ABSTRACT
Rickettsiae include diverse Gram-negative microbial species that exhibit obligatory intracellular lifecycles between mammalian hosts and arthropod vectors. Human infections with arthropod-borne Rickettsia continue to cause significant morbidity and mortality as recent environmental changes foster the proliferation of arthropod vectors and increased exposure to humans. However, the technical difficulties in working with Rickettsia have delayed our progress in understanding the molecular mechanisms involved in rickettsial pathogenesis and disease transmission. Recent advances in developing genetic tools for Rickettsia have enabled investigators to identify virulence genes, uncover molecular functions, and characterize host responses to rickettsial determinants. Therefore, continued efforts to determine virulence genes and their biological functions will help us understand the underlying mechanisms associated with arthropod-borne rickettsioses.
Rickettsioses: arthropod-borne diseases
Organisms belonging to the genus Rickettsia are obligate intracellular microorganisms with the ability to survive in both vertebrate and arthropod hosts. Rickettsiae are Gram-negative rod-shaped (0.3–0.5 × 0.8–1 µm) bacteria with genome sizes ranging between 1.1–1.5 Mbp (69–84% coding capacity) among different rickettsial species [Citation1–3]. Rickettsiae have evolved to downsize their genome and purged many genes involved in various metabolic pathways. Thus, rickettsiae are incapable of living extracellularly and pilfer various nutrient components for their replication from the hosts [Citation4]. Based on phylogeny, clinical symptoms, and antigenic properties, rickettsiae are classified into four groups: the spotted fever group (SFG), typhus group (TG), transitional group (TRG), and ancestral group (AG) [Citation5–7]. The TG consists of two species, R. typhi (murine typhus) and R. prowazekii (epidemic typhus), that are transmitted by faeces of infected fleas and lice, respectively [Citation8,Citation9]. Members of the SFG include tick-transmitted pathogens such as R. rickettsii (Rocky Mountain spotted fever), R. conorii (Mediterranean spotted fever), and R. parkeri (mild-moderate spotted fever), among others [Citation8,Citation10,Citation11]. With recent advances in molecular detection and identification, investigators continue to identify novel SFG Rickettsia species throughout the world [Citation8,Citation12]. The TRG includes the R. felis (flea-borne spotted fever), R. australis (tick-borne Queensland tick typhus), and R. akari (mite-borne rickettsialpox) [Citation5,Citation7,Citation8]. Species belonging to AG, such as R. bellii and R. canadensis, are considered non-pathogenic and comparatively less studied than rickettsial species with proven pathogenicity in the other groups [Citation5,Citation6,Citation8].
Humans are incidental hosts for Rickettsia and do not usually contribute to rickettsial transmission and maintenance. Instead, rickettsiae are maintained in mammalian hosts (e.g. small rodents) and/or their arthropod vectors (e.g. ticks and fleas). Rickettsioses occur when infected arthropod vectors feed on humans or contaminate skin openings or mucosal surfaces with Rickettsia-infected arthropod excrement. Unlike other Rickettsia species, humans serve as a reservoir for R. prowazekii [Citation13]. The human body louse, Pediculus humanus corporis, is the principal vector but not a reservoir for R. prowazekii because infected lice die within a week as the pathogen replicates and damages the gut epithelium of the infected lice [Citation13,Citation14]. While the exact mechanisms are poorly understood, R. prowazekii can latently persist in patients and cause recrudescent Brill – Zinsser disease, potentially disseminating R. prowazekii months to years after the primary infection [Citation15,Citation16]. Most rickettsiae preferentially target vascular endothelial cells within the bloodstream, leading to local and systemic vascular injury and inflammation with tissue infiltration of leukocytes and thrombosis [Citation17,Citation18]. The clinical spectrum of rickettsial infections changes significantly from mild to severe diseases, with varying case-fatality rates [Citation12,Citation19,Citation20]. Several factors, including age, prompt diagnosis and antibiotic treatment, underlying conditions, and infecting rickettsial agents, contribute to the variability in clinical outcomes [Citation21]. Patients with rickettsioses often present non-specific clinical symptoms, including fever, rash, headache, malaise, and vomiting [Citation21]. Some patients develop single or multiple eschars (tache noire, black spot) at the tick bite site, representing a dermal necrotic lesion associated with a primary inoculation site of Rickettsia and extensive rickettsial replication [Citation22]. Patients also display maculopapular rashes as rickettsiae cause vascular damage and inflammation, leading to fluid escape into interstitial spaces [Citation22]. As the rickettsial infections progress, patients appear delirious and exhibit neurological symptoms during the end stages of diseases [Citation21]. While uncommon, extensive necrosis and gangrene of the extremities occur in severe cases of rickettsioses, requiring surgical interventions [Citation21,Citation23]. In the absence of timely intervention, infections with highly pathogenic Rickettsia species can cause life-threatening complications, such as severe vasculitis, encephalitis, sepsis, interstitial pneumonia with noncardiogenic pulmonary oedema and acute respiratory distress syndrome, and multiorgan failure [Citation21,Citation24–26].
During major wars, such as the first and second World Wars, poor hygiene conditions contributed to the onset of epidemic typhus, claiming millions of lives [Citation27]. While the reported cases of epidemic typhus remain low nowadays, several environmental and circumstantial conditions (e.g. war, poverty, and natural disasters) associated with poor hygiene can trigger a new outbreak for R. prowazekii, as recently reported in Burundi (1997), Russia (1997–1998), Rwanda (2012), and Algeria (1998) [Citation28–32]. Because of its potential to cause vector-transmittable severe illnesses, R. prowazekii has been weaponized by the Soviet Union [Citation33]. On the other hand, flea-borne Rickettsia continues to cause local outbreaks in tropical and subtropical climates around the world, including the coastal states of California, Hawaii, and Southeast Texas [Citation34–37]. In addition, recent environmental changes have promoted the expansion and invasion of several tick species with aggressive and non-specific biting behaviours, contributing to the rise of tick-borne rickettsioses in several parts of the world [Citation19]. For all rickettsioses, prompt diagnosis and early antibiotic intervention are the keys to full recovery without complications [Citation21]. However, untrained clinicians may have difficulty making differential diagnoses as non-specific febrile clinical presentations mimic other bacterial and viral infections. Molecular diagnostic tests (e.g. nucleic acid amplification) are available at reference diagnostic laboratories for accurate detection and identification of Rickettsia during the early phase of the disease [Citation21,Citation38]. However, these tests often yield variable results and are not readily available in endemic settings due to the high costs and specific skill sets required for analysis [Citation39]. On the other hand, serological tests often identify the presence of non-specific and cross-reactive antibodies after the acute phase of the disease (7–10 days post-infection) and fail to reveal the causative agent [Citation21,Citation38]. The challenging nature of proper diagnosis for rickettsioses in the acute phase has promoted empirical antibiotic treatments without any subsequent confirmation, contributing to the delay in developing diagnostic tools and persistent underappreciation of clinical significance [Citation40].
Due to their intrinsic nature to survive intracellularly and the lack of axenic growth media, rickettsiae have historically been recalcitrant to genetic manipulation. Over the past decades, several investigators have pioneered and made significant achievements in developing genetic tools for Rickettsia, identifying virulence factors, revealing the underlying molecular mechanisms, and understanding rickettsial biology and pathogenesis (). In this review, we discuss the pathogenesis of Rickettsia with a focus on virulence determinants of Rickettsia, their contributions to obligate intracellular lifecycles, their interactions with host cells, and the immune evasion strategies they utilize. We highlight these recent advances and how they have provided significant insights into understanding the unique intracellular lifecycle of Rickettsia.
Table 1. List of putative rickettsial virulence genes.
Rickettsial intracellular lifecycle and virulence determinants
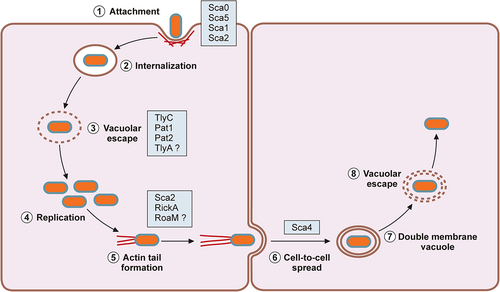
In mammalian hosts, pathogenic Rickettsia species target vascular endothelial cells and infiltrating immune cells, induce actin-mediated endocytosis, and escape into the cytoplasmic compartment (). Pathogenic Rickettsia species have evolved mechanisms to avoid intracellular detection and elimination of pathogens within the host cells. As a result, infections with pathogenic Rickettsia induce increased vascular permeability associated with rickettsial replication and disruption of vascular endothelial cells with perivascular infiltration of T cells and macrophages [Citation41]. Progressive endothelial cell injury leads to the generation of characteristic erythematous rash, disseminated vasculitis, cutaneous necrosis, pneumonitis, meningoencephalitis, and multiorgan failure [Citation13,Citation21,Citation39,Citation42]. On the other hand, previous investigations show that infection of vascular endothelial cells with Rickettsia activates a proinflammatory state and induces cytokine and chemokine responses [Citation43–47]. Thus, the molecular interactions between Rickettsia and endothelial cells have a significant role in rickettsioses. As rickettsiae continue to replicate and spread through the vasculature, perivascular neutrophilic and lymphohistiocytic inflammatory cells infiltrate into the site of infection to prevent further dissemination of the invading bacteria [Citation48,Citation49]. Recent investigations demonstrated that pathogenic Rickettsia species, such as R. rickettsii and R. conorii, have evolved to resist bactericidal mechanisms and establish a replicative niche within the cytosolic compartments of professional phagocytes, such as macrophages, suggesting that rickettsial survival in immune cells may contribute to rickettsial virulence and pathogenesis [Citation50,Citation51]. For instance, R. conorii replication in THP-1 macrophages induced unique proteome signatures and altered metabolic and lipid catabolic pathways, favouring anti-inflammatory M2 responses [Citation52,Citation53]. Here, we highlight recent investigations on the pathogenic mechanisms enabling Rickettsia to exploit and evade host immune protection mechanisms and establish an intracellular niche for their survival and transmission within mammalian hosts.
Figure 1. Rickettsial intracellular lifecycle.
Secretion Systems in Rickettsia
Bacterial surface and secreted proteins are responsible for numerous virulence mechanisms, such as sensing the environmental changes, protecting the microorganisms from stresses, adhering to and invading host cells, neutralizing innate immune components, and combating professional phagocytes. Similarly, Rickettsia utilizes diverse sets of surface-exposed proteins or secreted proteins to orchestrate its obligate intracellular lifecycles within target cells and establish a successful infection. Genome analyses identified five potential secretion systems in Rickettsia: Sec translocon, twin-arginine translocation pathway, Type I secretion system (T1SS), Type IV secretion system (T4SS), and Type V secretion system (T5SS) () [Citation54]. These analyses identified the presence of functional Sec translocon components in all Rickettsia species. Interestingly, rickettsiae lack two genes encoding LolE and LolB involved in the localization of the lipoprotein (Lol) pathway for the secretion of lipoproteins to the outer membrane [Citation54]. It is predicted that homodimer pairs of LolC and LolD make a functional unit of Lol transporters in Rickettsia. Several lines of experimental evidence suggest that Rickettsia employs a functional Sec secretion system [Citation54]. In R. rickettsii, RT-PCR analysis identified the presence of monocistronic transcript for secA gene [Citation55]. Further, heterologous expression of lepB from R. rickettsii and R. typhi and lspA from R. typhi complemented E. coli variants and confirmed signal peptidase activities [Citation56,Citation57]. While in silico secretion signal prediction tools successfully identified several Sec substrates in Rickettsia, recent studies suggest the presence of noncanonical secretion peptide sequences in Rickettsiales [Citation58].
Figure 2. Secretion systems in Rickettsia.
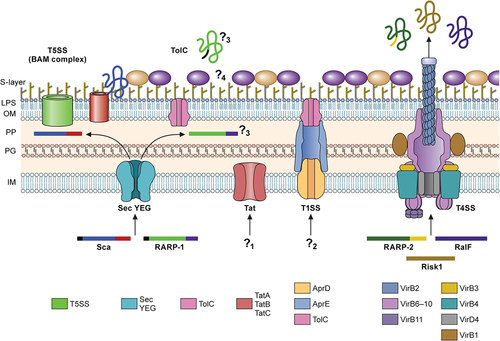
Rickettsia encodes seventeen outer membrane proteins (Omp), also known as surface cell antigens (Sca), which contain an N-terminal secretion signal for Sec-dependent secretion to the periplasmic space, followed by a passenger domain for virulence function, and a C-terminal β-barrel domain for protein translocation to outer-membrane via Type V secretion pathway (T5SS) [Citation54,Citation59–61]. Thus, the current model for the Rickettsia Sca secretion suggests that the Sec secretion machinery engages the N-terminal secretion signal of Sca proteins and translocates the polypeptides into the periplasmic space. Within the periplasmic area, the chaperone Skp and rickettsial β-barrel assembly machinery (BAM) assist the insertion of the C-terminal β-barrel domain in the outer membrane [Citation54,Citation62]. Then, conformational changes in the β-barrel domain facilitate the secretion of the passenger domain and dissociation from the BAM complex. The passenger domains of the Sca proteins are cleaved from the β-barrel domain by uncharacterized surface-associated enzymes or autocleavage reactions but remain non-covalently associated with the β-barrel domain on the surface of rickettsial cells [Citation63,Citation64]. It is interesting to postulate that the dissociated β-barrel domains may facilitate the translocation of Sca proteins lacking dedicated β-barrel domains or serve as outer-membrane channels for other molecules. On the other hand, the turnover rate of the β-barrel domains may dictate the release of the passenger domain to the extracellular spaces and facilitate the proper display of surface proteins to different environmental stimuli. However, the underlying molecular interactions between the passenger and β-barrel domains remain largely unresolved. As outer membrane proteins, Sca polypeptides constantly interact with host cytoplasmic or organelle proteins and are shown to be immunogenic, making them ideal candidates for understanding rickettsial biology and pathogenesis and developing vaccine antigens (see Attachment and Invasion).
TolC forms an outer-membrane channel in Gram-negative bacteria and functions as the exit tunnel for the Type I secretion system (T1SS) and drug efflux conduit associated with multidrug resistance. Thus, TolC plays a critical role in the virulence of Gram-negative bacterial pathogens. Current bioinformatics and experimental evidence support the conservation and presence of TolC in the outer membrane of Rickettsia [Citation5,Citation54,Citation65]. However, little is known about the molecular functions and TolC substrates in Rickettsia. A previous study suggested that Rickettsia ankyrin repeat protein 1 (RARP-1) is secreted and deposited in the cytosolic compartment of R. typhi-infected mammalian (Vero76 and HeLa) cells [Citation65]. Further analyses using a surrogate E. coli expression system determined that the secretion of recombinant RARP-1 is dependent on the Sec-TolC pathway [Citation65]. Interestingly, recent work demonstrated that, in contrast to R. typhi RARP-1, R. parkeri RARP-1 is not detected in the host cytoplasmic compartment [Citation66]. Rather, R. parkeri RARP-1 is secreted to the periplasmic space and interacts with other rickettsial factors that may mediate the secretion of other molecules or contribute to bacterial physiology [Citation66]. Ankyrin repeat proteins in Pseudomonas aeruginosa and Bdellovibrio bacteriovorus also reside within the periplasm and play various roles in maintaining bacterial physiology [Citation67,Citation68]. Given that RARP-1 is conserved across the genus, continued studies are necessary to determine the interacting partners in the periplasmic area and biological roles in the rickettsial physiology and virulence. It is also interesting to postulate that additional factors are involved in the secretion of RARP-1 in Rickettsia, contributing to different secretion profiles in R. typhi and R. parkeri.
The twin-arginine translocation (Tat) pathway in Gram-negative microorganisms is responsible for the secretion of fully folded substrates, including virulence factors, from the cytoplasmic area to the periplasmic space. Tat substrates are unique in that the N-terminal secretion signals contain a consensus “twin-arginine” motif at the distal end of the basic region [Citation69,Citation70]. Previous in silico analyses identified Tat genes (tatA, tatB, and tatC) dispersed throughout all rickettsial genomes but failed to reveal Tat substrates in Rickettsia, except PetA with a putative Tat signal peptide [Citation54]. Similar to the sequence variations in Sec secretion substrates, Tat substrates may vary substantially in Rickettsia, making it difficult to predict by the algorithms optimized for other bacterial organisms. Thus far, no substrates have been experimentally tested in Rickettsia. Future research is essential to determine the functionality of the Tat system and targeted substrates for their roles in rickettsial intracellular pathogenesis.
The T1SS is most well-characterized for a pore-forming toxin, haemolysin A (HlyA), in E. coli, illustrating the conserved and essential features of the T1SS. Additional cargos for T1SS include adhesins, iron-scavenger proteins, lipases, and proteases, with their sizes ranging from 20 kDa to 1,500 kDa, suggesting their diverse roles in bacterial pathogenesis [Citation71]. While the mechanism by which cargo molecules interact with the T1SS is ill-defined, the T1SS substrates are often defined by the presence of a GG repeat motif in the C-terminal end with varying degrees of conservation. In Rickettsia, the bioinformatic analysis identified a conserved operon encoding homolog proteins (e.g. Rc0427 – Rc0429 in R. conorii) capable of assembling into a functional T1SS: an inner membrane ATP binding cassette transporter, a periplasmic membrane fusion protein, and a multifunctional outer membrane protein of the TolC family (located in a separate location) [Citation54]. Unlike E. coli hly operon, Rickettsia does not have a gene encoding a HlyC homolog, which catalyzes post-translational acylation on two internal lysine residues of HlyA. Thus far, no known T1SS substrates have been characterized for Rickettsia. However, as recent studies characterized T1SS effector proteins in Ehrlichia and Orientia, it is highly probable that Rickettsia utilizes T1SS and TolC for the delivery of effector molecules that contribute to rickettsial pathogenesis and arthropod colonization and transmission. Additional studies are essential to characterize the T1SS substrate motif in Rickettsia and the underlying secretion mechanisms for T1SS.
The Type IV secretion system (T4SS) forms large protein complexes that traverse the cell envelope of bacterial organisms and deliver various molecules to bacterial or eukaryotic target cells, specifically for bacterial conjugation and secretion of effector molecules. The T4SS is the most versatile and divergent secretion system divided into subgroups based on genetic organization and evolution. For instance, the initial characterization of the incompatibility group of the representative conjugative plasmids subgrouped the T4SS into three major types: IncF, IncP, and IncI. Alternatively, T4SS that displays similarities to the Agrobacterium tumefaciens VirB/D4 system (IncF and IncP, comprised of 12 core proteins, VirB1 – VirB11 and VirD4) was designated as a Type IVA system, whereas genetic determinants closely related to the archetypal dot/icm (defective in organelle trafficking/intracellular multiplication) system of Legionella pneumophila was categorized as Type IVB system [Citation72]. The third group is comprised of those that display minimal homologies to IVA and IVB systems. Previous bioinformatic analyses suggest that Rickettsia encodes atypical P-T4SS (T4ASS), named as rickettsiales vir (virulence) homologs (rvh), with unprecedented gene duplication (rvhB9, rvhB8, and rvhB4) and proliferation (3–5 copies of rvhB6) dispersed throughout the genome [Citation54,Citation73]. Interestingly, the rvh T4SS encodes all known VirB/D4 components except for the VirB5, a minor component of the extracellular T pilis, corroborating current experimental evidence that Rickettsia lacks pili-like structures on the cell surface (with the exception of R. bellii and R. felis, potentially due to the presence of incomplete tra genes) [Citation54,Citation74,Citation75]. It is currently unknown how the anomalous rvh system assembles the secretion apparatus through which diverse substrate molecules translocate into host cells. The secretion of effector proteins and their biological roles during infection have been described for T4SS in Ehrlichia and Anaplasma [Citation76,Citation77]. For Rickettsia, a recent bacterial two-hybrid analysis demonstrated that RalF interacts with RvhD4, a homolog of VirD4 that recognizes target molecules and regulates substrate translocation in other P-T4SS, suggesting rvh-mediated secretion of RalF [Citation78]. The dot/icm T4BSS is known to secrete RalF comprised of an N-terminal Sec7 domain, which functions as a guanine nucleotide exchange factor of ADP-ribosylation factors (Arf), and a C-terminal Sec7-capping domain that regulates active site access to Arf [Citation79]. The L. pneumophila RalF recruits host Arf1 to the Legionella-containing vacuole (LCV) and facilitates the endoplasmic reticulum membrane fusion to LCV [Citation80]. Interestingly, RalF is pseudogenized in all SFG Rickettsia but present in TG, TRG, and AG Rickettsia with variable lengths of uncharacterized C-terminal domain with a conserved Pro-rich region and a rvh secretion signal. Experiments with in vitro tissue culture infections and ectopic expression analysis of RalF variants demonstrated that RalF is expressed early during host cell infection and modulates R. typhi invasion by activating Arf6, which subsequently recruits phosphatidylinositol 4-phosphate 5-kinase to the site of entry, enriching PI(4,5)P2 on the plasma membrane, and leading to host cytoskeleton remodelling [Citation81]. Another ankyrin repeat protein, RARP2, is recently shown to interact with RvhD4 and is predicted to be secreted by rvh T4SS [Citation82,Citation83]. RARP2 consists of a highly conserved N-terminal domain with putative cysteine protease activity, followed by a variable number of ankyrin repeats and a distinct C-terminal tail [Citation83]. Comparative genomic analysis suggests that the increasing number of ankyrin repeats is positively associated with rickettsial virulence and clear plaque formation in Rickettsia-infected tissue culture cells [Citation83]. During host cell infections, RARP2 is expressed early and shown to be deposited in the endoplasmic reticulum [Citation83]. However, the host cellular target for RARP2 is unknown. This work demonstrated that the cysteine protease activity and proper ankyrin repeats are necessary to disperse the trans-Golgi network by unknown mechanisms, potentially perturbing protein trafficking and sorting [Citation82]. Similar to other viral and bacterial infections, inhibiting protein trafficking to the plasma membrane may contribute to rickettsial pathogenesis and immune evasions by disrupting cellular barrier functions and downregulating immune recognition molecules, such as MHC-I [Citation82]. Additional genetic and functional studies are necessary to identify additional rvh substrates, determine their biological roles during the rickettsial obligate intracellular lifecycle, and define molecular functions of rvh components in forming the T4SS apparatus.
Attachment and invasion
In the absence of metabolic genes, rickettsiae must invade the host cytoplasm and acquire nutrients for survival and replication. Thus, rickettsial adherence and entrance into host cells are critical steps in initiating rickettsioses. Intracellular bacterial organisms utilize two general strategies, “zipper” or “trigger”-mechanisms, to enter host target cells [Citation1,Citation84]. The “zipper”-mechanism requires bacterial surface proteins, such as adhesins and invasins, to make direct physical contact with host receptors, initiating downstream signalling cascades for cytoskeletal rearrangement and bacterial internalization, as reported for Yersinia and Listeria species [Citation84]. On the other hand, the “trigger”-mechanism involves the injection of effector proteins by Type III and Type IV secretion systems and remodelling of host factors for the bacterial uptake, as described for Salmonella and Shigella species [Citation84]. While the rvh T4SS is present in all species of Rickettsia, additional studies are required to determine the role of rvh T4SS during the host cell invasion of Rickettsia (see Secretion Systems). Thus, it remains unknown whether Rickettsia utilizes the trigger mechanism for invasion. On the other hand, rickettsial surface proteins interact with host receptors and induce actin-mediated endocytosis (). Transmission electron microscopic analyses of non-phagocytic Vero cells interacting with R. conorii suggest that rickettsial entry is similar to L. monocytogenes and employs a “zipper”-mechanism also known as induced phagocytosis [Citation85–87].
Figure 3. Rickettsial determinants in host cell attachment and internalization.
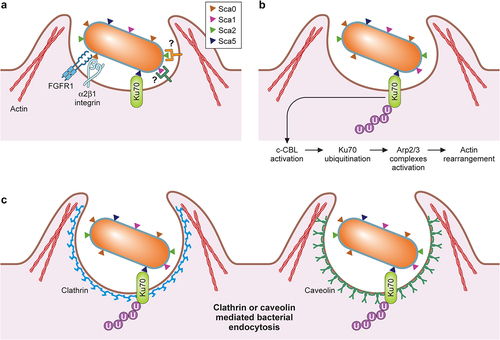
Sca5, also known as rickettsial outer membrane protein B (rOmpB), is conserved in all Rickettsia species [Citation59]. Sca5 is secreted via the Sec-T5SS pathway, abundantly expressed on the rickettsial surface, and predicted to form a paracrystalline surface layer (S-layer) [Citation54,Citation59,Citation88–90]. Previous biochemical studies identified Ku70 as a host receptor for Sca5 [Citation91]. Ku70 is predominantly expressed in the nucleus of most cells, forming a heterodimer with Ku80, which plays an essential role in the non-homologous end-joining pathway for DNA double-strand break repairs [Citation92]. In addition, Ku70 is also involved in cytosolic DNA-sensing and immune signalling pathways in various cell types [Citation93]. Interestingly, Ku70 has also been found in cholesterol-rich lipid rafts on the plasma membrane and facilitates interactions with metalloprotease-9 and fibronectin [Citation94,Citation95]. Following Sca5 interaction with Ku70, downstream signalling pathways activate Arp2/3 complexes, rearranging host actin filaments around invading Rickettsia, and induce clathrin- and caveolin-mediated endocytic pathways [Citation96,Citation97]. However, it remains unknown whether Sca5 interactions with Ku70 result in diminished levels of Ku70 and downregulation of cytoplasmic Ku70 activities for rickettsial immune evasion. The expression of recombinant Sca5 in E. coli was sufficient to increase the level of bacterial attachment and invasion into non-phagocytic tissue culture cells [Citation97–99]. Also, siRNA-based depletion of Ku70 and the pretreatment with Ku70-specific antibodies reduced R. conorii entry into Vero cells [Citation91,Citation97]. Both studies corroborate that Sca5 interactions with Ku70 mediate rickettsial uptake by host cells. Recent genetic studies also suggest that Sca5 facilitates rickettsial invasion as R. parkeri sca5 mutant displayed slower invasion kinetics than wild-type R. parkeri infecting HMEC (human microvascular endothelial cells) cells (30 minutes vs. 20 minutes for sca5 vs. wild-type) [Citation100]. Interestingly, the R. parkeri sca5 mutant ultimately reached the same invasion level in later time points, suggesting that additional rickettsial factors contribute to the attachment and invasion.
Sca0, also known as rOmpA, is another putative adhesin highly conserved in the SFG Rickettsia. It is currently postulated that Sca0, together with Sca5, constitute the rickettsial S-layer, but the exact molecular interactions between Sca0 and Sca5 for S-layer formations on Rickettsia are poorly understood [Citation101]. Initial work indicated that Sca0-specific monoclonal antibodies and purified Sca0 perturbed the R. rickettsii interactions with mouse fibroblast L929 cells [Citation102]. Furthermore, when expressed in E. coli, R. conorii Sca0 on the E. coli cellular surface increased E. coli attachment to and invasion into HeLa (human epithelial cells) and HMVEC-L (human lung microvascular endothelial cells) at a comparable level to E. coli expressing Sca5 [Citation103]. Affinity chromatography analysis with a truncated recombinant Sca0 (A.A. 954–1735) identified α2β1 integrin as a putative host receptor for Sca0 [Citation103]. Of note, the bioinformatic analysis identified an atypical conserved RNIGD motif within the recombinant Sca0, potentially involved in direct interaction with α2β1 integrin [Citation103]. Additional studies with α2β1 integrin-specific monoclonal antibodies and siRNA knockdown experiments in HMVEC-L have verified that Sca0 interacts with α2β1 integrin and mediates rickettsial attachment and invasion into endothelial cells [Citation103]. On the other hand, a recent study illustrated a potential interaction of the Sca0 β-barrel domain with fibroblast growth factor receptor-1 (FGFR1) and subsequent activation of FGFR1, which promoted caveolin-1-dependent uptake of R. rickettsii and R. conorii by endothelial cells [Citation104]. Inhibition of FGFR1 with a small molecule inhibitor (AZD4547) resulted in a substantial decrease in rickettsial burden in tissue culture endothelial cells and the lungs of R. conorii-infected C3H/HeN mice [Citation104]. In addition, siRNA knockdown of FGFR1, but not FGFR2, reduced the abundance of Rickettsia in endothelial cells, corroborating an important role for FGFR1-Sca0 interactions in the rickettsial invasion of endothelial cells [Citation104]. Interestingly, infections of endothelial cells with R. conorii significantly reduced the abundance of microRNA (miRNA)-424 and miRNA-503, leading to increased mRNA levels of FGFR1 and fibroblast growth factor 2 (FGF2), a primary ligand for FGFR1 [Citation105]. While it isn’t entirely clear how the increased levels of FGFR1 and FGF2 impact rickettsial pathogenesis, one can postulate that activated FGFR1 may facilitate the proliferation and prolonged survival of endothelial cells for Rickettsia. However, it remains unknown how the Sca0 β-barrel domain, which sits below the S-layer as an integral membrane protein, avoids steric hindrance and successfully interacts with FGFR1. It is possible that the interactions of the passenger domains of Sca0 and Sca5 with their respective host receptors induce conformational changes and allow the β-barrel domains to bind FGFR1 and other uncharacterized host receptors. Thus, continued studies are required to uncover molecular mechanisms by which Rickettsia activates FGFR1 and controls the miRNA levels in infected endothelial cells. It is important to note that another study addressed the role of Sca0 in rickettsial pathogenesis by generating a sca0 knockout by insertion of a premature stop codon in R. rickettsii using a group II intron system [Citation106]. In their study, Noriea et al. demonstrated that R. rickettsii sca0 mutant did not show any attachment or growth defects in Vero cells and displayed a similar capacity to cause spotted fever disease in guinea pigs compared to the wild-type R. rickettsii [Citation106]. Similarly, R. parkeri Sca0 was not required for invading endothelial cells [Citation100]. This data strongly argues that Rickettsia utilizes multiple redundant pathways to ensure their successful invasion into the nutrient-rich host cytoplasm ().
Other surface-exposed rickettsial proteins have been proposed to interact with host cells and facilitate rickettsial attachment and entry into the cytosolic compartment. For instance, E. coli expressing Sca1, a conserved polypeptide present in all Rickettsia species except for R. prowazekii and R. canadensis, enhanced E. coli attachment to non-phagocytic tissue culture cells but failed to facilitate E. coli invasion into the cytosol [Citation107,Citation108]. A similar experimental approach also determined that E. coli expressing Sca2, which is conserved in most SFG Rickettsia, displayed increased levels of attachment and invasion into multiple tissue culture cells [Citation109]. Corroborating these studies, preincubations of tissue culture cells with soluble recombinant passenger domains of Sca1 and Sca2 perturbed the interactions of R. conorii or E. coli expressing Sca2 with host cells, respectively [Citation108,Citation109]. However, R. rickettsii and R. parkeri without sca2 did not show significant defects in host cell invasion or growth [Citation110,Citation111]. Thus, additional genetic, biochemical, and molecular studies are necessary to determine the specific host receptors, identify involved protein domains, and reveal the subsequent molecular events facilitating rickettsial attachment and invasion into host cells. Recent advances in genetic analysis of Rickettsia permit investigators to study the biological roles of surface-exposed polypeptides, including conserved Sca proteins in rickettsial attachment and invasion into host cells, an essential step for rickettsial pathogenesis. Further, as demonstrated by Engström et al.,2019 future studies need to analyse the attachment and invasion kinetics to dissect multiple molecular mechanisms dedicated to the rickettsial invasion of host cells [Citation100].
Membranolytic factors of Rickettsia
Intracellular pathogens are primarily divided into two groups, depending on their survival strategies. The first group of intracellular pathogens have evolved diverse mechanisms to survive and replicate within vacuolar structures and subvert host cellular immunity pathways by actively modifying endocytic vacuoles and modulating host-derived membrane and nutrient trafficking. On the other hand, the second group of intracellular pathogens actively escape vacuolar structures and proliferate in nutrient-rich cytosol. Microscopic analyses of Rickettsia-infected cells suggest that Rickettsia readily exits endocytic vacuoles and resides in the cytosolic compartment. Previous bioinformatic analyses identified two different classes of rickettsial determinants that may allow rickettsial escape from endosomes: phospholipases (pat1, pat2, and pld) and haemolysins (tlyA and tlyC) [Citation54].
Many bacterial pathogens produce diverse sets of phospholipases to hydrolyse glycerophospholipids and modulate host cell membrane dynamics, signalling pathways, and cellular physiology. Previous work documented various molecular mechanisms of bacterial phospholipases involved in multiple host cellular processes. Rickettsia produces two patatin (Pat)-like proteins with phospholipase A2 (PLA2) activities (Pat1 and Pat2) and a phospholipase D (Pld) [Citation112–114]. Three genes responsible for rickettsial phospholipases share low sequence homologies and are found in different genomic locations. PLA2 enzymes catalyse the hydrolysis of the sn-2 acyl bond of membrane phospholipids, releasing free fatty acid and lysoglycero-phospholipids [Citation115]. On the other hand, Pld catalyzes the hydrolysis of the phosphodiester bond of glycerophospholipids to separate the head group from phosphatidic acid. Comparative genome sequence analysis determined that pat1 is present in all Rickettsia, but pat2 is often pseudogenized in many rickettsial species in the SFG, suggesting an ongoing reductive genome evolution process surrounding the pat2 gene [Citation114]. The vital role of PLA2 activities in rickettsial infections of tissue culture cells has been documented for various rickettsial pathogens, including R. typhi, R. prowazekii, R. rickettsii, and R. conorii [Citation116–119]. For instance, during Vero cell infections, R. typhi produced Pat1 and Pat2, deposited them on the bacterial surface, and secreted polypeptides into host cell cytosol by uncharacterized secretion pathways. Antibodies targeting Pat1 and Pat2 recognized the surface-associated molecules and delayed rickettsial escape from phagolysosome, preventing R. typhi exit into the cytosol of Vero cells [Citation114]. Interestingly, in the presence of host cell lysates, PLA2 activities of Pat1 and Pat2 increased significantly, implicating the existence of uncharacterized host cofactors altering the enzymatic kinetics [Citation114]. By characterizing R. parkeri pat1 mutant, a recent study confirmed that Pat1 plays a crucial role in facilitating R. parkeri escape from primary and secondary vacuoles and R. parkeri avoidance of autophagy killing [Citation120]. Interestingly, R. parkeri pat1 mutant did not show any growth defects in HMEC cells but generated smaller plaques than wild-type R. parkeri with diminished virulence in Ifnar1−/− Ifngr1−/− double knockout mice [Citation120].
Similar to pat1, pld is conserved in Rickettsia. The biological role of Rickettsia Pld was first described in a surrogate microorganism, Salmonella enterica serovar Typhimurium, a facultative intracellular bacterial organism that establishes replicative niche in Salmonella-containing vacuole (SCV) in host cells [Citation121]. Interestingly, introducing an expression vector that permits R. prowazekii pld expression under the R. prowazekii endogenous promoter in Salmonella allowed the pathogen to escape the SCV and relocate to the cytosolic compartment [Citation121]. Further, pretreatments of R. conorii and R. prowazekii with affinity-purified α-Pld mouse polyclonal antibodies reduced Vero cell cytotoxicity measured after seven days of infection [Citation113]. These results suggest an important role for Pld in rickettsial invasion into the cytosolic compartment. However, R. prowazekii pld mutant infecting mouse macrophage cells, RAW264.7, did not show defects in their escape into the cytosol and intracellular growth [Citation122]. It is undetermined whether the remaining phospholipases and haemolysins (described below) compensated for the loss of pld in R. prowazekii in macrophages. Regardless, R. prowazekii pld mutant failed to cause clinical diseases over the 14 days of infection in guinea pigs, suggesting that Pld is a putative virulence factor for rickettsioses [Citation122]. These studies demonstrate how important it is for Rickettsia to escape from the maturing endosomes and replicate in the cytoplasmic compartment. However, it is equally important for Rickettsia to remain in the cytosol and avoid host immune surveillance systems without breaking the integrity of the cellular membrane structures. Thus, future studies must reveal how Rickettsia regulates the cytotoxic activities of phospholipases, which may act as a double-edged sword for the rickettsial intracellular lifecycle [Citation112]. Additional experiments are also necessary to determine the biological roles of lipid byproducts generated by rickettsial phospholipases in regulating host immune signalling and vascular inflammation.
Pore-forming haemolysins serve as virulence factors for diverse human pathogens, including Escherichia coli, Listeria monocytogenes, and Staphylococcus aureus [Citation123–125]. The haemolytic activity of TG Rickettsia has been documented as early as 1948 by Clarke and Fox [Citation126]. Since then, the haemolytic principles of TG Rickettsia have been examined in various experimental conditions. These studies suggest that 1) TG Rickettsia presents slow haemolytic kinetics in lysing erythrocytes of multiple mammalian species, 2) metabolically inactive Rickettsia fails to cause haemolysis, and 3) Rickettsia may cause contact-dependent haemolysis [Citation127–129]. Recent whole-genome sequencing analyses revealed that Rickettsia has two conserved genes, tlyA and tlyC, encoding putative haemolysins. Introducing an expression vector harbouring R. typhi tlyC into two non-haemolytic microorganisms, E. coli and Proteus mirabilis, allowed the bacteria to confer haemolytic phenotypes with sheep erythrocytes [Citation130]. Similar to the experiment described for Pld, the expression of TlyC in Salmonella allowed the pathogen to escape the SCV and reside in the cytosolic compartment [Citation121]. However, the relative abundance of the bacteria in the cytosol was much higher for Salmonella expressing Pld [Citation121]. The molecular mechanisms associated with haemolysin secretion and haemolysis remain poorly defined. Plus, the significance of haemolysis for rickettsial pathogenesis is not well understood. Lastly, additional studies need to address whether rickettsial haemolysins provide synergistic activities with rickettsial phospholipases in altering host cellular membrane structures for the rickettsial intracellular lifecycle.
Intracellular immune evasion
Initially characterized as a survival strategy for nutrient-deficient cells, autophagy is a highly conserved cellular immune strategy to target and engulf invading microorganisms in double-membrane vesicles and subsequently eliminate them by fusing with lysosomes. For successful infections, intracellular pathogens have developed diverse immune escape strategies and mechanisms to avoid or exploit autophagosomes to establish intracellular niches for replication. Recent studies highlight how different Rickettsia species have evolved to interact with these complex processes for their survival and pathogenesis. For example, R. parkeri employs two protein-lysine methyltransferases (PKMT1 and PKMT2) that modify the surface proteins and prevent polyubiquitylations [Citation131]. Of the two enzymes, PKMT1 plays a significant role in the methylation of surface antigens, including Sca5. In the absence of PKMT1 or Sca5, R. parkeri is susceptible to ubiquitylation and subsequent elimination by Atg5 (autophagy-related gene 5)-dependent autophagy in mouse bone-marrow-derived macrophages. Both mutants were defective in spreading and causing diseases in mouse infection models for R. parkeri [Citation100,Citation131]. While the exact molecular mechanisms by which Sca5 prevents ubiquitylation of other surface molecules remain unresolved, the absence of Sca5 may have caused alterations to S-layers and exposed other susceptible residues in surface proteins [Citation100]. Similarly, R. parkeri mutants lacking the expression of wecA and rmlD, two genes involved in the biosynthesis of O-Ag, are susceptible to ubiquitylation, suggesting that O-Ag polysaccharides modulate the surface protein assembly and prevent ubiquitylation [Citation131,Citation132]. Homologous pkmt genes have been identified in most Rickettsia species, but low pathogenic Rickettsia species often contain a frameshift mutation within the pkmt2 gene, producing inactive PKMT2 [Citation133]. Nevertheless, previous work determined that R. typhi and R. australis undergo ubiquitination and induce Atg5-dependent autophagy for pathogenesis [Citation134,Citation135]. It remains unclear whether PKMT1 is inactive in R. typhi and R. australis. Other unknown mechanisms may also contribute to the immune subversion of these two pathogens exploiting autophagosomes. However, the interplay of autophagy with Rickettsia during infections and its impact on pathogenesis remains complex and not well understood.
Actin-based motility and rickettsial spread
Several intracellular bacterial pathogens, such as Listeria monocytogenes and Shigella flexneri, assemble actin filaments on their surface and propel themselves using actin-based motility (ABM) to avoid host immune detection and invade neighbouring cells [Citation136,Citation137]. Besides arthropod vectors infected by different Rickettsia groups, the ABM is described as one of the major differences between TG and SFG rickettsiae [Citation138,Citation139]. Two actin-polymerizing determinants, RickA and Sca2, have been characterized for ABM in Rickettsia. The rickA and sca2 genes are highly conserved in most SFG Rickettsia but missing in R. prowazekii [Citation59,Citation140,Citation141]. Another TG agent, R. typhi, lacks rickA and expresses a divergent Sca2, forming short actin tails [Citation60]. Unlike other bacterial pathogens, Rickettsia undergoes two different ABM phases regulated by unknown molecular mechanisms that alter the polar localization of each protein on the bacterial surface [Citation142]. By comparing genome sequences of virulent and avirulent strains of R. rickettsii, a recent study identified RoaM (regulator of actin-based motility) as a cytosolic factor that mediates the ABM in Rickettsia [Citation143]. However, it is unclear how RoaM interacts with other rickettsial determinants and modulates their functions in the ABM of Rickettsia. Once Rickettsia exits the endocytic vacuoles, the pathogen forms short actin tails with slow and curved motility using RickA, which is comprised of three domains: an uncharacterized N-terminal domain, a central proline-rich region with variable repeats, and a C-terminal Wiskott – Aldrich Syndrome (WASP)-like domain [Citation144]. Bioinformatics analysis failed to identify signal sequences and transmembrane domains. Thus, it remains unknown how RickA is translocated to and remains on the rickettsial surface. Biochemical actin polymerization assays determined that the C-terminal WASP-like domain is sufficient to activate the actin-related proteins-2/3 (Arp2/3) heterodimer complex, a eukaryotic actin nucleator that organizes branched actin filaments [Citation141,Citation144]. During R. parkeri infections of endothelial cells, R. parkeri utilized RickA and exhibited ABM in curved trajectories. On the other hand, RickA expression was insufficient to promote ABM for R. helvetica and R. raoultii [Citation145,Citation146]. In R. peacockii, a non-pathogenic member of the SFG, a rickettsial insertion sequence (IS) element, ISRpe1, disrupted several genes, including rickA, potentially contributing to its low virulence [Citation147]. Thus, the virulence functions of RickA for spotted fever rickettsioses remain unclear and require additional studies.
Following a replication phase with infrequent actin filament formation, SFG Rickettsia forms Arp2/3-independent long actin tails that provide fast motility and straight trajectories for intra- and inter-cellular spreads. Comparative genetic and insertional mutational analyses determined that Sca2 is responsible for the late-stage ABM of SFG Rickettsia. Sca2 is a rickettsial formin-like protein secreted via Sec-T5SS and harbours multiple noncanonical WASP-homology 2 (WH2) domains connected by folded structures of 45–55 amino acids in the passenger domain [Citation110,Citation148]. The WH2 domains are flanked by two proline-rich domains predicted to interact with the actin monomer binding protein called profilin. Interestingly, the N- and C-terminal repeat regions of Sca2 mediate intramolecular interactions and are predicted to form a circular shape to recruit two actin subunits for nucleation [Citation148]. Similar to eukaryotic formins, recombinant Sca2 passenger domain interacts with the fast-growing barbed ends of actin filaments and competes with capping protein to facilitate dose-dependent actin nucleation activity in the presence of profilin. Thus, Sca2 displays formin-like activities on the rickettsial surface and contributes to the ABM of SFG Rickettsia. R. rickettsii and R. parkeri sca2 transposon insertional mutants successfully evaded autophagic processes and displayed no growth defects in tissue culture cells. However, the mutants formed small plaques with minimal spread to adjacent cells and exhibited attenuated virulence in the animal infection models of spotted fever, suggesting that Sca2 is a virulence factor for Rickettsia [Citation110,Citation111]. In addition, R. helvetica, a member of the SFG Rickettsia predicted to be responsible for mild cases of spotted fever rickettsioses, possesses a full-length rickA, but has multiple premature stop codons in sca2 and exhibited no ABM and reduced cytopathogenesis, further emphasizing the role of Sca2 in spotted fever pathogenesis [Citation146].
While studies described above illustrate the biological significance of the ABM in rickettsial spread and pathogenesis, a recent study characterized another novel molecular mechanism that allows Rickettsia to spread to adjacent cells. Bioinformatic analysis identified that Sca4 is conserved in Rickettsia and contains two vinculin binding sites that share sequence homologies to mammalian talin-1 and S. flexneri IpaA2 [Citation149]. This molecular mimicry enabled R. rickettsii Sca4 to interact with and activate vinculin, as evidenced by co-sedimentation studies and co-crystal structures of Sca4 with vinculin [Citation149]. The Sca4 polypeptide sequence lacks the N-terminal secretion signal, but the C-terminal end of the protein was necessary for Sca4 secretion into host cells, suggesting that Sca4 may travel through T1SS or T4SS [Citation150]. Microscopic images of cells infected with R. parkeri revealed that, unlike L. monocytogenes utilizing the ActA-mediated ABM for spreading, most R. parkeri lacks actin filaments in vacuoles protruding into neighbouring cells [Citation150]. However, no significant differences were observed in the ABM between the R. parkeri wild-type and sca4 mutant [Citation150]. Instead, Sca4 is secreted into the host cytosol, inhibits the interaction between vinculin and α-catenin at the focal adhesion sites, and disturbs adherent junction complexes to reduce intercellular tension for efficient R. parkeri spread [Citation150]. Thus, R. parkeri sca4 transposon insertional mutant did not display abnormal attachment, invasion, or growth but formed small plaques in multiple cell types [Citation150]. Recent studies provide significant insights into the complex molecular interactions between rickettsial effector proteins and host target molecules involved in the cell-to-cell spread. As orchestrated exit and spread is an essential step for successful rickettsial survival and pathogenesis, it is plausible that additional factors are involved in each stage of rickettsial spread. Thus, continued genetic analysis of Rickettsia is critical to uncover novel rickettsial factors and their interactions with host target molecules to reveal underlying mechanisms that regulate rickettsial pathogenesis.
Undetermined roles of Rickettsial factors in arthropod colonization and transmission
The transmission of Rickettsia is mediated by various arthropod vectors (e.g. ticks, fleas, and lice). For successful disease transmission, pathogenic Rickettsia species must have evolved to adapt and survive in corresponding vectors, avoid immune surveillance systems, persist in key organ tissues, and utilize arthropod-derived molecules for survival strategies in diverse environments. However, the factors governing the susceptibility of any given arthropod species to Rickettsia infections are not well-defined. Comparative transcriptomic analysis of R. conorii infecting endothelial cells (HMEC) and AAE2 tick cells derived from Amblyomma americanum (lone star tick) strongly suggest that Rickettsia actively regulates different sets of genes during the infection of endothelium and tick cells [Citation151]. Further, R. rickettsii in A. aureolatum (yellow dog tick) exposed to a temperature shift or blood meal acquisition altered global gene expression profiles, including genes involved in T4SS [Citation152]. Similar observations have been documented for Borrelia burgdorferi, the causative spirochaetal agent of Lyme disease. Previous work extensively demonstrated that B. burgdorferi differentially regulates and expresses unique sets of genes for their successful infections of tick vectors and mammalian hosts [Citation153]. Thus, it is highly probable that Rickettsia secretes novel virulence determinants enabling their survival and persistent transmissions within arthropod vectors to increase their fitnesses to arthropod vector species. However, we have limited understanding of the biological roles of rickettsial determinants involved in the vertical (transovarial transmission: from adult female to offspring and transstadial transmission: from immature stages to subsequent growth stages) and horizontal transmissions (acquired during blood-feeding) between the arthropod vectors and mammalian hosts.
Molecular epidemiological studies determined that the infection rates of SFG Rickettsia species depend on the infecting Rickettsia organisms and target tick species. Recent tick surveillance studies documented that the number of ticks infected with highly virulent R. rickettsii is much lower (<1%) than those infected with Rickettsia species of mild to low virulence, such as R. parkeri or R. amblyommatis [Citation154–156]. Previous studies with artificial and in vivo tick feeding systems demonstrate that multiple factors, such as tick infection methods, Rickettsia and tick species, infection doses, and tick growth stages, contribute to the variability in rickettsial maintenance and transmission rates. For instance, A. aureolatum infected with R. rickettsii via animal feeding maintained the pathogen for multiple growth stages (larva, nymph, and adult) and generations [Citation157]. Interestingly, immature ticks did not suffer from R. rickettsii infections, but the infected ticks exhibited a reduced oviposition success rate [Citation157]. On the other hand, A. cajennense (Cayenne tick) failed to support stable R. rickettsii infections with inefficient vertical transmission rates [Citation158]. In another experiment, laboratory rearing of field-collected A. maculatum (Gulf Coast tick) determined that R. parkeri infections do not reduce tick reproduction or survival [Citation159]. By measuring multiple metrics, such as engorgement weight, nutrient conversion, egg production, and offspring viability, a recent study also determined that D. variabilis (American dog tick) and A. maculatum exhibit variable fitnesses and transmission rates for R. parkeri, R. amblyommatis, R. rickettsii, and R. montanaensis [Citation160]. Based on these studies, it is posited that SFG Rickettsia species with low virulence are best suited to colonize ticks by vertical transmission routes, whereas highly virulent SFG Rickettsia species rely on horizontal transmissions to overcome negative impacts on tick viability and reproduction.
A recent study examined ticks-to-host transmission dynamics by allowing R. rickettsii-infected D. variabilis ticks to feed on naïve guinea pigs [Citation161]. By PCR-detecting the presence of R. rickettsii at the bite site and distant tissue samples, this study determined that R. rickettsii transmission occurs as early as 30 minutes after tick biting [Citation161]. As expected, the abundance of R. rickettsii increased as ticks continued to feed on guinea pigs [Citation161]. However, it remains to be examined whether this rapid and time-dependent rickettsial transmission contributes to the disease severity in human patients. It has been posited that pathogenic Rickettsia species, including R. rickettsii, but not non-pathogenic Rickettsia species, can infect salivary glands, which permits their fast transmission along with immunosuppressive salivary molecules. For instance, R. peacockii and R. buchneri, non-pathogenic and endosymbiotic SFG Rickettsia, are known to infect ovarian tissues with limited tissue distribution in ticks [Citation162]. However, a recent study demonstrated that R. buchneri is also present in the salivary glands of I. scapularis ticks [Citation163]. Furthermore, comparative genome analyses identified multiple mutations within the coding sequences of putative virulence determinants, including RickA, Sca0, and Sca1 in R. peacockii and Sca2 and RickA in R. buchneri [Citation164,Citation165]. However, the absence of Sca2 or RickA did not alter the ability of R. parkeri to infect the midgut, salivary glands, and ovaries of A. maculatum, suggesting that Sca2 and RickA are dispensable for R. parkeri infection of tick organ tissues [Citation166].
R.felis belongs to the TRG Rickettsia and causes emerging flea-borne rickettsioses worldwide as the primary vector (Ctenocephalides felis) exhibits a broad spectrum of host range and natural habitats. While the vertical transmission of R. felis has been described, the efficiency and fitness costs have not been carefully examined [Citation167,Citation168]. Interestingly, R. felis has been identified in the salivary glands of cat fleas, inducing changes in global transcriptomic profiles of infected fleas [Citation169,Citation170]. The horizontal transmission of R. felis has also been described in laboratory-reared cat fleas fed on an artificial feeding system [Citation168]. Further, field studies determined the presence of R. felis DNA or R. felis-specific antibodies in small animals, suggesting their contributions to the horizontal transmission of R. felis [Citation171–175]. A recent study corroborates these findings by demonstrating that R. felis-infected dogs became rickettsiaemic and allowed uninfected cat fleas to become infected with R. felis [Citation176]. These studies suggest that domestic dogs, and other small animals, can serve as mammalian reservoirs for R. felis and contribute to the horizontal transmission of R. felis [Citation176]. Genome sequences of R. felis revealed the presence of putative virulence factors (e.g. Sca proteins, phospholipases, ankyrin repeat proteins, tetratricopeptide repeat proteins) that are uniquely associated with the pathogen, along with multiple copies of spoT and toxin-antitoxin systems [Citation75]. However, it remains unknown how these factors play out during the colonization of the cat fleas and transmission between the cat fleas and mammalian hosts.
The human body louse (Pediculus humanus humanus) serves as a primary vector for R. prowazekii [Citation13]. However, unlike ticks and fleas, the body louse fails to be a reservoir for R. prowazekii because the infection leads to the death of the body louse. Instead, humans and eastern flying squirrel, Glaucomys volans volans, in the United States, can be persistently infected with R. prowazekii [Citation177,Citation178]. During the R. prowazekii infection of the body louse, the pathogen targets the epithelial lining of the midgut, leading to severe disruption of the digestive tract, releasing the R. prowazekii-infected blood into the haemocoel (making the body louse turn to red) [Citation9,Citation179]. The body louse excretes R. prowazekii with the faecal contents as early as three days post-infection [Citation9]. R. prowazekii in the faeces is stable and remains infectious for several months; however, factors enabling this phenotype remain unknown [Citation179]. Thus, the transmission of epidemic typhus occurs through the contamination of the bite site or mucous membranes with louse faeces contaminated with R. prowazekii. An experimental body louse infection model largely corroborated the earlier findings and characterized the kinetics and outcomes of R. prowazekii infections in the body louse [Citation180]. Future studies with the body louse infection model may allow investigators to identify novel determinants contributing to the R. prowazekii infection of the epithelial cells of the body louse and R. prowazekii transmission to mammalian hosts.
Concluding remarks
Rickettsia causes arthropod-transmitted febrile illnesses with divergent clinical symptoms ranging from self-limited mild conditions to severe life-threatening diseases with debilitating consequences. Rickettsia continues to evolve by removing nonessential genes for survival and replication and adapting to diverse environmental conditions. With recent developments in genetic tools and genome sequencing, investigations have identified novel Rickettsia species and characterized associated arthropod vectors. Further, comparative genome sequence analyses and mutational studies have suggested the presence of core genes and virulence determinants that may exhibit essential functions in rickettsial pathogenesis and colonization of target vector species. Recent studies highlight the complex molecular interactions between multiple rickettsial effector proteins and host target molecules. On the other hand, genome sequencing determined the presence of numerous hypothetical genes with no significant homologies to other known proteins, suggesting that Rickettsia provides a unique platform to reveal previously uncharacterized molecular mechanisms for obligate intracellular pathogens. Therefore, continued investigation of the bacterial effectors and host targets that mediate rickettsial intracellular lifecycle in mammalian hosts and arthropod vectors will enhance our understanding of this crucial mechanism of virulence and reveal cellular pathways that are exploited by pathogens during infection.
Acknowledgments
The author appreciates the reviewers’ constructive criticisms and their effort in reviewing this review article and apologizes for any references that were not included due to space constraints. In addition, the authors thank the Kim laboratory members for their daily encouragement and insightful discussions. Finally, the authors express their sincere gratitude to Jorge Benach, David Thanassi, Erich Mackow, and Adrianus van der Velden for their continued support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Chan YG, Riley SP, Martinez JJ. Adherence to and invasion of host cells by spotted fever group Rickettsia Adherence to and invasion of host cells by spotted fever group Rickettsia species. Front Microbiol. 2010 December;1. DOI:10.3389/fmicb.2010.00139.
- Blanc G, Ogata H, Robert C, et al. Reductive genome evolution from the mother of Rickettsia. PLoS Genet. 2007;3(1):e14.
- Diop A, Raoult D, Fournier PE. Rickettsial genomics and the paradigm of genome reduction associated with increased virulence. Microbes Infect. 2018;20(7–8):401–409.
- Driscoll TP, Verhoeve VI, Guillotte ML, et al. Wholly rickettsia! reconstructed metabolic profile of the quintessential bacterial parasite of eukaryotic cells. MBio. 2017;8(5). DOI:10.1128/mBio.00859-17.
- Gillespie JJ, Williams K, Shukla M, et al. Rickettsia phylogenomics: unwinding the intricacies of obligate intracellular life. PLoS One. 2008;3(4):e2018. DOI:10.1371/journal.pone.0002018
- Gillespie JJ, Beier MS, Rahman MS, et al. Plasmids and Rickettsial Evolution: insight from Rickettsia felis. PLoS One. 2007;2(3):e266. DOI:10.1371/journal.pone.0000266
- Weinert LA, Werren JH, Aebi A, et al. Evolution and diversity of Rickettsiabacteria. BMC Biol. 2009;7(1):6.
- Salje J. Cells within cells: rickettsiales and the obligate intracellular bacterial lifestyle. Nat Rev Microbiol. 2021;19(6):375–390.
- Gillespie JJ, Ammerman NC, Beier-Sexton M, et al. Louse- and flea-borne rickettsioses: biological and genomic analyses. Vet Res. 2009;40(2):1–13.
- Paddock CD, Sumner JW, Comer JA, et al. Rickettsia parkeri: a newly recognized cause of spotted fever rickettsiosis in the United States. Clin Infect Dis an off Publ Infect Dis Soc Am. 2004;38(6):805–811.
- Walker DH. Rocky Mountain spotted fever: a disease in need of microbiological concern. Clin Microbiol Rev. 1989;2(3):227–240.
- Parola P, Paddock CD, Socolovschi C, et al. Update on tick-borne rickettsioses around the world: a geographic approach. Clin Microbiol Rev. 2013;26(4):657–702.
- Angelakis E, Bechah Y, Raoult D. The history of Epidemic Typhus. Am Soc Microbiol. 2016;4(4). DOI:10.1016/S0891-5520(03)00093-X
- Houhamdi L, Fournier PE, Fang R, et al. An experimental model of human body louse infection with Rickettsia prowazekii. J Infect Dis. 2002;186(11):1639–1646.
- BRILL NE. An acute infectious disease of unknown origin; a clinical study based on 221 cases. Am J Med. 1952;13(5):533–541.
- MURRAY ES, BAEHR G, SHWARTZMAN G, et al. BRILL’S DISEASE: i. Clinical and laboratory diagnosis. J Am Med Assoc. 1950;142(14):1059–1066.
- Bechah Y, Capo C, Raoult D, et al. Infection of endothelial cells with virulent Rickettsia prowazekii increases the transmigration of leukocytes. J Infect Dis. 2008;197(1):142–147.
- Otterdal K, Portillo A, Astrup E, et al. High serum CXCL10 in Rickettsia conorii infection is endothelial cell mediated subsequent to whole blood activation. Cytokine. 2016;83:269–274.
- Piotrowski M, Rymaszewska A. Expansion of tick-borne rickettsioses in the world. Microorganisms. 2020;8(12):1–28.
- Doppler JF, Newton PN. A systematic review of the untreated mortality of murine typhus. PLoS Negl Trop Dis. 2020;14(9):1–13.
- Biggs HM, Behravesh CB, Bradley KK, et al. Diagnosis and management of tickborne rickettsial diseases: rocky mountain spotted fever and other spotted fever group rickettsioses, ehrlichioses, and anaplasmosis - United States a practical guide for health care and public health professionals. MMWR Recomm Rep. 2016;65(2):1–44.
- Walker DH, Occhino C, Tringali GR, et al. Pathogenesis of rickettsial eschars: the tache noire of boutonneuse fever. Hum Pathol. 1988;19(12):1449–1454.
- Kirkland KB, Marcom PK, Sexton DJ, et al. Rocky mountain spotted fever complicated by gangrene: report of six cases and review. Clin Infect Dis. 1993;16(5):629–634.
- Kaplowitz LG, Fischer JJ, Sparling PF. Rocky Mountain spotted fever: a clinical dilemma. Curr Clin Top Infect Dis. 1981;2(89):108.
- Tran LT, Helms JL, Sierra-Hoffman M, et al. Rickettsia typhi infection presenting as severe ARDS. IDCases. 2019;18:e00645.
- Sekeyová Z, Danchenko M, Filipčík P, et al. Rickettsial infections of the central nervous system. PLoS Negl Trop Dis. 2019;13(8):1–18.
- Kelly DJ, Richards AL, Temenak J, et al. The past and present threat of Rickettsial diseases to military medicine and international public health. Clin Infect Dis. 2002;34(Suppl 4):S145–169.
- Raoult D, Woodward T, Dumler JS. The history of epidemic typhus. Infect Dis Clin North Am. 2004;18(1):127–140.
- Jensen G, Aurell M, Raoult D. Outbreak of epidemic typhus in Russia. Lancet. 1998;352(9134):1151.
- Mokrani K, Fournier PE, Dalichaouche M, et al. Reemerging threat of epidemic typhus in Algeria. J Clin Microbiol. 2004;42(8):3898–3900.
- Niang M, Brouqui P, Raoult D. Epidemic typhus imported from Algeria. Emerg Infect Dis. 1999;5(5):716–718.
- Umulisa I, Omolo J, Muldoon KA, et al. A mixed outbreak of epidemic typhus fever and trench fever in a youth rehabilitation center: risk factors for illness from a case-control study, Rwanda, 2012. Am J Trop Med Hyg. 2016;95(2):452–456.
- Azad AF. Pathogenic rickettsiae as bioterrorism agents. Clin Infect Dis. 2007;45(1):S52–5.
- Adjemian J, Parks S, McElroy K, et al. Murine typhus in Austin, TexRas, USA, 2008. Emerg Infect Dis. 2010;16(3):412–417.
- Murine typhus–Hawaii, 2002.MMWR Morb Mortal Wkly Rep. 2003:52(50):1224–1226.
- Abramowicz KF, Rood MP, Krueger L, et al. Urban focus of Rickettsia typhi and Rickettsia felis in Los Angeles, California. Vector-Borne Zoonotic Dis. 2011;11(7):979–984.
- Manea SJ, Sasaki DM, Ikeda JK, et al. Clinical and epidemiological observations regarding the 1998 Kauai murine typhus outbreak. Hawaii Med J. 2001;60(1):7–11.
- Abdad MY, Abdallah RA, Fournier P-E, et al. A Concise Review of the Epidemiology and Diagnostics of Rickettsiosis: rickettsia and Orientia spp. J Clin Microbiol. 2018;56(8):1–10.
- Robinson MT, Satjanadumrong J, Hughes T, et al. Diagnosis of spotted fever group Rickettsia infections: the Asian perspective. Epidemiol Infect. 2019;147:e286.
- Paddock CD, Holman RC, Krebs JW, et al. Assessing the magnitude of fatal Rocky mountain spotted fever in the United States: comparison of two national data sources. Am J Trop Med Hyg. 2002;67(4):349–354.
- Herrero-Herrero JI, Walker DH, Ruiz-Beltran R. Immunohistochemical evaluation of the cellular immune response to rickettsia conorii in taches noires. J Infect Dis. 1987;155(4):802–805.
- Walker DH, Gear JHS. Correlation of the distribution of Rickettsia conorii, microscopic lesions, and clinical features in South African tick bite fever. Am J Trop Med Hyg. 1985;34(2):361–371.
- Kaplanski G, Teysseire N, Farnarier C, et al. IL-6 and IL-8 production from cultured human endothelial cells stimulated by infection with Rickettsia conorii via a cell-associated IL-1α-dependent pathway. J Clin Invest. 1995;96(6):2839–2844.
- Sporn LA, Marder VJ. Interleukin-1α production during Rickettsia rickettsii infection of cultured endothelial cells: potential role in autocrine cell stimulation. Infect Immun. 1996;64(5):1609–1613.
- Damås JK, Davì G, Jensenius M, et al. Relative chemokine and adhesion molecule expression in Mediterranean spotted fever and African tick bite fever. J Infect. 2009;58(1):68–75.
- Dignat-George F, Teysseire N, Mutin M, et al. Rickettsia conorii infection enhances vascular cell adhesion molecule- 1- and intercellular adhesion molecule-1-dependent mononuclear cell adherence to endothelial cells. J Infect Dis. 1997;175(5):1142–1152.
- Rydkina E, Turpin LC, Sahni SK. Rickettsia rickettsii infection of human macrovascular and microvascular endothelial cells reveals activation of both common and cell type-specific host response mechanisms. Infect Immun. 2010;78(6):2599–2606.
- Shapiro MR, Fritz CL, Tait K, et al. Rickettsia 364D: a newly recognized cause of eschar-assodated illness in California. Clin Infect Dis. 2010;50(4):541–548.
- Denison AM, Amin BD, Nicholson WL, et al. Detection of Rickettsia rickettsii, Rickettsia parkeri, and Rickettsia akari in skin biopsy specimens using a multiplex real-time polymerase chain reaction assay. Clin Infect Dis. 2014;59(5):635–642.
- Kristof MN, Allen PE, Yutzy LD, et al. Significant growth by Rickettsia species within human macrophage-like cells is a phenotype correlated with the ability to cause disease in mammals. Pathogens. 2021;10(2):1–14.
- Curto P, Simões I, Riley SP, et al. Differences in intracellular fate of two spotted fever group Rickettsia in macrophage-like cells. Front Cell Infect Microbiol. 2016;6(JUL):1–14.
- Curto P, Santa C, Allen P, et al. A pathogen and a non-pathogen spotted fever group rickettsia trigger differential proteome signatures in macrophages. Front Cell Infect Microbiol. 2019;9(MAR):1–26.
- Allen PE, Noland RC, Martinez JJ. Rickettsia conorii survival in THP-1 macrophages involves host lipid droplet alterations and active rickettsial protein production. Cell Microbiol. 2021;23(11):1–18.
- Gillespie JJ, Kaur SJ, Sayeedur Rahman M, et al. Secretome of obligate intracellular Rickettsia. FEMS Microbiol Rev. 2015;39(1):47–80.
- Rahman MS, Simser JA, Macaluso KR, et al. Functional analysis of secA homologues from rickettsiae. Microbiology. 2005;151(2):589–596.
- Rahman MS, Simser JA, Macaluso KR, et al. Molecular and functional analysis of the lepB gene, encoding a type I signal peptidase from Rickettsia rickettsii and Rickettsia typhi. J Bacteriol. 2003;185(15):4578–4584.
- Rahman MS, Ceraul SM, Dreher-Lesnick SM, et al. The lspA gene, encoding the type II signal peptidase of Rickettsia typhi: transcriptional and functional analysis. J Bacteriol. 2007;189(2):336–341.
- Payne SH, Bonissone S, Wu S, et al. Unexpected diversity of signal peptides in prokaryotes. MBio. 2012;3(6):1–6. DOI:10.1128/mBio.00339-12
- Blanc G, Ngwamidiba M, Ogata H, et al. Molecular evolution of Rickettsia surface antigens: evidence of positive selection. Mol Biol Evol. 2005;22(10):2073–2083.
- McLeod MP, Qin X, Karpathy SE, et al. Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J Bacteriol. 2004;186(17):5842–5855.
- Sears KT, Ceraul SM, Gillespie JJ, et al. Surface proteome analysis and characterization of surface cell antigen (Sca) or autotransporter family of Rickettsia typhi. PLoS Pathog. 2012;8(8):e1002856.
- Ieva R, Tian P, Peterson JH, et al. Sequential and spatially restricted interactions of assembly factors with an autotransporter β domain. Proc Natl Acad Sci U S A. 2011;108(31). DOI:10.1073/pnas.1103827108
- Noriea NF, Clark TR, Mead D, et al. Proteolytic cleavage of the immunodominant outer membrane protein rOmpa in Rickettsia rickettsii. Schneewind O, ed. J Bacteriol. 2017; 199(6): e00826-16. DOI:10.1128/JB.00826-16.
- Hackstadt T, Messer R, Cieplak W, et al. Evidence for proteolytic cleavage of the 120-kilodalton outer membrane protein of rickettsiae: identification of an avirulent mutant deficient in processing. Infect Immun. 1992;60(1):159–165.
- Kaur SJ, Sayeedur Rahman M, Ammerman NC, et al. TolC-dependent secretion of an ankyrin repeat-containing protein of Rickettsia typhi. J Bacteriol. 2012;194(18):4920–4932.
- Sanderlin AG, Hanna RE, Lamason RL. The ankyrin repeat protein RARP-1 is a periplasmic factor that supports Rickettsia parkeri growth and host cell invasion. bioRxiv. Published online 2022:2022.02.23.481736
- Howell ML, Alsabbagh E, Ma JF, et al. AnkB, a periplasmic ankyrin-like protein in Pseudomonas aeruginosa, is required for optimal catalase B (KatB) activity and resistance to hydrogen peroxide. J Bacteriol. 2000;182(16):4545–4556.
- Lambert C, Cadby IT, Till R, et al. Ankyrin-mediated self-protection during cell invasion by the bacterial predator Bdellovibrio bacteriovorus. Nat Commun. 2015:6. DOI:10.1038/ncomms9884
- Bendtsen JD, Nielsen H, Widdick D, et al. Prediction of twin-arginine signal peptides. BMC Bioinformatics. 2005;6:1–9.
- Frain KM, van Dijl JM, Robinson C. The Twin-Arginine Pathway for Protein Secretion. EcoSal Plus. 2019;8(2):1–12.
- Spitz O, Erenburg IN, Beer T, et al. Type I Secretion Systems—One Mechanism for All? Microbiol Spectr. 2019;7(2). DOI:10.1128/microbiolspec.psib-0003-2018
- Grohmann E, Christie PJ, Waksman G, et al. Type IV secretion in Gram-negative and Gram-positive bacteria. Mol Microbiol. 2018;107(4):455–471.
- Gillespie JJ, Ammerman NC, Dreher-Lesnick SM, et al. An anomalous type IV secretion system in Rickettsia is evolutionarily conserved. PLoS One. 2009; 4(3):10.1371/journal.pone.0004833
- Ogata H, La Scola B, Audic S, et al. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2006;2(5):733–744.
- Ogata H, Robert C, Audic S, et al. Rickettsia felis, from culture to genome sequencing. Ann N Y Acad Sci. 2005;1063:26–34.
- Liu H, Bao W, Lin M, et al. Ehrlichia type IV secretion effector ECH0825 is translocated to mitochondria and curbs ROS and apoptosis by upregulating host MnSOD. Cell Microbiol. 2012;14(7):1037–1050.
- Lin M, den Dulk-Ras A, Hooykaas PJJ, et al. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell Microbiol. 2007;9(11):2644–2657.
- Rennoll-Bankert KE, Rahman MS, Gillespie JJ, et al. Which way in? The RalF Arf-GEF Orchestrates Rickettsia host cell invasion. PLoS Pathog. 2015;11(8): 1–28.
- Amor JC, Swails J, Zhu X, et al. The structure of RalF, an ADP-ribosylation factor guanine nucleotide exchange factor from Legionella pneumophila, reveals the presence of a cap over the active site. J Biol Chem. 2005;280(2):1392–1400.
- Nagai H, Kagan JC, Zhu X, et al. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science. 2002;295(5555):679–682. (80). DOI:10.1126/science.1067025
- Rennoll-Bankert KE, Rahman MS, Guillotte ML, et al. RalF-mediated activation of Arf6 controls Rickettsia typhi invasion by co-opting phosphoinositol metabolism. Infect Immun. 2016;84(12):3496–3506.
- Aistleitner K, Clark T, Dooley C, et al. Selective fragmentation of the trans-Golgi apparatus by Rickettsia rickettsii. PLoS Pathog. 2020;16(5):1–21.
- Lehman SS, Noriea NF, Aistleitner K, et al. The rickettsial ankyrin repeat protein 2 is a type IV secreted effector that associates with the endoplasmic reticulum. MBio. 2018;9(3):1–15.
- Alonso A, García-Del Portillo F. Hijacking of eukaryotic functions by intracellular bacterial pathogens. Int Microbiol off J Spanish Soc Microbiol. 2004;7(3):181–191.
- Teysseire N, Boudier JA, Raoult D. Rickettsia conorii entry into Vero cells. Infect Immun. 1995;63(1):366–374.
- Walker TS, Winkler HH. Penetration of cultured mouse fibroblasts (L cells) by Rickettsia prowazeki. Infect Immun. 1978;22(1):200–208.
- Walker TS. Rickettsial interactions with human endothelial cells in vitro: adherence and entry. Infect Immun. 1984;44(2):205–210.
- Ching W, Dasch GA, Carl M, et al. Structural Analyses of the 120‐kda serotype protein antigens of typhus group Rickettsiae: comparison with other s‐layer proteins. Ann N Y Acad Sci. 1990;590(1):334–351.
- Gilmore RD, Cieplak W, Policastro PF, et al. The 120 kilodalton outer membrane protein (rOmp B) of Rickettsia rickettsii is encoded by an unusually long open reading frame: evidence for protein processing from a large precursor. Mol Microbiol. 1991;5(10):2361–2370.
- Gilmore RD, Joste N, McDonald GA. Cloning, expression and sequence analysis of the gene encoding the 120kd surface‐exposed protein of Rickettsia rickettsii. Mol Microbiol. 1989;3(11):1579–1586.
- Martinez JJ, Seveau S, Veiga E, et al. Ku70, a component of DNA-dependent protein kinase, is a mammalian receptor for Rickettsia conorii. Cell. 2005;123(6):1013–1023.
- Koike M. Dimerization, translocation and localization of Ku70 and Ku80 proteins. J Radiat Res. 2002;43(3):223–236.
- Sui H, Hao M, Chang W, et al. The role of Ku70 as a cytosolic DNA sensor in innate immunity and beyond. Front Cell Infect Microbiol. 2021;11(October):1–18.
- Monferran S, Paupert J, Dauvillier S, et al. The membrane form of the DNA repair protein Ku interacts at the cell surface with metalloproteinase 9. Embo J. 2004;23(19):3758–3768.
- Monferran S, Muller C, Mourey L, et al. The Membrane-associated form of the DNA repair protein ku is involved in cell adhesion to fibronectin. J Mol Biol. 2004;337(3):503–511.
- Martinez JJ, Cossart P. Early signaling events involved in the entry of Rickettsia conorii into mammalian cells. Published online 2004. DOI:10.1242/jcs.01382
- Chan YGY, Cardwell MM, Hermanas TM, et al. Rickettsial outer-membrane protein B (rOmpb) mediates bacterial invasion through Ku70 in an actin, c-Cbl, clathrin and caveolin 2-dependent manner. Cell Microbiol. 2009;11(4):629–644.
- Uchiyama T. Adherence to and invasion of vero cells by recombinant Escherichia coli expressing the outer membrane protein rOmpb of Rickettsia japonica. Ann N Y Acad Sci. 2003;990:585–590.
- Uchiyama T, Kawano H, Kusuhara Y. The major outer membrane protein rOmpb of spotted fever group rickettsiae functions in the rickettsial adherence to and invasion of Vero cells. Microbes Infect. 2006;8(3):801–809.
- Engström P, Burke TP, Mitchell G, et al. Evasion of autophagy mediated by Rickettsia surface protein OmpB is critical for virulence. Nat Microbiol. 2019;4(12):2538–2551.
- Policastro PF, Hackstadt T. Differential activity of Rickettsia rickettsii ompA and ompB promoter regions in a heterologous reporter gene system. Microbiology. 1994;140(11):2941–2949.
- Li H, Walker DH. rOmpa is a critical protein for the adhesion of Rickettsia rickettsii to host cells. Microb Pathog. 1998;24(5):289–298.
- Hillman RD, Baktash YM, Martinez JJ. OmpA-mediated rickettsial adherence to and invasion of human endothelial cells is dependent upon interaction with α2β1 integrin. Cell Microbiol. 2013;15(5):727–741.
- Sahni A, Patel J, Narra HP, et al. Fibroblast growth factor receptor-1 mediates internalization of pathogenic spotted fever rickettsiae into host endothelium. PLoS One. 2017;12(8):1–16.
- Sahni A, Narra HP, Patel J, et al. MicroRNA-regulated rickettsial invasion into host endothelium via fibroblast growth factor 2 and its receptor FGFR1. Cells. 2018;7(12):1–15.
- Noriea NF, Clark TR, Hackstadt T. Targeted knockout of the Rickettsia rickettsii OmpA surface antigen does not diminish virulence in a mammalian model system. MBio. 2015;6(2). DOI:10.1128/mBio.00323-15
- Ngwamidiba M, Blanc G, Raoult D, et al. Sca 1, a previously undescribed paralog from autotransporter protein-encoding genes in Rickettsia species. BMC Microbiol. 2006;6(1):12.
- Riley SP, Goh KC, Hermanas TM, et al. The Rickettsia conorii autotransporter protein sca1 promotes adherence to nonphagocytic mammalian cells. Infect Immun. 2010;78(5):1895–1904.
- Cardwell MM, Martinez JJ. The Sca2 autotransporter protein from Rickettsia conorii is sufficient to mediate adherence to and invasion of cultured mammalian cells. Infect Immun. 2009;77(12):5272–5280.
- Kleba B, Clark TR, Lutter EI, et al. Disruption of the Rickettsia rickettsii Sca2 autotransporter inhibits actin-based motility. Infect Immun. 2010;78(5):2240–2247.
- Burke TP, Engström P, Tran CJ, et al. Interferon receptor-deficient mice are susceptible to eschar-associated rickettsiosis. Elife. 2021;10:e67029. DOI:10.7554/eLife.67029.
- Rahman MS, Ammerman NC, Sears KT, et al. Functional characterization of a phospholipase A2 homolog from Rickettsia typhi. J Bacteriol. 2010;192(13):3294–3303.
- Renesto P, Dehoux P, Gouin E, et al. Identification and characterization of a phospholipase D-superfamily gene in Rickettsiae. J Infect Dis. 2003;188(9):1276–1283.
- Rahman MS, Gillespie JJ, Kaur SJ, et al. Rickettsia typhi possesses phospholipase A2 enzymes that are involved in infection of host cells. PLoS Pathog. 2013; 9(6):10.1371/journal.ppat.1003399
- Burke JE, Dennis EA. Phospholipase A2 biochemistry. Cardiovasc Drugs Ther. 2009;23(1):49–59.
- Winkler HH, Miller ET. Phospholipase a and the interaction of Rickettsia prowazekii and mouse fibroblasts (L-929 cells). Infect Immun. 1982;38(1):109–113.
- Winkler HH, Daugherty RM. Phospholipase a activity associated with the growth of Rickettsia prowazekii in L929 cells. Infect Immun. 1989;57(1):36–40.
- Walker DH, Firth WT, Ballard JG, et al. Role of phospholipase-associated penetration mechanism in cell injury by Rickettsia rickettsii. Infect Immun. 1983;40(2):840–842.
- Walker DH, Feng HM, Popov VL. Rickettsial phospholipase a2 as a pathogenic mechanism in a model of cell injury by typhus and spotted fever group rickettsiae. Am J Trop Med Hyg. 2001;65(6):936–942.
- Borgo G, Burke T, Lo N, et al. A patatin-like phospholipase mediates Rickettsia parkeri escape from host 2 membranes. 1–66. Published online 2021.
- Whitworth T, Popov VL, Yu XJ, et al. Expression of the Rickettsia prowazekii pld or tlyC gene in Salmonella enterica serovar typhimurium mediates phagosomal escape. Infect Immun. 2005;73(10):6668–6673.
- Driskell LO, Yu XJ, Zhang L, et al. Directed mutagenesis of the Rickettsia prowazekii pld gene encoding phospholipase D. Infect Immun. 2009;77(8):3244–3248.
- Ristow LC, Welch RA. Hemolysin of uropathogenic Escherichia coli: a cloak or a dagger? Biochim Biophys Acta - Biomembr. 2016;1858(3):538–545.
- Oliveira D, Borges A, Simões M. Staphylococcus aureus toxins and their molecular activity in infectious diseases. Toxins (Basel). 2018;10(6). DOI:10.3390/toxins10060252
- Seveau S. Multifaceted activity of listeriolysin o, the cholesterol-dependent cytolysin of listeria monocytogenes. 2014;80. DOI:10.1007/978-94-017-8881-6_9.
- Clarke D, Fox J. The Phenomenon of in vitro hemolysis produced by the rickettsiae of typhus fever, with a note on the mechanism of rickettsial toxicity in mice. J Exp Med. 1948;88(1):25–41.
- Snyder J, Bovarnick M, Miller J, et al. Observations on the hemolytic properties of typhus rickettsiae. J Bacteriol. 1954;67(6):724–730.
- Ramm LE, Winkler HH. Rickettsial hemolysis: adsorption of rickettsiae to erythrocytes. Infect Immun. 1973;7(1):93–99.
- Winkler HH. Rickettsial hemolysis: adsorption, desorption, readsorption and hemagglutination. Infect Immun. 1977;17(3):607–612.
- Radulovic S, Troyer JM, Beier MS, et al. Identification and molecular analysis of the gene encoding Rickettsia typhi hemolysin. Infect Immun. 1999;67(11):6104–6108.
- Engström P, Burke TP, Tran CJ, et al. Lysine methylation shields an intracellular pathogen from ubiquitylation and autophagy. Sci Adv. 2021;7(26). DOI:10.1126/sciadv.abg2517
- Kim HK, Premaratna R, Missiakas DM, et al. Rickettsia conorii O antigen is the target of bactericidal Weil-Felix antibodies. Proc Natl Acad Sci U S A. 2019;116(39):19659–19664.
- Abeykoon AH, Noinaj N, Choi BE, et al. Structural insights into substrate recognition and catalysis in outer membrane protein B (OmpB) by Protein-lysine methyltransferases from rickettsia. J Biol Chem. 2016;291(38):19962–19974.
- Voss OH, Gillespie JJ, Lehman SS, et al. Risk1, a phosphatidylinositol 3-Kinase Effector, Promotes Rickettsia typhi Intracellular Survival. MBio. 2020;11(3):e00820–20.
- Bechelli J, Vergara L, Smalley C, et al. Atg5 supports rickettsia australis infection in macrophages in vitro and in vivo. MBio. 2019;87(1):1–19.
- Pizarro-Cerdá J, Cossart P. Listeria monocytogenes: cell biology of invasion and intracellular growth. Gram-Positive Pathog. 851–863. DOI:10.1128/9781683670131.ch53 Published online 2019.
- Agaisse H. Molecular and cellular mechanisms of Shigella flexneri dissemination. Front Cell Infect Microbiol. 2016;6(MAR):1–10.
- Teysseire N, Chiche-Portiche C, Raoult D. Intracellular movements of Rickettsia conorii adn R. typhi based on actin polymerization. Res Microbiol. 1992;143(9):821–829.
- Heinzen RA, Hayes SF, Peacock MG, et al. Directional actin polymerization associated with spotted fever group Rickettsia infection of Vero cells. Infect Immun. 1993;61(5):1926–1935.
- Walker DH, Yu XJ. Progress in rickettsial genome analysis from pioneering of Rickettsia prowazekii to the recent Rickettsia typhi. Ann N Y Acad Sci. 2005;1063:13–25.
- Gouin E, Egile C, Dehoux P, et al. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature. 2004;427(6973):457–461.
- Reed SCO, Lamason RL, Risca VI, et al. Rickettsia actin-based motility occurs in distinct phases mediated by different actin nucleators. Curr Biol. 2014;24(1):98–103.
- Nock AM, Clark TR, Hackstadt T. Regulator of actin-based motility (RoaM) downregulates actin tail formation by rickettsia rickettsii and is negatively selected in mammalian cell culture. MBio. 2022;13(2):e0035322.
- Jeng RL, Goley ED, D’Alessio JA, et al. A Rickettsia WASP-like protein activates the Arp2/3 complex and mediates actin-based motility. Cell Microbiol. 2004;6(8):761–769.
- Balraj P, El Karkouri K, Vestris G, et al. RickA expression is not sufficient to promote actin-based motility of Rickettsia raoultii. PLoS One. 2008;3(7). DOI:10.1371/journal.pone.0002582
- Speck S, Kern T, Aistleitner K, et al. In vitro studies of Rickettsia-host cell interactions: confocal laser scanning microscopy of Rickettsia helvetica-infected eukaryotic cell lines. PLoS Negl Trop Dis. 2018;12(2):1–17.
- Simser JA, Rahman MS, Dreher-Lesnick SM, et al. A novel and naturally occurring transposon, ISRpe1 in the Rickettsia peacockii genome disrupting the rickA gene involved in actin-based motility. Mol Microbiol. 2005;58(1):71–79.
- Alqassim SS, Lee IG, Dominguez R. Rickettsia Sca2 recruits two actin subunits for nucleation but lacks WH2 domains. Biophys J. 2019;116(3):540–550.
- Park HJ, Lee JH, Gouin E, et al. The Rickettsia surface cell antigen 4 applies mimicry to bind to and activate vinculin. J Biol Chem. 2011;286(40):35096–35103.
- Lamason RL, Bastounis E, Kafai NM, et al. Rickettsia Sca4 reduces vinculin-mediated intercellular tension to promote spread. Cell. 2016;167(3):670–683.e10.
- Narra HP, Sahni A, Alsing J, et al. Comparative transcriptomic analysis of Rickettsia conorii during in vitro infection of human and tick host cells. BMC Genomics. 2020;21(1):1–21.
- Galletti MFBM, Fujita A, Nishiyama MY, et al. Natural blood feeding and temperature shift modulate the global transcriptional profile of rickettsia rickettsii infecting its tick vector. PLoS One. 2013;8(10):1–12.
- Tokarz R, Anderton JM, Katona LI, et al. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect Immun. 2004;72(9):5419–5432.
- Hecht JA, Allerdice MEJ, Dykstra EA, et al. Multistate survey of American dog ticks (Dermacentor variabilis) for Rickettsia Species. Vector-Borne Zoonotic Dis. 2019;19(9):652–657.
- Hyochol AHN PhD, Weaver M PhD, Lyon D PhD, et al. Survey of Rickettsia parkeri and Amblyomma maculatum associated with small mammals in southeastern Virginia. Physiol Behav. 2017;176(1):139–148.
- Sanchez-Vicente S, Tagliafierro T, Coleman JL, et al. Polymicrobial nature of tick-borne diseases. MBio. 2019;10(5). DOI:10.1128/mBio.02055-19
- Labruna MB, Ogrzewalska M, Soares JF, et al. Experimental Infection of Amblyomma aureolatum Ticks with rickettsia rickettsii. Emerg Infect Dis. 2011;17(5):829–834.
- Soares JF, Soares HS, Barbieri AM, et al. Experimental infection of the tick Amblyomma cajennense, Cayenne tick, with Rickettsia rickettsii, the agent of Rocky Mountain spotted fever. Med Vet Entomol. 2012;26(2):139–151.
- Wright CL, Gaff HD, Sonenshine DE, et al. Experimental vertical transmission of Rickettsia parkeri in the Gulf Coast tick, Amblyomma maculatum. Ticks Tick Borne Dis. 2015;6(5):568–573.
- Harris EK, Verhoeve VI, Banajee KH, et al. Comparative vertical transmission of Rickettsia by Dermacentor variabilis and Amblyomma maculatum. Ticks Tick Borne Dis. 2017;8(4):598–604.
- Levin ML, Ford SL, Hartzer K, et al. Minimal Duration of tick attachment sufficient for transmission of infectious Rickettsia rickettsii (Rickettsiales: rickettsiaceae) by its primary vector dermacentor variabilis (Acari: ixodidae): duration of Rickettsial Reactivation in the Vector Revisite. J Med Entomol. 2020;57(2):585–594.
- Karim S, Kumar D, Budachetri K. Recent advances in understanding tick and rickettsiae interactions. Parasite Immunol. 2021;43(5):1–11.
- Al-Khafaji AM, Armstrong SD, Varotto Boccazzi I, et al. Rickettsia buchneri, symbiont of the deer tick Ixodes scapularis, can colonise the salivary glands of its host. Ticks Tick Borne Dis. 2020;11(1):101299.
- Felsheim RF, Kurtti TJ, Munderloh UG. Genome sequence of the endosymbiont Rickettsia peacockii and comparison with virulent Rickettsia rickettsii: identification of virulence factors. PLoS One. 2009;4(12). DOI:10.1371/journal.pone.0008361
- Kurtti TJ, Felsheim RF, Burkhardt NY, et al. Rickettsia buchneri sp. Nov., a rickettsial endosymbiont of the blacklegged tick ixodes scapularis. Int J Syst Evol Microbiol. 2015;65(3):965–970.
- Harris E, Jirakanwisal K, Verhoeve V, et al. Role of Sca2 and RickA in the Dissemination of Rickettsia parkeri in Amblyomma maculatum. Infect Immun. 2018;86(6):e00123–18.
- Azad AF, Sacci JB, Nelson WM, et al. Genetic characterization and transovarial transmission of a typhus-like rickettsia found in cat fleas. Proc Natl Acad Sci U S A. 1992;89(1):43–46.
- Hirunkanokpun S, Thepparit C, Foil LD, et al. Horizontal transmission of Rickettsia felis between cat fleas, Ctenocephalides felis. Mol Ecol. 2011;20(21):4577–4586.
- Macaluso KR, Pornwiroon W, Popov VL, et al. Identification of Rickettsia felis in the salivary glands of cat fleas. Vector-Borne Zoonotic Dis. 2008;8(3):391–396.
- Danchenko M, Laukaitis HJ, MacAluso KR. Dynamic gene expression in salivary glands of the cat flea during Rickettsia felis infection. Pathog Dis. 2021;79(5):1–8.
- Sashika M, Abe G, Matsumoto K, et al. Molecular survey of rickettsial agents in feral raccoons (Procyon lotor) in Hokkaido, Japan. Jpn J Infect Dis. 2010;63(5):353–354.
- Boostrom A, Beier MS, Macaluso JA, et al. Geographic association of Rickettsia felis-infected opossums with human murine typhus, Texas. Emerg Infect Dis. 2002;8(6):549–554.
- Tay ST, Mokhtar AS, Low KC, et al. Identification of rickettsiae from wild rats and cat fleas in Malaysia. Med Vet Entomol. 2014;28(SUPPL.1):104–108.
- Phoosangwalthong P, Hii SF, Kamyingkird K, et al. Cats as potential mammalian reservoirs for Rickettsia sp. genotype RF2125 in Bangkok, Thailand. Vet Parasitol Reg Stud Reports. 2018;13(June):188–192.
- Giudice E, Di Pietro S, Alaimo A, et al. A Molecular Survey of Rickettsia felis in fleas from cats and dogs in sicily (Southern Italy). PLoS One. 2014; 9(9):10.1371/journal.pone.0106820
- Ng-Nguyen D, Hii SF, Hoang MTT, et al. Domestic dogs are mammalian reservoirs for the emerging zoonosis flea-borne spotted fever, caused by Rickettsia felis. Sci Rep. 2020;10(1):1–10.
- Bozeman FM, Masiello SA, Williams MS, et al. Epidemic typhus rickettsiae isolated from flying squirrels. Nature. 1975;255(5509):545–547.
- Duma RJ, Mcgill TM, Sonenshine DE, et al. Epidemic Typhus in the United States Associated with Flying Squirrels. JAMA J Am Med Assoc. 1981;245(22):2318–2323.
- Laukaitis HJ, Macaluso KR. Unpacking the intricacies of Rickettsia–vector interactions. Trends Parasitol. 2021;37(8):734–746.
- Houhamdi L, Fournier PE, Fang R, et al. An experimental model of human body louse infection with Rickettsia typhi. Ann N Y Acad Sci. 2003;990:617–627.
