ABSTRACT
Accurate chromosome segregation in mammalian cells is guided by the centromere, a specialized chromosome region defined by the histone H3 variant centromere protein A (CENP-A). It is not well understood how cells maintain CENP-A levels at centromeres while continuously going through genome replications and cell divisions. A MgcRacGAP-dependent small GTPase molecular switch has been shown as essential for centromeric CENP-A maintenance. By using quantitative imaging, pulse-chase and live cell analysis, a recent work has suggested that the diaphanous formin mDia2, a well-established small GTPase effector, functions downstream of this small GTPase pathway to maintain CENP-A levels at centromeres. A constitutively active mDia2 construct is able to rescue the CENP-A loading defect caused by MgcRacGAP depletion. This study has uncovered an unsuspected role of the cytoskeleton protein mDia2 as an effector of the MgcRacGAP-dependent small GTPase signaling inside the nucleus to participate in the epigenetic regulation of centromere maintenance during cell cycle.
Faithful transmission of genetic materials during cell divisions requires 2 sets of replicated chromosomes being accurately segregated into 2 daughter cells. In mitosis, chromosome segregation is driven by the mitotic spindle, where microtubule polymers form mechanical linkages to the kinetochore—a proteinaceous structure assembled at a specialized chromosomal region called the centromere.Citation1 To ensure accurate chromosome segregation, it is important that the presence and the position of the centromere on each chromosome are stably maintained over many rounds of cell divisions. Through the course of evolution, many eukaryotes today use a conserved protein called centromere protein A (CENP-A) to mark the identity of centromeric chromatin. This is an epigenetic mark, as CENP-A is a histone H3 variant that forms nucleosomes composing centromeric chromatin regardless of the DNA sequences involved.Citation1 When DNA replicates in S-phase, however, the number of preexisting marker molecules per centromere will be diluted by half as they are redistributed to 2 daughter chromatin strands. Therefore, an outstanding question in the field is how CENP-A, as the marker molecule, maintains its level at centromeres. We now know that mammalian cells address this issue by replenishing the CENP-A level per centromere in early G1 phase of the cell cycle, right after the previous round of genome division and before the next round of DNA replication.Citation2 Although several important stages and molecular players have been identified to license, load and stabilize newly synthesized CENP-A proteins at centromeres labeled with preexisted and diluted CENP-A,Citation3-7 a complete understanding is missing regarding how new CENP-A proteins become stably incorporated into centromeric nucleosomes during early G1.
A Rho family small GTPase molecular switch is required for new CENP-A incorporation at centromeres
Rho family small GTPase proteins along with their cognate guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs) are generally distributed in the cytosol and participate in signaling regulation in membrane and cytoskeleton dynamics.Citation8,9 Recent studies have suggested nuclear functions of the Rho family small GTPase signaling. Net1, a RhoA specific GEF, and active RhoA have been found inside the nucleus.Citation10 A nuclear pool of RhoA is specifically activated in a Net1-dependent manner upon ionizing radiation (IR).Citation10 Loss of Ect2 and Net1 has been shown to suppress RhoB activation upon IR-induced DNA damage.Citation11 Rac1, another Rho family small GTPase protein, has a functional nuclear localization signal (NLS) for nuclear import mediated by karyopherin α2 and associates with numerous nuclear proteins.Citation12
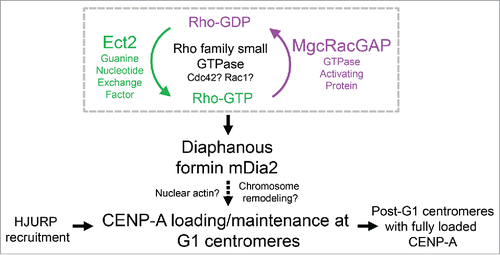
Leptomycin B (LMB) treatment results in a nuclear accumulation of the GTPase activating protein MgcRacGAP concurrent with a decrease in the cytoplasm.Citation13 The guanine exchange factor Ect2 contains a NLS and is preferentially localized to the nucleus at steady state.Citation14 Knocking down either MgcRacGAP or Ect2 for one cell cycle reduces the total CENP-A levels at the centromere by half, which is consistent with a defect in CENP-A replenishment.Citation15 Furthermore, depleting Cdc42 or Rac1, but not RhoA, results in a similar phenotype as MgcRacGAP or ECT2 depletion, indicating that Cdc42 and/or Rac1 are the small GTPases involved in new CENP-A loading/maintenance.Citation15 Thus, it is proposed that a molecular switch of Rho family small GTPase is essential in regulating epigenetic centromere maintenance by stabilizing newly loaded CENP-A.Citation15 However, it is difficult to know the exact functional role for the small GTPase pathway in this process without identifying the downstream effector(s).
The formin mDia2 functions downstream of the MgcRacGAP-dependent GTPase pathway to maintain CENP-A levels at the centromere
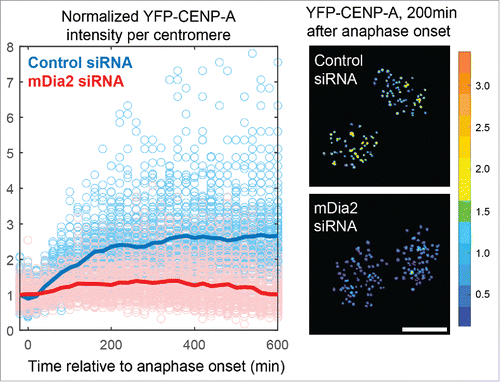
The diaphanous formin family proteins function as effectors of the small GTPase signaling in diverse aspects of the cell cycle and are well-established cytoskeleton regulators involved in both actin and microtubule dynamics.Citation16,17 Three members of the diaphanous formin family, mDia1-3, have conserved domain structures and a similar mode of action but differed choices of upstream GTPase signaling pathwaysCitation18-27 (). While investigating kinetochore functions of formin mDia3Citation28,29 using quantitative imaging, we have unexpectedly observed that depleting mDia2, but not mDia3, results in an almost 50% reduction of total CENP-A levels at centromeres after one round of cell cycle.Citation30 Furthermore, ratiometric live cell imaging studies () along with pulse chase approaches demonstrate that cells depleted of mDia2 are unable to maintain the continuous loading of new CENP-A onto centromeres.Citation30 The formin mDia2 have been shown to be the effector of either Rac or Cdc42,Citation20,21,23 2 small GTPases whose depletion results in a similar centromeric CENP-A reduction phenotype.Citation15 Unlike mDia1 or mDia3, mDia2 shuttles from cytoplasm to nucleoplasm evidenced by nuclear accumulation upon LMB treatmentCitation31 and a clear nuclear distribution in early G1 cells ().
Figure 1. Diaphanous formins are effectors of the small GTPase signaling. (A) Schematic diagram of the general structure and the mode of action of diaphanous formin proteins. GBD – GTPase binding domain; DID – Dia interacting domain; FH – Formin homology domain; DAD – Dia auto-inhibition domain. Small GTPase binding releases the auto-inhibitory interaction between the DID and the DAD domains. The FH1FH2 alone can be used as a constitutively active construct that functions without the GTPase signaling. (B) Comparison between the 3 members of diaphanous subfamily formins, with emphasis on the cognate upstream small GTPase signaling and the subcellular localization. Scale bar, 10 µm.
Figure 2. High resolution quantitative live-cell imaging for systematically measurement of total CENP-A levels at centromeres in real time. Left: overlaid plots showing ratiometric measurement of YFP-CENP-A levels from centromeres in multiple cells (light blue/red circles are raw data points, and dark blue/red lines are sample averages). Right: 2 representative frames of cells with YFP-CENP-A on the centromeres pseudocolored based on intensity levels. (Data were replotted from Liu and Mao., JCB, 2016Citation30 Scale bar, 10 µm.).
Diaphanous formins assume an auto-inhibited state resulted from an intra-molecular interaction between the DID and DAD domains. Upon small Rho GTPase binding at the N-terminus, the auto-inhibition of mDia formins is released to expose the formin homology domain (FH2) responsible for actin polymerization and microtubule stabilization.Citation32 A short-version of mDia proteins consisting of formin homology domains without regulatory regions required for auto-inhibition () is constitutively active even in the absence of small GTPase binding.Citation22 Such a constitutively active construct of mDia2 is able to rescue the CENP-A loading defect resulted from depletion of MgcRacGAP.Citation30 This epistatic analysis suggests that mDia2 is the effector downstream of the MgcRacGAP-mediated small GTPase switch in maintaining CENP-A levels at centromeres.
The identification of mDia2 as a small GTPase effector required for maintaining newly loaded CENP-A in early G1 has led to an obvious question: what exactly does mDia2 do in maintaining CENP-A levels at centromeres ()? Diaphanous formins are well known for their functions in polymerizing linear actin filaments and stabilizing microtubules.Citation33-35 The microtubule stabilization activity seems unlikely to be involved in mDia2's CENP-A function, as tubulins and microtubules are not normally found inside the nucleus.Citation36 On the contrary, recent works from multiple groups have implicated functional roles for the actin nucleation activity of formin proteins inside the nucleus.Citation36-38 As a potent actin nucleator, formin proteins promote actin polymerization inside the nucleus to participate in the regulation of serum response factor transcriptional activityCitation36 and the regulation of DNA damage response.Citation38 Consistent with the hypothesis that the actin nucleation activity of mDia2 is important for new CENP-A loading, constitutively active mDia2 constructs with point mutations that have been shown to be defective in polymerizing actin filaments in vitro and in vivo,Citation17 unlike the wild-type constitutively active construct, are not able to rescue centromeric CENP-A levels upon depleting endogenous mDia2.Citation30
Figure 3. Schematic summary showing mDia2 functions downstream of the MgcRacGAP-dependent GTPase pathway to maintain CENP-A levels at the centromere. Depending on the GEF (Ect2) and GAP (MgcRacGAP), the Rho family small GTPase cycles between active form and inactive form, which is important for CENP-A maintenance at the centromeres. Diaphanous formin mDia2 has an epistatic relationship downstream of the MgcRacGAP-dependent small GTPase molecular switch and promotes G1 CENP-A loading/maintenance.
A new model of epigenetic regulation of centromeric nucleosome assembly
The assembly of CENP-A nucleosomes underlying the centromeric chromatin includes several discernable steps. Upon anaphase onset, released from the CDK-based inhibition and promoted by the Polo kinase 1 (Plk1)-based stimulation, the Mis18 complex (consisting Mis18α, Mis18β and Mis18 binding protein 1) localizes to the centromere to license the process of new CENP-A recruitment.Citation7,39 The Mis18 complex is crucial for the recruitment of Holliday junction recognition protein (HJURP), a chaperon that binds to CENP-A in a pre-nucleosomal complex, to the centromere to execute its nucleosome assembly functions for new CENP-A.Citation5,6,40 Notably, the localization of HJURP at G1 centromeres appears to be transient, as HeLa cells yield a 2∼3 hr time window in early G1 in which HJURP can be detected on the centromere. Knocking down mDia2 does not affect HJURP recruitment onto the centromere.Citation30 However, upon mDia2 depletion, the percentage of cells with their centromeres decorated with HJURP is substantially increased.Citation30 A reasonable explanation for a prolonged centromeric localization of HJURP is that the transient HJURP turnover at early G1 centromeres is delayed in the absence of mDia2. A stochastic model with numerical simulation suggests that extended dwelling time of HJURP molecules at centromeres can indeed cause not only significantly higher percentage of cells with HJURP positive centromeres but also a defective loading process over time, which is consistent with observations from live-cell imaging studies.Citation30 Therefore, in the absence of mDia2, HJURP-bound CENP-A cannot be stably incorporated into the centromeric nucleosomes ().
Concluding remarks
That the constitutively active form of mDia2 is able to rescue the reduced levels of CENP-A at centromeres caused by depletion of MgcRacGAP strongly suggests the formin mDia2 as the downstream effector of the small GTPase-mediated molecular switch in new CENP-A loading at centromeres. It is proposed that the small GTPase activity modifies newly incorporated CENP-A at the correct place as a late-G1 surveillance step by either adding or removing a mark (modification or conformation change).Citation15 However, the identification of mDia2 as the downstream effector of this small GTPase pathway does not seem to support this hypothesis. First, ratiometric live-cell imaging analysis shows that depletion of mDia2 results in a failure in new CENP-A incorporation at centromeres in early G1.Citation30 And second, mDia2 depletion causes a prolonged centromeric localization of HJURP at G1,Citation30 which is consistent with a failed incorporation of CENP-A molecules into centromeric nucleosomes. Thus, one challenge for future studies will be to understand the exact functional roles for the small GTPase molecular switch and the formin mDia2 in regulating epigenetic centromere maintenance.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by a grant from the National Institute of Health (GM89768) to YM.
References
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and Kinetochores: From epigenetics to mitotic checkpoint signaling. Cell 2003; 112:407-21; PMID:12600307; https://doi.org/10.1016/S0092-8674(03)00115-6
- Jansen LET, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol 2007; 176:795-805; PMID:17339380; https://doi.org/10.1083/jcb.200701066
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, et al. Priming of centromere for CENP-A recruitment by human hMis18α, hMis18β, and M18BP1. Dev Cell 2007; 12:17-30; PMID:17199038; https://doi.org/10.1016/j.devcel.2006.11.002
- Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. Functional genomics identifies a Myb domain–containing protein family required for assembly of CENP-A chromatin. J Cell Biol 2007; 176:757-63; PMID:17339379; https://doi.org/10.1083/jcb.200701065
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP Is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 2009; 137:485-97; PMID:19410545; https://doi.org/10.1016/j.cell.2009.02.040
- Foltz DR, Jansen LET, Bailey AO, Yates Iii JR, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell 2009; 137:472-84; PMID:19410544; https://doi.org/10.1016/j.cell.2009.02.039
- McKinley Kara L, Cheeseman Iain M. Polo-like Kinase 1 Licenses CENP-A Deposition at Centromeres. Cell 2014; 158:397-411; PMID:25036634; https://doi.org/10.1016/j.cell.2014.06.016
- Ridley AJ. Rho GTPase signalling in cell migration. Curr Opin Cell Biol 2015; 36:103-12; PMID:26363959; https://doi.org/10.1016/j.ceb.2015.08.005
- Schaefer A, Reinhard NR, Hordijk PL. Toward understanding RhoGTPase specificity: structure, function and local activation. Small GTPases 2014; 5:6; PMID:25483298; https://doi.org/10.4161/21541248.2014.968004
- Dubash AD, Guilluy C, Srougi MC, Boulter E, Burridge K, Garcia-Mata R. The small GTPase RhoA localizes to the nucleus and is activated by Net1 and DNA damage signals. PloS one 2011; 6:e17380; PMID:21390328; https://doi.org/10.1371/journal.pone.0017380
- Srougi MC, Burridge K. The nuclear guanine nucleotide exchange factors Ect2 and Net1 regulate RhoB-mediated cell death after DNA damage. PloS one 2011; 6:e17108; PMID:21373644; https://doi.org/10.1371/journal.pone.0017108
- Sandrock K, Bielek H, Schradi K, Schmidt G, Klugbauer N. The nuclear import of the small GTPase Rac1 is mediated by the direct interaction with karyopherin alpha2. Traffic 2010; 11:198-209; PMID:19961560; https://doi.org/10.1111/j.1600-0854.2009.01015.x
- Cha K, Sen P, Raghunayakula S, Zhang XD. The cellular distribution of RanGAP1 is regulated by CRM1-mediated nuclear export in mammalian cells. PloS one 2015; 10:e0141309.
- Chalamalasetty RB, Hummer S, Nigg EA, Sillje HH. Influence of human Ect2 depletion and overexpression on cleavage furrow formation and abscission. J Cell Sci 2006; 119:3008-19; PMID:16803869; https://doi.org/10.1242/jcs.03032
- Lagana A, Dorn JF, De Rop V, Ladouceur AM, Maddox AS, Maddox PS. A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP-A. Nat Cell Biol 2010; 12:1186-93; PMID:21102442; https://doi.org/10.1038/ncb2129
- Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Ann Rev Biochem 2007; 76:593-627; PMID:17373907; https://doi.org/10.1146/annurev.biochem.75.103004.142647
- Bartolini F, Moseley JB, Schmoranzer J, Cassimeris L, Goode BL, Gundersen GG. The formin mDia2 stabilizes microtubules independently of its actin nucleation activity. J Cell Biol 2008; 181:523-36; PMID:18458159; https://doi.org/10.1083/jcb.200709029
- Alberts AS, Bouquin N, Johnston LH, Treisman R. Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein beta subunits and the yeast response regulator protein Skn7. J Biol Chem 1998; 273:8616-22; PMID:9535835; https://doi.org/10.1074/jbc.273.15.8616
- Gasman S, Kalaidzidis Y, Zerial M. RhoD regulates endosome dynamics through Diaphanous-related Formin and Src tyrosine kinase. Nat Cell Biol 2003; 5:195-204; PMID:12577064; https://doi.org/10.1038/ncb935
- Ji P, Jayapal SR, Lodish HF. Enucleation of cultured mouse fetal erythroblasts requires Rac GTPases and mDia2. Nat Cell Biol 2008; 10:314-21; PMID:18264091; https://doi.org/10.1038/ncb1693
- Lammers M, Meyer S, Kuhlmann D, Wittinghofer A. Specificity of interactions between mDia isoforms and Rho proteins. J Biol Chem 2008; 283:35236-46; PMID:18829452; https://doi.org/10.1074/jbc.M805634200
- Palazzo AF, Cook TA, Alberts AS, Gundersen GG. mDia mediates Rho-regulated formation and orientation of stable microtubules. Nat Cell Biol 2001; 3:723-9; PMID:11483957; https://doi.org/10.1038/35087035
- Peng J, Wallar BJ, Flanders A, Swiatek PJ, Alberts AS. Disruption of the diaphanous-related formin Drf1 gene encoding mDia1 reveals a role for Drf3 as an effector for Cdc42. Curr Biol 2003; 13:534-45; PMID:12676083; https://doi.org/10.1016/S0960-9822(03)00170-2
- Wallar BJ, Deward AD, Resau JH, Alberts AS. RhoB and the mammalian Diaphanous-related formin mDia2 in endosome trafficking. Exp Cell Res 2007; 313:560-71; PMID:17198702; https://doi.org/10.1016/j.yexcr.2006.10.033
- Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat Cell Biol 1999; 1:136-43; PMID:10559899; https://doi.org/10.1038/11056
- Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch BM, Narumiya S. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. Embo J 1997; 16:3044-56; PMID:9214622; https://doi.org/10.1093/emboj/16.11.3044
- Yasuda S, Oceguera-Yanez F, Kato T, Okamoto M, Yonemura S, Terada Y, Ishizaki T, Narumiya S. Cdc42 and mDia3 regulate microtubule attachment to kinetochores. Nature 2004; 428:767-71; PMID:15085137; http://dx.doi.org/10.1038/nature02452
- Cheng L, Zhang J, Ahmad S, Rozier L, Yu H, Deng H, Mao Y. Aurora B regulates formin mDia3 in achieving metaphase chromosome alignment. Dev Cell 2011; 20:342-52; PMID:21397845; https://doi.org/10.1016/j.devcel.2011.01.008
- Liu C, Chuang J-Z, Sung C-H, Mao Y. A dynein independent role of Tctex-1 at the kinetochore. Cell Cycle 2015; 14:1379-88; PMID:25928583; https://doi.org/10.1080/15384101.2014.1000217
- Liu C, Mao Y. Diaphanous formin mDia2 regulates CENP-A levels at centromeres. J Cell Biol 2016; 213:415-24; PMID:27185834; https://doi.org/10.1083/jcb.201512034
- Miki T, Okawa K, Sekimoto T, Yoneda Y, Watanabe S, Ishizaki T, Narumiya S. mDia2 shuttles between the nucleus and the cytoplasm through the importin-α/β- and CRM1-mediated nuclear transport mechanism. J Biol Chem 2009; 284:5753-62; PMID:19117945; http://dx.doi.org/10.1074/jbc.M806191200
- Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol 2010; 11:62-74; PMID:19997130; https://doi.org/10.1038/nrm2816
- Bartolini F, Gundersen GG. Formins and microtubules. Biochim Biophys Acta 2010; 1803:164-73; PMID:19631698; https://doi.org/10.1016/j.bbamcr.2009.07.006
- Mao Y. FORMIN a link between kinetochores and microtubule ends. Trends Cell Biol 2011; 21:625-9; PMID:21920754; https://doi.org/10.1016/j.tcb.2011.08.005
- Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Ann Rev Biochem 2007; 76:593-627; PMID:17373907; https://doi.org/10.1146/annurev.biochem.75.103004.142647
- Baarlink C, Wang H, Grosse R. Nuclear Actin Network Assembly by Formins Regulates the SRF Coactivator MAL. Science 2013; PMID:23558171
- Plessner M, Melak M, Chinchilla P, Baarlink C, Grosse R. Nuclear F-actin formation and reorganization upon cell spreading. J Biol Chem 2015; PMID:25759381
- Belin BJ, Lee T, Mullins RD. DNA damage induces nuclear actin filament assembly by Formin-2 and Spire-1/2 that promotes efficient DNA repair. eLife 2015; 4:e07735; PMID:AMBIGUOUS
- Silva Mariana CC, Bodor Dani L, Stellfox Madison E, Martins Nuno MC, Hochegger H, Foltz Daniel R, Jansen LE. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell 2012; 22:52-63; PMID:22169070; https://doi.org/10.1016/j.devcel.2011.10.014
- Barnhart MC, Kuich PHJL, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol 2011; 194:229-43; PMID:21768289; https://doi.org/10.1083/jcb.201012017
