ABSTRACT
The 20 members of the Rho GTPase family are key regulators of a wide-variety of biological activities. In response to activation, they signal via downstream effector proteins to induce dynamic alterations in the organization of the actomyosin cytoskeleton. In this review, post-translational modifications, mechanisms of dysregulation identified in human pathological conditions, and the ways that Rho GTPases might be targeted for chemotherapy will be discussed.
Introduction
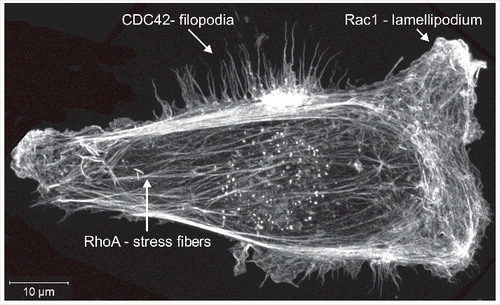
The Rho GTPases were initially discovered on the basis of their homology to the Ras GTPases,Citation1 and they were quickly discovered to have significant effects on cell morphology and actin cytoskeleton organization. RhoA activation leads to the formation of filamentous actin (F-actin) bundles called stress fibers,Citation2,3 Rac1 promotes the generation of F-actin-rich lamellipodia,Citation4 while CDC42 induces the extension of bundled F-actin projections called filopodiaCitation5,6 (). Since their initial discovery over 30 years ago, Rho GTPases have emerged as critically important and central regulators of the signaling pathways that influence the actomyosin cytoskeleton in processes including adhesion, migration and cell division,Citation7-9 as well as proliferation, differentiation and gene transcription.Citation10-12 This review will focus on the Rho proteins themselves, including recent findings regarding their post-translational modifications, and how this knowledge informs our understanding of disease-associated mutations and contributes to small molecule inhibitor development.
Figure 1. Filamentous actin structures. MDA MB 231 human breast cancer cells were fixed and stained with fluorescently labeled phalloidin to visualize filamentous actin structures. The image shows a maximum-projection assembled from 22 Z-plane images acquired with a Zeiss LSM 880 Airyscan microscope.
The Rho GTPase family
The Rho GTPase family is comprised of 20 members (), which act as molecular switch proteins that undergo cycles of activation and inactivation in response to GDP/GTP exchange and GTP hydrolysis (). Guanine nucleotide exchange factors (GEFs) promote the release of GDP, allowing GTP to associate with the Rho protein and induce conformational changes for downstream signal transduction. GTPase accelerating proteins (GAPs) increase the catalytic activity of Rho proteins to hydrolyse GTP to GDP by providing an “arginine finger” that enables catalysis.Citation13,14
Figure 2. Rho GTPases are molecular switches. When associated GEF molecules promote GDP release, GTP is bound which results in conformational changes in the Switch 1 and Switch 2 regions that enable interactions with downstream effector proteins and consequent signal transduction. Upon association with GAPs, GTP is hydrolysed to GDP and Pi is released to inactivate the Rho protein.
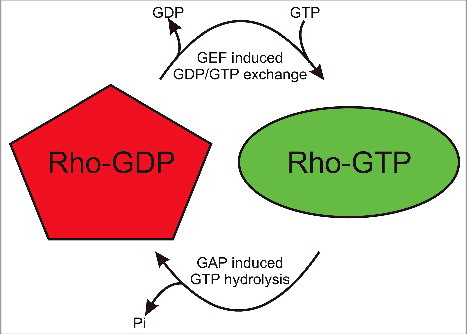
Table 1. Rho GTPases, post-translational modifications and mutations.
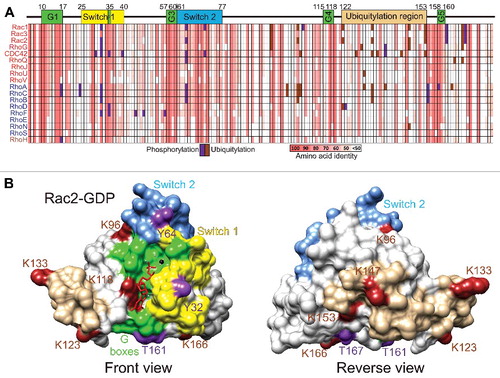
An amino acid identity matrix is shown in that indicates the percentage relatedness of each Rho GTPase protein to the other family members. In agreement with previous alignment studies,Citation15 grouping by hierarchical clusteringCitation16 indicated that there are 4 main protein clusters: Rac (red typeface) Rho (purple), RhoH (brown) and RhoBTB (green). The relationship between Rho GTPases is shown in by a phylogenetic tree where the separation between proteins is proportional to the calculated evolutionary distances.Citation16 Although clustering was based on amino acid identity, many of the proteins within clusters share overlapping biological functions. shows the amino acid conservation greater than 50% for the amino acids within the most well conserved segment of the 18 most related proteins, corresponding to residues 4 to 177 for the Rac1 reference sequence.
Figure 3. The Rho GTPase family. (A) An amino acid identity matrix was generated by comparing the primary sequence of 20 human Rho proteins. Grouping into Rac (red), Rho (purple), RhoH (brown) and RhoBTB (green) groups was by hierarchical clustering using the Multalin multiple sequence alignment program.Citation16 (B) A rooted phylogenetic tree of the 20 human Rho GTPases was generated by hierarchical clusing using the Multalin multiple sequence alignment program.Citation16 Distances between proteins are proportional to their PAM (point accepted mutation) score of molecular evolution.
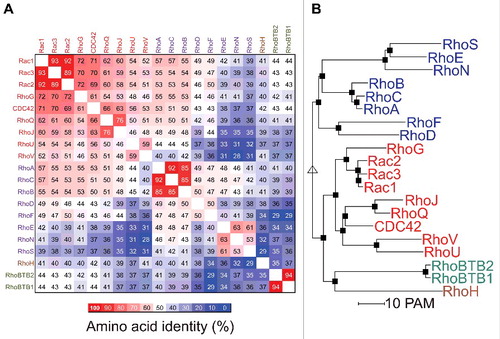
Figure 4. Amino acid conservation in the Rho GTPases. (A) The core conserved segments, with important features numbered by their positions within amino acids 4–177 of Rac1, of the 18 most related Rho GTPases are depicted with percentages of amino acid identity ranging from 100% (red) to less than 50% (white). Guanine nucleotide binding regions are shown as G1 to G5 boxes (green). Switch 1 (yellow) and Switch 2 (magenta) regions are also depicted. (B) Rac2-GDP (PDB ID: 2W2T) structure was rendered with Chimera,Citation112 with G boxes (green), Switch 1 (yellow) and Switch 2 (magenta) regions indicated. (C) Amino acid identity for 18 Rho GTPases was determined with Clustal Omega Citation113 and mapped onto Rac2-GDP with Chimera. Colors depicting amino acid identity from 100% (red) to 0% (blue) as indicated.
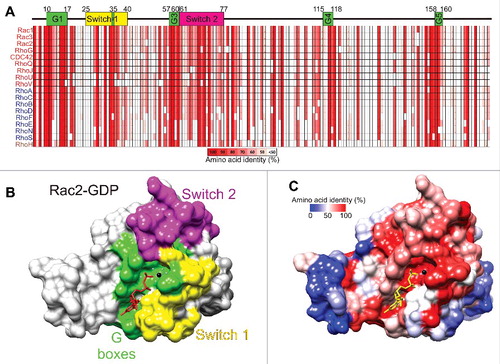
The most conserved regions of the Rho GTPases are the 5 G-boxes () that fold in the 3-dimensional protein to form the nucleotide binding region as shown for the Rac2-GDP structure (PDB ID: 2W2TCitation17) depicted in . The Switch 1 () and Switch 2 regions () change their conformations when GDP is exchanged for GTP to signal downstream to effector proteins. By mapping amino acid identity of the 18 most related Rho GTPases () onto the 3-dimensional Rac2-GDP structure, the conservation of the guanine nucleotide binding region as well as Switch 2 are clear (), while distal regions are less conserved (). The conservation of Switch 1 amino acids is more variable, with conserved and divergent residues. The amino acid diversity in Switch 1 allows each Rho protein to associate with different effectors to induce distinct responses and behaviors.
Regulatory post-translational modifications
In addition to the carboxyl-terminal modifications that have been reviewed elsewhereCitation18 which direct membrane localization, including prenylation (geranylgeranylation or farnesylation), endoproteolytic removal of the terminal 3 amino acids and carboxy-methylation, as well as palmitoylation of some family members, various Rho GTPases undergo further post-translational modifications that influence their activity and functions. Given the extensive homology within the Rho GTPase family, it is likely that comparable post-translational modifications may occur at conserved sites ( and ). Phosphorylations (purple) and ubiquitylations (brown) from the literature and from PhosphoSite (www.phosphosite.org) have been plotted on the amino acid identity grid in . In addition, phosphorylations (purple) and ubiquitylations (brown) identified for Rac2 have been mapped onto the front () and reverse () views of the 3-dimensional structure.
Figure 5. Phosphorylation and ubiquitylation of Rho GTPases. (A) Phosphorylation (purple) and ubiquitylation (brown) events reported in the literature or PhosphoSite (www.phosphosite.org) were mapped onto the amino acid identity grid depicting the core conserved segments, analogous to amino acids 4–177 of Rac1, of the 18 most related Rho GTPases, ranging from 100% identity (salmon) to less than 50% (white). (B) The phosphorylations (purple) and ubiquitylations (brown) of Rac2 were mapped onto the 3-dimensional structure, with G boxes (green), Switch 1 (yellow) and Switch 2 (magenta) regions depicted. (C) View rotated 180° of Rac2 phosphorylations and ubiquitylations.
The tyrosine analogous to RhoA Y34 (identical in 20/20 Rho proteins) in the Switch1 region is phosphorylated in RhoACitation19 and CDC42.Citation20 Nitration of RhoA Y34 in response to lipopolysaccharide treatment results in reduced GDP affinity to promote GDP/GTP exchange and consequent RhoA activation,Citation21 which suggests that phosphorylation at this conserved site might also be a mechanism of activation. Interestingly, AMPylation of RhoA Y34, Rac1 Y32 or CDC42 Y32, which could be catalyzed by the human HYPE protein,Citation22 blocked interactions with downstream effector proteins to inhibit signaling,Citation23 suggesting that modifications at this site could be a general mechanism for inactivation as well as activation. Phosphorylation of the tyrosine in the Switch 2 region analogous to RhoA Y66 (15/20 identical) occurs in RhoA,Citation19 Rac1,Citation24 and CDC42.Citation25 Rac1 Y64 phosphorylation inhibited cell spreading and lamellipodia formation, likely due to increased association with RhoGDIs and decreased association with GEFs and effectors,Citation24 while CDC42 Y64 phosphorylation also increased RhoGDI binding.Citation25 The conservation of these tyrosine residues, their accessibility on the Switch 1 and Switch 2 sites and the impact of their modifications on protein function suggest that they may be commonly targeted for post-translational modification as a regulatory mechanism.
RhoA can also be inhibited by Mst3-mediated phosphorylation on S26Citation26 which is serine or threonine at this analogous position in 12/20 Rho GTPases. Rac1 is phosphorylated on S71 (13/20 identical) by Akt to inhibit GTP binding,Citation27 similar to the CDC42 S71 phosphorylationCitation28 that is PI-3 kinase inhibitor sensitive.Citation29 The extensive conservation of many of the phosphorylation sites suggests that they may be general targets for post-translational modifications to provide additional levels of regulation. How these modifications may interact with disease-associated mutations (discussed below) is currently unknown, nor is it clear how these modifications would impact the efficacy of small molecule inhibitors. The consequence of these modifications on Rho GTPase activity and function illustrate the complexity and multi-layered nature of the regulatory mechanisms.
Ubiquitylation and regulation of protein stability
In addition to regulation through modulation of inactivation/activation cycling and subcellular localization, Rho GTPase signaling may also be influenced by varying protein levels. Expression of RhoB,Citation30 RhoC,Citation31 Rac3,Citation32 RhoG,Citation33 RhoU,Citation34 RhoE,Citation31,35 RhoSCitation36 and RhoHCitation37 can be induced by various stimuli. Conversely, ubiquitylation and subsequent proteasome-mediated degradation act to limit signaling and consequent effects on the actin cytoskeleton. RhoA is modified by ubiquitin ligase complexes directed by the E3 targeting proteins Smurf1 on K6 and K7,Citation38,39 by FBXL19 on K135,Citation40 and BACURD family proteins.Citation41 Rac3 is ubiquitylated on K166 by the E3 protein FBXL19,Citation42 while Rac1 is ubiquitylated by HACE on K147,Citation43,44 XIAP and c-IAP1 on K147,Citation45 and by FBXL19 on K166 dependent on S71 phosphorylation.Citation46 Unlike the conservation of several tyrosine residues that undergo phosphorylation in multiple Rho GTPases, the positions of ubiquitylated lysines are not conserved (), and most identified ubiquitylation events occur in a “Ubiquitylation region” on the opposite side of the protein from the GTP-binding and switch regions (). Many additional Rho GTPases have lysines scattered around this surface that are potentially ubiquitylated to regulate protein stability.
The significance of Rac1 regulation via ubiquitylation was revealed by the discovery of HACE1 ubiquitin ligase as a tumor suppressor in Wilm's tumor.Citation47 Subsequent investigations found HACE1 expression to be reduced in multiple cancers by several mechanisms including chromosomal translocation, deletions or loss-of-heterozygosity, as well as by epigenetic suppression.Citation47,48 Although reduced HACE1 expression would be predicted to have pleiotropic consequences due to its multiple targets, a key effect of HACE1 loss is increased Rac1 activity that induces cell migration and reactive oxygen species generationCitation43,44,49 that likely contribute to Rac-mediated tumor progression.Citation50
Rho GTPase mutations
Almost concurrent with their discovery, it was recognized that HRas, KRas and NRas GTPases had high frequencies of activating mutations in human cancers that work by reducing intrinsic and/or GAP-induced GTPase activity, thereby locking the protein in the on-state.Citation51 Similar GTPase deficient mutations have not been detected for Rho GTPases, and for many years it was believed that Rho proteins were not mutated in pathological conditions. However, the advent of large-scale gene sequencing has revealed both gain-of-function and loss-of-function mutations in several Rho GTPases. These findings have implications for how these proteins might be targeted by therapeutic agents, and suggest potential adverse effects that such treatments might evoke.
Immunodeficiency syndromes
Although infrequent, inactivating Rac2 mutations are associated with human immunodeficiency syndromes.Citation52 Rac2 has important roles in regulating the NADPH oxidase complex that generates superoxide in phagocytic cells of the immune system.Citation53 In addition, Rac2 also contributes to the chemotactic and phagocytic activities of immune cells such as neutrophils.Citation52 A Rac2 D57N mutation was identified in a human neutrophil immunodeficiency syndrome patient; the effect of this mutation was to decrease Rac2 GTP-binding, resulting in a dominant-negative acting protein that repressed endogenous Rac function.Citation54,55 Rac2 D57N was also identified in an additional patient screened for T-cell lymphopenia.Citation56 Homozygous Rac2 nonsense mutations at codon 56 (W56X) were identified in siblings with common variable immunodeficiency.Citation57 Unlike the manifestation of neutrophil dysfunction in patients bearing D57N mutations within weeks after birth, patients with W56X mutations did not present severe neonatal abnormalities. Instead, symptoms including recurrent infections did not emerge until the patients reached 6 months and 2 y of age,Citation57 suggesting that the effect of Rac2 protein absence was less potent than the dominant-inhibitory action of Rac2 D57N on endogenous wild-type Rac1.Citation54,55
RhoH is predominantly expressed in haematopoietic cells,Citation52 and is GTPase defective due to 2 differences at conserved sites analogous to Rac1 G12 and Q61 (similar to differences in RhoE, RhoN, RhoS, RhoBTB1 and RhoBTB2 at these positions) that would affect attacking water and GAP arginine finger co-ordination such that it remains constitutively GTP-bound.Citation37 RhoH deletion in mice revealed essential roles in T cell receptor signaling that are required for thymocyte selection and maturation.Citation58 Two adult human siblings with T cell defects that made them susceptible to infections by β-papilloma viruses were found to have homozygous nonsense RhoH mutations in codon 38 (Y38X) that resulted in loss of protein expression.Citation59 Consistent with the effects observed in RhoH−/− mice,Citation58,59 T cell receptor function was impaired and there were reduced numbers of tissue-homing integrin β7-positive T cells that would likely contribute to the observed susceptibility to β-papilloma viral infections.
Cancer
Although many of the first DBL-homology-containing guanine nucleotide exchange factors (GEFs) were discovered on the basis of their ability to oncogenically transform murine fibroblasts, including the prototype GEF Dbl,Citation60 GTPase-defective Rho proteins are inefficient oncogenes. Their poor transforming activity was attributed to a requirement for rapid GDP/GTP cycling, akin to the effect of activated GEFs, rather than persistent activation induced by GTPase-inactivating mutations. Consistent with this hypothesis, the rapid-cycling CDC42 F28L mutation, in which nucleotide-binding site and hydrogen bonding network disorder enhance spontaneous GDP/GTP exchange,Citation61 had potent transforming activity.Citation62
Recent high-throughput sequencing has led to the discovery of several significantly occurring Rac1 mutations (). In sun-exposed melanomas, Rac1 P29S substitutions were identifiedCitation63,64 that were proposed to alter Switch 1 conformation to destabilize the GDP-bound state and stabilize the GTP-bound form.Citation63,64 The Rac1 P29S mutation was also detected in a case of head and neck squamous cell carcinoma.Citation65 Analogous Rac2 P29LCitation63,66 and P29Q mutationsCitation66 have been identified, reinforcing the importance of this Proline residue for normal Switch I region function. Additional activating Rac1 mutations were identified in various cancer cell lines,Citation66 each of which were found to increase spontaneous GDP release to allow rapid GDP/GTP cycling that increases signal output.Citation66 Similarly, there is elevated expression of the rapidly GDP/GTP exchanging Rac1B splice variant in colorectal,Citation67 breast,Citation68 lung,Citation69 thyroid,Citation70 and pancreaticCitation71 cancers. These findings indicate that increased Rac signaling contributes to processes that promote tumorigenesis.
In contrast to the significant occurrence of Rac1 activation in cancer, frequent inactivating RhoA G17V mutations have been detected in T cell lymphomas.Citation72-74 The substitution of Valine for Glycine in the nucleotide binding pocket was predicted to introduce a bulky side-chainCitation72 that would result in reduced GTP binding.Citation73,74 In addition, RhoA G17V more effectively bound RhoGEFs than wild-type RhoA, and acted as a dominant-negative protein in cells to inhibit endogenous RhoA functions.Citation73,74 Sequencing RhoA in Burkitt lymphomas revealed additional mutations () that were predicted to reduce GEF binding and consequent GDP/GTP exchange.Citation75 In diffuse-type gastric cancer, further RhoA mutations were identified and found to confer growth promoting effects that wild-type RhoA did not.Citation76 Two additional mutations were found in RhoA in head and neck squamous cell carcinoma that mapped to the Switch 1 region.Citation65 Given that the mutations often clustered in regions important for GTP binding or effector interaction (including recurrent Y42 mutations),Citation77 these alterations may act as loss-of-function mutations that exert dominant-negative actions. The distribution of apparently inactivating RhoA mutations at varying amino acids also suggests that loss-of-function mutations could act via different mechanisms to achieve the same outcome. Since Rho signaling antagonizes Rac activity,Citation78,79 one possibility is that the effect of reduced RhoA signaling on tumorigenesis is mediated, at least in part, by enabling Rac functions.
In addition to the coding mutation described above, the RhoH gene is frequently altered by mutations in 5’ untranslated regions and by chromosomal translocations.Citation80 In fact, the intronless RhoH gene was first detected as part of a translocation between chromosomes 3 and 4 with the BCL6 gene in a non-Hodgkin lymphoma cell line, and was initially called TTF for translocation three four.Citation81 The RhoH gene was found to have undergone aberrant somatic hypermutation in germinal center-derived diffuse large-cell lymophomas, akin to the hypermutation of immunoglobulin variable region genes that normally takes place in B cell centroblasts to increase antibody diversity.Citation82 Since its initial discovery, RhoH gene mutations have been identified in lymphomas, mature B-cell neoplasms, multiple myeloma and lymphoproliferative disorders linked to immunodeficiencies associated with viral infections.Citation80,83 In addition, reduced RhoH expression has been described in hairy cell leukemiaCitation84 and acute myeloid leukemia,Citation85 although the causes for the low expression has not been determined. These alterations consistently lead to decreased RhoH expression, however the link between low RhoH protein levels and haematopoietic tumourigenesis is unclear. Although RhoH−/− mice have impaired T cell receptor signaling and defective thymocyte selection and maturation that lead to T cell deficiency, they did not develop lymphomas.Citation58,86 It was noted that basal Rac1 activity was higher in RhoH−/− mice,Citation86 while RhoH was also found to inhibit signaling downstream of active Rac1.Citation37 Therefore, one possibility is that, like inactivating RhoA mutants, reduced RhoH signaling may contribute to tumourigenesis by enhancing Rac functions.
In addition to their Rho GTPase related N-terminal region, RhoBTB1 and RhoBTB2 have a large C-terminal extension that includes a Proline-rich region, 2 BTB (broad-complex, tramtrack, bric à brac) domains and a conserved C-terminal region.Citation87 The additional domains associate with a number of proteins, several of which are components of ubiquitin ligase complexes that regulate the degradation of proteins involved with cell cycle progression and vesicle transport, as well as the influencing the stability of the RhoBTB proteins themselves. RhoBTB2 was identified as a putative tumor suppressor gene (Deleted in Breast Cancer 2; DBC2) that is located on chromosome 8p21 which is deleted or mutated in breast cancer.Citation88 Subsequently, it has been found that there is significant loss of heterozygosity (LOH) of the RhoBTB2 gene in bladderCitation89 and gastric cancers,Citation90 and LOH of the RhoBTB1 gene in head and neck squamous cell carcinoma.Citation91 In addition, rare somatic mutations have been identified in the RhoBTB1 and RhoBTB2 genes in these studies. However, more frequent are observations of reduced RhoBTB1 and RhoBTB2 expression, likely due to epigenetic factors including promoter methylation.Citation92 The tumor suppressor functions of RhoBTB1 and RhoBTB2, which may be lost as a result of gene deletion, mutation or reduced gene expression, are believed to be related to actions of the protein-associating C-terminal extension and not the Rho-homologous N-termini.Citation87 Although expression of RhoBTB1 or RhoBTB2 did not significantly affect actin cytoskeleton structures,Citation93 it has been suggested that the Rho-homologous region may interact with the BTB domain containing region to regulate associations with additional protein partners.Citation94 Therefore, it remains to be determined whether the rare mutations detected in the Rho-homolgous region (http://cancer.sanger.ac.uk/cosmic) may affect the tumor suppressive function of RhoBTB1 and RhoBTB2 by interfering with the formation of protein complexes with the C-terminal regions.
Large scale sequencing efforts have identified sporadic mutations in all 20 Rho GTPases in a large variety of cancers (http://cancer.sanger.ac.uk/cosmic). Some substitutions may be so conservative that they have little to no effect on protein function, while others may be adventitious mutations that are merely by-products of high mutation rates. Additional research will reveal whether specific mutations are passengers or drivers that contribute substantively to tumor initiation, growth and progression.
Small molecule inhibitor development
Given the initial associations of Rho GTPase signaling with cancer metastasisCitation95 and later discovery of activating mutations in tumors, there has been interest in developing small molecule inhibitors to inhibit Rho GTPase signaling.Citation96 One strategy employed has been to inhibit the interactions between GTPase and cognate GEF. The most widely-used GEF blocking compound is NSC23766, which was originally identified by in silico molecule docking and then validated as impairing the interaction of Rac1 with Trio and TIAM1 GEFs.Citation97 Although the potency of this compound is relatively low, limiting its potential for further development as a clinical candidate, it has been very useful as a tool compound and for providing proof-of-concept data. The inhibitor EHT1864,Citation98 which binds Rac1, Rac1b, Rac2 and Rac3, induces nucleotide release and can block TIAM1 interaction leading to inhibition of signalingCitation99 Additional small molecule Rac1/RhoG inhibitors that target GEF interactions have been identifiedCitation100,101 on the basis of blocking Trio GEF interactions, as well as Y16Citation102 and RhosinCitation103 that were discovered based on interference with LARG-RhoA binding. Cell-based screening for inhibitors of PIP2-induced actin polymerization in Xenopus laevis egg cytoplasmic extracts led to the identification of a series of Pirl1-related compounds that were found to block GEF-induced CDC42 activation,Citation104 latterly the Pirl1 related compound 2 was named CASIN.Citation105
An alternative strategy has been to directly target the GTPase. Compound library screening based on competition with fluorescent-GTP for bead-immobilized GTPase binding identified both broad-specificityCitation106 and CDC42-selective molecules.Citation107 In silico docking and validation by surface-plasmon resonance resulted in the identification of RhoA inhibitors that act by GTP-competition.Citation108 A similar virtual screen led to the discovery of Rhosin,Citation103 however in this case binding occurs in a shallow groove that forms part of the interface with GEFs including LARG, DBL and LBC.
Although these results validate the feasibility of discovering Rho GTPase inhibitors, there are several challenges that must be met if compounds are to be developed toward the ultimate goal of clinical use. GTP-competitive molecules would require very high affinities to overcome high cellular GTP concentrations.Citation109 Given that many of the activating mutations identified for Rac1 enable GEF-independent GDP/GTP exchangeCitation63,64 as does the Rac1B splice variant often elevated in cancers,Citation67-71,110 direct GTPase inhibition, including through GTP-competition, may be the most effective means to reduce signal output. Target selectivity is a significant issue, particularly since examining the selectivity of protein-protein interaction inhibitors is technically challenging due to the complexity of potential on-target and off-target protein binding partners. Indeed, by testing NSC23766 and EHT1864 in homozygous Rac1 knockout platelets, it was determined that both compounds have Rac1-independent effects including direct inhibition of the Rac1 effectors PAK1 and PAK2.Citation111 Many protein-protein inhibitors bind to surface pockets at the interaction interface, rather than deep pockets such as the nucleotide binding region. Although there may be sufficient differences in these relatively shallow surface pockets to confer selectivity, it is may be difficult for structure-activity relationship (SAR) medicinal chemistry to simultaneously increase potency and maintain selectivity.
The human mutation data suggests that selectivity would indeed be important for compounds to be used clinically. While Rac1 inhibition appears to be a potentially efficacious strategy, concomitant RhoA inhibition could have pro-tumorigenic effects.Citation65,72-74,76 In addition, the immunodeficiencies observed in patients with Rac2 mutationsCitation54-56 suggest that a Rac1 inhibitor might be immunosuppressive due to the likely difficulty in avoiding simultaneous inhibition of the highly-related (92% identity) Rac2 protein. If Rac1 inhibitors also blocked RhoH function, there might also be a risk of immunosuppressionCitation59 that could lead to susceptibility to viral infection and an increased risk of haematopoietic cancers.Citation80,83-85
Future directions
Despite the relative maturity of the Rho GTPase field, novel discoveries are continually made. Greatest attention has been paid to RhoA, Rac1 and CDC42, leaving considerable opportunity for details regarding the regulation and function of the remaining family members to be uncovered. Post-translational modifications at highly-conserved sites are suggestive that they may occur broadly. The occurrence of genetic mutations in the coding sequences of every Rho GTPase also suggests that there may be functional consequences for each protein. The recent emergence of gene-editing technologies will enable studies on the effect of these mutations in cellular contexts, and using in vivo models. Conditional genetically-modified in vivo models will continue to uncover the biological functions of the Rho GTPases in physiologically relevant environments as well as their contributions to pathological conditions, and may provide important clues about the potential utility, as well as adverse effects, of pharmacological inhibitors. Ultimately, the benefits and risks of potential Rho GTPase targeted therapeutics will be elucidated following rigorous testing in appropriate model systems.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
Thanks to Dominika Rudzka, Margaret O'Prey and Shehab Ismail (CRUK Beatson Institute) for assistance and advice with figure preparation.
Additional information
Funding
References
- Madaule P, Axel R. A novel ras-related gene family. Cell 1985; 41:31-40; PMID:3888408; https://doi.org/10.1016/0092-8674(85)90058-3
- Paterson HF, Self AJ, Garrett MD, Just I, Aktories K, Hall A. Microinjection of recombinant p21rho induces rapid changes in cell morphology. J Cell Biol 1990; 111:1001-7; PMID:2118140; https://doi.org/10.1083/jcb.111.3.1001
- Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 1992; 70:389-99; PMID:1643657; https://doi.org/10.1016/0092-8674(92)90163-7
- Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 1992; 70:401-10; PMID:1643658; https://doi.org/10.1016/0092-8674(92)90164-8
- Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 1995; 81:53-62; PMID:7536630; https://doi.org/10.1016/0092-8674(95)90370-4
- Kozma R, Ahmed S, Best A, Lim L. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol 1995; 15:1942-52; PMID:7891688; https://doi.org/10.1128/MCB.15.4.1942
- Zuo Y, Oh W, Frost JA. Controlling the switches: Rho GTPase regulation during animal cell mitosis. Cell Signal 2014; 26:2998-3006; PMID:25286227; https://doi.org/10.1016/j.cellsig.2014.09.022
- Sadok A, Marshall CJ. Rho GTPases: masters of cell migration. Small GTPases 2014; 5:e29710; PMID:24978113; https://doi.org/10.4161/sgtp.29710
- Takeichi M. Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling. Nat Rev Mol Cell Biol 2014; 15:397-410; PMID:24824068; https://doi.org/10.1038/nrm3802
- Rajakyla EK, Vartiainen MK. Rho, nuclear actin, and actin-binding proteins in the regulation of transcription and gene expression. Small GTPases 2014; 5:e27539; PMID:24603113; https://doi.org/10.4161/sgtp.27539
- Duquette PM, Lamarche-Vane N. Rho GTPases in embryonic development. Small GTPases 2014; 5:e972857; https://doi.org/10.4161/sgtp.29716
- Coleman ML, Marshall CJ, Olson MF. RAS and RHO GTPases in G1-phase cell-cycle regulation. Nat Rev Mol Cell Biol 2004; 5:355-66; PMID:15122349; https://doi.org/10.1038/nrm1365
- Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: Critical Elements in the Control of Small G Proteins. Cell 2007; 129:865-77; PMID:17540168; https://doi.org/10.1016/j.cell.2007.05.018
- Kötting C, Gerwert K. The dynamics of the catalytic site in small GTPases, variations on a common motif. FEBS Letters 2013; 587:2025-7; PMID:23684641; https://doi.org/10.1016/j.febslet.2013.05.021
- Boureux A, Vignal E, Faure S, Fort P. Evolution of the Rho family of ras-like GTPases in eukaryotes. Mol Biol Evol 2007; 24:203-16; PMID:17035353; https://doi.org/10.1093/molbev/msl145
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 1988; 16:10881-90; PMID:2849754; https://doi.org/10.1093/nar/16.22.10881
- Bunney TD, Opaleye O, Roe SM, Vatter P, Baxendale RW, Walliser C, Everett KL, Josephs MB, Christow C, Rodrigues-Lima F, et al. Structural insights into formation of an active signaling complex between Rac and phospholipase C gamma 2. Mol Cell 2009; 34:223-33; PMID:19394299; https://doi.org/10.1016/j.molcel.2009.02.023
- Liu M, Bi F, Zhou X, Zheng Y. Rho GTPase regulation by miRNAs and covalent modifications. Trends Cell Biol 2012; 22:365-73; PMID:22572609; https://doi.org/10.1016/j.tcb.2012.04.004
- Uezu A, Okada H, Murakoshi H, del Vescovo CD, Yasuda R, Diviani D, Soderling SH. Modified SH2 domain to phototrap and identify phosphotyrosine proteins from subcellular sites within cells. Proc Natl Acad Sci USA 2012; 109:E2929-E38; PMID:23027962; https://doi.org/10.1073/pnas.1207358109
- Trost M, Sauvageau M, Hérault O, Deleris P, Pomiès C, Chagraoui J, Mayotte N, Meloche S, Sauvageau G, Thibault P. Posttranslational regulation of self-renewal capacity: insights from proteome and phosphoproteome analyses of stem cell leukemia. Blood 2012; 120:e17-e27; PMID:22802335; https://doi.org/10.1182/blood-2011-12-397844
- Rafikov R, Dimitropoulou C, Aggarwal S, Kangath A, Gross C, Pardo D, Sharma S, Jezierska-Drutel A, Patel V, Snead C, et al. Lipopolysaccharide-induced Lung Injury Involves the Nitration-mediated Activation of RhoA. J Biol Chem 2014; 289:4710-22; PMID:24398689; https://doi.org/10.1074/jbc.M114.547596
- Faber PW, Barnes GT, Srinidhi J, Chen J, Gusella JF, MacDonald ME. Huntingtin Interacts with a Family of WW Domain Proteins. Human Mol Genet 1998; 7:1463-74; https://doi.org/10.1093/hmg/7.9.1463
- Worby CA, Mattoo S, Kruger RP, Corbeil LB, Koller A, Mendez JC, Zekarias B, Lazar C, Dixon JE. The Fic Domain: Regulation of Cell Signaling by Adenylylation. Mol Cell 2009; 34:93-103; PMID:19362538; https://doi.org/10.1016/j.molcel.2009.03.008
- Chang F, Lemmon C, Lietha D, Eck M, Romer L. Tyrosine phosphorylation of Rac1: a role in regulation of cell spreading. PLoS One 2011; 6:e28587; PMID:22163037; https://doi.org/10.1371/journal.pone.0028587
- Tu S, Wu WJ, Wang J, Cerione RA. Epidermal Growth Factor-dependent Regulation of Cdc42 Is Mediated by the Src Tyrosine Kinase. J Biol Chem 2003; 278:49293-300; PMID:14506284; https://doi.org/10.1074/jbc.M307021200
- Tang J, Ip JPK, Ye T, Ng Y-P, Yung W-H, Wu Z, Fang W, Fu AKY, Ip NY. Cdk5-Dependent Mst3 Phosphorylation and Activity Regulate Neuronal Migration through RhoA Inhibition. J Neurosci 2014; 34:7425-36; PMID:24872548; https://doi.org/10.1523/JNEUROSCI.5449-13.2014
- Kwon T, Kwon DY, Chun J, Kim JH, Kang SS. Akt Protein Kinase Inhibits Rac1-GTP Binding through Phosphorylation at Serine 71 of Rac1. J Biol Chem 2000; 275:423-8; PMID:10617634; https://doi.org/10.1074/jbc.275.1.423
- Schoentaube J, Olling A, Tatge H, Just I, Gerhard R. Serine-71 phosphorylation of Rac1/Cdc42 diminishes the pathogenic effect of Clostridium difficile toxin A. Cell Microbiol 2009; 11:1816-26; PMID:19709124; https://doi.org/10.1111/j.1462-5822.2009.01373.x
- Pothula S, Bazan HE, Chandrasekher G. Regulation of Cdc42 expression and signaling is critical for promoting corneal epithelial wound healing. Invest Ophthalmol Vis Sci 2013; 54:5343-52; PMID:23833064; https://doi.org/10.1167/iovs.13-11955
- Jahner D, Hunter T. The ras-related gene rhoB is an immediate-early gene inducible by v-Fps, epidermal growth factor, and platelet-derived growth factor in rat fibroblasts. Mol Cell Biol 1991; 11:3682-90; PMID:1710770; https://doi.org/10.1128/MCB.11.7.3682
- Croft DR, Crighton D, Samuel MS, Lourenco FC, Munro J, Wood J, Bensaad K, Vousden KH, Sansom OJ, Ryan KM, et al. p53-mediated transcriptional regulation and activation of the actin cytoskeleton regulatory RhoC to LIMK2 signaling pathway promotes cell survival. Cell Res 2011; 21:666-82; PMID:21079653; https://doi.org/10.1038/cr.2010.154
- Haataja L, Groffen J, Heisterkamp N. Characterization of RAC3, a Novel Member of the Rho Family. J Biol Chem 1997; 272:20384-8; PMID:9252344; https://doi.org/10.1074/jbc.272.33.20384
- Vincent S, Jeanteur P, Fort P. Growth-regulated expression of rhoG, a new member of the ras homolog gene family. Mol Cell Biol 1992; 12:3138-48; PMID:1620121; https://doi.org/10.1128/MCB.12.7.3138
- Tao W, Pennica D, Xu L, Kalejta RF, Levine AJ. Wrch-1, a novel member of the Rho gene family that is regulated by Wnt-1. Genes Dev 2001; 15:1796-807; PMID:11459829; https://doi.org/10.1101/gad.894301
- Hansen SH, Zegers MMP, Woodrow M, Rodriguez-Viciana P, Chardin P, Mostov KE, McMahon M. Induced Expression of Rnd3 Is Associated with Transformation of Polarized Epithelial Cells by the Raf–MEK–Extracellular Signal-Regulated Kinase Pathway. Mol Cell Biol 2000; 20:9364-75; PMID:11094087; https://doi.org/10.1128/MCB.20.24.9364-9375.2000
- Kim Y-S, Kim B, Karaki H, Hori M, Ozaki H. Up-regulation of Rnd1 during pregnancy serves as a negative-feedback control for Ca2+ sensitization of contractile elements in rat myometrium. Biochem Biophys Res Commun 2003; 311:972-8; PMID:14623277; https://doi.org/10.1016/j.bbrc.2003.10.100
- Li X, Bu X, Lu B, Avraham H, Flavell RA, Lim B. The hematopoiesis-specific GTP-binding protein RhoH is GTPase deficient and modulates activities of other Rho GTPases by an inhibitory function. Mol Cell Biol 2002; 22:1158-71; PMID:11809807; https://doi.org/10.1128/MCB.22.4.1158-1171.2002
- Wang H-R, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, Wrana JL. Regulation of Cell Polarity and Protrusion Formation by Targeting RhoA for Degradation. Science 2003; 302:1775-9; PMID:14657501; https://doi.org/10.1126/science.1090772
- Ozdamar B, Bose R, Barrios-Rodiles M, Wang H-R, Zhang Y, Wrana JL. Regulation of the Polarity Protein Par6 by TGFß Receptors Controls Epithelial Cell Plasticity. Science 2005; 307:1603-9; PMID:15761148; https://doi.org/10.1126/science.1105718
- Wei J, Mialki RK, Dong S, Khoo A, Mallampalli RK, Zhao Y, Zhao J. A new mechanism of RhoA ubiquitination and degradation: Roles of SCFFBXL19 E3 ligase and Erk2. Biochim Biophys Acta 2013; 1833:2757-64; PMID:23871831; https://doi.org/10.1016/j.bbamcr.2013.07.005
- Chen Y, Yang Z, Meng M, Zhao Y, Dong N, Yan H, Liu L, Ding M, Peng HB, Shao F. Cullin Mediates Degradation of RhoA through Evolutionarily Conserved BTB Adaptors to Control Actin Cytoskeleton Structure and Cell Movement. Mol Cell 2009; 35:841-55; PMID:19782033; https://doi.org/10.1016/j.molcel.2009.09.004
- Dong S, Zhao J, Wei J, Bowser RK, Khoo A, Liu Z, Luketich JD, Pennathur A, Ma H, Zhao Y. F-box protein complex FBXL19 regulates TGFbeta1-induced E-cadherin down-regulation by mediating Rac3 ubiquitination and degradation. Mol Cancer 2014; 13:76; PMID:24684802; https://doi.org/10.1186/1476-4598-13-76
- Castillo-Lluva S, Tan CT, Daugaard M, Sorensen PH, Malliri A. The tumour suppressor HACE1 controls cell migration by regulating Rac1 degradation. Oncogene 2013; 32:1735-42; PMID:22614015; https://doi.org/10.1038/onc.2012.189
- Torrino S, Visvikis O, Doye A, Boyer L, Stefani C, Munro P, Bertoglio J, Gacon G, Mettouchi A, Lemichez E. The E3 Ubiquitin-Ligase HACE1 Catalyzes the Ubiquitylation of Active Rac1. Dev Cell 2011; 21:959-65; PMID:22036506; https://doi.org/10.1016/j.devcel.2011.08.015
- Oberoi TK, Dogan T, Hocking JC, Scholz RP, Mooz J, Anderson CL, Karreman C, Meyer zu Heringdorf D, Schmidt G, Ruonala M, et al. IAPs regulate the plasticity of cell migration by directly targeting Rac1 for degradation. EMBO J 2012; 31:14-28; PMID:22117219; https://doi.org/10.1038/emboj.2011.423
- Zhao J, Mialki RK, Wei J, Coon TA, Zou C, Chen BB, Mallampalli RK, Zhao Y. SCF E3 ligase F-box protein complex SCF(FBXL19) regulates cell migration by mediating Rac1 ubiquitination and degradation. FASEB J 2013; 27:2611-9; PMID:23512198; https://doi.org/10.1096/fj.12-223099
- Anglesio MS, Evdokimova V, Melnyk N, Zhang L, Fernandez CV, Grundy PE, Leach S, Marra MA, Brooks-Wilson AR, Penninger J, et al. Differential expression of a novel ankyrin containing E3 ubiquitin-protein ligase, Hace1, in sporadic Wilms' tumor versus normal kidney. Human Mol Genet 2004; 13:2061-74; https://doi.org/10.1093/hmg/ddh215
- Zhang L, Anglesio MS, O'Sullivan M, Zhang F, Yang G, Sarao R, Nghiem MP, Cronin S, Hara H, Melnyk N, et al. The E3 ligase HACE1 is a critical chromosome 6q21 tumor suppressor involved in multiple cancers. Nat Med 2007; 13:1060-9; PMID:17694067; https://doi.org/10.1038/nm1621
- Daugaard M, Nitsch R, Razaghi B, McDonald L, Jarrar A, Torrino S, Castillo-Lluva S, Rotblat B, Li L, Malliri A, et al. Hace1 controls ROS generation of vertebrate Rac1-dependent NADPH oxidase complexes. Nature Commun 2013; 4; PMID:NOT_FOUND
- Goka ET, Lippman ME. Loss of the E3 ubiquitin ligase HACE1 results in enhanced Rac1 signaling contributing to breast cancer progression. Oncogene 2015; PMID:25659579
- Prior IA, Lewis PD, Mattos C. A Comprehensive Survey of Ras Mutations in Cancer. Cancer Res 2012; 72:2457-67; PMID:22589270; https://doi.org/10.1158/0008-5472.CAN-11-2612
- Troeger A, Williams DA. Hematopoietic-specific Rho GTPases Rac2 and RhoH and human blood disorders. Exp Cell Res 2013; 319:2375-83; PMID:23850828; https://doi.org/10.1016/j.yexcr.2013.07.002
- Hordijk PL. Regulation of NADPH Oxidases: The Role of Rac Proteins. Circ Res 2006; 98:453-62; PMID:16514078; https://doi.org/10.1161/01.RES.0000204727.46710.5e
- Williams DA, Tao W, Yang F, Kim C, Gu Y, Mansfield P, Levine JE, Petryniak B, Derrow CW, Harris C, et al. Dominant negative mutation of the hematopoietic-specific Rho GTPase, Rac2, is associated with a human phagocyte immunodeficiency. Blood 2000; 96:1646-54; PMID:10961859
- Ambruso DR, Knall C, Abell AN, Panepinto J, Kurkchubasche A, Thurman G, Gonzalez-Aller C, Hiester A, deBoer M, Harbeck RJ, et al. Human neutrophil immunodeficiency syndrome is associated with an inhibitory Rac2 mutation. Proc Natl Acad Sci USA 2000; 97:4654-9; PMID:10758162; https://doi.org/10.1073/pnas.080074897
- Accetta D, Syverson G, Bonacci B, Reddy S, Bengtson C, Surfus J, Harbeck R, Huttenlocher A, Grossman W, Routes J, et al. Human phagocyte defect caused by a Rac2 mutation detected by means of neonatal screening for T-cell lymphopenia. J Allergy Clin Immun 2011; 127:535-8.e2; PMID:21167572; https://doi.org/10.1016/j.jaci.2010.10.013
- Alkhairy OK, Rezaei N, Graham RR, Abolhassani H, Borte S, Hultenby K, Wu C, Aghamohammadi A, Williams DA, Behrens TW, et al. RAC2 loss-of-function mutation in 2 siblings with characteristics of common variable immunodeficiency. J Allergy Clin Immun 2015; 135:1380-4.e5; PMID:25512081; https://doi.org/10.1016/j.jaci.2014.10.039
- Gu Y, Chae H-D, Siefring JE, Jasti AC, Hildeman DA, Williams DA. RhoH GTPase recruits and activates Zap70 required for T cell receptor signaling and thymocyte development. Nat Immunol 2006; 7:1182-90; PMID:17028588; https://doi.org/10.1038/ni1396
- Crequer A, Troeger A, Patin E, Ma CS, Picard C, Pedergnana V, Fieschi C, Lim A, Abhyankar A, Gineau L, et al. Human RHOH deficiency causes T cell defects and susceptibility to EV-HPV infections. J Clin Invest 2012; 122:3239-47; PMID:22850876; https://doi.org/10.1172/JCI62949
- Srivastava SK, Wheelock RH, Aaronson SA, Eva A. Identification of the protein encoded by the human diffuse B-cell lymphoma (dbl) oncogene. Proc Natl Acad Sci USA 1986; 83:8868-72; PMID:3491366; https://doi.org/10.1073/pnas.83.23.8868
- Adams PD, Loh AP, Oswald RE. Backbone dynamics of an oncogenic mutant of Cdc42Hs shows increased flexibility at the nucleotide-binding site. Biochemistry 2004; 43:9968-77; PMID:15287724; https://doi.org/10.1021/bi0490901
- Lin R, Bagrodia S, Cerione R, Manor D. A novel Cdc42Hs mutant induces cellular transformation. Curr Biol 1997; 7:794-7; PMID:9368762; https://doi.org/10.1016/S0960-9822(06)00338-1
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al. A landscape of driver mutations in melanoma. Cell 2012; 150:251-63; PMID:22817889; https://doi.org/10.1016/j.cell.2012.06.024
- Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, Cheng E, Davis MJ, Goh G, Choi M, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet 2012; 44:1006-14; PMID:22842228; https://doi.org/10.1038/ng.2359
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. The Mutational Landscape of Head and Neck Squamous Cell Carcinoma. Science 2011; 333:1157-60; PMID:21798893; https://doi.org/10.1126/science.1208130
- Kawazu M, Ueno T, Kontani K, Ogita Y, Ando M, Fukumura K, Yamato A, Soda M, Takeuchi K, Miki Y, et al. Transforming mutations of RAC guanosine triphosphatases in human cancers. Proc Natl Acad Sci USA 2013; 110:3029-34; PMID:23382236; https://doi.org/10.1073/pnas.1216141110
- Jordan P, Brazao R, Boavida MG, Gespach C, Chastre E. Cloning of a novel human Rac1b splice variant with increased expression in colorectal tumors. Oncogene 1999; 18:6835-9; PMID:10597294; https://doi.org/10.1038/sj.onc.1203233
- Schnelzer A, Prechtel D, Knaus U, Dehne K, Gerhard M, Graeff H, Harbeck N, Schmitt M, Lengyel E. Rac1 in human breast cancer: overexpression, mutation analysis, and characterization of a new isoform, Rac1b. Oncogene 2000; 19:3013-20; PMID:10871853; https://doi.org/10.1038/sj.onc.1203621
- Liu J, Lee W, Jiang Z, Chen Z, Jhunjhunwala S, Haverty PM, Gnad F, Guan Y, Gilbert HN, Stinson J, et al. Genome and transcriptome sequencing of lung cancers reveal diverse mutational and splicing events. Genome Res 2012; 22:2315-27; PMID:23033341; https://doi.org/10.1101/gr.140988.112
- Silva AL, Carmo F, Bugalho MJ. RAC1b overexpression in papillary thyroid carcinoma: a role to unravel. Eur J Endocrinol 2013; 168:795-804; PMID:23482591; https://doi.org/10.1530/EJE-12-0960
- Mehner C, Miller E, Khauv D, Nassar A, Oberg AL, Bamlet WR, Zhang L, Waldmann J, Radisky ES, Crawford HC, et al. Tumor cell-derived MMP3 orchestrates Rac1b and tissue alterations that promote pancreatic adenocarcinoma. Mol Cancer Res 2014; 12:1430-9; PMID:24850902; https://doi.org/10.1158/1541-7786.MCR-13-0557-T
- Yoo HY, Sung MK, Lee SH, Kim S, Lee H, Park S, Kim SC, Lee B, Rho K, Lee J-E, et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat Genet 2014; 46:371-5; PMID:24584070; https://doi.org/10.1038/ng.2916
- Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y, Muto H, Tsuyama N, Sato-Otsubo A, Okuno Y, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet 2014; 46:171-5; PMID:24413737; https://doi.org/10.1038/ng.2872
- Palomero T, Couronne L, Khiabanian H, Kim MY, Ambesi-Impiombato A, Perez-Garcia A, Carpenter Z, Abate F, Allegretta M, Haydu JE, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet 2014; 46:166-70; PMID:24413734; https://doi.org/10.1038/ng.2873
- Rohde M, Richter J, Schlesner M, Betts MJ, Claviez A, Bonn BR, Zimmermann M, Damm-Welk C, Russell RB, Borkhardt A, et al. Recurrent RHOA mutations in pediatric Burkitt lymphoma treated according to the NHL-BFM protocols. Genes Chrom Cancer 2014; 53:911-6; PMID:25044415; https://doi.org/10.1002/gcc.22202
- Kakiuchi M, Nishizawa T, Ueda H, Gotoh K, Tanaka A, Hayashi A, Yamamoto S, Tatsuno K, Katoh H, Watanabe Y, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet 2014; 46:583-7; PMID:24816255; https://doi.org/10.1038/ng.2984
- Sahai E, Alberts AS, Treisman R. RhoA effector mutants reveal distinct effector pathways for cytoskeletal reorganization, SRF activation and transformation. EMBO J 1998; 17:1350-61; PMID:9482732; https://doi.org/10.1093/emboj/17.5.1350
- Yamaguchi Y, Katoh H, Yasui H, Mori K, Negishi M. RhoA inhibits the nerve growth factor-induced Rac1 activation through Rho-associated kinase-dependent pathway. J Biol Chem 2001; 276:18977-83; PMID:11279039; https://doi.org/10.1074/jbc.M100254200
- Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, Sahai E, Marshall CJ. Rac activation and inactivation control plasticity of tumor cell movement. Cell 2008; 135:510-23; PMID:18984162; https://doi.org/10.1016/j.cell.2008.09.043
- Fueller F, Kubatzky KF. The small GTPase RhoH is an atypical regulator of haematopoietic cells. Cell Commun Signal 2008; 6:6; PMID:18823547; https://doi.org/10.1186/1478-811X-6-6
- Dallery E, Galiegue-Zouitina S, Collyn-d'Hooghe M, Quief S, Denis C, Hildebrand MP, Lantoine D, Deweindt C, Tilly H, Bastard C, et al. TTF, a gene encoding a novel small G protein, fuses to the lymphoma- associated LAZ3 gene by t(3;4) chromosomal translocation. Oncogene 1995; 10:2171-8; PMID:7784061
- Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RSK, Kuppers R, Dalla-Favera R. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature 2001; 412:341-6; PMID:11460166; https://doi.org/10.1038/35085588
- Preudhomme C, Roumier C, Hildebrand MP, Dallery-Prudhomme E, Lantoine D, Lai JL, Daudignon A, Adenis C, Bauters F, Fenaux P, et al. Nonrandom 4p13 rearrangements of the RhoH/TTF gene, encoding a GTP- binding protein, in non-Hodgkin's lymphoma and multiple myeloma. Oncogene 2000; 19:2023-32; PMID:10803463; https://doi.org/10.1038/sj.onc.1203521
- Galiègue-Zouitina S, Delestré L, Dupont C, Troussard X, Shelley CS. Underexpression of RhoH in Hairy Cell Leukemia. Cancer Res 2008; 68:4531-40; PMID:Can't; https://doi.org/10.1158/0008-5472.CAN-07-5661
- Iwasaki T, Katsumi A, Kiyoi H, Tanizaki R, Ishikawa Y, Ozeki K, Kobayashi M, Abe A, Matsushita T, Watanabe T, et al. Prognostic implication and biological roles of RhoH in acute myeloid leukaemia. Eur J Haematol 2008; 81:454-60; PMID:18691253; https://doi.org/10.1111/j.1600-0609.2008.01132.x
- Dorn T, Kuhn U, Bungartz G, Stiller S, Bauer M, Ellwart J, Peters T, Scharffetter-Kochanek K, Semmrich M, Laschinger M, et al. RhoH is important for positive thymocyte selection and T-cell receptor signaling. Blood 2007; 109:2346-55; PMID:17119112; https://doi.org/10.1182/blood-2006-04-019034
- Ji W, Rivero F. Atypical Rho GTPases of the RhoBTB Subfamily: Roles in Vesicle Trafficking and Tumorigenesis. Cells 2016; 5; PMID:27314390; https://doi.org/10.3390/cells5020028
- Hamaguchi M, Meth JL, von Klitzing C, Wei W, Esposito D, Rodgers L, Walsh T, Welcsh P, King MC, Wigler MH. DBC2, a candidate for a tumor suppressor gene involved in breast cancer. Proc Natl Acad Sci USA 2002; 99:13647-52; PMID:12370419; https://doi.org/10.1073/pnas.212516099
- Knowles MA, Aveyard JS, Taylor CF, Harnden P, Bass S. Mutation analysis of the 8p candidate tumour suppressor genes DBC2 (RHOBTB2) and LZTS1 in bladder cancer. Cancer Lett 2005; 225:121-30; PMID:15922864; https://doi.org/10.1016/j.canlet.2004.10.047
- Cho YG, Choi BJ, Kim CJ, Song JH, Zhang C, Nam SW, Lee JY, Park WS. Genetic analysis of the DBC2 gene in gastric cancer. Acta Oncol 2008; 47:366-71; PMID:17906984; https://doi.org/10.1080/02841860701644094
- Beder LB, Gunduz M, Ouchida M, Gunduz E, Sakai A, Fukushima K, Nagatsuka H, Ito S, Honjo N, Nishizaki K, et al. Identification of a candidate tumor suppressor gene RHOBTB1 located at a novel allelic loss region 10q21 in head and neck cancer. J Cancer Res Clin Oncol 2006; 132:19-27; PMID:16170569; https://doi.org/10.1007/s00432-005-0033-0
- Berthold J, Schenkova K, Rivero F. Rho GTPases of the RhoBTB subfamily and tumorigenesis. Acta Pharmacol Sin 2008; 29:285-95; PMID:18298893; https://doi.org/10.1111/j.1745-7254.2008.00773.x
- Aspenström P, Fransson Å, Saras J. Rho GTPases have diverse effects on the organization of the actin filament system. Biochem J 2004; 377:327-37; PMID:14521508; https://doi.org/10.1042/bj20031041
- Berthold J, Schenkova K, Ramos S, Miura Y, Furukawa M, Aspenstrom P, Rivero F. Characterization of RhoBTB-dependent Cul3 ubiquitin ligase complexes–evidence for an autoregulatory mechanism. Exp Cell Res 2008; 314:3453-65; PMID:18835386; https://doi.org/10.1016/j.yexcr.2008.09.005
- Olson MF, Sahai E. The actin cytoskeleton in cancer cell motility. Clin Exp Metastasis 2009; 26:273-87; PMID:18498004; https://doi.org/10.1007/s10585-008-9174-2
- Mardilovich K, Olson MF, Baugh M. Targeting Rho GTPase signaling for cancer therapy. Future Oncol 2012; 8:165-77; PMID:22335581; https://doi.org/10.2217/fon.11.143
- Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci USA 2004; 101:7618-23; PMID:15128949; https://doi.org/10.1073/pnas.0307512101
- Désiré L, Bourdin J, Loiseau N, Peillon H, Picard V, De Oliveira C, Bachelot F, Leblond B, Taverne T, Beausoleil E, et al. RAC1 inhibition targets amyloid precursor protein processing by gamma-secretase and decreases Abeta production in vitro and in vivo. J Biol Chem 2005; 280:37516-25; PMID:Can't; https://doi.org/10.1074/jbc.M507913200
- Shutes A, Onesto C, Picard V, Leblond B, Schweighoffer F, Der CJ. Specificity and mechanism of action of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. J Biol Chem 2007; 282:35666-78; PMID:17932039; https://doi.org/10.1074/jbc.M703571200
- Blangy A, Bouquier N, Gauthier-Rouvière C, Schmidt S, Debant A, Leonetti JP, Fort P. Identification of TRIO-GEFD1 chemical inhibitors using the yeast exchange assay. Biol Cell 2006; 98:511-22; PMID:16686599; https://doi.org/10.1042/BC20060023
- Bouquier N, Vignal E, Charrasse S, Weill M, Schmidt S, Léonetti JP, Blangy A, Fort P. A cell active chemical GEF inhibitor selectively targets the Trio/RhoG/Rac1 signaling pathway. Chem Biol 2009; 16:657-66; PMID:19549603; https://doi.org/10.1016/j.chembiol.2009.04.012
- Shang X, Marchioni F, Evelyn CR, Sipes N, Zhou X, Seibel W, Wortman M, Zheng Y. Small-molecule inhibitors targeting G-protein–coupled Rho guanine nucleotide exchange factors. Proc Natl Acad Sci USA 2013; 110:3155-60; PMID:23382194; https://doi.org/10.1073/pnas.1212324110
- Shang X, Marchioni F, Sipes N, Evelyn Chris R, Jerabek-Willemsen M, Duhr S, Seibel W, Wortman M, Zheng Y. Rational Design of Small Molecule Inhibitors Targeting RhoA Subfamily Rho GTPases. Chem Biol 2012; 19:699-710; PMID:22726684; https://doi.org/10.1016/j.chembiol.2012.05.009
- Peterson JR, Lebensohn AM, Pelish HE, Kirschner MW. Biochemical Suppression of Small-Molecule Inhibitors: A Strategy to Identify Inhibitor Targets and Signaling Pathway Components. Chem Biol 2006; 13:443-52; PMID:16632257; https://doi.org/10.1016/j.chembiol.2006.02.009
- Sakamori R, Das S, Yu S, Feng S, Stypulkowski E, Guan Y, Douard V, Tang W, Ferraris RP, Harada A, et al. Cdc42 and Rab8a are critical for intestinal stem cell division, survival, and differentiation in mice. J Clin Invest 2012; 122:1052-65; PMID:22354172; https://doi.org/10.1172/JCI60282
- Surviladze Z, Waller A, Wu Y, Romero E, Edwards BS, Wandinger-Ness A, Sklar LA. Identification of a small GTPase inhibitor using a high-throughput flow cytometry bead-based multiplex assay. J Biomol Screen 2010; 15:10-20; PMID:20008126; https://doi.org/10.1177/1087057109352240
- Hong L, Kenney SR, Phillips GK, Simpson D, Schroeder CE, Nöth J, Romero E, Swanson S, Waller A, Strouse JJ, et al. Characterization of a Cdc42 Protein Inhibitor and Its Use as a Molecular Probe. J Biol Chem 2013; 288:8531-43; PMID:23382385; https://doi.org/10.1074/jbc.M112.435941
- Deng J, Feng E, Ma S, Zhang Y, Liu X, Li H, Huang H, Zhu J, Zhu W, Shen X, et al. Design and Synthesis of Small Molecule RhoA Inhibitors: A New Promising Therapy for Cardiovascular Diseases? J Med Chem 2011; 54:4508-22; PMID:21615130; https://doi.org/10.1021/jm200161c
- Traut T. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem 1994; 140:1-22; PMID:7877593; https://doi.org/10.1007/BF00928361
- Zhou C, Licciulli S, Avila JL, Cho M, Troutman S, Jiang P, Kossenkov AV, Showe LC, Liu Q, Vachani A, et al. The Rac1 splice form Rac1b promotes K-ras-induced lung tumorigenesis. Oncogene 2013; 32:903-9; PMID:22430205; https://doi.org/10.1038/onc.2012.99
- Dutting S, Heidenreich J, Cherpokova D, Amin E, Zhang SC, Ahmadian MR, Brakebusch C, Nieswandt B. Critical off-target effects of the widely used Rac1 inhibitors NSC23766 and EHT1864 in mouse platelets. J Thromb Haemost 2015; 13:827-38; PMID:25628054; https://doi.org/10.1111/jth.12861
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 2004; 25:1605-12; PMID:15264254; https://doi.org/10.1002/jcc.20084
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011; 7:539; PMID:21988835; https://doi.org/10.1038/msb.2011.75
- Okada S, Yamada E, Saito T, Ohshima K, Hashimoto K, Yamada M, Uehara Y, Tsuchiya T, Shimizu H, Tatei K, et al. CDK5-dependent Phosphorylation of the Rho Family GTPase TC10α Regulates Insulin-stimulated GLUT4 Translocation. J Biol Chem 2008; 283:35455-63; PMID:18948252; https://doi.org/10.1074/jbc.M806531200
- Alan JK, Berzat AC, Dewar BJ, Graves LM, Cox AD. Regulation of the Rho Family Small GTPase Wrch-1/RhoU by C-Terminal Tyrosine Phosphorylation Requires Src. Mol Cell Biol 2010; 30:4324-38; PMID:20547754; https://doi.org/10.1128/MCB.01646-09
