the serum IgG level increased significantly after booster immunization, with a peak at six weeks, indicating that the inserted mutant strain could stimulate a systemic immune response and further prevent the invasion of antigens.
Studies have shown that protective immunity could be triggered by activated B cells secreting sIgA and activated T cells differentiation [Citation55,Citation56]. The research of Gao et. al revealed that the application of LAB as a delivery vehicle for oral vaccines could induce the occurrence of cellular immune responses in oral immunization experimental with mice [Citation44]. In this study, to analyse the specific T-cell immune responses stimulated by the mutant strains in mice, T cell differentiation and cytokine production were tested. Flow cytometry data showed that the CD4+ and CD8+ T cells levels were significantly increased, and the cellular immunity of orally immunized mice was activated. This is a key method to assess the ability of vaccines to induce cellular immune responses [Citation57]. Studies have shown that an appropriate dose of LAB promotes the differentiation of Th cells and the production of sIgA by B lymphocytes [Citation58]. For analysing the ability of recombinant strains to trigger helper T cell differentiation (Th1 or Th2), the levels of cytokines induced by the mutant strains in mice were assessed [Citation57]. CD4+ Th lymphocytes secrete cytokines that could participate in the regulation of antibody production and cellular immune responses. In this study, we evaluated cytokine levels, including IFN-γ, IL-4, IL-17, IL-2, IL-10, and IL-12. We observed that oral immunization with VP4/△Alr W56, VP4/△Alr W56, and VP4/△Alr HLJ-27 could significantly induce cytokine production, revealing that these mutant strains could arouse Th1, Th2, and Th17-type cellular responses in vivo. The production of SIgA to fight pathogenic bacteria or foreign immunogens was thus via a T cell-dependent immune response [Citation59]. In this study, the heterologous protein VP4 expressed by the Lactobacillus-induced Th1 and Th2-type immune responses, indicating that T cell-mediated immunity is key. Furthermore, these mutant strains could elicit an efficient lymphocyte proliferation response against the PoRV VP4 antigen. Our results suggest that Lactobacillus expressing the PoRV VP4 antigen might serve as candidate vaccine vectors and provide a novel construction method to induce potent immune responses against viral infections. However, these findings need to be confirmed via further challenge-protection experiments in piglets.
Author contributions
Y.L. and X.W. conceived and designed the study. F.L., Z.M., L.S., X.F., X.L., Y.J., and W.C. performed the experiments. H.Z., L.W., X.Q., and L.T. analysed and interpreted the data. F.L. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Ethical statements
Animal experiments were carried out in accordance with the recommendations in the institutional and national guidelines for animal care and use. The protocol was approved by the Committee on the Ethics of Animal Experiments of Northeast Agricultural University, Harbin, China (2016NEFU–315, 13 April 2017). All procedures were performed under ether anaesthesia to minimize suffering.
Acknowledgements
We sincerely thank researcher Yang Sheng, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences for kindly providing the plasmid pLCNICK.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available from the corresponding author, Yijing Li, upon reasonable request.
Additional information
Funding
References
- Clark HF, Burke CJ, Volkin DB, et al. Safety, immunogenicity and efficacy in healthy infants of G1 and G2 human reassortant rotavirus vaccine in a new stabilizer/buffer liquid formulation. Pediatr Infect Dis J. 2003;22(10):914–920. DOI:10.1097/01.inf.0000091887.48999.77
- Hideaki S, Takashi W , Shinji F, et al. A novel mucosal vaccine targeting Peyer’s patch M cells induces protective antigen-specific IgA responses. A novel mucosal vaccine targeting Peyer’s patch M cells induces protective antigen-specific IgA responses. International Immunology. 2014;(11):619–625.
- Ho PS, Kwang J, Lee YKJV. Intragastric administration of Lactobacillus casei expressing transmissible gastroentritis coronavirus spike glycoprotein induced specific antibody production. Vaccine. 2005;23(11):1335–1342.
- Yin JY, Guo C-Q, Wang Z, et al. Directed chromosomal integration and expression of porcine rotavirus outer capsid protein VP4 in Lactobacillus casei ATCC393. Appl Microbiol Biotechnol. 2016;100(22):1–12. DOI:10.1007/s00253-016-7779-y
- Pouwels PH, Leer RJ, Boersma WJJOB. The potential of Lactobacillus as a carrier for oral immunization: Development and preliminary characterization of vector systems for targeted delivery of antigens. J Biotechnol. 1996;44(1–3):183.
- Gibson GR, Probert HM, Loo JV, et al. Dietary modulation of the human colonic microbiota: Updating the concept of prebiotics. Nutr Res Rev. 2004;17(2):259–275. DOI:10.1079/NRR200479
- Davies FL, Gasson MJJJODR. Genetics of lactic acid bacteria. J Dairy Res. 1981;48(2):363–376.
- Hugenholtz J, Smid EJJCOIB. Nutraceutical production with food-grade microorganisms. 2002;13(5):497–507. DOI:10.1016/s0958-1669(02)00367-1.
- Jingqi Zhao,Ting Yan,Yaqi Wang, et al. Lactobacillus plantarum BC299 can alleviate dextran sulphate sodium-induced colitis by regulating immune response and modulating gut microbiota. Int J Food Sci Technol. 2021; 56(11):5698–5707.
- Chen Y, Liu S, Yu C, et al. Evaluating the effectiveness of Lactobacillus zeae against enterotoxigenic Escherichia coli F4 infection in an in vitro porcine intestinal epithelial cell model. 2021.
- Dogi CA, Galdeano CM, Perdigón G. Gut immune stimulation by non pathogenic Gram(+) and Gram(-) bacteria. Comparison with a probiotic strain. Cytokine. 2008;41(3):223–231.
- Galdeano CM, De Moreno De Leblanc A, Vinderola G, et al. Proposed model: Mechanisms of immunomodulation induced by probiotic bacteria. Clin Vaccin Immunol. 2007;14(5):485–492. DOI:10.1128/CVI.00406-06
- LU Wenwei, Z Yang, L Ding, et al. Evaluation of intestinal microbiota in mice after oral administration of lactobacillus paracasei LC01. Food Sci. 2016.
- Vidal K, Benyacoub J, Moser M, et al. Effect of Lactobacillus paracasei NCC2461 on antigen-specific T-cell mediated immune responses in aged mice. Rejuvenation Res. 2008;11(5):957. DOI:10.1089/rej.2008.0780
- O“sullivan DJ, Klaenhammer TRJG. High- and low-copy-number Lactococcus shuttle cloning vectors with features for clone screening. Gene. 1993;137(2):227–231.
- Pouwels PHJAEM. Incompatibility of Lactobacillus vectors with replicons derived from small cryptic lactobacillus plasmids and segregational instability of the introduced vectors. Appl Environ Microbiol. 1991;19(1):28.
- Lu WW, Wang T, Wang Y, et al. A food-grade fimbrial adhesin FaeG expression system in Lactococcus lactis and Lactobacillus casei. Can J Microbiol. 2016;62(3):241–248. DOI:10.1139/cjm-2015-0596
- Bron PA Benchimol M G, Lambert J, et al. Use of the alr Gene as a Food-Grade Selection Marker in Lactic Acid Bacteria. Appl Environ Microbiol. 2002;68(11):5663–5670. DOI:10.1128/AEM.68.11.5663-5670.2002
- Zajdowicz SLW, Jones-Carson J, Vazquez-Torres A, et al. Alanine racemase mutants of Burkholderia pseudomallei and Burkholderia mallei and use of alanine racemase as a non-antibiotic-based selectable marker. PLoS One. 2011;6(6):e21523. DOI:10.1371/journal.pone.0021523
- Hols P, Defrenne C, Ferain T, et al. The alanine racemase gene is essential for growth of Lactobacillus plantarum. J Bacteriol. 1997;179(11):3804–3807. DOI:10.1128/jb.179.11.3804-3807.1997
- Liu Q, Jiang Y, and Yang W, et al. Protective effects of a food-grade recombinant Lactobacillus plantarum with surface displayed AMA1 and EtMIC2 proteins of Eimeria tenella in broiler chickens. Microbial Cell Factories. 2020;19(1):11-28.
- Pham ML, Tran A-M, and Mathiesen G, et al. Cell wall anchoring of a bacterial chitosanase in lactobacillus plantarum using a food-grade expression system and two versions of an LP TG anchor. International Journal of Molecular Sciences. 2020;21(11):3773. DOI:10.3390/ijms21113773
- Ehrlich SD, Bruand C, Sozhamannan S, et al. Plasmid replication and structural stability in Bacillus subtilis. Res Microbiol. 1991;142(7–8):869–873. DOI:10.1016/0923-2508(91)90067-K
- Bidart GN, Rodríguez-Díaz J, Monedero V, et al. A unique gene cluster for the utilization of the mucosal and human milk-associated glycans galacto- N -biose and lacto- N -biose in L actobacillus casei. Mol Microbiol. 2014;93(3):521–538. DOI:10.1111/mmi.12678
- Jan-Peter VP, and Britton RAJNAR. High efficiency recombineering in lactic acid bacteria. Nucleic Acids Research. 2012(10):e76.
- Pijkeren J, Britton RAJMCF. Precision genome engineering in lactic acid bacteria. Microb Cell Fact. 2014;13(1):S10.
- Hille, Hille F, Richter H, et al. The biology of CRISPR-cas: Backward and forward. Cell. 2018;172(6):1239–1259. DOI:10.1016/j.cell.2017.11.032
- Liu C, Zhang L, and Liu H, et al. Delivery strategies of the CRISPR-cas9 gene-editing system for therapeutic applications. Journal of Controlled Release. 2017;17–26.
- Eugene Koonin EV, Makarova KS, et al. Origins and evolution of CRISPR-cas systems. Philosophical Transactions of the Royal Society of; 2019.
- Jiang W, Bikard D, Cox D, et al. RNA-Guided editing of bacterial genomes using CRISPR-cas systems. Nat Biotechnol. 2013;31(3):233–239. DOI:10.1038/nbt.2508
- Jee-Hwan O, van P-J-P-J-N-A. Research, CRISPR–Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 2014(17):e131.
- Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. DOI:10.1126/science.1138140
- Xin Song, He Huang, Zhiqiang Xiong, et al. CRISPR-Cas9d10a nickase-assisted genome editing in Lactobacillus casei. Appl Environ Microbiol. 2017;83(22):01259–17.
- Diaz-Salinas MA, Romero P, Espinosa R, et al. The spike protein VP4 defines the endocytic pathway used by rotavirus to enter MA104 cells. J Virol. 2013;87(3):1658–63. doi:10.1128/JVI.02086-12.
- Liu X, Zhang GM, Li HZJCJOB. Passive immune protection of neonatal mice with recombinant adenovirus expressing rotavirus VP4 antigen. Vet Microbiol. 2007;20(1):33–36.
- YH Zhang, HU Huai-Rong, XY Xian, et al. Isolation and Identification of Lactic Acid Bacteria from Intestinal Tract of Rongchang Pig. J Southwest Univ(Natural). 2016.
- Wang L, Xia T, Guo T, et al. Recombinant lactobacillus casei expressing capsid protein vp60 can serve as vaccine against rabbit hemorrhagic disease virus in rabbits. Vet Microbiol. 2019;7(4).
- Meiling Y, Wang L, Ma S, et al. immunogenicity of egfp-marked recombinant lactobacillus casei against transmissible gastroenteritis virus and porcine epidemic diarrhea virus. Viruses. 2017;9(10):274. DOI:10.3390/v9100274
- Liu D, Wang X, Ge J, et al. Comparison of the immune responses induced by oral immunization of mice with Lactobacillus casei-expressing porcine parvovirus VP2 and VP2 fused to Escherichia coli heat-labile enterotoxin B subunit protein. Comp Immunol Microbiol. 2011;34(1):73–81. DOI:10.1016/j.cimid.2010.02.004
- Xiaona W, Wang L, Huang X, et al. Oral delivery of probiotics expressing dendritic cell-targeting peptide fused with porcine epidemic diarrhea virus coe antigen: A promising vaccine strategy against PEDV. Viruses. 2017;9(11):312. DOI:10.3390/v9110312
- Qiao X, Li G, Wang X, et al. Recombinant porcine rotavirus VP4 and VP4-LTB expressed in Lactobacillus casei induced mucosal and systemic antibody responses in mice. BMC Microbiol. 2009;9(1):249. DOI:10.1186/1471-2180-9-249
- Hou X, Jiang X, Jiang Y, et al. Oral immunization against PEDV with recombinant Lactobacillus casei expressing dendritic cell-targeting peptide fusing COE protein of PEDV in Piglets. BMC Microbiol. 2018;10(3):106.
- Gao X, Ma Y, Wang Z, et al. Oral immunization of mice with a probiotic Lactobacillus casei constitutively expressing the α-toxoid induces protective immunity against Clostridium perfringens α-toxin. Virulence. 2019;10(1):166–179. DOI:10.1080/21505594.2019.1582975
- Xu T, Li Y, Shi Z, et al. Efficient genome editing in clostridium cellulolyticum via CRISPR-cas9 nickase. Appl Environ Microbiol. 2015;81(13):4423–4431. DOI:10.1128/AEM.00873-15
- Ran F, Hsu P, Lin C-Y, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. DOI:10.1016/j.cell.2013.08.021
- Wild J, Hennig J, Lobocka M, et al. Identification of the dadX gene coding for the predominant isozyme of alanine racemase in Escherichia coli K12. Mol Gen Genet. 1985;198(2):315–322. DOI:10.1007/BF00383013
- Bron PA, Hoffer SM, Van Swam II, et al. Selection and characterization of conditionally active promoters in Lactobacillus plantarum, using alanine racemase as a promoter probe. Molecular & General Genetics Mgg. 2004;70(1):310–317.
- Guan Wang, Mingyue Hao, Qiong Liu, et al. Protective effect of recombinant Lactobacillus plantarum against H 2 O 2 -induced oxidative stress in HUVEC cells. J Zhejiang Univ Sci B. 2021;22(5):348–365.
- Ying L, Deng ZL, Zhang ZS, et al. A novel expression vector for Corynebacterium glutamicum with an auxotrophy complementation system. ScienceDirect. Chin Med J (Engl). 1994;107(3):171–175.
- Offit PA, Shaw RD, Greenberg HBJJOV. Passive protection against rotavirus-induced diarrhea by monoclonal antibodies to surface proteins vp3 and vp7. J Virol. 1986;58(2):700–703.
- Li YJ, Ma G-P, Li G-W, et al. Oral Vaccination with the porcine rotavirus VP4 outer capsid protein expressed by Lactococcus lactis induces specific antibody production. J Biomed Biotechnol. 2010;2010(3):708460. DOI:10.1155/2010/708460
- Jiang X, Yu M, Qiao X, et al. Up-Regulation of MDP and tuftsin gene expression in Th1 and Th17 cells as an adjuvant for an oral Lactobacillus casei vaccine against anti-transmissible gastroenteritis virus. Science. 1940;91(19):554–555. DOI:10.1126/science.91.2371.554
- Sutherland DB, Fagarasan SJCOII. IgA synthesis: A form of functional immune adaptation extending beyond gut. Appl Microbiol Biotechnol. 2012;24(3):261–268.
- Holmgren J, Czerkinsky CJNM. Mucosal immunity and vaccines. Nat Med. 2005;11(Suppl 4):S45–S53.
- Macpherson AJ, McCoy K D, and Johansen F-E, et al. The immune geography of IgA induction and function. Mucosal Immunology. 2008;1(1):11–22. DOI:10.1038/mi.2007.6
- Mantis NJ, Rol N, Corthésy BJMI. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011;4(6):603.
- SH Shi, WT Yang, GL Yang, et al. Lactobacillus plantarum vaccine vector expressing hemagglutinin provides protection against H9N2 challenge infection. Virus Research, 2016. 211.
- Ma S, Wang Li, Huang X, et al. Oral recombinant Lactobacillus vaccine targeting the intestinal microfold cells and dendritic cells for delivering the core neutralizing epitope of porcine epidemic diarrhea virus. Microb Cell Fact. 2018;17(1):20. DOI:10.1186/s12934-018-0861-7
- Fagarasan, Fagarasan S, Kawamoto S, et al. Adaptive immune regulation in the Gut: T cell–Dependent and T cell–Independent IgA synthesis. Annu Rev Immunol. 2010;28(1):243–273. DOI:10.1146/annurev-immunol-030409-101314