ABSTRACT
The Birt-Hoge-Dubé syndrome tumor suppressor Folliculin is a regulator of metabolism and has as a wide range of cellular and organismal phenotypes associated with its disruption. However, the molecular mechanisms which underlie its functions are poorly understood. Folliculin has been described to associate with lysosomes in response to nutrient depletion and form a key part of the signaling network that controls the activity of mTORC1. We recently reported that Folliculin can control the nutrient dependent cytoplasmic distribution of lysosomes by promoting the formation of a complex with the Golgi-associated small GTPase Rab34 and its effector RILP. We thus define a mechanistic connection between the lysosomal nutrient signaling network and the transport machinery that controls the distribution and dynamics of this organelle. Here we summarise the main conclusions from that study, attempt to integrate our findings with other recent studies on lysosome distribution/dynamics, and discuss the potential consequences of the dysregulation of this processes caused by Folliculin loss for Birt-Hoge-Dubé syndrome and normal cell function.
The tumor suppressor Folliculin (FLCN) is disrupted in the autosomal dominant Birt-Hoge-Dubé (BHD) syndrome. BHD syndrome patients may present with lung cysts that can cause spontaneous pneumothorax, skin tumors known as fibrofolliculomas as well as kidney cysts and kidney cancer.Citation1 It has been clear for some time that the 64kDa protein product of the FLCN gene is a regulator of cellular metabolism that is expressed in most cell types.Citation2 FLCN can form a complex with 2 larger (≈125–130 kDa) proteins FNIP1 and FNIP2 (Folliculin Interacting Proteins 1 and 2).Citation2,3 In yeast, Lst7 and Lst4 (lethal with Sec13 7, 4), are orthologues of FLCN and FNIPs respectively and also form a complex.Citation4,5 Recent structural studies have shown that FLCN and FNIP proteins are both comprised of a Longin and a DENN (differentially expressed in normal versus neoplastic cells) domain – both of which are protein folds that have been variously implicated in the regulation of small GTPases and membrane traffic.Citation6-8
Both on an organismal and cellular level, FLCN is pleiotropic, and many phenotypes appear to be highly context dependent. For example, FLCN has been proposed to both positively and negatively regulate cell-cell adhesion and migration,Citation9-11 and BHD renal tumors may have either increased or decreased mTORC1 (mechanistic target of rapamycin complex 1) activity.Citation12,13 The FLCN/FNIP complex interacts with AMPK (adenosine monophosphate-activated protein kinase) and dependent on the system studied appears to either promote, suppress or have little effect on its activity.Citation2,14,15 FLCN/FNIP have also been linked to diverse cell functions such as autophagy,Citation16,17 ciliogenesis,Citation18 exit of stem cells from pluripotencyCitation19 and lysosome biogenesis.Citation20,21 Together, these data point to a central role for FLCN/FNIP in metabolic homeostatsis, but also show that loss or disruption of FLCN impacts on a range of cellular functions.
On a mechanistic level, recent studies from the Sabatini and Ferguson groups have provided a particularly crucial insight; FLCN (which appears predominantly cytoplasmic under normal cell culture conditions), is recruited to lysosomes upon nutrient depravation.Citation21,22 At the lysosome, FLCN/FNIP can interact with the Rag (Ras related GTP binding protein) GTPases, and in vitro, the complex possesses GAP (GTPase activating protein) activity toward RagC.Citation22 The Lst7/Lst4 complex functions in a similar manner at the yeast vacuole.Citation5 It is suggested that RagC GAP activity is important for amino acid dependent recruitment and activity of mTORC1 on lysosomes. This in turn regulates the activity of the lysosome associated transcription factors TFEB (transcription factor EB) and TFE3 (Transcription Factor Binding To IGHM Enhancer 3), as loss of FLCN inhibits their phosphorylation, promotes their nuclear translocation and activity, and drives lysosome biogenesis.Citation20,21,23 Thus, the lysosome is a key site of action of FLCN. However, whether this pathway is sufficient to account for the broad and context dependent consequences of FLCN disruption is not clear.
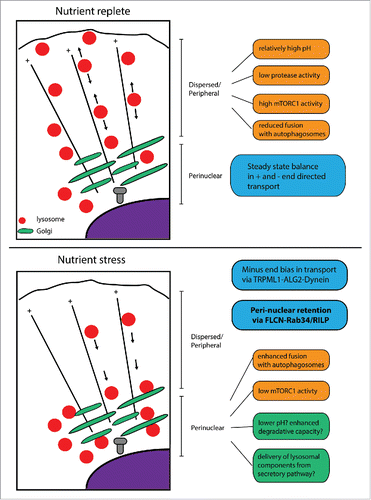
Regulation of lysosome distribution by FLCN
Consistent with work from the Rubinzstein lab, we noted that in HeLa cells, starvation not only promotes FLCN association with lysosomes, but also results in a shift in their distribution in the cytoplasm.Citation24,25 In many cell types, including HeLa, lysosomes are typically localized throughout the cell with some enrichment proximal to the microtubule organizing center(MTOC)/Golgi which is typically in a perinuclear position. There is dynamic exchange between these populations.Citation26 Starvation causes a centripetal shift in that distribution with lysosomes concentrating in the perinuclear region.Citation25 We sought to understand whether this correlation of FLCN-lysosome association and propensity toward a starvation-induced perinuclear localization were linked. Consistent with that proposition, depletion of FLCN or FNIP1/2 using RNAi suppressed starvation induced perinuclear clustering. Over-expression of FLCN and FNIP, while not strikingly impacting on steady state lysosome distribution, did promote the formation of dynamic tubules that extended from lysosomes, which have been linked to the activities of several small GTPases, their effectors and microtubule motor proteins.
We next considered the established pathways that control lysosome distribution and tubulation. To drive transport toward the plus end of microtubules that are typically located at the cell periphery, the lysosome associated small GTPase Arl(Arf-like)8b recruits the adaptor protein SKIP (SifA and kinesin interacting protein) which in turn recruits kinesin-1 for plus end directed microtubule transport.Citation27-29 The recently described BORC (BLOC-one-related complex) initiates this process by recruiting Arl8b.Citation30 Kinesin-1 can also be recruited by Rab7 though its effector FYCO1 (FYVE and coiled-coil domain containing 1), and a recent study has also suggested that kinesin-1 may also interact with phospholipids.Citation31-33 To promote transport toward the minus end of microtubules that are predominantly located in a perinuclear position at the microtubule organizing center, Rab7 can also recruit RILP (rab interacting lysosomal protein) which in turn recruits cytoplasmic dyneinCitation34,35 and indicates that lysosome-motor interactions may regulate bidirectional positioning upon microtubules. In addition to these lysosome intrinsic components, other reports have shown that the lysosome extrinsic, predominantly Golgi localized, Rab34 and Rab36 GTPases can also impact upon lysosome distribution by promoting their perinuclear clustering. For Rab34, this also requires interaction with RILP.Citation36-38 Overall, it is likely that the summation of these opposing centrifugal and centripetal directed activities defines the dynamic distribution of the organelle.
We found that FLCN interacts directly with RILP using its C-terminal DENN domain (a protein fold that in other contexts has been linked to Rab GEF (guanine nucleotide exchange factor) activityCitation8), suggesting the possibility that FLCN may control lysosome distribution via this Rab effector. We also found that depletion of FLCN suppressed Rab34 induced perinuclear aggregation of lysosomes. Moreover, using an effector pull down assay, we noted that starvation increased the amount of Rab34 retained on GST-RILP resin, suggesting that activation of Rab34 may be metabolically regulated.
How does a predominantly Golgi localized GTPase control the distribution of lysosomes? One could envisage either a direct or indirect mechanism. In the indirect scenario, Rab34, via RILP would in some way signal to affect the activity of the lysosome associated transport components. Alternatively, Golgi localized Rab34/RILP may itself interact with lysosomes. A series of super resolution and ultrastructural imaging experiments strongly favored the latter hypothesis – that Rab34 positive perinuclear membranes contact lysosomes. We showed that this association acts to limit lysosomal motility and so promotes their retention in this region of the cell.
We next sought to establish how FLCN could regulate these contacts, focusing on its DENN domain. Although many DENN domains act as Rab GEFs, we were unable to detect any Rab34 GEF activity from the FLCN-DENN in vitro. However, targeting of constitutively active Rab34 to mitochondria resulted in the DENN domain dependent re-localization of FLCN, suggesting that FLCN may interact with the active form of the GTPase. We went on to show that the FLCN-DENN domain is capable of promoting the formation of the active Rab34-RILP complex in vitro.
Thus, an attractive model presents itself - starvation‐induced FLCN association with lysosomes drives dynamic, Rab34/RILP driven interactions between lysosomes and Golgi membranes that result in the limitation of their motility, promoting perinuclear retention and thus contributes to control of their cytoplasmic distribution (). While it is clear there is much work to do to fully understand the basis of these contacts, our studies did reveal the expression of RILP can promote the association of Rab34 with Rab7. As RILP is a dimer,Citation39,40 we speculate that this could form the basis of these contacts and that one role of the FLCN/FNIP complex is to promote their formation under conditions of nutrient stress.
Potential impacts of the dysregulation of lysosome dynamics
Together, these data suggest that FLCN may couple the lysosomal nutrient signaling network to the cellular machinery that controls the intracellular distribution of the organelle itself. This new mechanistic insight into the role of FLCN at the lysosome may have the potential to explain some of the diverse and context dependent phenotypes associated with its loss. The starvation induced perinuclear clustering of lysosomes has been suggested to be important in autophagic flux by promoting the fusion of lysosomes with autophagosomes.Citation25 Moreover, the localization of lysosomes can affect mTORC1 activity, with higher activity associated with a more dispersed/peripheral localization.Citation25 Thus, the suppression of mTORC1 activity caused by loss of RagC GAP activityCitation22 may be counterbalanced by an opposing effect from a propensity toward peripheral distribution. Enhanced lysosomal biogenesis and/or exocytosis driven by the activation of TFEB/TFE3 would add a third variable.Citation21,41 The balance between these activities in various cell types could therefore give rise to the differing mTORC1 activity phenotypes found in various model systems studied.
Recently, the Grinstein lab has elegantly demonstrated that the position of lysosomes can directly control their luminal pH, with lysosomes at the cell periphery tending to be less acidic compared to the perinuclear population.Citation42 This may due to more limited access to the biosynthetic pathway, greater proton leakage and reduced vacuolar ATPase (proton pump) activity. This in turn impacts on the degradative capacity of the Cathepsin L protease. RILP was recently reported to interact directly with the vacuolar ATPase and regulate its activity, suggesting a potential mechanism that may couple positioning and acidity.Citation43
The distribution of lysosomes also affects their interaction with other organelles. A recent report suggested Rab34 may respond to extrinsic stimuli (lipopolysaccharide) to promote context‐dependent changes in lysosome distribution to modulate lysosome fusion with phagosomes and control antigen cross presentation in dendritic cells;Citation44 it would be interesting to determine whether FLCN may also contribute to this pathway. Similarly, elevation of intracellular calcium triggers lysosome exocytosis to participate in plasma membrane repairCitation45 and blocking lysosome transport to the cell periphery reduces cell spreading, migration and cancer cell invasion.Citation30,46
Thus, a lack of capacity to control organelle dynamics in response to metabolic cues has the potential to impact on a wide range of cellular functions. A better understanding of these effects could help to unify the often seemingly disparate phenotypic and mechanistic data on FLCN.
Future directions
As described above, a clear priority now is to understand more broadly the role of lysosome distribution/dynamics in normal cell function and in BHD syndrome. However, it is worth considering earlier work on the yeast homolog of FLCN, Lst7. Identified in a genetic screen as synthetically lethal with a temperature sensitive Sec13 mutant, Lst7 plays a key role in the transport of the Gap1p general amino acid permease to the yeast cell membrane.Citation47 Given the central role of lysosome associated amino acid transporters in nutrient signaling, further work should explore whether the FLCN/FNIP complex also regulates trafficking of these proteins in mammalian cells. One attractive hypothesis is that lysosome-Golgi contacts described in our study may form part of a trafficking pathway.Citation48 Another recent study has described a key role for the lysosomal calcium channel TRPML1 (transient receptor potential cation channel, mucolipin subfamily, member 1 / Mucolipin-1) in promoting the serum starvation dependent translocation of lysosomes to the perinuclear region via ALG-2 (apoptosis-linked gene-2) and cytoplasmic dynein, independently of Rab7.Citation49 It seems possible that the TRPML1-ALG-2-dynein axis may initiate a net minus end bias in MT transport of lysosomes to the perinuclear region and this positioning is subsequently stabilised by Rab34. FLCN/FNIP induced lysosomal tubules also appear not dissimilar to those tubules that emerge from autolysosomes that participate in lysosomal reformation.Citation31 Future studies should explore whether the FLCN/FNIP complex can also impact upon these pathways. Moreover, it is also clear that changes in cytosolic pH can affect lysosome transport, with extracellular acidification resulting in a transient decrease in intracellular pH promoting lysosome dispersion, whereas alkalinisation, which occurs during starvation, promoting perinuclear aggregation.Citation25,50 It may be the case that the lysosomal association of a number of these proteins may be regulated in part by cytosolic pH or lysosomal membrane potential. Finally, our study highlights how cells can make acute adaptations to their intracellular transport pathways in response to changes in metabolic state; it seems likely that other mechanisms remain to be discovered.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Work in the author's laboratory is supported by the Wellcome Trust (097316/Z/11/Z) and Biotechnology and Biological Sciences Research Council (BB/L006774/1).
References
- Schmidt LS, Linehan WM. Molecular genetics and clinical features of Birt–Hogg–Dubé syndrome. Nat Rev Urol 2015; 12:558–69; PMID:26334087; https://doi.org/10.1038/nrurol.2015.206
- Baba M, Hong S-B, Sharma N, Warren MB, Nickerson ML, Iwamatsu A, Esposito D, Gillette WK, Hopkins RF, Hartley JL, et al. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc Natl Acad Sci USA 2006; 103:15552–7; PMID:17028174; https://doi.org/10.1073/pnas.0603781103
- Hasumi H, Baba M, Hong S-B, Hasumi Y, Huang Y, Yao M, Valera VA, Linehan WM, Schmidt LS. Identification and characterization of a novel folliculin-interacting protein FNIP2. Gene 2008; 415:60–7; PMID:18403135; https://doi.org/10.1016/j.gene.2008.02.022
- Pacitto A, Ascher DB, Wong LH, Blaszczyk BK, Nookala RK, Zhang N, Dokudovskaya S, Levine TP, Blundell TL. Lst4, the yeast Fnip1/2 orthologue, is a DENN-family protein. Open Biol 2015; 5:150174; PMID:26631379; https://doi.org/10.1098/rsob.150174
- Péli-Gulli M-P, Sardu A, Panchaud N, Raucci S, De Virgilio C. Amino Acids Stimulate TORC1 through Lst4-Lst7, a GTPase-Activating Protein Complex for the Rag Family GTPase Gtr2. Cell Rep 2015; 13:1–7; https://doi.org/10.1016/j.celrep.2015.08.059
- Nookala RK, Langemeyer L, Pacitto A, Ochoa-Montano B, Donaldson JC, Blaszczyk BK, Chirgadze DY, Barr FA, Bazan JF, Blundell TL. Crystal structure of folliculin reveals a hidDENN function in genetically inherited renal cancer. Open Biol 2012; 2:120071–1; PMID:22977732; https://doi.org/10.1098/rsob.120071
- Dapeng Zhang LMIFHLA. Discovery of Novel DENN Proteins: Implications for the Evolution of Eukaryotic Intracellular Membrane Structures and Human Disease. Front Gene 2012; 3:283
- Marat AL, Dokainish H, McPherson PS. DENN Domain Proteins: Regulators of Rab GTPases. J Biol Chem 2011; 286:13791–800; PMID:21330364; https://doi.org/10.1074/jbc.R110.217067
- Khabibullin D, Medvetz DA, Pinilla M, Hariharan V, Li C, Hergrueter A, Laucho Contreras M, Zhang E, Parkhitko A, Yu JJ, et al. Folliculin regulates cell-cell adhesion, AMPK, and mTORC1 in a cell-type-specific manner in lung-derived cells. Physiol Rep 2014; 2:e12107-7; PMID:25121506; https://doi.org/10.14814/phy2.12107
- Nahorski MS, Seabra L, Straatman-Iwanowska A, Wingenfeld A, Reiman A, Lu X, Klomp JA, Teh BT, Hatzfeld M, Gissen P, et al. Folliculin interacts with p0071 (plakophilin-4) and deficiency is associated with disordered RhoA signalling, epithelial polarization and cytokinesis. Hum Mol Genet 2012; 21:5268–79; PMID:22965878; https://doi.org/10.1093/hmg/dds378
- Medvetz DA, Khabibullin D, Hariharan V, Ongusaha PP, Goncharova EA, Schlechter T, Darling TN, Hofmann I, Krymskaya VP, Liao JK, et al. Folliculin, the Product of the Birt-Hogg-Dube Tumor Suppressor Gene, Interacts with the Adherens Junction Protein p0071 to Regulate Cell-Cell Adhesion. PLoS ONE 2012; 7:e47842; PMID:23139756; https://doi.org/10.1371/journal.pone.0047842
- Hasumi Y, Baba M, Ajima R, Hasumi H, Valera VA, Klein ME, Haines DC, Merino MJ, Hong S-B, Yamaguchi TP, et al. Homozygous loss of BHD causes early embryonic lethality and kidney tumor development with activation of mTORC1 and mTORC2. Proc Natl Acad Sci USA 2009; 106:18722–7; PMID:19850877; https://doi.org/10.1073/pnas.0908853106
- Hudon V, Sabourin S, Dydensborg AB, Kottis V, Ghazi A, Paquet M, Crosby K, Pomerleau V, Uetani N, Pause A. Renal tumour suppressor function of the Birt-Hogg-Dubé syndrome gene product folliculin. J Med Genet 2010; 47:182–9; PMID:19843504; https://doi.org/10.1136/jmg.2009.072009
- Yan M, Gingras M-C, Dunlop EA, Nouët Y, Dupuy F, Jalali Z, Possik E, Coull BJ, Kharitidi D, Dydensborg AB, et al. The tumor suppressor folliculin regulates AMPK-dependent metabolic transformation. J Clin Invest 2014; 124:2640–50; PMID:24762438; https://doi.org/10.1172/JCI71749
- Hasumi H, Baba M, Hasumi Y, Lang M, Huang Y, Oh HF, Matsuo M, Merino MJ, Yao M, Ito Y, et al. Folliculin-interacting proteins Fnip1 and Fnip2 play critical roles in kidney tumor suppression in cooperation with Flcn. Proc Natl Acad Sci USA 2015; 112:E1624–31; PMID:25775561; https://doi.org/10.1073/pnas.1419502112
- Dunlop EA, Seifan S, Claessens T, Behrends C, Kamps MA, Rozycka E, Kemp AJ, Nookala RK, Blenis J, Coull BJ, et al. FLCN, a novel autophagy component, interacts with GABARAP and is regulated by ULK1 phosphorylation. Autophagy 2014; 10:1749–60; PMID:25126726; https://doi.org/10.4161/auto.29640
- Bastola P, Stratton Y, Kellner E, Mikhaylova O, Yi Y, Sartor MA, Medvedovic M, Biesiada J, Meller J, Czyzyk-Krzeska MF. Folliculin Contributes to VHL Tumor Suppressing Activity in Renal Cancer through Regulation of Autophagy. PLoS ONE 2013; 8:e70030; PMID:23922894; https://doi.org/10.1371/journal.pone.0070030
- Luijten MNH, Basten SG, Claessens T, Vernooij M, Scott CL, Janssen R, Easton JA, Kamps MAF, Vreeburg M, Broers JLV, et al. Birt-Hogg-Dube syndrome is a novel ciliopathy. Hum Mol Genet 2013; 22:4383–97; PMID:23784378; https://doi.org/10.1093/hmg/ddt288
- Betschinger J, Nichols J, Dietmann S, Corrin PD, Paddison PJ, Smith A. Exit from Pluripotency Is Gated by Intracellular Redistribution of the bHLH Transcription Factor Tfe3. Cell 2013; 153:335–47; PMID:23582324; https://doi.org/10.1016/j.cell.2013.03.012
- Martina JA, Diab HI, Lishu L, Jeong-A L, Patange S, Raben N, Puertollano R. The nutrient-responsive transcription factor TFE3 Promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal 2014; 7:ra9-ra9; PMID:24448649; https://doi.org/10.1126/scisignal.2004754
- Petit CS, Roczniak-Ferguson A, Ferguson SM. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol 2013; 202:1107–22; PMID:24081491; https://doi.org/10.1083/jcb.201307084
- Tsun Z-Y, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell 2013; 52(4):495–505; PMID:24095279
- Hong S-B, Oh H, Valera VA, Baba M, Schmidt LS, Linehan WM. Inactivation of the FLCN tumor suppressor gene induces TFE3 transcriptional activity by increasing its nuclear localization. PLoS ONE 2010; 5:e15793; PMID:21209915; https://doi.org/10.1371/journal.pone.0015793
- Starling GP, Yip YY, Sanger A, Morton PE, Eden ER, Dodding MP. Folliculin directs the formation of a Rab34–RILP complex to control the nutrient‐dependent dynamic distribution of lysosomes. EMBO Rep 2016; 17:e201541382-841; https://doi.org/10.15252/embr.201541382
- Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, Jahreiss L, Sarkar S, Futter M, Menzies FM, et al. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol 2011; 13:453–60; PMID:21394080; https://doi.org/10.1038/ncb2204
- Matteoni R, Kreis TE. Translocation and clustering of endosomes and lysosomes depends on microtubules. J Cell Biol 1987; 105:1253–65; PMID:3308906; https://doi.org/10.1083/jcb.105.3.1253
- Rosa-Ferreira C, Munro S. Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev Cell 2011; 21:1171–8; PMID:22172677; https://doi.org/10.1016/j.devcel.2011.10.007
- Pernigo S, Lamprecht A, Steiner RA, Dodding MP. Structural basis for kinesin-1:cargo recognition. Science 2013; 340:356–9; PMID:23519214; https://doi.org/10.1126/science.1234264
- Boucrot E, Henry T, Borg J-P, Gorvel J-P, Méresse S. The intracellular fate of Salmonella depends on the recruitment of kinesin. Science 2005; 308:1174–8; PMID:15905402; https://doi.org/10.1126/science.1110225
- Pu J, Schindler C, Jia R, Jarnik M, Backlund P, Bonifacino JS. BORC, a Multisubunit Complex that Regulates Lysosome Positioning. Dev Cell 2015; 33:176–88; PMID:25898167; https://doi.org/10.1016/j.devcel.2015.02.011
- Du W, Su QP, Chen Y, Zhu Y, Jiang D, Rong Y, Zhang S, Zhang Y, Ren H, Zhang C, et al. Kinesin 1 drives autolysosome tubulation. Dev Cell 2016; 37:326–36; PMID:27219061; https://doi.org/10.1016/j.devcel.2016.04.014
- Raiborg C, Wenzel EM, Pedersen NM, Olsvik H, Schink KO, Schultz SW, Vietri M, Nisi V, Bucci C, Brech A, et al. Repeated ER–endosome contacts promote endosome translocation and neurite outgrowth. Nature 2015; 520:234–8; PMID:25855459; https://doi.org/10.1038/nature14359
- Pankiv S, Alemu EA, Brech A, Bruun J-A, Lamark T, Øvervatn A, Bjørkøy G, Johansen T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol 2010; 188:253–69; PMID:20100911; https://doi.org/10.1083/jcb.200907015
- Jordens I, Fernandez-Borja M, Marsman M, Dusseljee S, Janssen L, Calafat J, Janssen H, Wubbolts R, Neefjes J. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol 2001; 11:1680–5; PMID:11696325; https://doi.org/10.1016/S0960-9822(01)00531-0
- Cantalupo G. Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J 2001; 20:683–93; PMID:11179213; https://doi.org/10.1093/emboj/20.4.683
- Wang T, Hong W. Interorganellar regulation of lysosome positioning by the Golgi apparatus through Rab34 interaction with Rab-interacting lysosomal protein. Mol Biol Cell 2002; 13:4317–32; PMID:12475955; https://doi.org/10.1091/mbc.E02-05-0280
- Chen L, Hu J, Yun Y, Wang T. Rab36 regulates the spatial distribution of late endosomes and lysosomes through a similar mechanism to Rab34. Mol Membr Biol 2010; 27:23–30; PMID:19961360; https://doi.org/10.3109/09687680903417470
- Goldenberg NM, Grinstein S, Silverman M. Golgi-bound Rab34 Is a Novel Member of the Secretory Pathway. Mol Biol Cell 2007; 18:4762–71; PMID:17881736; https://doi.org/10.1091/mbc.E06-11-0991
- Wu M, Wang T, Loh E, Hong W, Song H. Structural basis for recruitment of RILP by small GTPase Rab7. EMBO J 2005; 24:1491–501; PMID:15933719; https://doi.org/10.1038/sj.emboj.7600643
- Colucci AMR, Campana MC, Bellopede M, Bucci C. The Rab-interacting lysosomal protein, a Rab7 and Rab34 effector, is capable of self-interaction. Biochem Biophys Res Commun 2005; 334:128–33; PMID:15996637; https://doi.org/10.1016/j.bbrc.2005.06.067
- Medina DL, Fraldi A, Bouche V, Annunziata F, Mansueto G, Spampanato C, Puri C, Pignata A, Martina JA, Sardiello M, et al. Transcriptional Activation of Lysosomal Exocytosis Promotes Cellular Clearance. Dev Cell 2011; 21:421–30; PMID:21889421; https://doi.org/10.1016/j.devcel.2011.07.016
- Johnson DE, Ostrowski P, Jaumouillé V, Grinstein S. The position of lysosomes within the cell determines their luminal pH. J Cell Biol 2016; 212:677–92; PMID:26975849; https://doi.org/10.1083/jcb.201507112
- De Luca M, Cogli L, Progida C, Nisi V, Pascolutti R, Sigismund S, Di Fiore PP, Bucci C. RILP regulates vacuolar ATPase through interaction with the V1G1 subunit. J Cell Sci 2014; 127:2697–708; PMID:24762812; https://doi.org/10.1242/jcs.142604
- Alloatti A, Kotsias F, Pauwels A-M, Carpier J-M, Jouve M, Timmerman E, Pace L, Vargas P, Maurin M, Gehrmann U, et al. Toll-like Receptor 4 Engagement on Dendritic Cells Restrains Phago-Lysosome Fusion and Promotes Cross-Presentation of Antigens. Immunity 2015; 43:1087–100; PMID:26682983; https://doi.org/10.1016/j.immuni.2015.11.006
- Reddy A, Caler EV, Andrews NW. Plasma Membrane Repair Is Mediated by Ca2+-Regulated Exocytosis of Lysosomes. Cell 2001; 106:157–69; PMID:11511344; https://doi.org/10.1016/S0092-8674(01)00421-4
- Steffan JJ, Williams BC, Welbourne T, Cardelli JA. HGF-induced invasion by prostate tumor cells requires anterograde lysosome trafficking and activity of Na+-H+ exchangers. J Cell Sci 2010; 123:1151–9; PMID:20215403; https://doi.org/10.1242/jcs.063644
- Roberg KJ, Bickel S, Rowley N, Kaiser CA. Control of Amino Acid Permease Sorting in the Late Secretory Pathway of Saccharomyces cerevisiae by SEC13, LST4, LST7 and LST78. Genetics 1997; 147:1569–84; PMID:9409822
- Milkereit R, Persaud A, Vanoaica L, Guetg A, Verrey F, Rotin D. LAPTM4b recruits the LAT1-4F2hc Leu transporter to lysosomes and promotes mTORC1 activation. Nat Comms 2015; 6:7250; https://doi.org/10.1038/ncomms8250
- Li X, Rydzewski N, Hider A, Zhang X, Yang J, Wang W, Gao Q, Cheng X, Xu H. A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. - PubMed - NCBI. Nat Cell Biol 2016; 18:404–17; PMID:26950892; https://doi.org/10.1038/ncb3324
- Heuser J. Changes in lysosome shape and distribution correlated with changes in cytoplasmic pH. J Cell Biol 1989; 108:855–64; PMID:2921284; https://doi.org/10.1083/jcb.108.3.855