ABSTRACT
CagA is a significant oncogenic factor injected into host cells by Helicobacter pylori, which is divided into two subtypes: East Asian type (CagAE), characterized by the EPIYA-D motif, and western type (CagAW), harboring the EPIYA-C motif. CagAE has been reported to have higher carcinogenicity than CagAW, although the underlying reason is not fully understood. SHIP2 is an intracellular phosphatase that can be recruited by CagA to perturb the homeostasis of intracellular signaling pathways. In this study, we found that SHIP2 contributes to the higher oncogenicity of CagAE. Co-Immunoprecipitation and Pull-down assays showed that CagAE bind more SHIP2 than CagAW. Immunofluorescence staining showed that a higher amount of SHIP2 recruited by CagAE to the plasma membrane catalyzes the conversion of PI(3,4,5)P3 into PI(3,4)P2. This alteration causes higher activation of Akt signaling, which results in enhanced IL-8 secretion, migration, and invasion of the infected cells. SPR analysis showed that this stronger interaction between CagAE and SHIP2 stems from the higher affinity between the EPIYA-D motif of CagAE and the SH2 domain of SHIP2. Structural analysis revealed the crucial role of the Phe residue at the Y + 5 position in EPIYA-D. After mutating Phe of CagAE into Asp (the corresponding residue in the EPIYA-C motif) or Ala, the activation of downstream Akt signaling was reduced and the malignant transformation of infected cells was alleviated. These findings revealed that CagAE hijacks SHIP2 through its EPIYA-D motif to enhance its carcinogenicity, which provides a better understanding of the higher oncogenic risk of H. pylori CagAE.
Introduction
Helicobacter pylori is a Gram-negative microaerophilic bacillus that colonizes the stomach [Citation1]. More than 440 million people carry this bacterium [Citation2]. Persistent infection with H. pylori is a precipitating factor in the development of gastric diseases, including chronic gastritis, peptic ulcer, gastric mucosa-associated lymphoid tissue (MALT) lymphoma, and gastric cancer (GC) [Citation3]. Numerous studies have revealed a variety of virulence factors in H. pylori determining its pathogenicity, including cytotoxin-associated antigen A (CagA), vacuolating cytotoxin (VacA), duodenal ulcer promoting gene A protein (DupA), outer inflammatory protein (OipA), and gamma-glutamyl transpeptidase (GGT) [Citation4]. Among these, CagA is closely associated with carcinogenesis [Citation5].
The molecular weight of CagA ranges from 125 to 145 kDa in different strains [Citation6]. This variation is mainly attributed to the different repetition numbers of the glutamic acid-proline-isoleucine-tyrosine-alanine (EPIYA) motif and the CagA polymeric motif (CM) at its carboxyl-terminal (C-terminal) [Citation5]. Owing to the diversity of its flanking sequence, the EPIYA motif can be categorized into four types: EPIYA-A, -B, -C, and -D [Citation7]. H. pylori strains popular in East Asian countries such as China, Japan, and Korea possess EPIYA-D, and the arrangement of EPIYA motifs is EPIYA-A-B-D. Prevalent strains in the rest of the world predominantly harbor EPIYA-A-B-C (varying numbers of EPIYA-C, mostly 1–3) [Citation7]. Thus, CagA can be divided into two categories: Western type (CagAW) and East Asian type (CagAE). Epidemiological studies have shown a closer association between CagAE and GC. However, the underlying cause is not yet fully understood.
In H. pylori infection, CagA is delivered into gastric epithelial cells via the bacterial type IV secretion system and is located on the inner surface of the plasma membrane [Citation8]. Intracellular CagA undergoes tyrosine phosphorylation at EPIYA motifs by cellular kinases, such as Csk, c-Src, and c-Abl [Citation9]. CagA acts as a non-physiological scaffold/hub protein by interacting with multiple host signaling molecules, including pro-oncogenic phosphatase SHP2, growth factor receptor-bound protein 2 (Grb2), and inositol polyphosphate 5-phosphatase (SHIP2), in a tyrosine phosphorylation-dependent or phosphorylation-independent manner [Citation10]. The CagA-mediated manipulation of intracellular signaling promotes the neoplastic transformation of gastric epithelial cells. Among these intracellular signaling molecules, SHP2 has been reported to have a higher affinity for binding the EPIYA-D motif in CagAE than that with the EPIYA-C motif in CagAW, thus contributing to the stronger pathogenesis of CagAE [Citation10]. This study opens up a window to resolve the pathogenic differences between the two CagA subtypes from the perspective of the intracellular regulatory network.
SHIP2, also known as inositol polyphosphate 5-phosphatase-like protein-1 (INPPL1), is a 142 kDa protein containing an SH2 domain in the amino (N) terminus [Citation11]. The major role of this lipid phosphatase is to dephosphorylate phosphatidylinositol(3,4,5)trisphosphate (PI(3,4,5)P3) at the 5’-position and convert it to PI(3,4)P2 [Citation12]. By activating the PI3K/Akt signaling pathway, SHIP2 plays an oncogenic role in several tumors, including colorectal cancer, hepatocellular carcinoma, and non-small cell lung cancer [Citation13]. While, it was found to play a tumor-suppressive role in GC by inhibiting PI3K/Akt signaling [Citation14]. This suggests that the role of SHIP2 in carcinogenesis is intricate. A recent study showed that SHIP2 interacts with the EPIYA motifs of CagA through its SH2 domain, thereby strengthening the attachment of H. pylori and potentiating the delivery of CagA [Citation15]. Our study also confirmed the binding of SHIP2 to CagA and found a difference in its affinity for CagAW and CagAE. We wondered whether this binding difference influenced the pathogenesis of CagAE and CagAW.
In this study, we investigated the effect of the SHIP2-CagA interaction on some transformation indications of gastric cells, explored the mechanism underlying the affinity difference between SHIP2-CagAE and SHIP2-CagAW, and clarified the biological effects of this difference in H. pylori pathogenesis.
Materials and methods
Cell lines, bacterial strains and cultivation
Human immortalized gastric epithelial cell (GES-1) obtained from Beyotime Biotechnology (C6268, Shanghai, China) was cultured in DMEM supplemented with 10% (v/v) fetal bovine serum (FBS) (ExCell, Shanghai, China), and gastric cancer cell line AGS was cultured in RPMI-1640 medium supplemented with the same amount of FBS. The cells were incubated in a humidified atmosphere containing 5% (v/v) CO2 at 37°C. H. pylori strains SHCH30 (encoding CagAE, EPIYA-A-B-D), 26695 (encoding CagAW, EPIYA-A-B-C) and G27 were preserved in the laboratory. All H. pylori strains were cultured on Brucella agar plates (OXOID, Basingstoke, UK) containing 5% (v/v) defibrinated sheep blood at 37°C under microaerobic conditions (5% O2, 10% CO2 and 85% N2). Escherichia coli DH5α used for plasmid cloning, and BL21 (DE3) strain for heterologous expression were cultured routinely at 37°C in Luria-Bertani medium.
Construction of CagA variant strains
H. pylori strain G27 was used to knockout cagA gene, and then to construct mutants expressing CagAE or CagAW. The original cagA gene in G27 was deleted by homologous recombination method as previously described [Citation16]. The resulting CagA-deleted strain (ΔCagA) exhibits kanamycin resistance. The coding sequence of CagAW or CagAE was amplified from the genome DNA of H. pylori 26695 or SHCH30, which was transformed into ΔCagA mutant via H. pylori-E. coli shuttle plasmid pCHFHP [Citation17] to generate H. pylori G27 mutants expressing CagAW or CagAE. The point mutation of CagAE with substitution of Phe in EPIYA-D to Asp or Ala was generated by overlapping PCR, and resulting sequence was also transformed into ΔCagA mutant via shuttle vector.
Infection assay
AGS and GES-1 cells were seeded into 6-well cell culture plates at a density of 3 × 105 cells per well and cultured overnight. H. pylori in the logarithmic phase was collected, suspended in PBS, and added to the cell cultures at an MOI of 100. When necessary, SHP099 (Aladdin, Shanghai, China) was added to the cell cultures 48 h before infection at a concentration of 10 µM to specifically inhibit the activity of SHP2. The morphology of infected cells was examined using a light microscope (Olympus, Tokyo, Japan). Elongated cells with the longest axis exceeding the shortest axis by more than 2-fold were counted as the hummingbird phenotype. Cell diameters were measured using ImageJ software (https://imagej.nih.gov/ij/). Three independent experiments were performed, and ten different observation fields in the culture dishes were counted in each repeat.
Silencing of SHIP2
Lentivirus-mediated SHIP2 short hairpin RNA (shRNA) silencing vector was constructed by GenePharma Biotech Company (Shanghai, China). AGS and GES-1 cells were infected with the virus according to the manufacturer’s protocol. Fluorescence-activated cell sorting (FACS) and puromycin were used to screen stable clones.
Co-immunoprecipitation assay
Co-immunoprecipitation of SHIP2 with p-CagA was conducted in GES-1 and AGS cell lines. After 6 h of infection with H. pylori, cells were lysed and centrifuged. Antibody against SHIP2 was added into the supernate for an overnight incubation at 4°C. Pierce protein A/G magnetic beads from the Immunoprecipitation Kit (Beyotime Biotechnology) were then added to precipitate the antigen-antibody complexes for 2 h at 4°C. The beads were washed three times with PBS buffer and boiled in SDS loading buffer. The co-precipitates were analysed by Western blotting.
Expression and purification of GST-tag or His-tag fused SHIP2-SH2 domain
The DNA fragments encoding the SH2 domain of SHIP2 were synthesized by Beijing Tsingke Biotech (Beijing, China), and then cloned into the vector pGEX-4T-1 for GST-tagged or pET-22b for His-tagged recombinant protein expression. The resulting plasmids were transformed into E. coli BL21 (DE3) strain, which was inoculated in LB medium with shaking at 37°C until Abs. 600 nm reached 0.6. Isopropyl-β-D-thiogalactopyranoside (IPTG) (0.1 mM) was added to induce protein expression at 16°C for 16 h. The cells were harvested and suspended in 20 mM Tris-HCl (pH 8.0) containing 500 mM NaCl. After lysis by sonication, the cell lysate was centrifuged at 11,000 g for 30 min at 4°C. GST-tag or His-tag proteins in the supernatant were subsequently purified by affinity chromatography [Citation18] and gel filtration chromatography using an AKTA purification apparatus (GE Healthcare, Stockholm, Sweden) according to the manufacturer’s instructions.
Transwell assays
Cell invasion and migration were detected using 24-well transwell plates with 8 μm pore filters (Corning, New York, NY, USA) with or without Matrigel. For cell invasion detection, cells at a density of 1 × 105 per well were seeded with serum-free medium into the upper chambers pretreated with Matrigel at 37°C for 4 h. Medium plus 20% (v/v) FBS was added to the lower chambers. After incubation for 48 h, the cells and Matrigel above the chamber membranes were wiped off. Cells that invaded the other side were fixed, stained with 0.1% crystal violet, and counted. Cell migration was detected using the same procedure, except that without Matrigel pretreatment, and the incubation time was 24 h. Each assay was repeated at least three times.
Western blot analysis
The cells were collected and lysed in cold RIPA buffer (Beyotime, Shanghai, China) supplemented with 1% (v/v) phenylmethanesulfonyl fluoride (PMSF) (Beyotime). The protein concentration was measured using a BCA kit (Beyotime). These proteins were then separated on SDS polyacrylamide gels and subsequently transferred onto a polyvinylidene fluoride (PVDF) membrane. The membrane was blocked with 5% nonfat milk for 2 h at room temperature and incubated with primary antibodies overnight at 4°C. After washing off the primary antibody with TBST buffer, the secondary antibodies were added and incubated for another 1 h at room temperature. The signals were detected and imaged using an image analyzer (Tanon, Shanghai, China). ImageJ software was used to analyse the data obtained. Experiments were independently repeated at least three times.
Antibodies against pSer473-Akt (AP1208), Akt (A17909), NF-κB p65 (A19653), and phospho-NF-κB p65 (AP0125) were purchased from Abcam (Cambridge, UK). Anti-SHIP2 (C76A7) antibody was purchased from Cell Signaling Technology (Danvers, MA, USA). Monoclonal anti-PI(3,4)P2 (Z-P034) was purchased from Echelon Biosciences (San Jose, CA, USA). Anti-CagA monoclonal antibody (sc -28368) and anti-phosphotyrosine antibody (sc-7020) used for phosphorylated CagA detection were both purchased from Santa Cruz Biotechnology (Texas, USA). Anti-GAPDH polyclonal antibody (60004–1-Ig) and β-actin (81115–1-RR) monoclonal antybody purchased from Proteintech Group (Wuhan, China) were used to detect the internal reference proteins. Secondary antibodies, including anti-rabbit (PR30011) and anti-mouse IgG (PR30012), produced in goats were obtained from Proteintech Group.
Immunofluorescence staining
The cells were seeded onto 12-well chamber slides (Solarbio, Beijing, China) overnight before infection with H. pylori. After 6 h of co-incubation with H. pylori, the cells were fixed in 4% paraformaldehyde for 30 min, permeabilized with 0.01% Triton X-100 for 20 min, and incubated in 5% BSA for 1 h. The fixed samples were incubated with the primary antibody overnight at 4°C and secondary antibodies (FITC Goat anti-mouse IgG and TRITC Goat anti-rabbit IgG, AS001 and AS040, ABclonal) at room temperature for 1 h. Cellular nuclei were stained with DAPI for 15 min. Images were captured using a confocal laser microscope (Carl Zeiss; Oberkochen, Germany). Co-localization analysis was performed by using “Colocalization Finder” in ImageJ. It was performed in triplicate.
Molecular dynamic simulation
The structure of SHIP2 was downloaded from the PDB database (PDB ID: 2MK2). Structures of two short peptides of tyrosine-phosphorylated EPIYA-D (ASPEPIpYATIDFD, EPIpYA-D; pY, phosphotyrosine) and EPIYA-C (VSPEPIpYATIDDL, EPIpYA-C) were obtained from Hayashi’s data (PDB: 5X94 and PDB: 5X7B) [Citation10] with mild modifications using Alphafold2 [Citation19]. Based on these primary structures, we constructed SHIP2-EPIYA molecular dynamics (MD) simulations using OpenMM v7.5.1 [Citation20]. The Amber14SB force field [Citation21] was adopted for the treatment of standard residues in SHIP2 and the peptides, whereas the phosphorylated tyrosine was prepared by Acpype [Citation22] based on the Amber GAFF2 force field [Citation23]. The simulations were performed under periodic boundary conditions with transferable intermolecular potential with 3 points (TIP3P) water molecules in a cubic box with a minimum distance of 1.2 nm to establish the complex structures. Salt ions Na+ and Cl− were added to neutralize the system, and the salt concentration was set to 0.15 M. After 1000 steps of steepest decent energy minimization, a 1 ns NVT ensemble based equilibration (temperature = 300 K) was conducted by freezing the heavy atoms (non-hydrogen atoms) of the protein-peptide complexes and allowing free movement of the water molecules and ion atoms. Another round of 2 ns equilibration was performed by allowing fully flexible relaxation for all atoms. Starting from the obtained state, a random initial velocity was set, and 10 independent production simulations with a constant temperature (300 K) and pressure (1 bar) were carried out for 200 ns using a 2 fs time step. During the production simulations (NPT ensemble), the hydrogen atom related bonds and covalent bonds between non-hydrogen atoms are constrained by LINCS [Citation24] and SHAKE [Citation25] algorithms. The distance cutoff for the non-bonding interaction was 1.2 nm and the long-range electrostatic interaction was computed using the Particle Mesh Ewald (PME) method.
Based on the above simulation trajectory, we used the protein-peptide binding energy calculation tool MMGB/PBSA to evaluate the binding energy and affinity between SHIP2 and the two CagA-derived peptides.
Surface plasmon resonance (SPR) analysis
The peptides EPIpYA-D and EPIpYA-C were synthesized by NovoPep Limited (Shanghai, China). Affinity analysis of the SHIP2-SH2 domain and CagA-EPIYA peptides was conducted using a BIAcore T100 instrument (GE Healthcare). Briefly, recombinant SHIP2-SH2 was immobilized on a BIAcore CM5 sensor chip (GE Healthcare) as the receptor in phosphate-buffered saline (PBS, pH 5.0) to achieve a response of 1000/1500 RU. The analytes (90 µL of EPIpYA-D or EPIpYA-C at different concentrations in PBS buffer) were injected at a flow rate of 30 µL/min at 25°C. Sensorgrams were recorded and analysed using the Biacore T100 Evaluation Software.
Pull-down assay
Equal amounts of purified recombinant GST-SH2 or GST-tag were immobilized on GST bind resin. Cells infected with H. pylori were harvested, suspended in Tris-HCl buffer (20 mM, pH 8.0), and lysed by sonication. The supernatants were collected and incubated with resins immobilized with GST-tagged proteins for 2 h at 4°C with rotation. Finally, the resins were washed three times with buffer, and cellular components pulled down by the resins were eluted using 20 mM reduced glutathione and detected by western blotting. The assay was repeated at least three times.
IL-8 detection
The cell culture supernatants of AGS and GES-1 cells incubated with H. pylori were collected for IL-8 detection using a human IL-8 ELISA Kit (Elabscience Biotechnology, Wuhan, China) according to the manufacturer’s instructions. Three independent detections were performed.
Statistical analyses
For every result requiring statistical analysis, at least three independent experiments were conducted. The values are expressed as mean ± standard deviation (SD). Statistical analysis was performed using Student’s t-test or two-way analysis of variance (ANOVA) in GraphPad Prism version 7.0. A p-value less than 0.1 was considered statistically significant.
Results
SHIP2 acts in CagA inducing cellular oncogenic transformation and contributes to the higher oncogenicity of CagAE
It has been reported that H. pylori CagA can recruit SHIP2 through its EPIYA motif, interacting with the SHIP2-SH2 domain to strengthen its delivery and pathogenesis [Citation15]. However, the actions of SHIP2 in this process and whether there is a difference between its interaction with CagAE and CagAW have not fully been understood. Here, we silenced SHIP2 and detected several cell transformation traits associated with CagA using infection assays. To exclusively compare the differences between CagAE and CagAW, two complementary H. pylori strains with identical genetic backgrounds, except for CagA types, were generated. The sequence alignment of the two CagA subtypes is shown in Figure S1. Before the cell traits examination, we measured the amounts of phosphorylated CagA (p-CagA, which only exists in host cells) in the lysates of cells infected with different strain variants to examine whether comparable CagA was injected into cells. From and Fig. S2A, we can see that there is no significant difference between groups in the amount of intracellular p-CagA, indicating that the influence of strain variants can be ignored in subsequent cell infection experiments.
Figure 1. Pathogenic effect of H. pylori CagAE and CagAW mediated by SHIP2 in AGS cells.
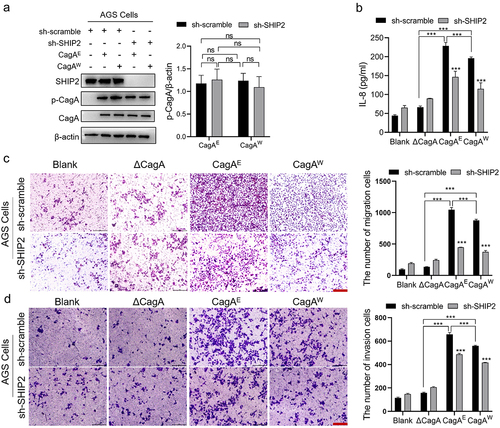
Secretion of the inflammatory factor IL-8 is thought to be helpful in the pathogenesis of H. pylori and is correlated with CagA [Citation26]. Our results showed that for the CagA-deleted strain, the secretion of IL-8 induced by infection was not obvious. But for either CagAE or CagAW harboring strains, IL-8 levels in cell cultures increased significantly after infection, and the former induced more. However, knockdown of SHIP2 dramatically reduced IL-8 release and reduced the difference between the two types of CagA: for AGS cells, CagAE-caused IL-8 secretion was 23% higher than CagAW in wild cell lines, while it was only 12% higher after knockdown of SHIP2; for GES-1 cells, SHIP2 knockdown decreased the discrepancy from 33% higher for CagAE to 11% ( and Fig. S2B). This suggests that SHIP2 is involved in the increased pathogenicity of CagAE.
Transwell assays were performed to examine cell migration and invasion. As shown in , incubation with the CagA-positive strains enhanced the migration and invasion of AGS cells, and CagAE appeared to be more effective. Silencing SHIP2 also reduced these cell properties. Moreover, the descending range in the CagAE group was sharper (40% decrease in motility and 24% decrease in invasion) than that of CagAW (31% decrease in motility and 9% decrease in invasion) in AGS-1 cells. The same trend was also detected in the GES-1 cell lines (Fig. S2C and D).
These data revealed that SHIP2 is involved in cell properties alteration induced by H. pylori CagA, including IL-8 secretion, migration, and invasion, and contributes to the higher oncogenicity of CagAE.
CagAE tethered more SHIP2 to the plasma membrane and enhanced the activation of Akt signaling
To explain how SHIP2 mediates the pathogenesis of CagAE and CagAW, the interactions between CagA and SHIP2 were explored intracellularly. CagA is known to localize to the inner membrane after translocation into the host cells [Citation27]. SHIP2 is broadly distributed in the cytoplasm [Citation28]. Because of this interaction, SHIP2 is tethered to the inner leaflet of the plasma membrane. Therefore, immunofluorescence staining was firstly used to determine their distribution in the cells. As expected, when cells were infected with H. pylori, either East Asian strain SHCH30 or Western strain 26-695, injected CagA was mainly observed on the inner membrane. In addition, more SHIP2 was transferred to the membrane (), which is consistent with the report by Fujii et al. [Citation15]. Co-localization analysis using ImageJ showed that the Rcoloc values of CagAE and SHIP2 were significantly higher than those of CagAW, suggesting the formation of a greater number of CagAE-SHIP2 complex.
Figure 2. The effect of H. pylori infection on the sub-localization of SHIP2, production of PI(3,4)P2 and activation of Akt signaling.
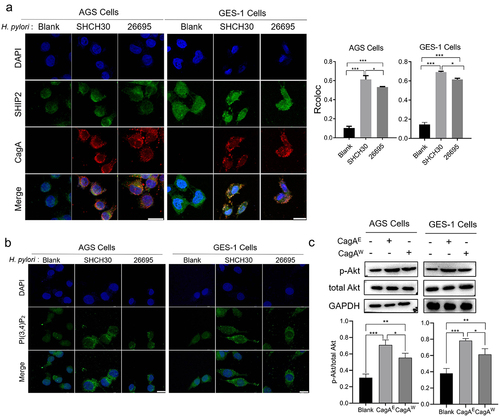
Considering that SHIP2 recruited to the membrane hydrolyzes PI(3,4,5)P3 to PI(3,4)P2 because of its lipid phosphatase activity [Citation12], we further detected the proportion of PI(3,4)P2 and PI(3,4,5)P3 in the cell membrane. Based on the fluorescence intensity, the amount of PI(3,4)P2 was higher in CagAE injected cells than that of CagAW (). The membrane accumulation of PI(3,4)P2 in CagAE injected cells confirmed the result of co-localization that more SHIP2 was recruited to the membrane by CagAE, thereby exhibiting its catalytic activity toward PI(3,4,5)P3.
As proposed in the “two PIP hypothesis” [Citation29], PI(3,4)P2 and PI(3,4,5)P3 act as two major activators of the Akt pathway, which then triggers cell survival and proliferation, thereby increasing the oncogenic potential of cells. PI(3,4)P2 has been reported to lead to higher levels of Akt activation than PI(3,4,5)P3 through phosphorylating its residue Ser473 [Citation29]. This means that more SHIP2 on the membrane produces more PI(3,4)P2 and activates the Akt pathway at a higher level. Therefore, the amount of pSer473-Akt was further detected in the infected cells (). Because some other H. pylori virulence factors may also be involved in the activation of Akt pathway, we used the isogenic background mutants we constructed for infection in this experiment. We found that H. pylori infection significantly increased the amount of phosphorylated Akt, and the CagAE harboring strain induced more phosphorylation than CagAW. This result revealed that CagAE leads to higher activation of Akt signaling than CagAW through its interaction with SHIP2, thereby enhancing its oncogenic activity.
CagAE exhibits higher affinity to SHIP2 than CagAW
The reason for the formation of different amounts of the CagAE-SHIP2 and CagAW-SHIP2 complexes during infection was further explored. First, an immunoprecipitation assay was performed on the infected cells. Since we only focused on the interaction of intracellular CagA with SHIP2 (not involving the influence of other factors on cells), East Asian strains SHCH30 and Western strain 26695 were used. First, we confirmed that these two strains inject comparable CagA into host cells, either for p-CagA or for total CagA ( and Fig. S3). The results also showed that SHIP2 was more abundantly co-precipitated with CagAE than with CagAW (), indicating a higher affinity of CagAE for SHIP2.
Figure 3. Detection of the interaction difference between CagAE or CagAW with SHIP2.
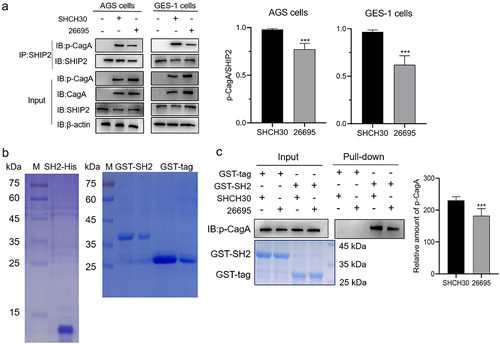
Previous studies have shown that SHIP2 interacts with CagA via its SH2 domain [Citation15]. We then compared the affinity of CagAE and CagAW to the heterogeneously expressed SHIP2-SH2 domain infused with the GST tag (, right). An equal amount of SH2 recombinant protein was immobilized on the resin and mixed with lysates of infected cells. As shown in , a significantly higher amount of CagAE was pulled down than that of CagAW. This confirmed the results of the co-immunoprecipitation assays.
SPR assay was performed to quantify the binding force of the SHIP2-SH2 domain to CagAE and CagAW. Because only EPIYA motifs in CagA are involved in interacting with the SH2 domain, and EPIYA-C and EPIYA-D motifs represent the specific sequences of CagAW and CagAE respectively, we synthesized tyrosine-phosphorylated peptides EPIpYA-D and EPIpYA-C to refer to CagAE and CagAW. When peptide analytes flowed through the chip immobilizing the SH2 domain, the sensorgrams recorded by SPR Evaluation Software confirmed the interaction of SH2 with EPIYA-C/D motifs (). Based on the association and dissociation processes of several concentration gradients of analytes to and from receptors, the affinity constant (KD) values were calculated by steady-state affinity analysis. A higher KD value indicates a weaker affinity, as it requires a greater amount of analytes for interaction. The KD value of EPIpYA-D interacting with SH2 was 8.903 × 10−7 M, indicating that binding 50% of SH2 on the chip requires only 0.8903 µM of the EPIpYA-D peptide. This was approximately 10 times lower than that of EPIpYA-C (KD = 8.347 × 10−6 M, 8.347 µM) (), demonstrating the higher affinity of EPIYA-D for SHIP2-SH2.
Figure 4. Surface plasmon resonance (SPR) analysis of the interactions between SHIP2-SH2 domain with EPIpYA-D or EPIpYA-C peptides. Recombinant His tagged SHIP2-SH2 (Figure3(b)) was immobilized on a BIAcore CM5 sensor chip as the receptor. EPIpYA-C (a) or EPIpYA-D (b) at different concentrations flow through the chip. Sensorgrams were recorded by a BIAcore T100 instrument. For each peptide interaction, the response values at a fixed time point for different concentrations of peptides are marked and fitted into a stoichiometric reaction curve (c for EPIpYA-c and d for EPIpYA-D). The values of KD were then calculated according to the curves with Biacore T100 Evaluation Software.
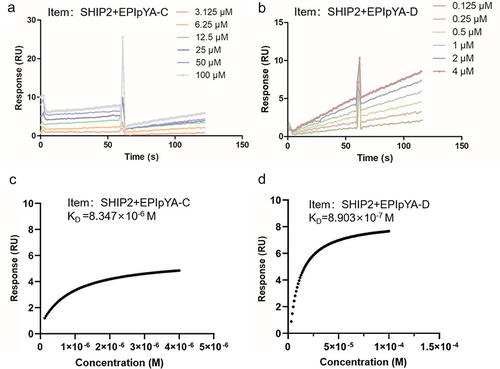
All these data revealed that the EPIYA-D motif had a higher affinity for the SHIP2-SH2 domain, which lays the foundation for the higher affinity of CagAE for SHIP2 than CagAW.
Structural analysis of the interaction between EPIYA-C/D peptide and SHIP2-SH2 domain
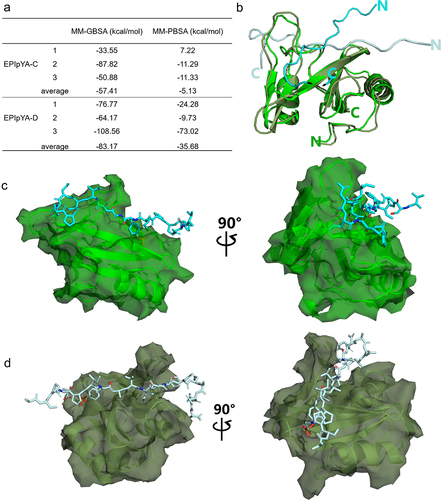
Molecular dynamic simulations were carried out to calculate the possible interaction complexes between the CagA EPIpYA-C/D peptide and SHIP2. First, we calculated the binding energies of the two complexes using the MMGB/PBSA tool (). We found that the average binding energy of SHIP2-SH2 and EPIpYA-C, −57.41 kcal/mol, is significantly higher than that of EPIpYA-D (−83.17 kcal/mol) based on MMGBSA. The calculation results from MMPBSA are similar. This also indicated that SHIP2-SH2 binds more easily to the EPIpYA-D motif. The two simulated complexes with the highest probability are superimposed in . From these complexes structures, it can be observed that both EPIpYA-C and EPIpYA-D, located on the surface of the SH2 domain, formed relatively tight interactions with SHIP2. The EPIpYA-C polypeptide forms a curled conformation for the interaction and its N-terminal is more free (), while the EPIpYA-D polypeptide is relatively more extended and close to the SH2 domain ().
Figure 5. Comparative analysis of the interaction of SHIP2-SH2 domain with EPIpYA-C and EPIpYA-D peptides. (a) The binding energies of two peptides with SHIP2-SH2 domain were calculated with two algorithms, MMGBSA and MMPBSA. (b) Superimposition of SHIP2-SH2 domain interacting with EPIpYA-C and EPIpYA-D motifs. (c) Overall structure of the SHIP2-SH2-EPIpYA-C complex in two views. (d) Overall structure of the SHIP2-SH2-EPIpYA-D complex in two views. Cyan and light gray indicate EPIpYA-C and EPIpYA-D peptides respectively where their Tyr residues were both phosphorylated. Green and dark gray indicate SH2 domains interacting with EPIpYA-C and EPIpYA-D peptides respectively.
As shown in the detailed diagram in , Tyr (pY-115) in the middle of the EPIpYA-C polypeptide formed tight bonds with several Arg residues (Arg-19, Arg-38, Arg-61) in SH2 domain. However, the Ala-116 and Ile-118 residues at the Y + 1 and Y + 3 locations were completely exposed, without obvious interaction (). In the EPIpYA-D and SHIP2-SH2 complex model, the central Tyr (pY-115) formed stable salt-bridge interactions with some polar amino acids of SHIP2-SH2 (Ser-42, Arg-61), similar to that of EPIpYA-C. The obvious difference is that the C-terminus of EPIpYA-D forms a tight bond with SHIP2-SH2 through multiple forces, including hydrogen bonds between Thr-117 (Y + 2) and Ser75 in SHIP2, and nonpolar interactions between Ile-118 (Y + 3) and Val-72, Leu-89, and Tyr-93 of SH2. Notably, the Phe-120 residue at the Y + 5 position of EPIpYA-D, which is absent in EPIpYA-C, formed stable π-π stacking with Arg-81 of SH2 (), representing a specific feature of the SHIP2-EPIpYA-D interaction. These nonpolar interactions and hydrogen bonds result in lower binding energies for the interaction between EPIYA-D and SHIP2-SH2.
Figure 6. Key sites analysis in complexes of SHIP2-SH2-EPIpYA-C and SHIP2-SH2-EPIpYA-D. (a) Complex of SHIP2-SH2-EPIpYA-C. (b) Details of interaction surface of SHIP2-SH2-EPIpYA-C complex. (c) Complex of SHIP2-SH2-EPIpYA-D. (d) Details of interaction surface of SHIP2-SH2-EPIpYA-D. pY-115 indicates the phosphorylated tyrosine residue in synthesized peptides. Other residues on peptides are marked in black. The amino acids in green represent residues on SHIP2-SH2 domain those interact with EPIpYA-C motif, and in dark grey represent residues on SHIP2-SH2 domain those interact with EPIpYA-D peptide.
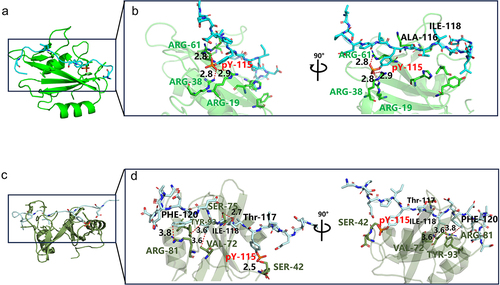
This interaction analysis provided an explanation for the greater affinity of EPIYA-D to SHIP2 at the atomic level, and suggested that Phe-120 (Y + 5) of the peptide plays an important role in the strong binding of CagAE with SHIP2.
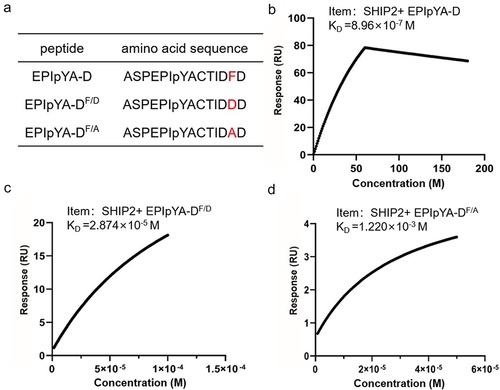
Confirmation of the key role of residue Phe-120 in the interaction between EPIpYA-D and SHIP2-SH2
Structural analysis highlighted the critical role of Phe-120 (Y + 5) in EPIYA-D in its strong interaction with SH2. To validate this analysis, we synthesized mutated EPIpYA-D peptides with Phe replaced by Ala (a simple residue without a side chain) (EPIpYA-DF/A) or Asp (the corresponding residue in EPIpYA-C) (EPIpYA-DF/D) () and tested their binding strength to SHIP2-SH2 using SPR. As shown in , the KD value of EPIpYA-D to SHIP2-SH2 is 8.96 × 10−7 M, which shows very good repeatability with . However, it increased to 2.874 × 10−5 M when EPIpYA-DF/D interacted with SHIP2-SH2 (), indicating a 32-fold reduction in affinity. This is close to the KD value of EPIYA-C for SHIP2-SH2. The substitution of Asp with Ala in EPIpYA-D led to a greater reduction in its binding to SHIP2-SH2 (1.220 × 10−3 M) (), suggesting that Asp at the Y + 5 position in EPIYA-C also plays a role in the interaction. From these data, the effect of the residue at the Y + 5 position in EPIYA-C/D on affinity to SHIP2 is F>D>A.
Role of residue Phe in EPIYA-D motif in affecting the pathogenicity of CagAE
In vitro structural analysis demonstrated the significance of Phe-120 at the Y + 5 position in the EPIpYA-D peptide interaction with SHIP2. Its role in the pathogenesis of CagAE was further investigated by generating two H. pylori mutant strains, CagAF/A and CagAF/D, based on the CagAE complementary strain by substitution of Phe with Asp or Ala. The virulence changes in the infected cells were then detected.
After infecting GES-1 or AGS cells with these strains, we also found that intracellular p-CagA was at roughly the same level in the input cell lysate (). However, the co-precipitation of SHIP2 with both CagAF/D and CagAF/A was lower than that of CagAE (), which was consistent with the SPR results.
Figure 8. Verification of role of residue Phe in CagAE-SHIP2 interaction by cell infection assays. (a) Immunoprecipitation analysis of CagAE and its mutants (CagAF/D and CagAF/A) with SHIP2 in infected cells. Antibody against CagA was used as IP antibody, antibody against SHIP2 was used to reveal the amount of SHIP2 interacted with CagA. (b) Western blotting indicating activation of Akt pathway in infected cells. (c) Western blotting detecting the phosphorylation level of p-65 to indicate the activation of NF-κB pathway in infected cells. (d) Secretion of IL-8 by infected cells. Error bars represent means ± SD, **indicates p < 0.01, ***indicates p < 0.001. The other notes represent the same meaning as above.
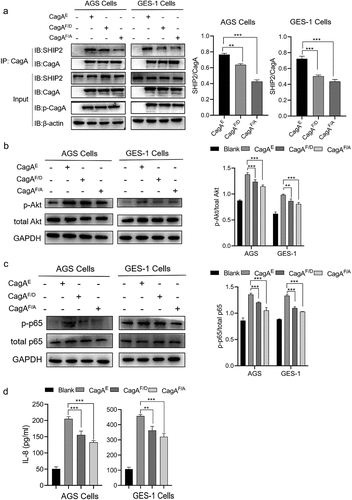
The activation of Akt and NF-κB signaling was detected by western blotting. Akt regulates many cellular processes including transcription, cell migration and autophagy [Citation30]. As shown in , the CagAF/D and CagAF/A mutants caused less phosphorylation of Akt than CagAE, implying that the alteration of Phe in CagAE affects the recruitment of SHIP2, thereby affecting the activation of the Akt pathway. CagA is a bacterial T4SS effector protein that is capable of activating NF-κB and inducing NF-κB-mediated IL-8 production [Citation31]. Furthermore, p65 is one of the components of the NF-κB transcription factor family, and its phosphorylation also indicates the activation of this pathway. As shown in , the CagAF/D and CagAF/A mutants induced less phosphorylation of p65. The production of IL-8 also decreased in mutant-infected cells (). These results show that the alteration of Phe in CagAE also has an impact on the activation of the NF-κB pathway.
Oncogenic transformations of infected cells treated with CagAE mutants were then measured. It has been reported that Phe-120 of the EPIYA-D motif is also crucial for its interaction with SHP2, another intracellular receptor of CagA involved in cell oncogenic and morphological transformation. Therefore, a specific inhibitor of SHP2, SHP099, was employed to limit the influence of SHP2 in this study. It has been reported that CagA-SHP2 interaction can activate the Ras-Erk pathway and subsequently induce a specific hummingbird phenotype of host cells. Therefore, the alteration of cell shape was first examined. Morphological observations showed that CagAE induced many hummingbird alterations, while fewer elongated cells was formed in CagA mutant strains (CagAF/A and CagAF/D) infecting cells (). However, when SHP099 was added, the differences in the morphological changes between the groups were not obvious. These results confirmed the role of Phe-120 in its interaction with SHP2 and that hummingbird-like changes are only associated with SHP2 and its downstream Ras-Erk pathway.
Figure 9. Role of residue Phe in EPIYA-D motif in affecting the oncogenicity of CagAE. SHP099 was used as an inhibitor to exclude the effect of interaction between SHP2 and CagA, as Phe in EPIYA-D motif was also reported to play roles in SHP2-CagAE interaction to improve the cytotoxicity of CagAE. (a) Examination of CagA induced specific hummingbird phenotype. Elongated cells with the longest axis exceeding the shortest axis by more than 2-fold were counted as the hummingbird phenotype. (b) Migration and (c) invasion of infected AGS and GES-1 cells examined by transwell assays. Bars in images represent 100 µm. Left, representative images; Right, statistical data. Error bars represent means ± SD, ns indicates no statistical significance, ***indicates p < 0.001.
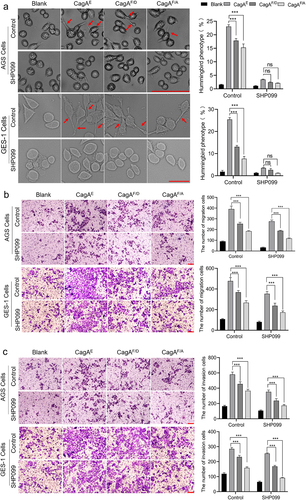
Then, the ability of cells to migrate and invade was examined, which is associated with both the SHP2-Erk and SHIP2-Akt pathways. From , we found that for blank cell lines without treatment with SHP099, cell migration and invasion were both reduced by infection with mutant strains (F/D or F/A) compared to infection with the CagAE harboring strain. After inhibitor treatment, there was still a relatively significant decrease between cells infected with F/D or F/A mutants and the wild-type strain. These results implied that, in addition to interacting with SHP2, Phe-120 in EPIYA-D could also enhance the pathogenicity of CagAE through its interaction with other receptors, such as SHIP2.
Discussion
Chronic infection with CagA-positive H. pylori is strongly associated with the development of GC, and epidemiological studies have shown that CagAE is a higher risk factor than CagAW [Citation32]. However, the mechanism underlying the higher carcinogenicity of CagAE remains unclear. During infection, CagA can be injected into host gastric epithelial cells through T4SS [Citation8]. Intracellular CagA binds to the inner leaflet of the plasma membrane where it is phosphorylated by host kinases at the Tyr residue in the Glu-Pro-Ile-Tyr-Ala (EPIYA) motif [Citation33]. This phosphorylation enables CagA to interact with host SH2 domain-containing proteins, including SHP2, PI3K, SHIP2, and Crk, thereby perturbing their functions to promote neoplastic transformation of the cell [Citation15,Citation34]. Owing to the sequence differences between EPIYA-C and EPIYA-D, CagAE might have a different affinity spectrum with its intracellular receptor proteins from CagAW, which will inevitably affect the regulation of intracellular signals. This speculation was verified by Hayashi, who studied the interaction between CagAE/W and SHP2 and found that the CagAE-specific EPIYA-D segment binds to SHP2 more strongly than the EPIYA-C segment, contributing to the higher occurrence of the “hummingbird phenotype” in infected cells [Citation10]. This finding opens a window to explain the difference between the pathogenicities of CagAE and CagAW. However, additional differential receptor proteins of CagAE/W require further investigation.
SHIP2 is a member of CagA-binding SH2-containing proteins [Citation15]. Studies on the oncogenic functions of this molecule have been inconsistent. Evidence suggests that its protumorigenic or tumor suppressor function is largely dependent on cell models [Citation35]. In GC, SHIP2 is frequently expressed depressedly and has an antagonistic effect on tumorigenesis and proliferation in gastric cancer [Citation14]. However, H. pylori can hijack SHIP2 through intracellular CagA to induce gastric cell carcinogenesis [Citation36]. In this study, we found that by interacting with CagA, SHIP2 induces cell migration, invasion, and IL-8 secretion, thereby contributing to the higher oncogenicity of CagAE than that of CagAW. This is consistent with the results of Fujii [Citation15].
However, unlike Fujii’s observation that SHIP2 binds slightly strongly to EPIYA-C than EPIYA-D [Citation15], our data demonstrated that CagAE recruits more SHIP2 than CagAW. This discrepancy arises from the higher affinity between the EPIYA-D motif in the CagAE and the SH2 domain of SHIP2. Investigation of the CagA-SHIP2 complexes revealed the critical role of the Phe residue at the Y + 5 position in EPIYA-D, which binds to Arg-81 in the SHIP2-SH2 domain with strong strength. By mutating Phe to Asp or Ala, the strength of CagAE interaction with SHIP2 was significantly reduced, subsequently diminishing its capacity to promote the oncogenic transformation of cells. From the binding force, we found that SHIP2-SH2 displays selectivity for the benzene-containing residue at the pY + 5 position in the EPIYA-D motif. The preference order was Phe > Asp > Ala, indicating that the cyclic spatial structure at this site facilitates a higher affinity for SHIP2-SH2.
The essential role of Phe in EPIYA-D has also been previously reported in the interaction between CagAE and SHP2 [Citation10]. Both SHIP2 and SHP2 interact with CagA through their SH2 domains, which exhibit sequence and structural similarities. It is reasonable that the presence of Phe in EPIYA-D crucially contributed to the high affinity of CagAE for both SHIP2 and SHP2. Based on these observations, we speculate that Phe at position Y + 5 in EPIYA-D is crucial for reinforcing the interaction between CagAE and other SH2-containing receptors. However, this inference requires further validation.
In addition, Fujii et al. found that CagA increased its delivery into gastric epithelial cells by interacting with SHIP2 [Citation15]. However, in our study, the injection of CagA into the cells was not reduced after silencing SHIP2. Further experiments are needed to confirm this issue.
The findings in this study expand our understanding of the differential regulation of intracellular signals between CagAE and CagAW and are helpful in elucidating the higher carcinogenic effect of CagAE.
Conclusions
In this study, we found that SHIP2 is an intracellular receptor hijacked by CagA that enhances the carcinogenicity of H. pylori. CagAE exhibited a higher affinity for SHIP2 than CagAW. Depending on this higher affinity, CagAE tethers more SHIP2 to the plasma membrane, produces more PI(3,4)P2, and causes higher activation of Akt signaling, thereby enhancing the neoplastic traits of infected cells. Structural analysis revealed the crucial role of Phe at the Y + 5 position in the EPIYA-D motif, which interacted with the Arg-81 residue of SHIP2-SH2 to establish this higher affinity.
Author contributions
X. F. J., Q.W.W., and X.Y.C. contributed equally to the study. H.L.Z. designed and supervised this study. X. F. J., J.H.Z., and S.C. searched the literature. Q.W.W. and X.Y.C. performed experiments. X. F. J. wrote the original manuscript. H.L.Z., S.Z.L., J.F.S., B.Q.L., and Y. Z. supervised the study and contributed resources. All authors contributed to and edited the manuscript.
EPIYA C peptide percentage.jpg
Download JPEG Image (1.7 MB)EPIYA C peptide MS confirmation.jpg
Download JPEG Image (614.4 KB)EPIYA D peptide MS confirmation.jpg
Download JPEG Image (1.5 MB)Supplemental Material.docx
Download MS Word (1 MB)EPIYA D percentage.jpg
Download JPEG Image (1.8 MB)Acknowledgements
The computation in this work was provided by Shanghai ZELIXIR BIOTECH through the ZCloud platform (https://cloud.zelixir.com).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data Availability statement
The data generated during the study is available at figshare database at https://figshare.com doi.org/[10.6084/m9.figshare.25356829].
Supplemental data
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21505594.2024.2375549
Additional information
Funding
References
- Warren JR, Marshall B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;1(8336):1273–17. doi: 10.1016/S0140-6736(83)92719-8
- Hooi JKY, Lai WY, Ng WK, et al. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology. 2017;153(2):420–429. doi: 10.1053/j.gastro.2017.04.022
- Jiang J, Chen Y, Shi J, et al. Population attributable burden of Helicobacter pylori-related gastric cancer, coronary heart disease, and ischemic stroke in China. Eur J Clin Microbiol Infect Dis. 2017;36(2):199–212. doi: 10.1007/s10096-016-2810-x
- de Brito BB, Silva FAFD, Soares AS, et al. Pathogenesis and clinical management of Helicobacter pylori gastric infection. World J Gastroenterol. 2019;25(37):5578–5589. doi: 10.3748/wjg.v25.i37.5578
- Ansari S, Yamaoka Y. Helicobacter pylori virulence factors exploiting gastric colonization and its pathogenicity. Toxins (Basel). 2019;11(11):677. doi: 10.3390/toxins11110677
- Backert S, Blaser MJ. The Role of CagA in the gastric biology of helicobacter pylori. Cancer Res. 2016;76(14):4028–4031. doi: 10.1158/0008-5472.CAN-16-1680
- Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer. 2004;4(9):688–694. doi: 10.1038/nrc1433
- Backert S, Tegtmeyer N, Fischer W. Composition, structure and function of the Helicobacter pylori cag pathogenicity island encoded type IV secretion system. Future Microbiol. 2015;10(6):955–965. doi: 10.2217/fmb.15.32
- Backert S, Tegtmeyer N, Selbach M. The versatility of Helicobacter pylori CagA effector protein functions: The master key hypothesis. Helicobacter. 2010;15(3):163–176. doi: 10.1111/j.1523-5378.2010.00759.x
- Hayashi T, Senda M, Suzuki N, et al. Differential mechanisms for SHP2 Binding and activation are exploited by geographically distinct Helicobacter pylori CagA Oncoproteins. Cell Rep. 2017;20(12):2876–2890. doi: 10.1016/j.celrep.2017.08.080
- Pesesse X, Deleu S, De Smedt F, et al. Identification of a second SH2-domain-containing protein closely related to the phosphatidylinositol polyphosphate 5-phosphatase SHIP. Biochem Biophys Res Commun. 1997;239(3):697–700. doi: 10.1006/bbrc.1997.7538
- Thomas MP, Erneux C, Potter BV. SHIP2: Structure, function and inhibition. Chembiochem. 2017;18(3):233–247. doi: 10.1002/cbic.201600541
- Fu M, Fan W, Pu X, et al. Elevated expression of SHIP2 correlates with poor prognosis in non-small cell lung cancer. Int J Clin Exp Pathol. 2013;6(10):2185–2191.
- Ye Y, Ge YM, Xiao MM, et al. Suppression of SHIP2 contributes to tumorigenesis and proliferation of gastric cancer cells via activation of Akt. J Gastroenterol. 2016;51(3):230–240. doi: 10.1007/s00535-015-1101-0
- Fujii Y, Murata-Kamiya N, Hatakeyama M. Helicobacter pylori CagA oncoprotein interacts with SHIP2 to increase its delivery into gastric epithelial cells. Cancer Sci. 2020;111(5):1596–1606. doi: 10.1111/cas.14391
- Ji X, Zhao H, Zhang Y, et al. Construction of novel plasmid vectors for gene knockout in Helicobacter pylori. Curr Microbiol. 2016;73(6):897–903. doi: 10.1007/s00284-016-1140-7
- Ji X, Wang Y, Li J, et al. Application of FLP-FRT System to construct unmarked deletion in helicobacter pylori and functional study of gene hp0788 in pathogenesis. Front Microbiol. 2017;8:2357. doi: 10.3389/fmicb.2017.02357
- Galindo-Murillo R, Robertson JC, Zgarbová M, et al. Assessing the current state of amber force field modifications for DNA. J Chem Theory Comput. 2016;12(8):4114–4127. doi: 10.1021/acs.jctc.6b00186
- Bryant P, Pozzati G, Elofsson A. Improved prediction of protein-protein interactions using AlphaFold2. Nat Commun. 2022;13(1):1265. doi: 10.1038/s41467-022-28865-w
- Friedrichs MS, Eastman P, Vaidyanathan V, et al. Accelerating molecular dynamic simulation on graphics processing units. J Comput Chem. 2009;30(6):864–872. doi: 10.1002/jcc.21209
- Zhao H, Ji X, Chen X, et al. Functional study of gene hp0169 in Helicobacter pylori pathogenesis. Microb Pathog. 2017;104:225–231. doi: 10.1016/j.micpath.2017.01.039
- Sousa da Silva AW, Vranken WF. ACPYPE - AnteChamber python parser interface. BMC Res. 2016;5:367. doi: 10.1186/1756-0500-5-367
- Wang J, Wolf RM, Caldwell JW, Kollman PA, Case D A. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035
- Hess B, Bekker H, Berendsen HJC, et al. LINCS: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18(12):1463–1472. doi: 10.1002/(SICI)1096-987X(199709)18:12<1463:AID-JCC4>3.0.CO;2-H
- Kräutler V, Gunsteren WFV, Hünenberger PH. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J Comput Chem. 2001;22(5):501–508. doi: 10.1002/1096-987X(20010415)22:5<501:AID-JCC1021>3.0.CO;2-V
- Lu H, Hsu PI, Graham DY, et al. Duodenal ulcer promoting gene of Helicobacter pylori. Gastroenterology. 2005;128(4):833–848. doi: 10.1053/j.gastro.2005.01.009
- Zhao Q, Busch B, Jiménez-Soto LF, et al. Integrin but not CEACAM receptors are dispensable for Helicobacter pylori CagA translocation. PLOS Pathog. 2018;14(10):e1007359. doi: 10.1371/journal.ppat.1007359
- Xu L, Shao Y, Ren L, et al. IQGAP2 inhibits migration and invasion of gastric cancer cells via elevating SHIP2 phosphatase activity. Int J Mol Sci. 2020;21(6):1968. doi: 10.3390/ijms21061968
- Pedicone C, Meyer ST, Chisholm JD, et al. Targeting SHIP1 and SHIP2 in Cancer. Cancers (Basel). 2021;13(4):890. doi: 10.3390/cancers13040890
- Zhou X, Li T-M, Luo J-Z, et al. CYP2C8 suppress proliferation, migration, invasion and sorafenib resistance of hepatocellular carcinoma via PI3K/Akt/p27(kip1) Axis. J Hepatocell Carcinoma. 2021;8:1323–1338. doi: 10.2147/JHC.S335425
- Brandt S, Kwok T, Hartig R, et al. NF-κB activation and potentiation of proinflammatory responses by the Helicobacter pylori CagA protein. Proc Natl Acad Sci USA. 2005;102(26):9300–9305. doi: 10.1073/pnas.0409873102
- Azuma T, Yamazaki S, Yamakawa A, et al. Association between diversity in the Src homology 2 domain–containing tyrosine phosphatase binding site of Helicobacter pylori CagA protein and gastric atrophy and cancer. J Infect Dis. 2004;189(5):820–827. doi: 10.1086/381782
- Lang BJ, Gorrell RJ, Tafreshi M, et al. The Helicobacter pylori cytotoxin CagA is essential for suppressing host heat shock protein expression. Cell Stress Chaperones. 2016;21(3):523–533. doi: 10.1007/s12192-016-0680-x
- Higashi H, Tsutsumi R, Muto S, et al. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science. 2002;295(5555):683–686. doi: 10.1126/science.1067147
- Elong Edimo W, Schurmans S, Roger PP, et al. SHIP2 signaling in normal and pathological situations: Its impact on cell proliferation. Adv Biol Regul. 2014;54:142–151. doi: 10.1016/j.jbior.2013.09.002
- Wang Z, Shan Y, Wang R, et al. Structural Insights into the Binding Propensity of Human SHIP2 SH2 to Oncogenic CagA Isoforms from Helicobacter pylori. Int J Mol Sci. 2022;23(19):11299. doi: 10.3390/ijms231911299