ABSTRACT
The genus Shewanella consists of Gram-negative proteobacteria that are ubiquitously distributed in environment. As the members of this genus have rapidly increased within the past decade, several species have become emerging pathogens worldwide, attracting the attention of the medical community. These species are also associated with severe community- and hospital-acquired infections. Patients infected with Shewanella spp. had experiences of occupational or recreational exposure; meanwhile, the process of infection is complex and the pathogenicity is influenced by a variety of factors. Here, an exhaustive internet-based literature search was carried out in PUBMED using terms “Achromobacter putrefaciens,” “Pseudomonas putrefaciens,” “Alteromonas putrefaciens” and “Shewanella” to search literatures published between 1978 and June 2022. We provided a comprehensive review on the epidemiology, clinical features and pathogenicity of Shewanella, which will contribute a better understanding of its clinical aetiology, and facilitate the timely diagnosis and effective treatment of Shewanella infection for clinicians and public health professionals.
Introduction
The genus Shewanella comprises Gram-negative, facultative anaerobic, oxidase-positive and motile bacteria [Citation1,Citation2]. Due to the unique physiological and respiratory versatility, Shewanella spp. can survive in a wide range of ecological niches (for example, suboptimal environmental conditions with extreme salinity and high barometric pressure, spoilt foods and clinical specimens) and have been applied in environmental protection and industrial development [Citation2]. Since the first identification of Shewanella putrefaciens in 1931 [Citation3], several Shewanella species have recently emerged as worldwide pathogens, attracting the attention of the medical community [Citation4]. Species like Shewanella algae, Shewanella putrefaciens, and Shewanella xiamenensis [Citation5,Citation6] have been proven to associate with human [Citation4,Citation7] and aquatic livestock diseases [Citation8–11]. Also, Shewanella spp. can be found in food processing and storage [Citation7]. Nowadays, members of the genus Shewanella are more than 70 (http://www.bacterio.net/shewanella.html). In order to identify the clinical features and to evaluate resistance pattern of Shewanella species, Wincy et al. retrospectively analyzed demographics, antibiotics, microbiology, and outcomes of the 128 patients who has been admitted to a regional hospital in Hong Kong with Shewanella species infection from 1st April 2010 to 31th December 2020 [Citation4]. In this review, we searched for case reports of Shewanella spp. infections in PUBMED from 1978 to June 2022 and provided an overview of the epidemiological features, clinical manifestations and pathogenicity of Shewanella infection, which may be helpful in guiding treatment strategy determinations and providing responsive therapy.
Epidemiology
In sharp contrast to the beneficial effects of Shewanella, occupational or recreational exposure is the two most common routes of infection [Citation7]. The physiological versatility of the genus Shewanella allows for its wide distribution. As mentioned above, S. putrefaciens was originally isolated from rotten butter, and since then, it has been identified as a food spoiler in several foods, including poultry, beef and seafood [Citation7]. Other Shewanella species are mainly found in marine environments [Citation12], and S. algae, S. putrefaciens, and S. xiamenensis have become emerging opportunistic pathogens. The Shewanella-related infections are sporadically reported and many cases are being documented. All published literatures about the case reports of Shewanella infection have been systematically reviewed to summarize the demographic information and clinical characteristics [Citation3,Citation7,Citation13–18]. A total of 125 studies involving 273 patients were included for final analysis.
Geographical distribution
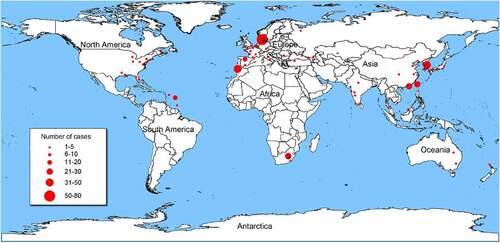
Cases of Shewanella infection have been reported around the world. Places with hot summer weather have the highest number of reported cases (), e.g. Southern Europe (n = 71), Southeast Asia (n = 48), and Southern Africa (n = 30). Most reported cases were from tropical, subtropical, and temperate countries such as Australia, Belgium, Denmark, Israel, Spain, and Turkey [Citation12]. Coastal cities and regions suitable for tourism and living, like Taiwan of China, Martinique, Barbados, Canary Islands, also have many cases. The prevalence of Shewanella infection varies greatly in different geographical locations, being correlated with the temperature of seawater and the frequency of strain occurrence [Citation19].
Figure 1. Geographical distribution of 273 Shewanella infectious cases. The geographical distribution of cases are Denmark (n = 71), Spain (n = 39), Africa (n = 30), China (n = 19), U.S.A (n = 17), Martinique (n = 16), India (n = 12), France (n = 9), Japan (n = 7), Korea (n = 5), Turkey (n = 6), Australia (n = 5), Caucasian (n = 3), Croatia (n = 2), Malaysia (n = 2), Thailand (n = 2), Belgium (n = 2), Italy (n = 2), Panama (n = 1), Mexico (n = 1), Moroccan (n = 1), Belize (n = 1), Wakefield (n = 1), Virginia (n = 1), Bahamas (n = 1), Romania (n = 1), Madagascar (n = 1), Germany (n = 1), Caribbean (n = 1), Cyprus (n = 1), UK (n = 1), Brunei Darussalam (n = 1), Puerto Rico (n = 1), Russia (n = 1), New Zealand (n = 1), Côte d’Ivoire (n = 1), and Israel (n = 1). Information on the geographical location of 5 cases is not available.
Population distribution and species composition
The ages of people infected with Shewanella ranged from neonates to 92 years old (). Among all the cases, the elderly (over 60 years old) accounted for the largest proportion (34.43%) compared to other age groups. Apart from patients with undisclosed sex (39.56%), the ratio of male to female was 2.84:1(122/43). In the study of Wincy et al., 61.7% of the 128 patients were male, with an average age of 78 [Citation4]. In terms of age and gender composition, we have reached the same conclusion. The reason for this may be that men are more likely to engage in occupations or activities related to marine habitats, including fishing and diving. Clinical strains of Shewanella can be isolated from samples such as blood, sputum, urine, and intra-abdominal. The species compositions of clinical Shewanella infection were S. algae (35.16%), S. putrefaciens (28.94%), and S. xiamenensis (0.37%). Some studies failed to provided definitive information for accurate species identification. In most of the cases (35.16%), Shewanella was a member of the multi-microbial infections, making it difficult to explain its exact role in pathogenicity and disease progression.
Table 1. Clinical characteristics associated with Shewanella infection.
According to the available case reports, S. algae and S. putrefaciens were most frequently isolated from blood samples and skin injury swabs, which is consistent with the conclusion drown by Janda [Citation7]. It is worth noting that in Denmark, S. algae has also been isolated in the pure culture of the ear swabs of 33 patients, suggesting its important role in causing ear infections [Citation15]. Moreover, S. putrefaciens has been isolated from patients suffered from peritonitis [Citation20–24] and meningitis [Citation25], and S. xiamenensis has been isolated from intestinal specimen [Citation6].
Infectious pathways and influencing factors
The infection caused by the genus of Shewanella is complex and can be influenced by a variety of factors. Firstly, Shewanella spp. are abundantly distributed in water environments, providing ample opportunities for these bacteria to come into close contact with humans [Citation7]. Recreation (e.g. diving, playing on the beach), occupational exposure (e.g. crabbing, fishing), seafood ingestion, puncture wounds caused by marine organisms (sea urchins, fish), or the direct exposure of a wound to aquatic environments can increase the risk of Shewanella infection. The percentage of exposure to marine environments was reported to be 43.59% among patients with Shewanella infection (). Secondly, Shewanella infections are commonly found in patients with immunocompromised state, including malignancies, severe heart failure, renal failure, hepato-biliary disease, neutropenia, and chronic ulcerations on the lower extremities [Citation7,Citation26], although infections in healthy individuals with no medical history had also been reported [Citation13,Citation27–30]. This kind of microorganisms can also be cultured from the clinical samples of burned or multiply traumatized patients, and patients with diabetes, leukaemia or immunosuppressive therapy. Among the above mentioned, liver diseases appeared to be a strong risk factor [Citation23,Citation31–34]. Thirdly, Shewanella spp. have been found to associate with cases of nosocomial infection, leading to the outbreaks of healthcare-associated infections [Citation5,Citation24,Citation35–37]. Invasive procedures like catheterization and intubation are also an important source of infection [Citation22,Citation24,Citation38]. An outbreak of 31 cases of abdominal and biliary tract infections or bacteraemia of Shewanella, caused by the exposure to a shared measuring cup, was reported in a general surgery unit in South Korea [Citation35]. In addition, the differences of development, living styles and environmental pollution conditions result in different incidence rates of Shewanella infection. Brink et al. reported 28 cases of bacteraemia caused by S. putrefaciens in South Africa, and almost all cases were related to poor hygiene [Citation16].
Clinical features and treatment
Symptom classification
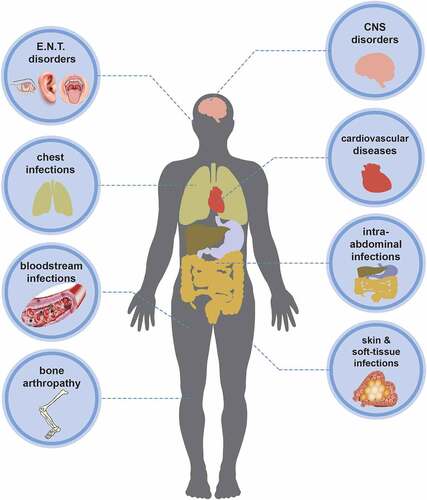
As opportunistic pathogens, Shewanella spp. can cause a wide spectrum of clinical diseases in human. Documented illnesses by Janda et al. showed that Shewanella-related syndromes can be divided into five categories, including skin and soft-tissue infections (SSTIs), invasive diseases (such as sepsis), hepatobiliary diseases (hepatocirrhosis, liver cancer, cholangitis), otitis media and associated sequelae, and other infection [Citation3]. SSTIs, including cellulitis, abscess, or necrotizing fasciitis, were considered as the most common clinical manifestation of infection. We divided Shewanella-related diseases into eight categories according to the infection site (), including ear, nose, and throat (E.N.T) disorders, central nervous system (CNS) disorders, chest infections, cardiovascular diseases, bloodstream infections (bacteraemia, septicaemia), intra-abdominal infections, bone arthropathy, skin and soft-tissue infections (SSTIs). The first four common clinical manifestations consist of bloodstream infections, SSTIs, E.N.T disorders, and intra-abdominal infections (). Certain bone or joint diseases such as arthritis [Citation13,Citation38,Citation39], osteomyelitis [Citation40–43], and discitis [Citation44] can also be caused by Shewanella. SSTIs and bloodstream infections were reported to be predominate in the 16 cases of Shewanella infection in hospitals in Taiwan, followed by biliary tract infections [Citation14]. Consistent with the conclusion of Vignier et al., bloodstream infection was the most common complication caused by Shewanella infection [Citation13]. Bacteraemia was not associated with the ear infection of Shewanella in any case published to date.
Underlying diseases
Among the patients with Shewanella infection, 70.70% of them (n = 193) had underlying diseases (), some even had multiple diseases. As Wincy et al. concluded, hepatobiliary diseases, malignancy, chronic kidney disease or end-stage renal failure, and diabetes mellitus are important underlying disease [Citation4] (). In our study, 31 patients (11.36%) had hepatobiliary, spleen and pancreatic related diseases, like hepatitis, cirrhosis, liver abscess, and gallstones. It needs to be acknowledged that, with the increase in the number of included cases, the status of E.N.T disorders in the underlying diseases cannot be ignored [Citation15]. Cancer infiltration, tumour block compression and other mechanical compression bring about neurological lesions and then result in joint diseases. Shewanella spp. are often found in bile as a part of mixed flora [Citation35,Citation45]. Biliary tract infection and cholelithiasis are mutually causal. Biliary tract obstruction caused by cholelithiasis will lead to cholestasis and bacterial reproduction. Diabetic microvascular lesions can affect the healing of foot ulcers, leaving the wound exposed and prone to bacterial infection [Citation46]. Among the patients with Shewanella infection, 30 (10.99%) were diabetics with the complication of lower limb soft tissue ulcer(). In addition to malignant tumours, kidney disease, diabetes and hepatobiliary diseases, respiratory tract-related diseases like pneumonia, tuberculosis, and chronic obstructive pulmonary disease are also considered to associate with Shewanella infection. Erfanmanesh et al. reported a mixed infection of Streptococcus dolphins and S. algae in 2019. They concluded that the systemic Streptococcosis may trigger the formation of ulceration by Shewanella and highlighted the potential significance of Shewanella as a pathogen to cause pulmonary oedema and concomitant infection [Citation47].
Table 2. Summary report of the underlying diseases of Shewanella infection.
Identification
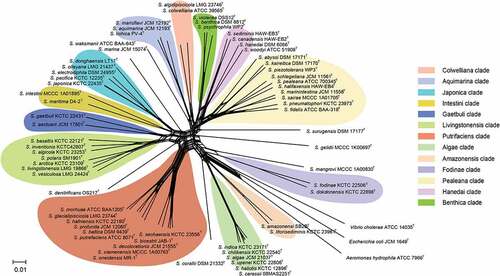
Most species in the genus of Shewanella are non-fermentable, with the same phenotypic characteristics; as a result, there are limited biochemical indexes to distinguish them at the species level. Meanwhile, existing clinical commercial analysis systems, such as API 20E, API 20 NE, Vitek 2 GN card (bioMérieux, France) and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), cannot correctly identify Shewanella at the species level [Citation7,Citation48] due to the limited information of species (like S. algae and S. putrefaciens) in databases. Our previous study, which based on the newly constructed peptide mass reference spectra database, containing 36 Shewanella species, demonstrated that Shewanella isolates can be effectively distinguished according to their different MS fingerprintings [Citation49], indicating that MALDI-TOF MS is a reliable and powerful tool for the rapid identification of Shewanella strains at the species level. Homology analysis of 16S rRNA gene is the most commonly used molecular identification method in prokaryotic systematics, known as the “molecular clock” of evolution [Citation50]. However, due to the low evolution rate and high sequence conservation of the 16S rRNA gene, it is difficult to distinguish closely related species in the Shewanella genus [Citation51,Citation52]. Although the gene gyrB has higher resolution than 16S rRNA, there are limitations to the accurate identification of Shewanella species at the species level due to the lack of a unified species cutoff value and the uneven quality of sequences submitted in public databases [Citation53–55]. Fang et al. established the method of multilocus sequence analysis (MLSA) based on several housekeeping genes [Citation56]. The type strains of fifty-nine species were used to describe the phylogenetic relationships and taxonomy of Shewanella, 12 distinct monophyletic clades were defined by using MLSA to clarify the evolutionary relationships of Shewanella spp. Among them, the Shewnaella pathogens isolated from humans are concentrated in two clades, Algae (S. algae, S. indica, S. haliotis) and Putrefaciens (S. putrefaciens, S. xiamenensis). The Shewanella spp. in the same clade are more closely related, showing <4 mol% GC variation and >84% concatenated similarity [Citation56]. Here, after updating the MLSA data of Shewanella type strains, split network tree has been constructed by using SplitsTree 4 program (version: 4.14.4) based on concatenated sequences of 66 Shewanella species type strains (). Thirteen monophyletic clades have been clarified, and the conclusion of GC variation and concatenated similarity within the same clade consistent with that of Fang et al.
Figure 3. Concatenated split network tree based on six genes loci. The gyrA, gyrB, infB, recN, rpoA, and topA gene sequences from 66 Shewanella species were concatenated and reconstructed using SplitsTree program (version: 4.14.4), based on Jukes-Cantor model.
By the end of 2021, over 300 genome sequences, which belong to 63 Shewanella species, had been collected and collated in GenBank. Genomic data revealed that the average genome size of Shewanella is 4.83Mbp and the average GC content is 46.84 mol%. The genomic sizes of marine strains were significantly correlated with water depth [Citation57]. Typically, deep-sea Shewanella species exhibited larger genomic sizes than their shallow-sea counterparts [Citation57]. As the number of genomic disclosures increases, researchers can achieve accurate identification of strains using conditions of average nucleotide identity (ANI) >95% and digital DNA – DNA hybridization (dDDH) >70% [Citation58]. Thorell et al. quantitatively assessed the genomic diversity within Shewanella using dDDH, and reconstructed the phylogenomic associations among Shewanella strains. The resulting phylogenetic reconstruction revealed two major species cluster show a big difference in G+C content [Citation59]. At the same time, it was emphasized that as new whole genome sequences from Shewanella type strains become available, the complexity of the taxonomic relationships within the genus Shewanella will be clearly realized.
Treatment
Shewanella-related infections can involve multiple parts of the human body. Based on the case reports, we summarized the treatment regimen and the resistance of the strains (). Most cases recover completely with treatment. However, there was no guideline of the antimicrobial therapy for Shewanella spp. infections in humans. Due to the existence of the patient’s underlying disease and the complexity of the disease spectrum, it cannot be clearly summarized what type of diseases were treated with particular antibiotic. It is certain that empirical therapy was initiated with ceftazidime, meropenem, and vancomycin et al. broad-spectrum coverage antibiotics, which could effectively control the growth of most pathogens () [Citation69]. Piperacillin-tazobactam was used in the initial empiric therapy for sepsis [Citation60]. Usually, targeted treatment is according to the results of in vitro antimicrobial susceptibility testing on isolates. In the majority of cases, β-lactams, aminoglycosides, and quinolones were given to patients, which mostly had effective results. Moreover, there were great differences in drug resistance among different Shewanella strains. The frequent emergence of multidrug resistant (MDR) strains under the selective pressure of antibiotics makes the treatment more difficult, and clinicians need to be more cautious in antibiotic selection.
Table 3. The treatment regimen and drug resistance of strains summerized in the case reports.
Pathogenicity
Morphological, physiological, and biochemical characteristics
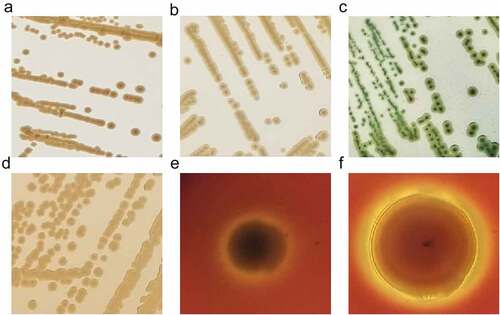
There is no uniform biochemical or physiological profile in the genus of Shewanella [Citation2]. As marine microorganisms, Shewanella spp. were originally isolated with marine agar 2216E, forming round, smooth, medium-sized, and raised colonies [Citation56]. On nutrient agar, the colonies are light brown, round and raised with intact margins. Selective media are also used for the initial isolation of Shewanella strains, such as thiosulphate citrate bile salts (TCBS) sucrose agar, MacConkey agar, and blood plates. Colonies on TCBS are medium-sized, smooth, raised, rounded, with colourless edges and black centres. On MacConkey agar, 1–2 mm yellow-brown colonies can be seen, while on sheep’s blood agar, some Shewanella species such as S. algae and S. putrefaciens can produce conspicuous β-haemolysis, and some strains can also form mucoid colonies. As researchers studied that β-haemolysis was detected in all S. algae strains but only in a couple of S. putrefaciens isolates [Citation111,Citation112] ().
Figure 4. Colony morphology of Shewanella strains on different plates. Figure 4a, 4b, 4c and d showed the colonies on marine agar 2216, nutrient agar, TCBS and MacConkey agar, respectively, after incubating at 37 °C for 18–24 h. Figure e and f represent the growth of S. algae JCM 21037T and S. putrefaciens ATCC 8071T on sheep blood agar, after incubating at 37 °C for 72 h, respectively.
Derby and Hammer described the following general biochemical features of Shewanella spp.: Gram-negative, indole-negative, and gelatinase-, sucrose-, and maltose-positive [Citation113]. This genus is positive for catalase and can reduce nitrate (NO3−) to nitrite (NO2−). Members of this genus are aerobic or facultative anaerobic, and some species can ferment D-glucose and N-acetylglucosamine to produce acid, usually without gas. Most of the Shewanella strains also present H2S-producing, urea-, arginine hydrolase-, lysine decarboxylase-negative phenotypes [Citation12,Citation114,Citation115]. This article summarizes physiological and biochemical properties of seven common Shewanella spp. () [Citation12,Citation116–120].
Table 4. Summary of physiological and biochemical characteristics of seven common species of the genus Shewanella.
Potential virulence factors
Pathogens must overcome several innate host defenses to attach to and colonize intestinal epithelium. Virulence factors of pathogens include the ability to invade human intestinal epithelial cells through virulence determinates, including flagella, homoserine lactones signal molecules, gelatinases, proteases and biofilm formation, which are essential for the first stage in infectious diseases [Citation121]. Carla et al. characterized the attachment and internalization ability of S. putrefaciens by using human colonic carcinoma (Caco-2) cells, which shared morphological and functional features with normal small intestinal cells in post-confluent stage in vitro. The results showed that S. putrefaciens showed ability to attach and internalized into Caco-2 cells [Citation121]. Members of the genus Shewanella have flagella, which promote motility, adhesion and biofilm formation. The mannose-sensitive haem agglutination (MSHA), and extracellular DNA (eDNA) play important roles in microbial attachment [Citation122,Citation123]. β-haemolysins has been found to involve in haemolytic activity, which is responsible for making S. algae more virulent [Citation111]. Also, quorum sensing (QS), a cell-to-cell communication process in bacteria, has been studied in different Shewanella spp. [Citation124]. In Gram-negative bacteria, the LuxI/R sensing acylhomoserine lactone (AHLs) was the most well-studied QS system [Citation125]. S. putrefaciens were also able to produce AHL molecules, which could coordinate the expression of multiple virulence factors, such as biofilm development [Citation121]. The homologous gene of LuxR has been predicted in the genomes of S. baltica, and the loss of this gene relates to the failure to produce AHLs. However, S. baltica can sense AHLs produced by other bacteria to assist QS-mediated cellular behaviours through LuxR receptor system [Citation126]. Chitinase, lipase, protease, elastin, and alkyl sulphate enzymes produced by certain species of Shewanella may also associate with damage host tissues, being responsible for disruption and depletion of the mucus barrier. The genus Shewanella also has O-side chains or capsules with thickness varying from 20 to 130 nm, depending on different species [Citation38]. The polysaccharide polymers on the surface of Shewanella are beneficial for its adhesion to solid substances and thus its infectivity [Citation127]. In S. oneidensis, the biofilm facilitates microbial aggregation and then matures from a monolayer state to a three-dimensional structure, mediating by mxdABCD gene cluster and biofilm-promoting factor A (BpfA) [Citation128,Citation129]. Moreover, for tetrodotoxin (TTX), there are evidences that it associates with the pathogenicity of Shewanella [Citation130].
Multiple virulence-specific genes, such as capsular polysaccharide biosynthesis, O‑antigen and lasB (vibriolysin related gene) et al. have been identified through genomes scan [Citation131]. In the genomes of S. algae, some putative genes, homologous to hlyA, hlyB, hlyD, and tolC genes encoding haemolysin operon, have been identified. RTX pore-forming toxin α-haemolysin, as the product encoded by hlyA, could change membrane permeability and lead to cell lysis [Citation132]. The genes related to type IV pili, which are regulated by external stimuli for directed movement, and the gene vasF related to VAS T6SS, provide a fresh perspective on the pathogenicity of S. algae. Almost all strains of S. algae contain katA gene, the product of which can decompose hydrogen peroxide, protect cells from the toxic effects of hydrogen peroxide, and enhance the bacterial colonization ability in hosts. Also, the identification of genomic islands (GIs) harbouring a suite of virulence genes and mobile elements implied that the GIs may help cross-species gene transfer and contribute to the independent acquisitions of virulence factors in S. algae [Citation133]. Genes related to the spoilage metabolic pathways (including trimethylamine metabolism, sulphur metabolism, putrescine metabolism, biofilm formation and serine protease production) and to illustrate the specific QS systems were identified, providing additional evidences for its metamorphic potential and pathogenicity to aquatic animals [Citation134]. The study by Tamez et al. proposed new candidate virulence factors including the Fur protein, OmpA, T6SS, type VI secretion effector Phospholipase A1, microbial collagenase, DNase, type IV pili, curli, twin-arginine translocation system, and ClpP [Citation135]. Alex A et al. detected genes encoding core components of type III secretion system (T3SS) and type VI secretion system (T6SS) gene cluster, as well as the homologs of T3SS effector molecules, which aid to penetrate host mucous barriers. All structural components of T6SS were detected [Citation136]. Virulence of pathogens depends on the activity of the T3SS injector and the effector proteins delivered to the host cell [Citation137,Citation138]. Previous studies revealed that VPA1328 and VopG are members of a larger family of T3SS2 effector proteins, linking to disruption of host cell survival and suppression of innate immunity in infected cells [Citation139–141]. The VopG-like proteins have been found in S. baltica [Citation142].
Antibiotic resistance
The antimicrobial susceptibility testing panel for aerobic Gram-negative bacilli could be used to perform the antimicrobial susceptibility testing of Shewanella by the micro-broth dilution method. Huang et al. determined the results of susceptible (S), intermediate (I), and drug resistance (R) according to the Enterobacteriaceae standards of the American Committee for Clinical Laboratory Standards Institute (CLSI) guidelines [Citation133,Citation143]. Kang et al. used a disk-diffusion technique to determine the susceptibility to several antimicrobial agents [Citation144]. The diameter of the zone of inhibition around each disk was measured and the interpretation of results was acquired according to the guidelines of the CLSI. It should be emphasized that there are no unambiguous criteria for interpretation of antibiotic resistance of Shewanella spp., therefore it is impossible to unequivocally compare the results obtained by different authors. Although the drug susceptibility of the genus Shewanella varies among different species, these bacteria were usually susceptible to the third- and fourth-generation cephalosporins, carbapenems, β-lactamase inhibitor combinations, aminoglycosides, chloramphenicol, erythromycin, aztreonam, and quinolones [Citation3,Citation13]. This is consistent with our findings from the summary of case reports (). Shewanella algae and S. putrefaciens are mostly susceptible to aminoglycosides, carbapenems, erythromycin, and quinolones and usually resistant to penicillin [Citation3]. Their susceptibility to ampicillin and cephalosporins is variable [Citation12]. It has been shown that the strains of S. algae strains harbouring eptA gene were colistin resistant [Citation15,Citation18,Citation145,Citation146], while those of S. putrefaciens were colistin sensitive [Citation111]. Antibiotic resistance is on the rise in the genus of Shewanella. Zhao et al. investigated multidrug resistance in S. xiamenensis isolated from an estuarine water sample in China [Citation147]. The results showed that the strain displayed resistance to ampicillin, aztreonam, cefepime, cefotaxime, chloramphenicol, ciprofloxacin, erythromycin, kanamycin and trimethoprim-sulfamethoxazole. As an important Shewanella pathogenic species, multi-drug resistant S. xiamenensis strains in the aquatic environment may become important reservoirs for resistance genes. Once infected in humans, it may lead to clinical treatment failure.
Bacteria of this genus are possible progenitors of many antibiotic resistance genes. The OXA-54 oxacillinase detected in S. oneidensis MR-1 was the progenitor of OXA-48 identified in Klebsiella pneumoniae [Citation148]. S. xiamenensis was the progenitor of blaOXA-181 found in K. pneumonia [Citation149]. S. algae was considered to be the origin of plasmid-mediated quinolone resistance determinant qnrA [Citation150]. It was also suggested that the genus Shewanella is a reservoir for the MCR-4 mobile colistin resistance genes [Citation151].
In Shewanella spp., the resistant genes were detected on the chromosomal regions surrounding by mobile genetic elements (MGEs) [Citation3,Citation45,Citation152,Citation153]. Numerous multidrug-resistant (MDR) and extensively drug-resistant (XDR) Shewanella strains have been reported in recent years [Citation18,Citation147,Citation152]. Most of the blaOXA-55 genes and their variants detected in S. algae are located on the chromosomes, indicating intrinsic resistance to β-lactams. However, the blaOXA-55 gene identified in the MDR S. algae strain MARS 14 is close to a MGEs (transposon) encoding gene, suggesting S. algae may be a possible reservoir of this gene [Citation18]. In the MDR S. xiamenensis strains, MGEs (transposon) or plasmid have been detected, containing a variety of resistance genes and conferring resistance to trimethoprim, aminoglycosides, quaternary ammonium compounds, β-lactams, chloramphenicol, sulphonamides, and tetracycline [Citation152]. Not all Shewanella strains carrying β-lactam resistance genes develop a corresponding phenotype [Citation154]. Jianhua Yin et al. discovered that ampicillin with high concentrations (>12.5 µg/ml) could induce the blaA gene, while ampicillin with low concentrations (<5 µg/ml) could not [Citation155].
Outstanding questions
First of all, the diagnostic ability of Shewanella-related infections needs to be strengthened. Shewanella-related infections were easily overlooked in clinical sets. Physicians should take care to diagnose if there exists Shewanella infection when patients had a history of aquatic exposure [Citation7]. Also, clinicians need to be aware of the ability of Shewanella spp. to cause invasive disease, and to understand the clinical and epidemiological characteristics of Shewanella infections, so as to provide an earlier microbiological presumptive diagnosis and to achieve greater controllability and predictability in clinical treatment.
Secondly, methods with more rapidity and accuracy need to be established for Shewanella spp. identification. Considering the complex and variable biochemical characteristics of the existing Shewanella species as well as the discovery of new species, the traditional commercial biochemical identification systems, such as API 20E, API 20 NE, Vitek 2 GN card (bioMérieux, France), cannot correctly identify Shewanella strains at the species level. Therefore, it is necessary to take enough representative strains into account in experimental verification. MALDI-TOF MS, despite the most commonly used instrument in clinical bacterial detection, sometimes can cause inaccurate species-level identification due to its dependence on databases. The development of modern sequencing technology has effectively revised the phylogenetic relationships of bacterial species and improved the accuracy of species identification. However, the 16S rRNA gene or single housekeeping gene lacks discriminatory value for Shewanella identification at the species level. MLSA, which needs PCR of several housekeeping genes, followed by truncation and concatenation of the gene sequences, still cannot meet the requirements of clinical detection timeliness. Researchers can also achieve accurate identification of strains based on whole-genome sequence information. However, it takes a long time to obtain the genome and then to analyze it. Therefore, it is particularly important to establish rapid and accurate detection methods, such as Shewanella species-specific gene PCR. Besides, standard procedures for the isolation of Shewanella strains in food and environmental samples should also be established.
Thirdly, considering the increasing antimicrobial resistance, research on effective and judicious use of antimicrobials as well as clarification of the duration of antibiotic use in clinical treatment is needed. As an important vehicle and a reservoir of antibiotic resistance genes, the genus of Shewanella contains a variety of drug-resistant genetic elements, showing resistance to many drugs, including β-lactams, quinolones, aminoglycosides, macrolides and carbapenems. The use of broad-spectrum antibiotics provides continuous selection pressure to Shewanella strains, allowing them to evolve and become more resistant. Therefore, it is urgent to conduct drug resistance monitoring for Shewanella spp. and to analyze the resistance characteristics at the species level.
Conclusion
From a clinical and aetiological perspective, this review summarized our present knowledge of the epidemiology features, clinical manifestations and pathogenic characteristics of Shewanella infection, highlighting that some species are associated with a wide spectrum of clinical diseases. Physicians should take care to diagnose Shewanella infection when patients had a history of aquatic exposure. Studies of the clinical and epidemiological characteristics of Shewanella-induced diseases have helped to achieve greater controllability and predictability in clinical treatment. Clinicians need to be aware of the potential of Shewanella spp. to cause invasive disease and should be able to provide earlier microbiological presumptive diagnosis. With the increasing incidences of Shewanella infection and the emerging drug resistance of Shewanella strains, further research is needed on how to use antibiotics effectively and judiciously, as well as on clarification of the duration of antibiotic use.
Authors contributors
KY, ZH and YX drafted the manuscript; DW edited the manuscript and supervised the project. All authors read and approved the final version of the manuscript. All figures are original creations for this manuscript, and no additional permissions are required for inclusion into the manuscript.
Acknowledgments
We apologize to those colleagues whose papers could not becited due to space limitations.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Mukherjee M, Zaiden N, Teng A, et al. Shewanella biofilm development and engineering for environmental and bioenergy applications. Curr Opin Chem Biol. 2020;59:84–92.
- Lemaire ON, Méjean V, Iobbi-Nivol C. The Shewanella genus: ubiquitous organisms sustaining and preserving aquatic ecosystems. FEMS Microbiol Rev. 2020;44(2):155–170.
- Yousfi K, Bekal S, Usongo V, et al. Current trends of human infections and antibiotic resistance of the genus Shewanella. Eur J Clin Microbiol Infect Dis. 2017;36(8):1353–1362. DOI:10.1007/s10096-017-2962-3
- Ng WW, Shum HP, To KK, et al. Emerging infections due to Shewanella spp.: a case series of 128 cases over 10 years. Front Med (Lausanne). 2022;9:850938.
- Zong Z. Nosocomial peripancreatic infection associated with Shewanella xiamenensis. J Med Microbiol. 2011;60(9):1387–1390.
- Antonelli A, Di Palo DM, Galano A, et al. Intestinal carriage of Shewanella xiamenensis simulating carriage of OXA-48–producing Enterobacteriaceae. Diagn Microbiol Infect Dis. 2015;82(1):1–3. DOI:10.1016/j.diagmicrobio.2015.02.008
- Janda JM, Abbott SL. The genus Shewanella : from the briny depths below to human pathogen. Crit Rev Microbiol. 2014;40(4):293–312.
- Cai J, Chen H, Thompson KD, et al. Isolation and identification of Shewanella alga and its pathogenic effects on post-larvae of abalone Haliotis diversicolor supertexta. J Fish Dis. 2006;29(8):505–508. DOI:10.1111/j.1365-2761.2006.00732.x
- Gram L, Huss HH. Microbiological spoilage of fish and fish products. Int J Food Microbiol. 1996;33(1):121–137.
- Sterniša M, Bucar F, Kunert O, et al. Targeting fish spoilers Pseudomonas and Shewanella with oregano and nettle extracts. Int J Food Microbiol. 2020;328:108664.
- Prachumwat A, Wechprasit P, Srisala J, et al. Shewanella khirikhana sp. nov. – a shrimp pathogen isolated from a cultivation pond exhibiting early mortality syndrome. Microb Biotechnol. 2020;13(3):781–795. DOI:10.1111/1751-7915.13538
- Holt HM, Gahrn-Hansen B, Bruun B. Shewanella algae and Shewanella putrefaciens: clinical and microbiological characteristics. Clin Microbiol Infect. 2005;11(5):347–352.
- Vignier N, Barreau M, Olive C, et al. Human infection with Shewanella putrefaciens and S. algae: report of 16 cases in martinique and review of the literature. Am J Trop Med Hyg. 2013;89(1):151–156. DOI:10.4269/ajtmh.13-0055
- Chen YS, Liu YC, Yen MY, et al. Skin and soft-tissue manifestations of Shewanella putrefaciens Infection. Clin Infect Dis. 1997;25(2):225–229. DOI:10.1086/514537
- Holt HM, Søgaard P, Gahrn-Hansen B. Ear infections with Shewanella alga: a bacteriologic, clinical and epidemiologic study of 67 cases. Clin Microbiol Infect. 1997;3(3):329–334.
- Brink AJ, van Straten A, van Rensburg AJ. Shewanella (Pseudomonas) putrefaciens bacteremia. Clin Infect Dis. 1995;20(5):1327–1332.
- Rajchgot J, Glicksman R, Bogoch II. Shewanella algae bacteremia from a foot ulcer exposed to seawater during a Caribbean vacation. J Travel Med. 2016;23(3):taw014.
- Cimmino T, Olaitan AO, Rolain JM. Whole genome sequence to decipher the resistome of Shewanella algae, a multidrug-resistant bacterium responsible for pneumonia, Marseille, France. Expert Rev Anti Infect Ther. 2016;14(2):269–275.
- Gram L, Bundvad A, Melchiorsen J, et al. Occurrence of shewanella algae in danish coastal water and effects of water temperature and culture conditions on its survival. Appl Environ Microbiol. 1999;65(9):3896–3900. DOI:10.1128/AEM.65.9.3896-3900.1999
- Bulat O, Bulat C, Blaj M, et al. A rare colonization in peritoneum after blunt abdominal trauma: s. putrefaciens and S. cerevisiae. Balkan Med J. 2018;35(4):333–335. DOI:10.4274/balkanmedj.2017.0773
- López Aperador C, Benitez-Parodi E B, Díaz N, et al. Peritonitis por Shewanella putrefaciens: a propósito de un caso. Nefrología. 2016;36(4):444–445. DOI:10.1016/j.nefro.2016.01.005
- Yim SY, Kang YS, Cha DR, et al. Fatal PD Peritonitis, Necrotizing Fasciitis, and Bacteremia Due to Shewanella Putrefaciens. Perit Dial Int. 2010;30(6):667–669. DOI:10.3747/pdi.2010.00084
- Ahmed N, Casey K, Liu E, et al. Necrotizing Fasciitis of the Lower Extremity Caused by Shewanella algae. Surg Infect (Larchmt). 2013;14(1):165–166. DOI:10.1089/sur.2012.068
- Bhandari S, Pan TL, Horvath J, et al. CAPD, swimming in Shewanella. Nephrol Dial Transplant. 2000;15(9):1484–1485. DOI:10.1093/ndt/15.9.1484
- Laudat P, Audurier A, Loulergue F, et al. Pseudomonas putrefaciens meningitis. J Infect. 1983;7(3):281–283. DOI:10.1016/S0163-4453(83)97412-1
- Tsai MS, You HL, Tang YF, et al. Shewanella soft tissue infection: case report and literature review. Int J Infect Dis. 2008;12(6):e119–124. DOI:10.1016/j.ijid.2008.03.020
- Bryant T, Ellenwood S, Butters O, et al. An uncommon cause of soft tissue and knee infection after penetrating injury in a non-immunocompromised adolescent male. SAGE Open Med Case Rep. 2021;9:2050313x211034683.
- Duan M, Wang D, Wang J, et al. A case report of intracranial infection caused by Shewanella putrefaciens. Neurol Sci. 2015;36(4):625–629. DOI:10.1007/s10072-014-1956-5
- Wang IK, Lee MH, Chen YM, et al. Polymicrobial bacteremia caused by Escherichia coli, Edwardsiella tarda, and Shewanella putrefaciens. Chang Gung Med J. 2004;27(9):701–705.
- Leong J, Mirkazemi M, Kimble F. Shewanella putrefaciens hand infection. Aust NZ J Surg. 2000;70(11):816–817.
- Fernández-Fernández E, Martín-Rodríguez AJ, Hernández M, et al. First clinical isolation report of Shewanella algae from the stools of a patient with acute enteritis in Spain. Rev Esp Quimioter. 2018;31(2):160–163.
- Poovorawan K, Chatsuwan T, Lakananurak N, et al. Shewanella haliotis associated with severe soft tissue infection, Thailand, 2012. Emerg Infect Dis. 2013;19(6):1019–1021. DOI:10.3201/eid1906.121607
- Schmidt U, Kapila R, Kaminski Z, et al. Pseudomonas putrefaciens as a cause of septicemia in humans. J Clin Microbiol. 1979;10(3):385–387. DOI:10.1128/jcm.10.3.385-387.1979
- Otsuka T, Noda T, Noguchi A, et al. Shewanella infection in decompensated liver disease: a septic case. J Gastroenterol. 2007;42:87–90.
- Oh HS, Kum KA, Kim EC, et al. Outbreak of Shewanella algae and Shewanella putrefaciens infections caused by a shared measuring cup in a general surgery unit in Korea. Infect Control Hosp Epidemiol. 2008;29:742–748.
- Shrishrimal K. Recurrent Ochrobactrum anthropi and Shewanella putrefaciens bloodstream infection complicating hemodialysis. Hemodial Int. 2012;16:113–115.
- Lee WS, Ou TY, Chen FL, et al. Shewanella putrefaciens bacteremia in a uremic patient receiving hemodialysis. J Microbiol Immunol Infect. 2016;49:159–160.
- Martín-Rodríguez AJ, Martín-Pujol O, Artiles-Campelo F, et al. Shewanella spp. infections in Gran canaria, Spain: retrospective analysis of 31 cases and a literature review. JMM Case Rep. 2017;4:e005131.
- Levy PY, Tessier JL. Arthritis due to Shewanella putrefaciens. Clin Infect Dis. 1998;26:536.
- Guinetti-Ortiz K, Bocanegra-Jesús A, Gómez de la Torre-Del Carpio A. Osteomyelitis due to Shewanella putrefaciens: case report and literature review. Medwave. 2016;16:e6642.
- Carlson RM, Dux K. Shewanella putrefaciens, a rare cause of osteomyelitis. Int J Low Extrem Wounds. 2013;12:231–233.
- Shimizu T, Matsumura Y. A case of bacteremia and suppurative vertebral osteomyelitis/discitis due to Shewanella algae occurring after raw-fish consumption. Kansenshogaku Zasshi. 2009;83:553–556.
- Botelho-Nevers E, Gouriet F, Rovery C, et al. First case of osteomyelitis due to Shewanella algae. J Clin Microbiol. 2005;43:5388–5390.
- Gressier M, Mbayo D, Deramond H, et al. First case of human spondylodiscitis due to Shewanella algae. Int J Infect Dis. 2010;14(3):e261–264. DOI:10.1016/j.ijid.2009.11.007
- Jousset AB, Dabos L, Bonnin RA, et al. CTX-M-15-Producing Shewanella species clinical isolate expressing OXA-535, a chromosome-encoded OXA-48 variant, putative progenitor of the Plasmid-Encoded OXA-436. Antimicrob Agents Chemother. 2018;62(1):e01879–17.
- Simonsen JR, Järvinen A, Hietala K, et al. Bacterial infections as novel risk factors of severe diabetic retinopathy in individuals with type 1 diabetes. Br J Ophthalmol. 2021;105:1104–1110.
- Erfanmanesh A, Beikzadeh B, Aziz Mohseni F, et al. Ulcerative dermatitis in barramundi due to coinfection with Streptococcus iniae and Shewanella algae. Dis Aquat Organ. 2019;134:89–97.
- Byun JH, Park H, Kim S. The phantom menace for patients with hepatobiliary diseases: Shewanella haliotis, often misidentified as Shewanella algae in biochemical tests and MALDI-TOF analysis. Jpn J Infect Dis. 2017;70:177–180.
- Yu K, Huang Z, Li Y, et al. Establishment and application of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for detection of Shewanella genus. Front Microbiol. 2021;12:625821.
- Yarza P, Yilmaz P, Pruesse E, et al. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol. 2014;12:635–645.
- Glaeser SP, Kämpfer P. Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst Appl Microbiol. 2015;38:237–245.
- Kim M, Oh HS, Park SC, et al. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351.
- Sung HR, Yoon JH, Ghim SY. Shewanella dokdonensis sp. nov., isolated from seawater. Int J Syst Evol Microbiol. 2012;62:1636–1643.
- Bozal N, Montes MJ, Tudela E, et al. Shewanella frigidimarina and Shewanella livingstonensis sp. nov. isolated from Antarctic coastal areas. Int J Syst Evol Microbiol. 2002;52:195–205.
- Miyazaki M, Nogi Y, Usami R, et al. Shewanella surugensis sp. nov., Shewanella kaireitica sp. nov. And Shewanella abyssi sp. nov., isolated from deep-sea sediments of Suruga Bay, Japan. Int J Syst Evol Microbiol. 2006;56:1607–1613.
- Fang Y, Wang Y, Liu Z, et al. Multilocus sequence analysis, a rapid and accurate tool for taxonomic classification, evolutionary relationship determination, and population biology studies of the genus Shewanella. Appl Environ Microbiol. 2019;85. DOI:10.1128/AEM.03126-18.
- Tang X, Yu L, Yi Y, et al. Phylogenomic analysis reveals a two-stage process of the evolutionary transition of Shewanella from the upper ocean to the hadal zone. Environ Microbiol. 2021;23:744–756.
- Sentausa E, Fournier PE. Advantages and limitations of genomics in prokaryotic taxonomy. Clin Microbiol Infect. 2013;19:790–795.
- Thorell K, Meier-Kolthoff JP, Sjöling Å, et al. Whole-Genome sequencing redefines Shewanella taxonomy. Front Microbiol. 2019;10:1861.
- Yan Y, Chai X, Chen Y, et al. The fulminating course of infection caused by Shewanella algae: a case report. Infect Drug Resist. 2022;15:1645–1650.
- Song JE, Kim S, Kang HK, et al. A case of bacterial keratitis caused by multi-drug-resistant Shewanella algae without marine exposure. Oxf Med Case Reports. 2021; omab131.
- Benaissa E, Abassor T, Oucharqui S, et al. Shewanella putrefaciens: a cause of bacteremia not to neglect. Idcases. 2021;26:e01294.
- Weiss TJ, Barranco-Trabi JJ, Brown A, et al. Case report: Shewanella algae pneumonia and bacteremia in an elderly male living at a long-term care facility. Am J Trop Med Hyg. 2021;106:60–61.
- Ojha N, Walsh K, Hess MJ. Non-necrotizing bullous cellulitis and bacteremia: a rare presentation of the Shewanella algae infection. Cureus. 2021;13:e12776.
- Rathish B, Mohammed SM, Ullal K, et al. Tropical aquatic skin and soft tissue infections: a series of three cases. Cureus. 2021;13:e13170.
- Patel A, Ascha M, Punjabi A, et al. Pyogenic flexor tenosynovitis caused by Shewanella putrefaciens. Cureus. 2020;12:e8113.
- Hong J, Steen C, Wong E, et al. Shewanella: an important, emerging and lethal pathogen in a patient with recurrent presentations of cholangitis. BMJ Case Rep. 2020;13:e237655.
- Ding CH, Wahab AA, Muttaqillah NAS, et al. Cellulitis due to Shewanella algae: crucial diagnostic clues from basic microbiological tests. Trop Biomed. 2019;36:883–887.
- Zhang F, Fang Y, Pang F, et al. Rare Shewanella spp. associated with pulmonary and bloodstream infections of cancer patients, China: a case report. BMC Infect Dis. 2018;18:454.
- Ignak S, Unay Demirel O, Soydan S, et al. Shewanella algae in a chronic suppurative otitis media patient with cholesteatoma. Drug Discov Ther. 2018;12:108–110.
- Dretler AW, Jacob JT, Rouphael NG. Swimming with the Pigs: a Case of Severe Soft Tissue Infection during a Caribbean Vacation. Case Rep Infect Dis. 2018;2018:4092609.
- Brulliard C, Traversier N, Allyn J, et al. Case report: disseminated Shewanella algae infection with meningoencephalitis in a traveler secondary to marine injury in madagascar. Am J Trop Med Hyg. 2017;97:1043–1044.
- Giroux PA, Sinna R, Mercut R, et al. Shewanella putrefaciens necrotizing fasciitis of the lower limb. Med Mal Infect. 2017;47:436–438.
- Tang TH, Cheng NH, Ho RT, et al. Shewanella-Related Bacteremia and Fournier’s Gangrene: a Case Report. Open Forum Infect Dis. 2016;3:ofw148.
- Shanmuganathan M, Goh BL, Lim C, et al. Shewanella algae Peritonitis in Patients on Peritoneal Dialysis. Perit Dial Int. 2016;36:574–575.
- Fluke EC, Carayannopoulos NL, Lindsey RW. Pyogenic Flexor Tenosynovitis Caused by Shewanella algae. J Hand Surg Am. 2016;41:e203–206.
- Mohr M, Köstler J, Salzberger B, et al. Polymicrobial soft tissue infection including Shewanella putrefaciens. Infection. 2016;44:563–564.
- Charles MV, Srirangaraj S, Kali A. Neonatal sepsis caused by Shewanella algae: a case report. Australas Med J. 2015;8:64–66.
- Baruah FK, Grover RK. Case report and literature review of carbapenem resistant shewanella putrefaciens isolated from ascitic fluid. J Clin Diagn Res. 2014;8:Dd01–02.
- Ananth AL, Nassiri N, Pamoukian VN. Shewanella algae: a rare cause of necrotizing fasciitis. Surg Infect (Larchmt). 2014;15:336–338.
- Liu PY, Shi ZY, Shyu CL, et al. Cobra bite wound infection caused by Shewanella algae. Int J Infect Dis. 2014;20:11–12.
- Smith JR, Morgan M, Palmer JH. Shewanella algae infection complicating an open lower limb fracture. J Plast Reconstr Aesthet Surg. 2014;67:e99–e100.
- Lee GC, Lee JY, Kim DM, et al. A fatal Shewanella algae infection after an open tibial fracture following a tropical storm: a case report. JBJS Case Connect. 2013;3:e92.
- Wagner N, Otto L, Podda M, et al. Travel-Related chronic hemorrhagic leg ulcer infection by Shewanella algae. J Travel Med. 2013;20:262–264.
- Mohan N, Sharma S, Padhi TR, et al. Traumatic endophthalmitis caused by Shewanella putrefaciens associated with an open globe fishhook injury. Eye (Lond). 2014;28:235.
- Prinja A, Singh J, Davis N, et al. A rare cause of wound infection after an open fracture: shewanella putrefaciens. BMJ Case Rep. 2013;2013: bcr2012008537-bcr2012008537. DOI:10.1136/bcr-2012-008537.
- Basir N, Yong AM, Chong VH. Shewanella putrefaciens, a rare cause of splenic abscess. J Microbiol Immunol Infect. 2012;45:151–153.
- Patel R, Abraham A, Thomas J, et al. A Rare Case of Pneumonia Caused by Shewanella putrefaciens. Case Rep Med. 2012;2012:597301.
- García-Fragoso L, García-García I, Rivera A. Shewanella algae bacteremia in a preterm newborn. Pediatr Infect Dis J. 2012;31:104–105.
- Jover-Díaz F, Gracia-Ruiz de Alda M, Cuadrado-Pastor JM, et al. Shewanella algae bacteremia after contact with seawater in an immunocompromised patient. Rev Clin Esp. 2011;211:489–490.
- Tucker C, Baroso G, Tan P. Ventilator-Associated pneumonia due to Shewanella putrefaciens. Am J Health Syst Pharm. 2010;67:1007–1009.
- Sardelic S, Karanovic J, Rubic Z, et al. Late ventriculoperitoneal shunt infection caused by Shewanella algae. Pediatr Infect Dis J. 2010;29:475–477.
- Ternavasio-de-la-Vega HG, Angel-Moreno A, Hernández-Cabrera M, et al. Skin and soft tissue infections (patera foot) in immigrants, Spain. Emerg Infect Dis. 2009;15:598–600.
- Debois J, Degreef H, Vandepitte J, et al. Pseudomonas putrefaciens as a cause of infection in humans. J Clin Pathol. 1975;28:993–996.
- Appelbaum PC, Bowen AJ. Opportunistic infection of chronic skin ulcers with Pseudomonas putrefaciens. Br J Dermatol. 1978;98:229–231.
- Heller HM, Tortora G, Burger H. Pseudomonas putrefaciens bacteremia associated with shellfish contact. Am J Med. 1990;88:85–86.
- Chen SC, Lawrence RH, Packham DR, et al. Cellulitis due to Pseudomonas putrefaciens: possible production of exotoxins. Rev Infect Dis. 1991;13:642–643.
- Domínguez H, Vogel BF, Gram L, et al. Shewanella alga bacteremia in two patients with lower leg ulcers. Clin Infect Dis. 1996;22:1036–1039.
- Yohe S, Fishbain JT, Andrews M. Shewanella putrefaciens abscess of the lower extremity. J Clin Microbiol. 1997;35:3363.
- Papanaoum K, Marshmann G, Gordon LA, et al. Concurrent infection due to Shewanella putrefaciens and Mycobacterium marinum acquired at the beach. Australas J Dermatol. 1998;39:92–95.
- Krsnik I, Arribalzaga K, Romanyk J. Shewanella alga bacteremia and associated cellulitis in a patient with multiple myeloma. Haematologia (Budap). 2002;32:79–80.
- Bulut C, Ertem GT, Gökcek C, et al. A rare cause of wound infection: shewanella putrefaciens. Scand J Infect Dis. 2004;36:692–694.
- Tan CK, Lai CC, Kuar WK, et al. Purulent pericarditis with greenish pericardial effusion caused by Shewanella algae. J Clin Microbiol. 2008;46:2817–2819.
- Saidel-Odes L, Borer A, Riesenberg K, et al. Shewanella spp. infection following treatment for upper gastrointestinal bleeding. Scand J Infect Dis. 2007;39:360–361.
- Yilmaz G, Aydin K, Bektas D, et al. Cerebellar abscess and meningitis, caused by Shewanella putrefaciens and Klebsiella pneumoniae, associated with chronic otitis media. J Med Microbiol. 2007;56:1558–1560.
- Liu MC, Gau SJ, Wu HC. Acute exudative tonsillitis caused by Shewanella algae in a healthy child. Scand J Infect Dis. 2006;38:1104–1105.
- Kim DM, Kang CI, Lee CS, et al. Treatment failure due to emergence of resistance to carbapenem during therapy for Shewanella algae bacteremia. J Clin Microbiol. 2006;44:1172–1174.
- Süzük S, Yetener V, Ergüngör F, et al. Cerebellar abscess caused by Shewanella putrefaciens. Scand J Infect Dis. 2004;36:621–622.
- Iwata M, Tateda K, Matsumoto T, et al. Primary Shewanella alga septicemia in a patient on hemodialysis. J Clin Microbiol. 1999;37:2104–2105.
- Dhawan B, Chaudhry R, Mishra BM, et al. Isolation of Shewanella putrefaciens from a rheumatic heart disease patient with infective endocarditis. J Clin Microbiol. 1998;36:2394.
- Khashe S, Janda JM. Biochemical and pathogenic properties of Shewanella alga and Shewanella putrefaciens. J Clin Microbiol. 1998;36:783–787.
- Nozue H, Hayashi T, Hashimoto Y, et al. Isolation and characterization of Shewanella alga from human clinical specimens and emendation of the description of S. alga Simidu et al., 1990, 335. Int J Syst Bacteriol. 1992;42:628–634.
- Ha D, Bw H. Bacteriology of butter. IV. Bacteriological studies on surface taint butter. Iowa Agric Exp Sta Res Bull. 1931;145:389–416.
- Pagniez H, Berche P. Opportunistic infections caused by Shewanella, new emergent bacteria. Med Mal Infect. 2005;35:186–191.
- Karpinets TV, Obraztsova AY, Wang Y, et al. Conserved synteny at the protein family level reveals genes underlying Shewanella species’ cold tolerance and predicts their novel phenotypes. Funct Integr Genomics. 2010;10:97–110.
- Verma P, Pandey PK, Gupta AK, et al. Shewanella indica sp. nov., isolated from sediment of the Arabian Sea. Int J Syst Evol Microbiol. 2011;61(9):2058–2064. DOI:10.1099/ijs.0.026310-0
- Sucharita K, Sasikala C, Park SC, et al. Shewanella chilikensis sp. nov., a moderately alkaliphilic gammaproteobacterium isolated from a lagoon. Int J Syst Evol Microbiol. 2009;59:3111–3115.
- Huang J, Sun B, Zhang X. Shewanella xiamenensis sp. nov., isolated from coastal sea sediment. Int J Syst Evol Microbiol. 2010;60:1585–1589.
- Fang Y, Wang Y, Liu Z, et al. Shewanella carassii sp. nov., isolated from surface swabs of crucian carp and faeces of a diarrhoea patient. Int J Syst Evol Microbiol. 2017;67:5284–5289.
- Yoon JH, Park S, Jung YT, et al. Shewanella seohaensis sp. nov., isolated from a tidal flat sediment. Antonie Van Leeuwenhoek. 2012;102:149–156.
- Dias C, Ribeiro M, Correia-Branco A, et al. Virulence, attachment and invasion of Caco-2 cells by multidrug-resistant bacteria isolated from wild animals. Microb Pathog. 2019;128:230–235.
- Gödeke J, Binnenkade L, Thormann KM. Transcriptome analysis of early surface-associated growth of Shewanella oneidensis MR-1. PLoS One. 2012;7:e42160.
- Thormann KM, Saville RM, Shukla S, et al. Initial Phases of biofilm formation in Shewanella oneidensis MR-1. J Bacteriol. 2004;186:8096–8104.
- Eickhoff MJ, Bassler BL, SnapShot: bacterial Quorum Sensing. Cell. 2018;174: 1328-1328.e1321. DOI:10.1016/j.cell.2018.08.003.
- Papenfort K, Bassler BL. Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Microbiol. 2016;14:576–588.
- Jie J, Yu H, Han Y, et al. Acyl-Homoserine-Lactones receptor LuxR of Shewanella baltica involved in the development of microbiota and spoilage of refrigerated shrimp. J Food Sci Technol. 2018;55:2795–2800.
- Korenevsky AA, Vinogradov E, Gorby Y, et al. Characterization of the lipopolysaccharides and capsules of Shewanella spp. Appl Environ Microbiol. 2002;68:4653–4657.
- Thormann KM, Duttler S, Saville RM, et al. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J Bacteriol. 2006;188:2681–2691.
- Zhou G, Yuan J, Gao H. Regulation of biofilm formation by BpfA, BpfD, and BpfG in Shewanella oneidensis. Front Microbiol. 2015;6:790.
- Wang D, Wang Y, Huang H, et al. Identification of tetrodotoxin-producing Shewanella spp. From feces of food poisoning patients and food samples. Gut Pathog. 2013;5:15.
- Leangapichart T, Hadjadj L, Gautret P, et al. Comparative genomics of two Shewanella xiamenensis strains isolated from a pilgrim before and during travels to the Hajj. Gut Pathog. 2021;13:9.
- Wu ZY, Ho SP, Cheng JF, et al. Whole-Genome characterization of Shewanella algae strain SYT3 isolated from seawater reveals insight into hemolysis. Future Microbiol. 2018;13:1709–1717.
- Huang Z, Yu K, Fu S, et al. Genomic analysis reveals high intra-species diversity of Shewanella algae. Microb Genom. 2022;8. DOI:10.1099/mgen.0.000786.
- Li J, Yu H, Yang X, et al. Complete genome sequence provides insights into the quorum sensing-related spoilage potential of Shewanella baltica 128 isolated from spoiled shrimp. Genomics. 2020;112:736–748.
- Tamez AM, McLaughlin RW, Li J, et al. Searching for putative virulence factors in the genomes of Shewanella indica and Shewanella algae. Arch Microbiol. 2021;203:683–692.
- Alex A, Antunes A. Whole-Genome Comparisons Among the Genus Shewanella Reveal the Enrichment of Genes Encoding Ankyrin-Repeats Containing Proteins in Sponge-Associated Bacteria. Front Microbiol. 2019;10:5.
- Lara-Tejero M, Galán JE. The Injectisome, a Complex Nanomachine for Protein Injection into Mammalian Cells. EcoSal Plus. 2019;8. DOI:10.1128/ecosalplus.ESP-0039-2018
- Galán JE, Lara-Tejero M, Marlovits TC, et al. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol. 2014;68:415–438.
- Gao X, Wan F, Mateo K, et al. Bacterial effector binding to ribosomal protein s3 subverts NF-kappaB function. PLoS Pathog. 2009;5:e1000708.
- Hemrajani C, Berger CN, Robinson KS, et al. NleH effectors interact with Bax inhibitor-1 to block apoptosis during enteropathogenic Escherichia coli infection. Proc Natl Acad Sci U S A. 2010;107:3129–3134.
- Fookes M, Schroeder GN, Langridge GC, et al. Salmonella bongori provides insights into the evolution of the Salmonellae. PLoS Pathog. 2011;7:e1002191.
- Plaza N, Urrutia IM, Garcia K, et al. Identification of a family of vibrio type III secretion system effectors that contain a conserved serine/threonine kinase domain. mSphere. 2021;6:e0059921.
- CLSIPerformance standards for antimicrobial susceptibility testing. 28th. ( CLSI supplement M100). Wayne, PA:Clinical and Laboratory Standards Institute; 2018.
- Kang CH, Shin Y, Jeon H, et al. Antibiotic resistance of Shewanella putrefaciens isolated from shellfish collected from the West Sea in Korea. Mar Pollut Bull. 2013;76:85–88.
- Telke AA, Rolain JM. Functional genomics to discover antibiotic resistance genes: the paradigm of resistance to colistin mediated by ethanolamine phosphotransferase in Shewanella algae MARS 14. Int J Antimicrob Agents. 2015;46:648–652.
- Zago V, Veschetti L, Patuzzo C, et al. Shewanella algae and Vibrio spp. strains isolated in Italian aquaculture farms are reservoirs of antibiotic resistant genes that might constitute a risk for human health. Mar Pollut Bull. 2020;154:111057.
- Zhao JY, Mu XD, Zhu YQ, et al. Identification of an integron containing the quinolone resistance gene qnrA1 in Shewanella xiamenensis. FEMS Microbiol Lett. 2015;362:fnv146.
- Poirel L, Héritier C, Nordmann P. Chromosome-Encoded ambler class D beta-lactamase of Shewanella oneidensis as a progenitor of carbapenem-hydrolyzing oxacillinase. Antimicrob Agents Chemother. 2004;48:348–351.
- Potron A, Poirel L, Nordmann P. Origin of OXA-181, an emerging carbapenem-hydrolyzing oxacillinase, as a chromosomal gene in Shewanella xiamenensis. Antimicrob Agents Chemother. 2011;55:4405–4407.
- Nordmann P, Poirel L. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J Antimicrob Chemother. 2005;56:463–469.
- Zhang H, Wei W, Huang M, et al. Definition of a family of nonmobile colistin resistance (Nmcr-1) determinants suggests aquatic reservoirs for MCR-4. Adv Sci (Weinh). 2019;6:1900038.
- Yousfi K, Touati A, Lefebvre B, et al. A novel plasmid, psx1, harboring a new tn1696 derivative from extensively drug-resistant shewanella xiamenensis encoding Oxa-416. Microb Drug Resist. 2017;23:429–436.
- Fang Y, Wang Y, Li Z, et al. Distribution and genetic characteristics of SXT/R391 integrative conjugative elements in Shewanella spp. From China. Front Microbiol. 2018;9:920.
- Ohama Y, Aoki K, Harada S, et al. Genetic environment surrounding bla(oxa-55-like) in clinical isolates of Shewanella algae clade and enhanced expression of bla(oxa-55-like) in a carbapenem-resistant isolate. mSphere. 2021;6:e0059321.
- Yin J, Sun Y, Mao Y, et al. Pbp1a/lpoa but not PBP1b/LpoB are involved in regulation of the major β-lactamase gene blaA in Shewanella oneidensis. Antimicrob Agents Chemother. 2015;59:3357–3364.