ABSTRACT
Resistance of bacteria to antibiotics is a major concern in medicine and veterinary science. The bacterial biofilm structures not only prevent the penetration of drugs into cells within the biofilm’s interior but also aid in evasion of the host immune system. Hence, there is an urgent need to develop novel therapeutic approaches against bacterial biofilms. One potential strategy to counter biofilms is to use phage depolymerases that degrade the matrix structure of the bacteria and enable access to bacterial cells. This review mainly discusses the methods by which phage depolymerases enhance the efficacy of the human immune system and the therapeutic applications of some phage depolymerases, such as single phage depolymerase application, combined therapy with phage depolymerase and antibiotics, and phage depolymerase cocktails, for treating bacterial biofilms. This review also summarizes the relationship between bacterial biofilms and antibiotic resistance.
Introduction
It is estimated that antimicrobial-resistant bacterial infections will become the leading cause of death by 2050 [Citation1]. The formation of bacterial biofilms is considered to be one of the major causes of antibiotic resistance in bacteria [Citation2]. Bacterial biofilm acts as a virulence factor. Biofilms can prevent the penetration of drugs through their matrix and reduce the ability of antibiotics to reach the surface of bacterial cells. Furthermore, biofilms enable bacteria to evade the host immune system [Citation3]. Some pathogenic bacteria, such as E. coli, K. pneumoniae, P. aeruginosa, S. aureus, and E. faecalis, can form biofilms on the surfaces of medical instruments and human and animal tissues [Citation4]. In fact, it has been estimated that bacterial biofilms are related to approximately 80% of chronic and recurrent bacterial infections in humans, including cystic fibrosis, endocarditis, meningitis, osteomyelitis, rhinosinusitis, and periodontitis, as well as kidney and prosthesis infections [Citation5]. Bacteria that are susceptible to antibiotics when in planktonic form can develop increased antibiotic resistance after biofilm formation on a suitable surface [Citation6]. The possible reasons may be bacterial biofilms reducing the penetration of antibiotics into the deeper layers, biofilm bacteria growing slowly in the deeper layers or biofilm bacteria developing molecular mechanisms of antibiotic resistance. There is, thus, an urgent need to develop novel therapeutic approaches, besides the conventional antibiotics used against bacterial biofilms.
The use of phage depolymerases is a promising therapeutic strategy for preventing and controlling bacterial biofilm-associated infections [Citation7]. Some phages have developed the ability to degrade the polysaccharide-based structures produced by biofilm-forming bacteria to gain entry into bacterial cells and replicate their genetic information [Citation8,Citation9]. Not all phages can encode depolymerases with exopolysaccharide-degrading activity. The depolymerases are encoded by some phages that infect the encapsulated bacteria. Phage depolymerases are encoded in the same open reading frames as phage structural proteins or in close proximity to these genes, which are located mainly on tail fibres, base plates, and neck [Citation10]. Phage depolymerases can be divided into two main groups: hydrolases (EC 3.2.1.-) and lyases (EC 4.2.2.-), based on the degradation mode of the carbohydrate polymers on the surface of the bacteria [Citation11]. These depolymerases can degrade polysaccharides by recognizing specific ligands on the bacterial surface. The specific binding to capsular polysaccharides (CPSs) and lipopolysaccharides (LPSs) results in the destruction of repeating units of the polysaccharide [Citation12]. Phage depolymerases do not directly kill the bacteria; instead, they strip the protective polysaccharide layers from the bacterial cells, which exposes and sensitizes them to components of the immune system or to antibacterial agents. Moreover, the degradation of polysaccharides by phage depolymerases can increase the penetration of antibiotics into the biofilm. Thus, phage depolymerases demonstrate a synergistic effect with some antibiotics against biofilm-forming pathogens [Citation13–16]. In the treatment of bacterial infections, phage depolymerase is compared with phage endolysin and phage holin in .
Table 1. Comparison of depolymerase, endolysin and holin derived from phages in the treatment of bacterial infections.
This review discusses the enhancing effect of phage depolymerase on the human immune system and the therapeutic applications of some phage depolymerases, such as single phage depolymerase application, combination therapy with phage depolymerase and antibiotics, and phage depolymerase cocktails, for the treatment of bacterial biofilms. Additionally, the relationships between bacterial biofilms and antibiotic resistance have been examined.
Relationship between bacterial biofilms and antibiotic resistence
Bacterial biofilms are multicellular communities that consist of water (up to 97%), microbial cells (2%–5%), exopolysaccharides (EPSs) (1%–2%), proteins (<1%–2%), and DNA and RNA (<1%–2%) [Citation17]. Biofilms maintain bacterial structural integrity, help them to adhere to biotic and abiotic surfaces, and protect them against antibiotics or the host’s immune system.
Conventional antibiotics can kill growing and dividing bacterial cells, such as planktonic bacteria, that is, those in free-living form [Citation18]. However, the minimum inhibitory concentrations (MICs) of conventional antibiotics for biofilm bacteria are 100–1000 times higher than those for planktonic bacteria [Citation19].
Formation of the bacterial biofilm
Biofilm formation can be influenced by numerous factors, such as the condition of the surface on which the biofilm form is formed, cellular structures, and chemical and physical growth factors [Citation20]. Hence, the formation of bacterial biofilm is a complex process requiring quorum sensing and different sets of genes for transcription and translation.
Bacterial biofilm formation can be classified into five stages (). The first stage is reversible attachment, wherein the cells newly and loosely attach to the surface via electrostatic forces, van der Waals forces, and hydrophobic interactions [Citation24]. At this stage the attachment is very weak and reversible. The bacteria either commit to the biofilm mode or return to the planktonic lifestyle. Fimbriae, pili, and flagella contribute to their attachment to rough and hydrophobic substances. The second stage of bacterial biofilm formation is monolayer formation, wherein the loose bacteria begin to produce extracellular polymeric substances and consolidate the attachment process [Citation21]. After monolayer formation, irreversible adhesion occurs. The third stage is microcolony formation, wherein various microbial cells continue to accumulate and grow into multilayered cell clusters surrounding EPS, thus leading to the formation of microcolonies and three-dimensional structures [Citation22,Citation24]. In the fourth stage, the microcolonies develop into mature biofilms that may be mushroom- or tower-like in shape with fluid-filled channels. These channels ensure the diffusion and circulation of nutrients, oxygen, and essential substances within the microenvironment [Citation23]. According to Marchabas, matured biofilm structure includes bulk of biofilm, linking film, conditioning film, and the substratum from the outside to the inside [Citation25]. Biofilm dispersal, the last stage of biofilm formation, involves detachment of the bacteria from the mature biofilm and their conversion into a planktonic state. This is a cyclical process, with the detached bacterial cells potentially being able to colonize new surfaces [Citation24]. Different saccharolytic enzymes, such as N-acetyl-heparosan lyase from E. coli [Citation26], alginate lyase from P. aeruginosa [Citation27], beta-N-acetyl-glucosaminidase (DspB) from A. actinomycetemcomitans [Citation28], and hyaluronidase from Streptococcus equi [Citation29], are produced during the final stage to facilitate bacterial release from the biofilm and the modulation of biofilm structure.
Figure 1. Schematic overview of bacterial biofilm formation and development stages. Biofilm formation starts with reversible adhesion of planktonic cells, and then with irreversible adhesion to the surface. The microbial cells continue to multiply and form micro-colonies and eventually develop into the mature biofilm. In the last stage, biofilm bacteria detach from the mature biofilm and disperse as planktonic state [Citation21–23].
![Figure 1. Schematic overview of bacterial biofilm formation and development stages. Biofilm formation starts with reversible adhesion of planktonic cells, and then with irreversible adhesion to the surface. The microbial cells continue to multiply and form micro-colonies and eventually develop into the mature biofilm. In the last stage, biofilm bacteria detach from the mature biofilm and disperse as planktonic state [Citation21–23].](/cms/asset/8ff23155-a293-4de6-8011-dde983a8969d/kvir_a_2273567_f0001_oc.jpg)
Bacterial biofilms reduce the penetration of antibiotics into deeper layers
Bacterial biofilm is a complex three-dimensional structure comprising bacteria, EPSs, metabolites, and nutrients. EPS forms the skeleton of the biofilm [Citation30], which maintains the integrity and persistence of the biofilm architecture. EPS can prevent or retard the penetration of antibiotics into the deeper layers of the biofilm independently or in combination with eDNAs [Citation5].
The ability of antibiotics to penetrate a bacterial biofilm varies among the various classes of antibiotics and bacterial genera. Singh et al. [Citation31] found that the average percentage reductions in the penetration of vancomycin, chloramphenicol, amikacin, ciprofloxacin, imipenem, cefotaxime, and tetracycline were 57%, 34%, 22%, 18%, 14%, 11%, and 9%, respectively, in S. epidermidis, S. aureus, K. pneumoniae, and E. coli biofilms. In another study, polysaccharide intercellular adhesin (PIA) was shown to reduce the penetration of many antibiotics, such as oxacillin, cefotaxime, teicoplanin, and vancomycin, via the biofilm [Citation32]. PIA, also known as poly-β-1–6-N-acetylglucosamine (PNAG), is the only EPS produced by staphylococci [Citation33]. PIA determines the cell surface hydrophobicity of S. epidermidis and mediates the initial adherence of the biofilms to some extent [Citation34]. Different bacterial strains might have different types of EPSs, and while some bacteria produce several different types, others produce only one dominant EPS molecule. Anderl et al [Citation35]. found that ampicillin did not penetrate the biofilm of K. pneumoniae, which indicates the resistance of this species to ampicillin when in biofilm form. Hoyle et al [Citation36]. reported a decrease in the diffusion of piperacillin through P. aeruginosa biofilms formed on dialysis membranes. P. aeruginosa biofilms prevented the entry of antibiotics into bacterial cells, and imipenem or ceftazidime at a concentration of 2560 μg/mL could not eradicate these biofilms. It was also reported that the permeation rates of macrolides, fluoroquinolones, beta-lactams, gentamicin, and amikacin through the alginate of P. aeruginosa strain 214 were reported to be 100%, >75%, >75%, 73%, and 59%, respectively, which indicates that biofilms can limit the permeation rates of antibiotics to various degrees [Citation37].
Biofilm bacteria grow slowly in deeper layers
The bacteria in the deeper layers of biofilms lack oxygen, as reported by Wu et al [Citation38]. In one study, the oxygen concentration within the gel biofilm was measured using microelectrodes. A decrease in the oxygen concentration to < 3% of air saturation at a depth of 500 μm was reported in S. aureus biofilm [Citation39]. Elsewhere [Citation40], it was documented that the strains exhibited low oxidoreductase activities, which included reductions in the expression levels of pyruvate dehydrogenase, ethanol dehydrogenase, glycerol-3-phosphate dehydrogenase, succinate dehydrogenase, and cytochromes bo and aa3 under anaerobic conditions. In another study on a P. aeruginosa clinical isolate, bottom-up proteomic analysis showed that the levels of L-arginine and polyamine metabolism were higher in anoxic regions of biofilms than in oxic ones [Citation41].
Besides oxygen limitations, biofilms also suffer from nutrient limitations that affect bacterial growth. Several nutrients exert multiple effects on the metabolism of the biofilm bacteria, with L-arginine being particularly prominent. Indeed, arginine and aspartic acid were reported to exert opposite effects on biofilm formation in P. putida KT2440 and Rup4959 [Citation42]. Similarly, Mills et al [Citation43]. observed that low concentrations of L-arginine induced an increase in the concentration of c-di-GMP in Salmonella typhimurium. Thus, L-arginine may be used to cope with biofilms based on the nutrient defining solution. Anderl et al [Citation44]. found that bacteria in stationary phase could be protected from antibiotics in the absence of carbon and nitrogen within the culture medium. The pH of the biofilm is a key factor determining bacterial metabolism and ranges from 5.6 in deeper layers of biofilms to 7.0 in superficial layers [Citation45]. Low pH can directly reduce the activities of oxacillin [Citation46]. A low nutrient level influences the metabolic state of biofilm bacteria.
These slow-growing bacteria deep in biofilms are more resistant to antimicrobial agents than those growing at an intermediate rate [Citation47]. For example, Zheng and Stewart [Citation48] noted that rifampin penetrates the biofilm of S. epidermidis but does not kill these bacteria. The reason for this failure to achieve bacterial killing was not the inadequate penetration of rifampin through the biofilm, but the slow or absent bacterial growth. Results showed that the average growth rate of biofilm bacteria was 0.035 ± 0.004 h−1, whereas the average growth rates of the stationary-phase and exponential-phase bacteria were 0.15 ± 0.06 h−1 and 0.82 ± 0.34 h−1, respectively. Other studies also showed that the slow growth of biofilm bacteria enable them to evade the effect of rifampicin [Citation49,Citation50]. These studies establish that metabolically inactive bacteria can escape the effects of conventional antibiotics.
Biofilm bacteria develop molecular mechanisms of antibiotic resistance
Biofilms provide an the ideal environment for horizontal gene transfer and the development of multidrug resistance [Citation51]. Conjugation is the most common mechanism by which the horizontal transfer of resistance genes occurs within biofilms. Indeed, a study reported that the transfer rate of the conjugative plasmid pGO1 in S. aureus biofilms was nearly 16,000 times higher than that observed in the planktonic state [Citation52]. Moreover, Kouzel et al [Citation53]. quantified the acquisition and spread of multidrug resistance and observed that the transfer efficiencies of ermC and aadA were higher during the early stages of N. gonorrhoeae biofilm formation. Resistance genes can also be transferred between different bacterial species in biofilms. For instance, an in vitro biofilm experiment demonstrated the transfer of the blaNDM-1 gene from Enterobacteriaceae into P. aeruginosa and A. baumannii via conjugation [Citation54]. Transduction, which occurs via temperate phages, is another mechanism by which resistance genes can be exchanged between bacteria. ϕ731 is a Shiga-toxin-encoding phage with a chloramphenicol resistance gene, which was reported to be transferred to pathogenic E. coli in biofilms at both 20°C and 37°C. It was also found that S. epidermidis temperate phages could spread antimicrobial resistance genes via transduction [Citation55]. Transformation in the biofilm is the third mechanism of horizontal gene transfer. For example, S. pneumoniae can acquire genes conferring resistance to streptomycin or trimethoprim resistance genes by recombination via the transformation of DNA from the environment. Two pneumococcal strains, S2Tet and S4Str, were incubated together, and the emergence of double-resistant pneumococci was observed at a recombination frequency of 2.5 × 10−4 4 h post-inoculation [Citation56].
In addition to gene transfer, gene mutations in biofilm bacteria can lead to resistance. The spontaneous mutation rates of several bacteria were reported to range from approximately 10−10 to 10−9 per nucleotide per generation [Citation57]. Furthermore, in biofilms, the occurrence rates of mutations are higher than the spontaneous mutation rates. This kind of mutation is usually called high mutability or hypermutability. In biofilm bacteria, mutations can confer an evolutionary advantage, especially under the pressure of nonlethal-dose antibiotics and growth restrictions. The presence of bacteria with hypermutability can lead to antibiotic resistance. P. aeruginosa is one of the most prone to producing biofilms bacteria. Thus, its hypermutation is considered to be the main driver for the development of antimicrobial resistance in this species in patients with chronic infections. One study reported a 105-fold increase in mutation frequency in biofilm bacteria compared with that in planktonic bacteria [Citation58]. Furthermore, several enzymes that protect DNA from oxidative damage were found to be downregulated in P. aeruginosa biofilms. For instance, the major pseudomonal antioxidant catalase encoded by katA was found to be downregulated by 7.7 folds. The downregulation of antioxidant enzymes leads to the accumulation of DNA damage, which accelerates the rate of mutagenic events [Citation58]. Sultan et al. [Citation59] observed the coexistence of qnrA, qnrB, qnrS, and gyrA gene mutations and biofilm production in almost 40% of quinolone-resistant uropathogenic E. coli. These findings indicate the relationship between gene mutations and biofilm production.
It is worth mentioning that some researchers believe that efflux pumps are not associated with biofilm formation. For example, Türkel et al [Citation60]. explored the relationship between efflux pump-associated gene expression levels and biofilm formation by collecting 100 clinical extended-spectrum β-lactamase-producing K. pneumoniae isolates and examining their biofilm-forming capabilities [Citation60]. The expression levels of AcrA, ketM, kdeA, kpnEF, and kexD, which are related to efflux pumps, were measured. Interestingly, no correlations were observed between the expression levels of these efflux pump genes and biofilm formation. In another study, Knight et al. reported that mutation of the major efflux pump gene ΔadeJ led to a minor decrease in biofilm formation [Citation61]. Moreover, Li et al [Citation62]. documented significant correlations between the efflux pump MexAB-OprM phenotype and biofilm formation in 110 carbapenem-resistant P. aeruginosa strains. Thus, there is a need for additional studies to determine the relationship between efflux pump genes and biofilm formation.
Applications of phage depolymerase for the treating of bacterial biofilms
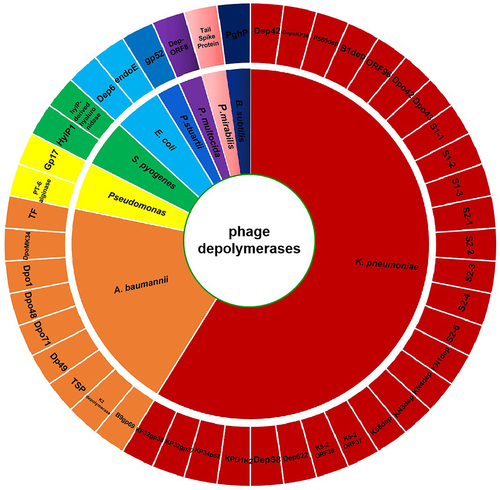
Antibiotic treatment often fails in clinical medicine owing to the impermeability of bacterial biofilms. Thus, other methods, such as the use of phage depolymerases, have been attempted to counter the resistance conferred by such biofilms. Phages are natural predators of bacteria, but not all phages can encode depolymerases with exopolysaccharide-degrading activity. Depolymerases are encoded by some phages that infect the encapsulated bacteria, such as E. coli K1 [Citation63], E. coli K20 [Citation64], K. pneumoniae K22 [Citation65], K. pneumoniae K23 [Citation66], K. pneumoniae K64 [Citation67], A. baumannii K26 [Citation68], and A. baumannii K92 [Citation69]. Although the use of a single phage depolymerase can combat bacterial biofilms, the complete eradication of bacterial biofilms may require the application of multi-phage depolymerase cocktails or their combined use with antibiotics. An overall classification of phage depolymerases reported in the treatment of bacterial biofilm and their corresponding bacterium genus targets are summarized in .
Figure 2. Wheel diagram summary depicting the classification of phage depolymerases reported in the treatment of bacterial biofilm and their corresponding bacterium genus targets.
The structure of phage depolymerase may be differ among different species of depolymerases. Whereas, phage depolymerase is a trimer crystal state in general [Citation70,Citation71]. According to the crystal structure of KP32gp38 and K1 CPS depolymerase, each monomer has three distinct domains: the N-terminal particle-binding domain, the central receptor-binding domain, the C-terminal β sandwich domain. The N-terminal domain consists of β-sheet and α-helix. Three monomers fold into a barrel-like structure. The central receptor-binding domain mainly features β-helix and contains at least one distinct carbohydrate‑binding site. The C-terminal β sandwich domain has a lectin-like fold formed by β strands. Three parallel chains tightly packed together to form a highly stable screw-like trimeric structure [Citation70,Citation71]. The protein sequences of phage depolymerases are compared in .
Application of a single phage depolymerase
To eliminate polysaccharide-producing bacteria, phages have evolved several species of depolymerases to overcome the capsular structure surrounding the bacteria. Phage depolymerases can be categorized into two main groups, namely, hydrolases (EC 3.2.1.-) and lyases (EC 4.2.2.-), based on the mode of degradation of carbohydrate polymers on the bacterial surface [Citation11]. The hydrolases include sialidases (EC 3.2.1.18) [Citation72], poly-γ-glutamate (γ-PGA) hydrolases (EC 3.4.19.9) [Citation73], xylosidases (EC 3.2.1.37) [Citation74], levanases (EC 3.2.1.65) [Citation75], dextranases (EC 3.2.1.11) [Citation76], lipases (EC 3.1.1.3) [Citation77], and rhamnosidases (EC 3.2.1.40) [Citation10]. Hydrolases are a group of enzymes that catalyse the cleavage of 1,4- glycosidic bonds in the glycosidic linkage, thereby degrading CPSs, EPSs, LPSs, and O-polysaccharides. In contrast, lyases cleave the glycosidic linkage between monosaccharides and the C4 position in uronic acid and introduce a double bond via the β-elimination mechanism between the C4 and C5 positions in nonreducing uronic acid. Some phage depolymerases are listed in according to those with defined classes and those yet to be defined.
Table 2. Therapy of phage depolymerase in the treatment of bacterial biofilm.
Sialidases (EC 3.2.1.18), also called neuraminidases, are a group of enzymes that hydrolyse the α-linkage of terminal sialic acids in glycans. Endosialidase E (endoE), derived from the E. coli K1 strain phage A192PP, selectively degrades sialic acid. However, endoE cannot kill pathogens or inhibit their growth, so its potential therapeutic efficacy remains unclear. Mushtaq et al [Citation72,Citation78]. conducted two studies to determine whether endoE could improve the outcome of E. coli K1 systemic infections in bacteraemia and meningitis rat models. The findings showed that the intraperitoneal administration of 20 µg of endoE can protected 3-day-old rats from systemic infection. The enzyme hydrolysed α-2,8-linked sialic acid and removed the capsular polysaccharides from the E. coli surfaces. Sensitization to the bactericidal effect of the complement system also occurred, and the phagocytic activity of the macrophages was enhanced when the capsular polysaccharides were removed.
Poly-γ-glutamate (γ-PGA) hydrolase (EC 3.4.19.9) is a type of peptidase that hydrolyzes γ-PGA into oligo-γ-glutamates. Kimura et al [Citation73]. discovered a novel γ-glutamyl hydrolase PghP from Bacillus subtilis phage ΦNIT1, which randomly hydrolysed γ-PGA into oligo-γ-glutamates.
Hyaluronate (HA) lyases (4.2.99.1) and hyaluronidases (EC 4.2.2.1) are classes of enzymes that digest hyaluronate. Baker et al [Citation80]. purified hylP-derived hyaluronidase from the S. pyogenes phage H4489A and reported that the phage HA lyase cleaved the N-acetylglucosaminidic bonds of hyaluronan and belonged to the category of hyaluronate lyases. The researchers further tested the substrate specificity and found that the phage HA lyase specifically cleaved hyaluronan, but not dermatan sulphate, keratan sulphate, chondroitin 4-sulphate, heparan sulphate, or heparin. This study may provide an alternative reagent to digest the S. pyogenes hyaluronan capsule. Another hyaluronate lyase named HylP1 from Streptococcus pyogenes prophage SF370.1 was tested to catalyse a β-elimination reaction of hyaluronan [Citation79]. Although phage HA lyases and hyaluronidases play important roles in removing the streptococcal hyaluronan capsule, they can transform the nonvirulent strains into virulent strains [Citation96].
Alginate lyases (EC 4.2.2) are a class of enzymes that catalyse the degradation of alginic acids, including mannuronate lyase (EC 4.2.2.3) and guluronate lyases (EC 4.2.2.11) [Citation97]. To date, only P. aeruginosa and A. vinelandii phages are known to encode alginate lyases [Citation97]. Glonti et al [Citation81]. reported that P. aeruginosa phage PT-6 could rapidly reduce the viscosity of the alginic acid capsule by 62%–66% within 15 min. Furthermore, PT-6 alginase was purified from A. vinelandii phage suspensions. The enzyme corresponding to a 37 kDa band was shown to degrade polysaccharides to a series of oligouronides. By analysing these oligouronides and together with kinetic information, the authors concluded that PT-6 alginase exhibited polyuronide-degrading activity. The A. vinelandii phage is another example of a phage with alginate lyase activity [Citation98].
Pectate lyases (EC 4.2.2.2) and pectin lyases (EC 4.2.2.10) are classes of pectolytic enzymes that degrade galacturonic acid. Gp17 is a pectate lyase obtained from the tail spikes of the P. putida phage φ15. Recombinantly expressed and purified Gp17 was reported to form opaque halo zones on the lawns of the host bacteria, which exhibited EPS-, CPS- or LPS-degrading properties [Citation82]. Similarly, Liu et al [Citation83]. reported that the phage IME200 exhibited polysaccharide depolymerase activity against A. baumannii. Based on the analysis of the complete genome sequences, open reading frame 48 was predicted to encode a polysaccharide depolymerase with pectate lyase activity (Dpo48), which was subsequently expressed, purified, and characterized. Dpo48 demonstrated high efficacy at a wide range of temperatures (20°C–70°C) and pH (5.0–9.0). It also exerted a synergistic effect with 50% serum on A. baumannii strains. Interestingly, another tail spike protein (TSP) with pectate lyase activity was derived from the P. mirabilis strain BB2000 phage (vB_PmiS_PM-CJR), which not only degraded the biofilms in vitro and reduced the adherence of bacteria to plastic pegs, but also improved the survival rates of Galleria mellonella larvae infected with the host cells [Citation84].
Xylosidases (EC 3.2.1.37), levanases (EC 3.2.1.65), dextranases (EC 3.2.1.11), lipases (EC 3.1.1.3), and rhamnosidases (EC 3.2.1.40) catalyse the hydrolysis of xylan, levan, dextran, triacylglycerols, and rhamnogalacturonan, respectively. Some phages such as the C. crescentus transducing phage Cr30 [Citation74], the L. fermentum phage phiPYB5 [Citation75], the B. subtilis phage SP10 [Citation76], and the Shigella phage SF6 [Citation77] contain xylosidases, levanases, dextranases, lipases, respectively. These phage-derived enzymes are extremely rare [Citation10], and have not been used for the treatment of bacterial biofilm until recently.
Apart from the defined classes of phage depolymerases, several undefined classes of phage depolymerases can inhibit the formation of bacterial biofilms or degrade those already formed. Shahed-Al-Mahmud et al [Citation7]. reported that the TSP from the A. baumannii φAB6 phage inhibited the colonization of host cells on the surface of Foley catheters. The researchers also evaluated the therapeutic effect of TSP in zebrafish infected with A. baumannii 54149 and reported that the survival rate of zebrafish administered TSP (80%) was significantly higher than that of zebrafish administered PBS (10%). Dp49 is another capsule depolymerase identified from the A. baumannii phage vB_AbaM_IME285. Positivity for the depolymerase activity of Dp49 was found in 25 out of 49 A. baumannii clinical isolates [Citation85]. In addition, Dp49 increased the serum killing ability against the strains Ab387 and Ab220 in vitro, whereas Dp49 the administration improved the survival rates of mice infected with Ab387 in vivo. Moreover, Oliveira et al. [Citation86] identified a 604-amino-acid virion protein, gp52, with depolymerase activity. The tail spike gp52 purified from the P. stuartii phage vB_PstP_Stuart made the host bacteria susceptible to serum killing by degrading the exopolysaccharide. In another study, the K. pneumoniae phage P560 depolymerase P560dep was shown to inhibit biofilm formation. Intraperitoneal administration of a 50 μg dose of P560dep protected 90%–100% of mice infected with the KL47 carbapenem-resistant K. pneumoniae from mortality. Although the depolymerase was not related to the bacterial killing, the authors considered it an attractive and promising agent to combat infectious diseases [Citation8]. The mechanisms by which parts of phage depolymerases degrade polysaccharides have been illustrated in certain studies focusing on the protein structures. A recent study by Squeglia et al. [Citation70] revealed the crystal structure of the Klebsiella phage capsule depolymerase KP32gp38. It presented as a trimer in solution and in the crystal state. The monomer comprised four protein domains, a flexible N-terminal domain, a catalytic domain, a carbohydrate-binding domain, and a lectin-like fold C-terminal domain.
Phage depolymerase can also be used to treat Shiga toxin-producing E. coli (STEC) infections. The STEC strain HB10 O91 phage PHB19 was isolated, and the whole genome was sequenced and annotated by a phage study group. The authors [Citation99] identified a novel phage depolymerase, Dep6, from the PHB19 TSP. In vitro, Dep6 effectively removed STEC biofilms and enhanced the susceptibility of host bacteria to serum killing. Furthermore, no toxic effects of Dep6 were observed in human red blood cells, lung carcinoma cells, or embryonic kidney cells in vitro and in vivo. In an STEC infection mouse model, pretreatment with Dep6 resulted in 100% survival compared with that in mice in the control group. Delayed treatment (3 h post infection) resulted in only 33% survival, whereas mice that were simultaneously treated with infection presented with a survival rate of 83%. In addition, the levels of proinflammatory cytokines, such as tumour necrosis factor-alpha, gamma interferon, and IL-1β, were reduced 24 h post infection in the Dep6-treated mice. However, the levels of IL-6 were not reduced. Thus, Dep6 appeared to be safe for mice based on the in vivo and in vitro assays. P. aeruginosa LPS can be degraded by phage depolymerase. LKA1gp49 is from P. aeruginosa phage LKA1 and cleaves β-band of LPS [Citation100]. K. pneumoniae K63 capsule can also be degraded by phage depolymerase KP36gp50 [Citation101].
These phage depolymerases possess the exopolysaccharide-degrading activity and glycoside hydrolases domains. One of the possible inhibitory mechanisms of a single phage depolymerase to bacterial biofilm may be the enzymes hydrolyse the glycosidic bonds of polysaccharide. There are four glycosidic bonds, including alpha-C-glycosidic bond, alpha-O-glycosidic bond, beta-C-glycosidic bond and beta-O-glycosidic bond. Each phage depolymerase may hydrolyse at least one kind of glycosidic bonds. However, phage depolymerases have specific and only one or a few types of capsular polysaccharide. We suspect that another possible inhibitory mechanisms may quench quorum sensing. Further studies will be done to explore whether phage depolymerases can disturb quorum sensing. And more phage depolymerases will be found to hydrolyse the glycosidic bonds of polysaccharide.
Phage depolymerase cocktail application
The narrow specificity of phage depolymerases is one of the major obstacles in their use for removing bacterial biofilms and has considerably restricted their application. Several studies have attempted to broaden the specificity via protein engineering and combining several different phage depolymerases to form cocktails ().
Few phage depolymerases have been tested in terms of their ability to degrade different types of bacterial capsules in vivo. Whether phage depolymerases can exhibit generalized therapeutic efficacy towards different kinds of capsules has remained unclear. For example, more than 80 different types of capsules were discovered based on the serological, biochemical, and genetic properties of E. coli [Citation102]. Lin et al [Citation63]. used five phage depolymerases to combat three capsule types in mouse infection models. The phage depolymerases K1E, K1F, K1H, K5, and K30 (gp41 and gp42) were cloned and purified in vitro. In a mouse thigh model, the majority of the mice were rescued by treatment with all of the phage depolymerases, except K1E, when the enzyme (20 µg per mouse) was injected within 0.5 h after the bacterial injection. Preliminary trials showed that the effective doses of K1F and K1H ranged between 2 and 5 µg per mouse. The effective dose of K5 ranged between 2 and 20 µg per mouse, whereas the effective dose of K30 gp41 was 20 µg per mouse. K30 gp42 did not appear to improve the survival rate, and K30 gp41 was found to be less effective than K1F, K1H, and K5. At a dose of 20 µg per mouse, the mixture of K30 gp41 and K30 gp42 rescued all three mice (3/3), whereas K30 gp41 rescued three out of eleven (3/11) mice, and K30 gp42 did not rescue any of the mice (0/3). Although the survival outcome appeared to be somewhat better in the mixture group than that in the two individually treated groups, the sample size was too small to draw any definitive conclusions. The potential acute toxicity of the five phage depolymerases (K1E, K1F, K1H, K5, and K30 gp41) was evaluated by injecting 100 µg of the different phage depolymerases into the right thigh and monitoring the survival, body weight gain, and behaviour of the mice. All of the mice appeared healthy without any change in behaviour over an observation period of 5 days. Statistical analysis indicated no significant differences in body weight gain between the phage depolymerase- and PBS-treated mouse groups. Nonetheless, further studies are required to confirm the safety and efficacy of applying phage depolymerase cocktails.
A cocktail of some phage depolymerases can be used to treat infection with K. pneumoniae. Recently, Pan et al [Citation93]. reported that the multi-host Klebsiella phage ϕK64–1 can infect the capsules of Klebsiella K1, K11, KN4, K21, KN5, K25, K35, K64, K30, and K69. To analyse the wide capsular-type – specific host spectrum, the authors expressed and purified putative proteins (S2–4, S1–1, S1–2, S1–3, S2–1, S2–2, S2–3, S2–5, and S2–6) that exhibited similarity with tail fiber/spike or lyase proteins. In terms of capsule depolymerization activities of these proteins, opaque halo zones were observed using a through spot test of capsule depolymerase on the individual bacterial lawns of K1, K11, KN4, K21, KN5, K25, K35, K64, and K30/K69. S2–6 could digest the capsules of both type K30 and type K69 bacteria. In another study, the same group detected other Klebsiella phage depolymerases, such as KN1dep, KN4dep, KN3dep, and K56dep, with high specificities for capsular serotypes KN1, KN4, KN3, and K56, respectively [Citation94]. Moreover, Majkowska-Skrobek et al [Citation103]. discovered a novel Klebsiella phage KP32, which produced two capsule depolymerases, KP32gp37 and KP32gp38, and infected the K3 and K21 capsular serotypes, respectively. Both depolymerases increased the lifespan of the Galleria mellonella larvae, which were infected with K. pneumoniae in a time-dependent manner. In another study, Liu et al [Citation92]. identified a new K. pneumoniae phage IME205 with two phage depolymerases (Dpo42 and Dpo43), which were stable across a relatively broad temperature range (20°C–70°C). The pH levels of Dpo42 and Dpo43 were in the ranges of ranged between 5.0–8.0 and 4.0–8.0, respectively. The two polymerases could strip the K47 CPSs and sensitize the serum complement to kill the host bacteria but did not exhibit any haemolytic activity against erythrocytes. Thus, Dpo42 and Dpo43 could be used to combat the K47 capsule in K. pneumoniae infections. Moreover, Klebsiella phage K5–2 was found to encode two capsule depolymerases K5–2 ORF37 and K5–2 ORF38. K5–2 ORF37 exhibited K8, K30, and K69 depolymerase activities, whereas K5–2 ORF38 exhibited K5 depolymerase activity [Citation95]. Owing the clear capsular types of K. pneumoniae that phage depolymerases can degrade, phage depolymerases have the potential to combat different capsular types when used as a cocktail.
Although a cocktail can broaden the antibacterial biofilm spectrum, no studies have focused on the different phage depolymerases used in cocktails to combat the bacteria capsules. Phage cocktail therapy has been used to treat M. abscessus infection [Citation104], P. aeruginosa respiratory infection [Citation105], and E. coli urinary tract infection in a mouse model [Citation106]. In addition, some phage cocktails have been used in clinical trials, such as P. aeruginosa phage cocktail to treat burn wound infection (phase 1/2 trial) [Citation107], oral E. coli phage cocktail to treat acute bacterial diarrhoea [Citation108], and topical C. acnes phage cocktail to treat skin acne (phase 1 trial) [Citation109], have been used in clinical trials. Additional studies are required to explore the effect and safety of phage depolymerase cocktails.
Phage depolymerase cocktail application mainly use the mechanism and effect of single depolymerase. Due to the high specificity, a single phage depolymerase can degrade only one or a few types of capsular polysaccharide. This kind of single phage depolymerase application limits the antibiofilm spectrum. In order to enlarge the antibiofilm spectrum, phage depolymerase cocktail application is one of the solutions. Two or more different phage depolymerase may hydrolyse different kinds of glycosidic bonds of polysaccharide. This cocktail application can improve the antibiofilm effect. In addition, phage depolymerase cocktail application can also deduce the resistance issue. Although the probability of polysaccharide mutation is low, the resistant mutation may generate when phage and the host (pathogenic bacteria) have coevolved over time.
Combination therapy with phage depolymerase and antibiotics
Although a single phage depolymerase tends to be somewhat effective in controlling bacterial biofilms, the high specificity of depolymerases limits the complete removal of the biofilm owing to variations in the EPS. In some cases, phage depolymerases might exert anti-polysaccharide activity against a small set of bacterial strains. To address the limitation of the narrow host spectrum, combination therapy with phage depolymerases and antibiotics, an approach widely adopted in humans, may prove useful in combating infections caused by biofilm-forming pathogens. The degradation of the biofilm matrix by phage depolymerases can increase the penetration of antibiotics to exert synergistic effects ().
Table 3. Combination therapy of phage depolymerase and some antibiotics in the treatment of bacterial biofilm.
Ciprofloxacin is one of the most commonly used antibiotics in clinic [Citation113]. Some phage capsule depolymerases exhibit synergy with ciprofloxacin, whereas others demonstrate no synergistic effects with ciprofloxacin. Verma et al [Citation110]. discovered a depolymerase derived from K. pneumoniae phage KPO1K2. The depolymerase and ciprofloxacin could reduce the bacterial numbers in mature 3-day (72 h) biofilms. The antibiofilm effect was reported to be significantly greater in the combination treatment group than in the group treated with ciprofloxacin alone (P > 0.05). Interestingly, the concomitant application of depolymerase and ciprofloxacin produced different effects on the biofilm. When applied concomitantly for 6 h, an insignificant reduction of 1.21 + 0.62 logs was observed in the biofilm bacterial count. However, when treated with depolymerase for 60 min followed by 6 h treatment with ciprofloxacin, a significant reduction of 3.72 + 1.2 logs was observed in the bacterial count. These findings indicate that the combined treatment with phage depolymerase and ciprofloxacin is effective in mature biofilms. However, some studies have reported contradictory findings. For example, Latka and Drulis-Kawa [Citation111] identified a K. pneumoniae phage depolymerase called KP34p57, which had no impact on ciprofloxacin activity. Their findings showed that the The combination of KP34p57 and ciprofloxacin did not improve the antibiotic activity. Moreover, the phage KP36 capsule depolymerase DepoKP36 did not affect the susceptibility of biofilm-forming K. pneumoniae strains to antibiotics such as ciprofloxacin, oxytetracycline, and chloramphenicol [Citation13].
Colistin is the last-resort antibiotic for clinical multidrug-resistant Gram-negative bacterial infections. The combination of phage depolymerase and colistin may produce a synergistic effect to combat A. baumannii infections. Chen et al [Citation16]. expressed a novel depolymerase Dpo71 from the A. baumannii phage IME-AB2 in vitro, which inhibited biofilm formation and interfered with the preformed biofilm. In addition, Dpo71 enhanced the bactericidal effect of colistin. Single-dose colistin (1 µg/mL, 1/2 MIC) alone or with 5% serum did not influence its antibacterial effect against the host cells. However, the application of 10 µg/mL Dpo71 + 1 µg/mL of colistin along with 5% serum resulted in a significant reduction in the counts of A. baumannii. This combination treatment resulted in a 7-log reduction in the bacterial count, and the antibacterial effect was boosted to nearly complete eradication. In comparison to 1 µg/mL colistin alone and 10 µg/mL dpo71 alone treatment, bacterial reduction was a 1-log and 0-log, respectively. The authors also revealed the underlying mechanisms for the superior action of the combination therapy compared with the individual treatments. Scanning electron microscopy showed that the bacterial capsule was stripped by Dpo71 and that the host cell surface did not contain any pilus-shaped protrusions. In contrast, the surfaces of the bacterial cells in the untreated group presented with both capsules and pilus-shaped protrusions. The removal of the capsule by phage depolymerase Dpo71 significantly enhanced the outer membrane destabilization ability of colistin, which promoted the interaction between antibiotics and the bacteria and facilitated the entry of the drug into the bacterial host. In infection models, the combination of Dpo71 and colistin could improve the survival rate of A. baumannii-infected Galleria mellonella. Although Dpo71 itself had no bactericidal efficacy, treatment with Dpo71 alone rescued 40% of the infected Galleria mellonella over a period of 72 h. However, the combined treatment of Dpo71 and colistin rescued 80% of the infected worms during the same observation period. Approximately 70% of the infected worms died within 18 h, and the mortality rate increased to 90% after 48 h. These results indicate that phage depolymerases can act as adjuvants with some antibiotics to enhance antibacterial activity. Nevertheless, the combination of phage depolymerase and colistin may not produce a synergistic effect. Luo et al [Citation112]. identified another A. baumannii phage depolymerase called TF, which did not exhibit any additive or synergistic effects with colistin on the host bacteria. Surprisingly, a temporary increase in the resistance of A. baumannii to colistin was observed after the EPS was peeled by phage depolymerase TF from the bacteria. Thus, it was considered that the loss of EPS may have reduced the colistin attachment, causing a temporary increase in resistance.
Polymyxin has been shown to exert the synergistic effects with phage depolymerase. K. pneumoniae phage SH-KP152226 depolymerase Dep42 increased the antibacterial activity of polymyxin when they were used together [Citation15]. The average bacterial count of the Dep42 + polymyxin treatment group was 5.260 ± 0.05 log, whereas those of the groups treated with Dep42 or polymyxin alone were 6.317 ± 0.01 and 6.013 ± 0.125 log, respectively, which demonstrates a significant reduction of 0.743 ± 0.05 log compared with that in the polymyxin group. Collectively, the findings of these studies strongly indicate that phage depolymerase Dep42 and polymyxin have a synergistic effect on multidrug-resistant K. pneumoniae.
Some antibiotics is neither synergistic nor antagonistic in combination with phage depolymerases. A. baumannii MK34 phage vB_AbaP_PMK34 capsule depolymerase DpoMK34 was neither synergistic nor antagonistic in combination with different antibiotics, such as colistin, imipenem, and amikacin [Citation14]. However, the addition of 500 µg/mL of DpoMK34 did not change the MIC values of the three tested antibiotics upon pretreatment or cotreatment with MK34 and DpoMK34. It was, thus, the authors concluded that DpoMK34 did not produce synergistic or antagonistic effects in combination with colistin, imipenem, and amikacin.
Some antibiotics, such as ciprofloxacin, colistin, polymyxin, have a synergistic effect with a certain phage depolymerase. While some antibiotics, such as imipenem, amikacin, have no synergistic effect with phage depolymerases. The most likely reason for the synergistic effects of phage depolymerases and some antibiotics is that the removal of CPS by phage depolymerases helps some antibiotics to access the bacterial surface. This changes the arrangement of bacteria in the biofilm to a dispersed state, which enhances the ability of antibiotics to penetrate into the biofilm. Eventually, the intensity of the antimicrobial attack is enhanced with the help of the phage depolymerase.
Phage depolymerase enhances the effects of human and mouse immune systems
CPSs are important virulence factors that help bacteria to escape the human immune system. Surface polysaccharides provide shields against components of the host immune system, such as the complement system and phagocytosis. Loss or alteration of CPSs can make bacteria more susceptible to clearance by the human immune system. lists some studies on how phage depolymerases enhance the susceptibility to human serum and kill bacteria.
Table 4. Phage depolymerases enhance the effect of human immune system in the treatment of bacterial biofilm.
Phage-derived capsule depolymerases can remove bacterial CPSs, and some phage depolymerases are reported to make the pathogens susceptible to serum killing. In recent years, A. baumannii has been considered to be an important nosocomial pathogen in intensive care units [Citation116]. Most clinical isolates are resistant to all available antibiotics and the current treatment methods are becoming less effective. One reason for this resistance is that the CPSs protect the bacteria from antibiotics via the polysaccharide structure and aid in their evasion of the host immune system. In other words, CPSs are a major virulence factor. Oliveira et al. [Citation87] purified a K2 capsule-specific depolymerase from the tail spike C-terminus of the A. baumannii phage vB_AbaP_B3 and assessed its activity in vivo. It was found to protect caterpillar larva and mice against bacterial infections. In a mouse sepsis model, the intraperitoneal injection of K2 depolymerase (dose, 50 μg) resulted in 60% of mice avoiding mortality due to infection. Additionally, significant reductions in the expression levels of tumour necrosis factor-alpha and interleukin-6 were observed. K2 depolymerase enhanced the ability of human serum to kill the host cells and reduced the number of bacteria to < 10 colony-forming units (CFU)/mL. The NIPH 2061 strain was not susceptible to serum killing following the inactivation of K2 depolymerase. It was, thus, concluded that the human complement system was activated to control the infection via the action of K2 depolymerase, which degraded the CPS and, consequently, affected the bacterial virulence.
DpoMK34 is another A. baumannii phage depolymerase, which was reported to increase the ability of the serum to kill A. baumannii MK34 in a concentration-dependent manner [Citation14]. Cells treated with 100 µg/mL DpoMK34 presented with a 1.8 ± 0.34 log reduction in 25% (v/v) human serum and a 5.05 log unit reduction in both 50% (v/v) and 75% (v/v) human sera when compared with that in PBS-treated cells. Moreover, heat inactivation of DpoMK34 did not cause any reductions in bacterial cell numbers, even in the 75% (v/v) human serum. Similar findings were reported in another study, wherein a 5 log reduction in Dpo48-treated bacteria was observed following treatment with a 50% volume of serum. The inactivation of the complement in the serum did not result in any reduction in the serum-dependent bacterial count [Citation83]. Oliveira et al [Citation88]. identified another A. baumannii phage depolymerase called B9gp69, which rendered K45 strains susceptible to serum killing in vitro. The number of depolymerase-pretreated bacteria incubated with serum was reduced to below the detection limit (<10 CFU/mL). Notably, B9gp69 digested the capsule polysaccharides of both K30 and K45 strains. Furthermore, the optimal activity of the enzyme could be maintained at temperatures ranging from 20°C to 80°C and pH values ranging from 5 to 9. These are clear indications of the effect of phage depolymerase on the bacterial susceptibility of bacteria to human serum killing.
The use of other phage depolymerases, such as Dep-ORF8 from P. multocida phage PHB02, which specifically degrades the serogroup A capsule, has been reported. The purified Dep-ORF8 significantly increased the survival of mice infected with P. multocida. Additionally, it did not increase the eosinophil and basophil counts or cause any other pathological changes when compared with the control group. Human serum, mouse serum, and mouse whole blood alone exhibited minor bactericidal effects of 1.2–1.7 log CFU reductions in P. multocida strain HB03 cell counts. Treatment with Dep-ORF8 + serum further reduced the cell counts (by 3.5–4.5 log CFU). However, no significant difference in viable cell counts was observed between the Dep-ORF8 + whole blood and the Dep-ORF8 + serum treatment groups. Heat inactivation of the serum resulted in an increase in the survival counts of the bacterial cells to levels equal to those in the PBS control group [Citation114].
pneumoniae phage depolymerases can sensitize cells to serum complement-mediated killing. KP32gp37 and KP32gp38 obtained from Klebsiella phage KP32 increased the sensitivity of serum-resistant cells to complement-mediated host bacterial killing. Decapsulation of the strains by depolymerases resulted in the exposure of the ligands to phagocytic cell attachment [Citation103]. Thus, phage depolymerases could combat the resistance of biofilm bacteria and increase the phagocytic activity of the cells, thereby killing the biofilm bacteria. KP32gp37 increased the phagocytic cell uptake of strain 271 by approximately two folds. Similarly, the geometric mean fluorescence intensity (gMFI) value of the KP32gp38-treated strain 358 (146.7 ± 13.6 gMFI) was higher than that of the untreated bacteria (83.9 ± 5.7 gMFI). In the case of strain 968, the gMFI value in the KP32gp38-treated group (102.3 ± 6.8) was lower than that in the untreated group (293.6 ± 24.4). Evaluation of the time-dependent bactericidal effect of depolymerases + serum against K. pneumoniae strains 271, 45, and 358 revealed that 0.1 µg/mL of KP32gp37 could lead to a 1.6 log decrease in the bacterial number after 3 h of exposure and a 4 log decrease after 7 h of exposure in the case of strain 271. For the K21-type strains, including strains 45 and 358, 100 µg/mL of KP32gp38 caused only a 1.7–4.6 log reduction in the bacterial number compared with that in the initial inoculum after 3 h of exposure. After 7 h of exposure, only minor reductions were observed in the depolymerase-treated Klebsiella strains 358 and 45 [Citation103]. Similarly, Dep6 derived from the STEC phage PHB19 TSP enhanced the serum sensitivity of the host strain. Dep6 at 30 µg/mL resulted in a 4.2 log reduction in the number of host bacteria, whereas that in the PBS-treated group remained at 8 × 108 CFU/mL [Citation99].
aeruginosa phage depolymerase DP is another example of enhancing bactericidal activity mediated by serum in vitro [Citation115]. The authors have isolated a lytic P. aeruginosa phage named IME180 from the sewage of a hospital. Through genomic sequence analysis of IME180 phage genome, DP has two catalytic regions, the Pectate lyase_3 super family and Glycosyl hydrolase_28 super family. The phage depolymerase DP can degrade exopolysaccharide of P. aeruginosa and enhance serum bactericidal activity in vitro. In the bactericidal assay, the bacterial enumeration reduces by two orders of magnitude in the serum and DP group. However, when either serum or DP is applied to bacteria individually, the bacterial enumeration is no obvious decrease or increase. In other words, the P. aeruginosa phage depolymerase DP has the potential to be an anti-microbial agent targeting P. aeruginosa.
The human immune system can not recognize pathogens when bacteria generate CPSs and encase themselves. CPSs help bacteria to escape the human immune system and bacteria fail to induce human immune response. When phage depolymerases strip the protective polysaccharide layers from the pathogen cells, which exposes them to the immune system. And the uncapsulate bacteria expose the lipoteichoic acids of the Gram-positive bacterial cell wall, lipopolysaccharide and outer membrane protein of the Gram-negative bacterial outer membrane. These structural component of the bacterial cell wall induce and activate the immune response, especially the complement system and macrophagocytes.
Conclusions
The resistance of bacterial biofilms to antibiotics and the human immune system prompted scientists to search for alternative methods to counter antibiotic-resistant strains. Studies have characterized several phage depolymerases that act against biofilm-forming bacteria, such as K. pneumoniae, E. coli, A. baumannii, P. mirabilis, P. aeruginosa, S. pyogenes, B. subtilis, and P. stuartii. As natural antimicrobial agents, phage depolymerases clearly have a potential role in preventing and treating bacterial biofilm-associated infections. Although phage depolymerases have been demonstrated to possess potential antibiofilm activity in vitro, no clinical trials on this approach have so far been conducted thus far. There is, thus, a need for detailed exploration of the use of phage depolymerases as potential therapeutic drugs. Fortunately, some phage endolysins have entered clinical trials. Considering the current biosafety standards and regulations, the clinical application of phage depolymerases is much easier than that of the phage itself. Nonetheless, there is a need for further studies confirming the therapeutic use of phage depolymerases in humans. Besides, the use of phage depolymerases in combinations with some innovative treatments such as antibacterial peptide, nano particle, silver, copper and zinc will be explored in the future. As it acts as a kind of enzymes, phage depolymerase can be synthesized with nanomaterials to form phage depolymerase-incorporated nanomaterials. Or liposome-coupled phage depolymerases were prepared. Then, the potential therapeutic effect of innovative treatments will be evaluated in future.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.
Additional information
Funding
References
- No authors listed. United Nations meeting on antimicrobial resistance. Bull World Health Organ. 2016;94(9):638–19. doi: 10.2471/BLT.16.020916
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–1322. doi: 10.1126/science.284.5418.1318
- Wang S, Ding Q, Zhang Y, et al. Evolution of virulence, fitness, and carbapenem resistance transmission in ST23 hypervirulent Klebsiella pneumoniae with the capsular polysaccharide synthesis gene wcaJ Inserted via insertion sequence elements. Microbiol Spectr. 2022;10(6):e0240022. doi: 10.1128/spectrum.02400-22
- Caldara M, Belgiovine C, Secchi E, et al. Environmental, microbiological, and immunological features of bacterial biofilms associated with implanted medical devices. Clin Microbiol Rev. 2022;35(2):e0022120. doi: 10.1128/cmr.00221-20
- Singh A, Amod A, Pandey P, et al. Bacterial biofilm infections, their resistance to antibiotics therapy and current treatment strategies. Biomed Mater. 2022;17(2):17. doi: 10.1088/1748-605X/ac50f6
- Rasool FN, Saavedra MA, Pamba S, et al. Isolation and characterization of human pathogenic multidrug resistant bacteria associated with plastic litter collected in Zanzibar. J Hazard Mater. 2021;405:124591. doi: 10.1016/j.jhazmat.2020.124591
- Shahed-Al-Mahmud M, Roy R, Sugiokto FG, et al. Phage φAB6-borne depolymerase combats Acinetobacter baumannii biofilm formation and infection. Antibiotics. 2021;10(3):10. doi: 10.3390/antibiotics10030279
- Li M, Wang H, Chen L, et al. Identification of a phage-derived depolymerase specific for KL47 capsule of Klebsiella pneumoniae and its therapeutic potential in mice. Virol Sin. 2022;37(4):538–546. doi: 10.1016/j.virs.2022.04.005
- Timoshina OY, Kasimova AA, Shneider MM, et al. Loss of a branch sugar in the Acinetobacter baumannii K3-type capsular polysaccharide due to Frameshifts in the gtr6 glycosyltransferase gene leads to susceptibility to phage APK37.1. Microbiol Spectr. 2023;11(1):e0363122. doi: 10.1128/spectrum.03631-22
- Pires DP, Oliveira H, Melo LD, et al. Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl Microbiol Biotechnol. 2016;100(5):2141–2151. doi: 10.1007/s00253-015-7247-0
- Drulis-Kawa Z, Majkowska-Skrobek G, Maciejewska B. Bacteriophages and phage-derived proteins–application approaches. Curr Med Chem. 2015;22(14):1757–1773. doi: 10.2174/0929867322666150209152851
- Topka-Bielecka G, Dydecka A, Necel A, et al. Bacteriophage-derived depolymerases against bacterial biofilm. Antibiotics. 2021;10(2):10. doi: 10.3390/antibiotics10020175
- Majkowska-Skrobek G, Latka A, Berisio R, et al. Capsule-Targeting Depolymerase, derived from Klebsiella KP36 phage, as a tool for the development of anti-virulent strategy. Viruses. 2016;8(12):324. doi: 10.3390/v8120324
- Abdelkader K, Gutierrez D, Latka A, et al. The specific capsule depolymerase of phage PMK34 sensitizes Acinetobacter baumannii to serum killing. Antibiotics. 2022;11(5):11. doi: 10.3390/antibiotics11050677
- Wu Y, Wang R, Xu M, et al. A novel polysaccharide depolymerase encoded by the phage SH-KP152226 confers specific activity against multidrug-resistant Klebsiella pneumoniae via biofilm degradation. Front Microbiol. 2019;10:2768. doi: 10.3389/fmicb.2019.02768
- Chen X, Liu M, Zhang P, et al. Phage-Derived Depolymerase as an antibiotic adjuvant against Multidrug-Resistant Acinetobacter baumannii. Front Microbiol. 2022;13:845500. doi: 10.3389/fmicb.2022.845500
- Sutherland IW. The biofilm matrix–an immobilized but dynamic microbial environment. Trends Microbiol. 2001;9(5):222–227. doi: 10.1016/S0966-842X(01)02012-1
- Belfield K, Bayston R, Birchall JP, et al. Do orally administered antibiotics reach concentrations in the middle ear sufficient to eradicate planktonic and biofilm bacteria? A review. Int J Pediatr Otorhinolaryngol. 2015;79(3):296–300. doi: 10.1016/j.ijporl.2015.01.003
- Hoiby N, Bjarnsholt T, Givskov M, et al. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35(4):322–332. doi: 10.1016/j.ijantimicag.2009.12.011
- Kostakioti M, Hadjifrangiskou M, Hultgren SJ. Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb Perspect Med. 2013;3(4):a010306. doi: 10.1101/cshperspect.a010306
- Jang H, Rusconi R, Stocker R. Biofilm disruption by an air bubble reveals heterogeneous age-dependent detachment patterns dictated by initial extracellular matrix distribution. NPJ Biofilms Microbiomes. 2017;3(1):6. doi: 10.1038/s41522-017-0014-5
- Abebe GM. The role of bacterial biofilm in antibiotic resistance and food contamination. Int J Microbiol. 2020;2020:1705814. doi: 10.1155/2020/1705814
- Petrova OE, Sauer K. Sticky situations: key components that control bacterial surface attachment. J Bacteriol. 2012;194(10):2413–2425. doi: 10.1128/JB.00003-12
- Jamal M, Ahmad W, Andleeb S, et al. Bacterial biofilm and associated infections. J Chin Med Assoc. 2018;81(1):7–11. doi: 10.1016/j.jcma.2017.07.012
- Habash M, Reid G. Microbial biofilms: their development and significance for medical device-related infections. J Clin Pharmacol. 1999;39(9):887–898. doi: 10.1177/00912709922008506
- Legoux R, Lelong P, Jourde C, et al. N-acetyl-heparosan lyase of Escherichia coli K5: gene cloning and expression. J Bacteriol. 1996;178(24):7260–7264. doi: 10.1128/jb.178.24.7260-7264.1996
- Blanco-Cabra N, Paetzold B, Ferrar T, et al. Characterization of different alginate lyases for dissolving Pseudomonas aeruginosa biofilms. Sci Rep. 2020;10(1):9390. doi: 10.1038/s41598-020-66293-2
- Lahiri D, Nag M, Dey A, et al. Immobilized enzymes as potent antibiofilm agent. Biotechnol Prog. 2022;38(5):e3281. doi: 10.1002/btpr.3281
- Woolcock JB. The capsule of Streptococcus equi. J Gen Microbiol. 1974;85(2):372–375. doi: 10.1099/00221287-85-2-372
- Latka A, Maciejewska B, Majkowska-Skrobek G, et al. Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl Microbiol Biotechnol. 2017;101(8):3103–3119. doi: 10.1007/s00253-017-8224-6
- Singh R, Sahore S, Kaur P, et al. Penetration barrier contributes to bacterial biofilm-associated resistance against only select antibiotics, and exhibits genus-, strain- and antibiotic-specific differences. Pathog Dis. 2016;74(6):ftw056. doi: 10.1093/femspd/ftw056
- Singh R, Ray P, Das A, et al. Penetration of antibiotics through Staphylococcus aureus and Staphylococcus epidermidis biofilms. J Antimicrob Chemother. 2010;65(9):1955–1958. doi: 10.1093/jac/dkq257
- Nguyen H, Nguyen TH, Otto M. The staphylococcal exopolysaccharide PIA - Biosynthesis and role in biofilm formation, colonization, and infection. Comput Struct Biotechnol J. 2020;18:3324–3334. doi: 10.1016/j.csbj.2020.10.027
- Nuryastuti T, Krom BP. Ica-status of clinical Staphylococcus epidermidis strains affects adhesion and aggregation: a thermodynamic analysis. Antonie Van Leeuwenhoek. 2017;110(11):1467–1474. doi: 10.1007/s10482-017-0899-2
- Anderl JN, Franklin MJ, Stewart PS. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother. 2000;44(7):1818–1824. doi: 10.1128/AAC.44.7.1818-1824.2000
- Hoyle BD, Alcantara J, Costerton JW. Pseudomonas aeruginosa biofilm as a diffusion barrier to piperacillin. Antimicrob Agents Chemother. 1992;36(9):2054–2056. doi: 10.1128/AAC.36.9.2054
- Abdi-Ali A, Mohammadi-Mehr M, Agha AY. Bactericidal activity of various antibiotics against biofilm-producing Pseudomonas aeruginosa. Int J Antimicrob Agents. 2006;27(3):196–200. doi: 10.1016/j.ijantimicag.2005.10.007
- Wu Y, Klapper I, Stewart PS. Hypoxia arising from concerted oxygen consumption by neutrophils and microorganisms in biofilms. Pathog Dis. 2018;76(4). doi: 10.1093/femspd/fty043
- Pabst B, Pitts B, Lauchnor E, et al. Gel-entrapped Staphylococcus aureus bacteria as models of biofilm infection exhibit growth in dense aggregates, oxygen limitation, antibiotic tolerance, and heterogeneous gene expression. Antimicrob Agents Chemother. 2016;60(10):6294–6301. doi: 10.1128/AAC.01336-16
- Uribe-Alvarez C, Chiquete-Felix N, Contreras-Zentella M, et al. Staphylococcus epidermidis: metabolic adaptation and biofilm formation in response to different oxygen concentrations. Pathog Dis. 2016;74(1):ftv111. doi: 10.1093/femspd/ftv111
- Pulukkody AC, Yung YP, Donnarumma F, et al. Spatially resolved analysis of Pseudomonas aeruginosa biofilm proteomes measured by laser ablation sample transfer. PLoS One. 2021;16(7):e0250911. doi: 10.1371/journal.pone.0250911
- Ramos-Gonzalez MI, Travieso ML, Soriano MI, et al. Genetic dissection of the regulatory network associated with high c-di-GMP levels in Pseudomonas putida KT2440. Front Microbiol. 2016;7:1093. doi: 10.3389/fmicb.2016.01093
- Mills E, Petersen E, Kulasekara BR, et al. A direct screen for c-di-GMP modulators reveals a salmonella typhimurium periplasmic ʟ-arginine–sensing pathway. Sci Signal. 2015;8(380):ra57. doi: 10.1126/scisignal.aaa1796
- Anderl JN, Zahller J, Roe F, et al. Role of nutrient limitation and stationary-phase existence in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother. 2003;47(4):1251–1256. doi: 10.1128/AAC.47.4.1251-1256.2003
- Hunter RC, Beveridge TJ. Application of a pH-sensitive fluoroprobe (C-SNARF-4) for pH microenvironment analysis in Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2005;71(5):2501–2510. doi: 10.1128/AEM.71.5.2501-2510.2005
- Venglarcik JR, Blair LL, Dunkle LM. pH-dependent oxacillin tolerance of Staphylococcus aureus. Antimicrob Agents Chemother. 1983;23(2):232–235. doi: 10.1128/AAC.23.2.232
- Xu KD, McFeters GA, Stewart PS. Biofilm resistance to antimicrobial agents. Microbiology (Reading). 2000;146(Pt 3):547–549. doi: 10.1099/00221287-146-3-547
- Zheng Z, Stewart PS. Penetration of rifampin through Staphylococcus epidermidis biofilms. Antimicrob Agents Chemother. 2002;46(3):900–903. doi: 10.1128/AAC.46.3.900-903.2002
- Schierholz JM, Beuth J, Pulverer G. Killing effects of antibiotics and two-fold antimicrobial combinations on proliferating and non growing staphylococci. Zentralbl Bakteriol. 1998;288(4):527–539. doi: 10.1016/S0934-8840(98)80072-8
- Zimmerli W, Frei R, Widmer AF, et al. Microbiological tests to predict treatment outcome in experimental device-related infections due to Staphylococcus aureus. J Antimicrob Chemother. 1994;33:959–967.
- Olsen I. Biofilm-specific antibiotic tolerance and resistance. Eur J Clin Microbiol Infect Dis. 2015;34(5):877–886. doi: 10.1007/s10096-015-2323-z
- Savage VJ, Chopra I, O’Neill AJ. Staphylococcus aureus biofilms promote horizontal transfer of antibiotic resistance. Antimicrob Agents Chemother. 2013;57(4):1968–1970. doi: 10.1128/AAC.02008-12
- Kouzel N, Oldewurtel ER, Maier B, et al. Gene transfer efficiency in gonococcal biofilms: role of biofilm Age, architecture, and pilin antigenic variation. J Bacteriol. 2015;197(14):2422–2431. doi: 10.1128/JB.00171-15
- Tanner WD, Atkinson RM, Goel RK, et al. Horizontal transfer of the blaNDM-1 gene to Pseudomonas aeruginosa and Acinetobacter baumannii in biofilms. FEMS Microbiol Lett. 2017;364(8). doi: 10.1093/femsle/fnx048
- Fisarova L, Botka T, Du X, et al. Staphylococcus epidermidis phages transduce antimicrobial resistance plasmids and mobilize chromosomal islands. mSphere. 2021;6(3):6. doi: 10.1128/mSphere.00223-21
- Lattar SM, Wu X, Brophy J, et al. A mechanism of unidirectional Transformation, leading to antibiotic resistance, occurs within nasopharyngeal pneumococcal biofilm consortia. MBio. 2018;9(3). doi: 10.1128/mBio.00561-18
- Uruen C, Chopo-Escuin G, Tommassen J, et al. Biofilms as promoters of bacterial antibiotic resistance and tolerance. Antibiotics. 2020;10(1):10. doi: 10.3390/antibiotics10010003
- Driffield K, Miller K, Bostock JM, et al. Increased mutability of Pseudomonas aeruginosa in biofilms. J Antimicrob Chemother. 2008;61(5):1053–1056. doi: 10.1093/jac/dkn044
- Sultan AM, Amer GF, Nabiel Y. Quinolone-resistant uropathogenic E. coli: is there a relation between qnr genes, gyrA gene target site mutation and biofilm formation? J Med Microbiol. 2021;70(10). doi: 10.1099/jmm.0.001432
- Turkel I, Yildirim T, Yazgan B, et al. Relationship between antibiotic resistance, efflux pumps, and biofilm formation in extended-spectrum β-lactamase producing Klebsiella pneumoniae. J Chemother. 2018;30(6–8):354–363. doi: 10.1080/1120009X.2018.1521773
- Knight DB, Rudin SD, Bonomo RA, et al. Acinetobacter nosocomialis: defining the role of efflux pumps in resistance to antimicrobial therapy, surface motility, and biofilm formation. Front Microbiol. 2018;9:1902. doi: 10.3389/fmicb.2018.01902
- Li XF, Shi HQ, Liang Y, et al. Interaction of biofilm and efflux pump in clinical isolates of carbapenem resistant P. aeruginosa. Eur Rev Med Pharmacol Sci. 2022;26(5):1729–1737. doi: 10.26355/eurrev_202203_28242
- Lin H, Paff ML, Molineux IJ, et al. Therapeutic application of phage capsule depolymerases against K1, K5, and K30 capsulated E. coli in mice. Front Microbiol. 2017;8:2257. doi: 10.3389/fmicb.2017.02257
- Nimmich W, Schmidt G, Krallmann-Wenzel U. Two different Escherichia coli capsular polysaccharide depolymerases each associated with one of the coliphage φK5 and φK20. FEMS Microbiol Lett. 1991;66(2):137–141. doi: 10.1016/0378-1097(91)90322-2
- Domingo-Calap P, Beamud B, Vienne J, et al. Isolation of four lytic phages infecting Klebsiella pneumoniae K22 clinical isolates from Spain. Int J Mol Sci. 2020;21(2):21. doi: 10.3390/ijms21020425
- Gorodnichev RB, Volozhantsev NV, Krasilnikova VM, et al. Novel Klebsiella pneumoniae K23-specific bacteriophages from different families: similarity of depolymerases and their therapeutic potential. Front Microbiol. 2021;12:669618. doi: 10.3389/fmicb.2021.669618
- Eckstein S, Stender J, Mzoughi S, et al. Isolation and characterization of lytic phage TUN1 specific for Klebsiella pneumoniae K64 clinical isolates from Tunisia. BMC Microbiol. 2021;21(1):186. doi: 10.1186/s12866-021-02251-w
- Kasimova AA, Arbatsky NP, Timoshina OY, et al. The K26 capsular polysaccharide from Acinetobacter baumannii KZ-1098: structure and cleavage by a specific phage depolymerase. Int j biol macromol. 2021;191:182–191. doi: 10.1016/j.ijbiomac.2021.09.073
- Drobiazko AY, Kasimova AA, Evseev PV, et al. Capsule-targeting depolymerases derived from Acinetobacter baumannii prophage regions. Int J Mol Sci. 2022;23(9):23. doi: 10.3390/ijms23094971
- Squeglia F, Maciejewska B, Latka A, et al. Structural and functional studies of a Klebsiella phage capsule Depolymerase Tailspike: mechanistic insights into capsular degradation. Structure. 2020;28(6):613–624.e4. doi: 10.1016/j.str.2020.04.015
- Tu IF, Lin TL, Yang FL, et al. Structural and biological insights into Klebsiella pneumoniae surface polysaccharide degradation by a bacteriophage K1 lyase: implications for clinical use. J Biomed Sci. 2022;29(1):9. doi: 10.1186/s12929-022-00792-4
- Mushtaq N, Redpath MB, Luzio JP, et al. Treatment of experimental Escherichia coli infection with recombinant bacteriophage-derived capsule depolymerase. J Antimicrob Chemother. 2005;56(1):160–165. doi: 10.1093/jac/dki177
- Kimura K, Itoh Y. Characterization of poly-γ-glutamate hydrolase encoded by a bacteriophage genome: possible role in phage infection of Bacillus subtilis encapsulated with poly-γ-glutamate. Appl Environ Microbiol. 2003;69(5):2491–2497. doi: 10.1128/AEM.69.5.2491-2497.2003
- Ely B, Gibbs W, Diez S, et al. The caulobacter crescentus transducing phage Cr30 is a unique member of the T4-like family of myophages. Curr Microbiol. 2015;70(6):854–858. doi: 10.1007/s00284-015-0799-5
- Yoon BH, Chang HI. Complete genomic sequence of the lactobacillus temperate phage LF1. Arch Virol. 2011;156(10):1909–1912. doi: 10.1007/s00705-011-1082-0
- Maaroufi H, Levesque RC. Glycoside hydrolase family 32 is present in Bacillus subtilis phages. Virol J. 2015;12(1):157. doi: 10.1186/s12985-015-0373-6
- Chua J, Manning PA, Morona R. The shigella flexneri bacteriophage Sf6 tailspike protein (Tsp)/endorhamnosidase is related to the bacteriophage P22 TSP and has a motif common to exo- and endoglycanases, and C-5 epimerases. Microbiology (Reading). 1999;145(7):1649–1659. doi: 10.1099/13500872-145-7-1649
- Mushtaq N, Redpath MB, Luzio JP, et al. Prevention and cure of systemic Escherichia coli K1 infection by modification of the bacterial phenotype. Antimicrob Agents Chemother. 2004;48(5):1503–1508. doi: 10.1128/AAC.48.5.1503-1508.2004
- Smith NL, Taylor EJ, Lindsay AM, et al. Structure of a group a streptococcal phage-encoded virulence factor reveals a catalytically active triple-stranded β-helix. Proc Natl Acad Sci U S A. 2005;102(49):17652–17657. doi: 10.1073/pnas.0504782102
- Baker JR, Dong S, Pritchard DG. The hyaluronan lyase of Streptococcus pyogenes bacteriophage H4489A. Biochem J. 2002;365(1):317–322. doi: 10.1042/bj20020149
- Glonti T, Chanishvili N, Taylor PW. Bacteriophage-derived enzyme that depolymerizes the alginic acid capsule associated with cystic fibrosis isolates of Pseudomonas aeruginosa. J Appl Microbiol. 2010;108(2):695–702. doi: 10.1111/j.1365-2672.2009.04469.x
- Cornelissen A, Ceyssens PJ, T’Syen J, et al. The T7-related Pseudomonas putida phage phi15 displays virion-associated biofilm degradation properties. PLoS One. 2011;6:e18597. doi: 10.1371/journal.pone.0018597
- Liu Y, Mi Z, Mi L, et al. Identification and characterization of capsule depolymerase Dpo48 from Acinetobacter baumannii phage IME200. PeerJ. 2019;7:e6173. doi: 10.7717/peerj.6173
- Rice CJ, Kelly SA, O’Brien SC, et al. Novel phage-derived depolymerase with activity against Proteus mirabilis biofilms. Microorganisms. 2021;9(10):2172. doi: 10.3390/microorganisms9102172
- Wang C, Li P, Zhu Y, et al. Identification of a novel Acinetobacter baumannii phage-derived depolymerase and its therapeutic application in mice. Front Microbiol. 2020;11:1407. doi: 10.3389/fmicb.2020.01407
- Oliveira H, Pinto G, Mendes B, et al. A tailspike with exopolysaccharide depolymerase activity from a New Providencia stuartii phage makes multidrug-resistant bacteria susceptible to serum-mediated killing. Appl Environ Microbiol. 2020;86(13). doi: 10.1128/AEM.00073-20
- Oliveira H, Mendes A, Fraga AG, et al. K2 capsule depolymerase is highly stable, is refractory to resistance, and protects larvae and mice from Acinetobacter baumannii sepsis. Appl Environ Microbiol. 2019;85(17). doi: 10.1128/AEM.00934-19
- Oliveira H, Costa AR, Ferreira A, et al. Functional analysis and antivirulence properties of a New depolymerase from a myovirus that infects Acinetobacter baumannii capsule K45. J Virol. 2019;93(4). doi: 10.1128/JVI.01163-18
- Pertics BZ, Cox A, Nyul A, et al. Isolation and characterization of a novel lytic bacteriophage against the K2 capsule-Expressing Hypervirulent Klebsiella pneumoniae strain 52145, and Identification of its functional depolymerase. Microorganisms. 2021;9(3):650. doi: 10.3390/microorganisms9030650
- Hernandez-Morales AC, Lessor LL, Wood TL, et al. Genomic and biochemical characterization of Acinetobacter podophage petty reveals a novel lysis mechanism and tail-associated depolymerase activity. J Virol. 2018;92(6). doi: 10.1128/JVI.01064-17
- Hsu CR, Lin TL, Pan YJ, et al. Isolation of a bacteriophage specific for a new capsular type of Klebsiella pneumoniae and characterization of its polysaccharide depolymerase. PLoS One. 2013;8(8):e70092. doi: 10.1371/journal.pone.0070092
- Liu Y, Leung S, Huang Y, et al. Identification of two depolymerases from phage IME205 and their antivirulent functions on K47 capsule of Klebsiella pneumoniae. Front Microbiol. 2020;11:218. doi: 10.3389/fmicb.2020.00218
- Pan YJ, Lin TL, Chen CC, et al. Klebsiella phage PhiK64-1 encodes multiple depolymerases for multiple host capsular types. J Virol. 2017;91(6):e02457–16. doi: 10.1128/JVI.02457-16
- Pan YJ, Lin TL, Chen YY, et al. Identification of three podoviruses infecting Klebsiella encoding capsule depolymerases that digest specific capsular types. Microbiol Biotechnol. 2019;12(3):472–486. doi: 10.1111/1751-7915.13370
- Hsieh PF, Lin HH, Lin TL, et al. Two T7-like Bacteriophages, K5-2 and K5-4, each encodes two capsule depolymerases: isolation and functional characterization. Sci Rep. 2017;7(1):4624. doi: 10.1038/s41598-017-04644-2
- Broudy TB, Pancholi V, Fischetti VA, et al. Induction of lysogenic bacteriophage and phage-associated toxin from group a streptococci during coculture with human pharyngeal cells. Infect Immun. 2001;69(3):1440–1443. doi: 10.1128/IAI.69.3.1440-1443.2001
- Wong TY, Preston LA, Schiller NL. ALGINATE LYASE: review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu Rev Microbiol. 2000;54(1):289–340. doi: 10.1146/annurev.micro.54.1.289
- Davidson IW, Lawson CJ, Sutherland IW. An alginate lysate from Azotobacter vinelandii phage. Journal Of General Microbiology. 1977;98(1):223–229. doi: 10.1099/00221287-98-1-223
- Chen Y, Li X, Wang S, et al. A novel tail-associated O91-specific polysaccharide depolymerase from a podophage reveals lytic efficacy of Shiga toxin-producing Escherichia coli. Appl Environ Microbiol. 2020;86(9). doi: 10.1128/AEM.00145-20
- Olszak T, Shneider MM, Latka A, et al. The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence. Sci Rep. 2017;7(1):16302. doi: 10.1038/s41598-017-16411-4
- Kaszowska M, Majkowska-Skrobek G, Markwitz P, et al. The mutation in wbaP cps gene cluster selected by phage-borne depolymerase abolishes capsule production and diminishes the virulence of Klebsiella pneumoniae. Int J Mol Sci. 2021;22(21):22. doi: 10.3390/ijms222111562
- Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 2006;75(1):39–68. doi: 10.1146/annurev.biochem.75.103004.142545
- Majkowska-Skrobek G, Latka A, Berisio R, et al. Phage-borne depolymerases decrease Klebsiella pneumoniae resistance to innate defense mechanisms. Front Microbiol. 2018;9:2517. doi: 10.3389/fmicb.2018.02517
- Guerrero-Bustamante CA, Dedrick RM, Garlena RA, et al. Toward a phage cocktail for tuberculosis: susceptibility and tuberculocidal action of mycobacteriophages against diverse mycobacterium tuberculosis strains. MBio. 2021;12(3):12. doi: 10.1128/mBio.00973-21
- Li M, Chang R, Lin Y, et al. Phage cocktail powder for Pseudomonas aeruginosa respiratory infections. Int J Pharm. 2021;596:120200. doi: 10.1016/j.ijpharm.2021.120200
- Mijbel AB, Gatea KS, Al-Bayati MA, et al. A novel phage cocktail therapy of the urinary tract infection in a mouse model. Arch Razi Inst. 2021;76(5):1229–1236. doi: 10.22092/ari.2021.356004.1762
- Jault P, Leclerc T, Jennes S, et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. Lancet Infect Dis. 2019;19(1):35–45. doi: 10.1016/S1473-3099(18)30482-1
- Sarker SA, Sultana S, Reuteler G, et al. Oral phage therapy of acute bacterial diarrhea with two coliphage preparations: a randomized trial in children from Bangladesh. EBioMedicine. 2016;4:124–137. doi: 10.1016/j.ebiom.2015.12.023
- Golembo M, Puttagunta S, Rappo U, et al. Development of a topical bacteriophage gel targeting Cutibacterium acnes for acne prone skin and results of a phase 1 cosmetic randomized clinical trial. Skin Health Dis. 2022;2(2):e93. doi: 10.1002/ski2.93
- Verma V, Harjai K, Chhibber S. Structural changes induced by a lytic bacteriophage make ciprofloxacin effective against older biofilm of Klebsiella pneumoniae. Biofouling. 2010;26(6):729–737. doi: 10.1080/08927014.2010.511196
- Latka A, Drulis-Kawa Z. Advantages and limitations of microtiter biofilm assays in the model of antibiofilm activity of Klebsiella phage KP34 and its depolymerase. Sci Rep. 2020;10(1):20338. doi: 10.1038/s41598-020-77198-5
- Luo CH, Hsu YH, Wu WJ, et al. Phage digestion of a bacterial capsule imparts resistance to two antibiotic Agents. Microorganisms. 2021;9(4):794. doi: 10.3390/microorganisms9040794
- Dunne MW, Aronin SI, Das AF, et al. Sulopenem or ciprofloxacin for the treatment of uncomplicated urinary tract infections in women: a phase 3, randomized trial. Clin Infect Dis. 2023;76(1):66–77. doi: 10.1093/cid/ciac738
- Chen Y, Sun E, Yang L, et al. Therapeutic application of bacteriophage PHB02 and its putative depolymerase against Pasteurella multocida capsular type a in mice. Front Microbiol. 2018;9:1678. doi: 10.3389/fmicb.2018.01678
- Mi L, Liu Y, Wang C, et al. Identification of a lytic Pseudomonas aeruginosa phage depolymerase and its anti-biofilm effect and bactericidal contribution to serum. Vir Gen. 2019;55(3):394–405. doi: 10.1007/s11262-019-01660-4
- Russo A, Gavaruzzi F, Ceccarelli G, et al. Multidrug-resistant Acinetobacter baumannii infections in COVID-19 patients hospitalized in intensive care unit. Infection. 2022;50(1):83–92. doi: 10.1007/s15010-021-01643-4

