ABSTRACT
Intracellular pathogens often exploit RAB functions to establish a safe haven in which to survive and proliferate. Ehrlichia chaffeensis, an obligatory intracellular bacterium, resides in specialized membrane-bound inclusions that have early endosome–like characteristics, e.g., resident RAB5 GTPase and RAB5 effectors, including VPS34 (the catalytic subunit of class III phosphatidylinositol 3-kinase), but the inclusions lack late endosomal or lysosomal markers. Within inclusions, Ehrlichia obtains host-derived nutrients by inducing RAB5-regulated autophagy using Ehrlichia translocated factor-1 deployed by its type IV secretion system. This manipulation of RAB5 by a bacterial molecule offers a simple strategy for Ehrlichia to avoid destruction in lysosomes and obtain nutrients, membrane components, and a homeostatic intra-host-cell environment in which to grow.
Introduction
RAB GTPases localize to distinct cellular membrane compartments and regulate multiple steps in eukaryotic vesicle trafficking including vesicle budding and invagination, vesicle tethering, and membrane fusion.Citation1,Citation2 RAB5 is found in nascent phagosomes and early endosomes that lack the microbicidal capacity required to kill invading pathogens; this requires subsequent maturation to late endosomes and subsequent fusion with lysosomes. Consequently, several intracellular pathogens subvert RAB5 functions. For example, Mycobacterium tuberculosis localizes to the RAB5-positive endocytic compartment by reducing the amount of phosphatidylinositol 3-phosphate (PtdIns3P) in the phagosomal membrane, thereby interfering with phagosome maturation.Citation3,Citation4 Listeria monocytogenes GAPDH binds and ADP-ribosylates RAB5A and impairs GDP/GTP exchange on this GTPase, thereby blocking phagosome maturation.Citation5 The Salmonella type III secretion system effector SopE (a GDP/GTP exchange factor mimic) activates RAB5 to trigger entry into host cells and promote fusion with early endosomes,Citation6 whereas SopB (a phosphoinositide phosphatase) recruits RAB5 and its effector VPS34 to promote maturation of Salmonella-containing vacuoles.Citation7
Ehrlichia chaffeensis is a tick-borne obligatory intracellular bacterium that causes human monocytic ehrlichiosis, an emerging life-threatening infectious disease in the US and other parts of the world.Citation8 Upon entry into host cells via receptor-mediated endocytosis,Citation9 Ehrlichia resides in specialized membrane-bound inclusions that provide sanctuary against host innate-immune microbicidal mechanisms and a means to acquire host cell–derived nutrients. These intracellular inclusions have early endosome–like characteristics, including transferrin receptor, transferrin, vacuolar-type H+-ATPase, and the small GTPase RAB5 and its effectors EEA1 (early endosome antigen 1), VPS34, and Rabankyrin-5, but they lack late endosomal or lysosomal markers or NADPH oxidase components.Citation8,Citation10,Citation11 E. chaffeensis infection is dependent on RAB5 and RAB5-regulated trafficking to establish E. chaffeensis inclusions, indicating that the recruitment and retention of RAB5-containing endosomes are necessary for subsequent expansion of the inclusions and pathogen replication.Citation11 RABs cycle between GTP-bound “active” and GDP-bound “inactive” conformational states. The exchange of GDP for GTP in RABs is catalyzed by a guanine nucleotide exchange factor (GEF), whereas GTP hydrolysis is facilitated by a GTPase-activating protein (GAP).Citation12 Overexpression of RAB5AS34N (dominant-negative RAB5A/RAB5A-DN, a GDP-bound form of RAB5 that sequesters GEF for RAB5 and thus prevents RAB5 activation) or the RAB5-specific GAP prevents E. chaffeensis infection,Citation11 indicating that GTP-bound RAB5 is required for E. chaffeensis infection.Citation11
One fundamental aspect of microbial pathogen virulence is “nutritional virulence,"Citation13 i.e., the ability to acquire nutrients for pathogen proliferation in competition with host cells and possibly other microbes. Because E. chaffeensis has a small genome (1.176 Mb) and relatively few protein-coding genes that are needed for biosynthesis of amino acids and intermediary metabolites,Citation8 the pathogen relies heavily on host-derived nutrients. Although the host-cell cytoplasm is rich in these nutrients, the ehrlichial inclusion membrane is essentially impermeable to nutrient flow, considering the fact that the inclusion maintains a weakly acidic intraluminal pH.Citation10 It is also unlikely that varieties of active transporters are synthesized and assembled on the inclusion membrane to import host nutrients during ehrlichial replication. Because Ehrlichia cannot replicate or even survive outside of a eukaryotic cell, this mechanism must ensure host-cell survival until Ehrlichia have undergone sufficient replication. Similar limitation is also found in a tick-borne obligatory intracellular bacterium, Anaplasma phagocytophilum, in the family Anaplasmataceae to which E. chaffeensis belong. A. phagocytophilum replicates in neutrophils in MAP1LC3/LC3 (microtubule associated protein 1 light chain 3; a mammalian ortholog family of yeast Atg8)-decorated early autophagosomes that are segregated from the endosomal and lysosomal pathway, thereby gaining access to host cytosolic nutrients while escaping autolysosomal degradation.Citation14-Citation16 Because E. chaffeensis resides in the intracellular compartment distinct from A. phagocytophilum,Citation8,Citation10,Citation11 and the compartment lacks LC3, out hypothesis was that E. chaffeensis species have evolved a unique molecular mechanism to acquire host-cell nutrients.
Autophagy is required for Ehrlichia proliferation
Autophagy is an essential and highly regulated eukaryotic cellular homeostatic process that sequesters and digests/recycles intracellular components,Citation17,Citation18 and autophagy is an important innate immune response to kill intracellular bacteria such as Salmonella, Shigella, Listeria, and MycobacteriumCitation19-Citation23 Induction of autophagy by rapamycin promotes intracellular killing of Salmonella and Mycobacterium,Citation23,Citation24 and inhibition of autophagy by the PtdIns3K inhibitor 3-methyladenine promotes the survival of intracellular mycobacteria.Citation23 In contrast to these bacteria, the intracellular replication of E. chaffeensis is enhanced by rapamycin and strongly inhibited by 3-methyladenine.Citation11 BECN1 (beclin 1; mammalian ortholog of yeast Vps30/Atg6) is an essential component of the class III PtdIns3K complexes that activate canonical autophagy.Citation25 A study using the cell-permeable and potent inhibitor of autophagy, spautin-1, BECN1 siRNA, or mouse bone marrow–derived macrophages from atg5flox/flox-Lyz2-Cre mice (in which Lyz2 promoter-driven Cre is used for myeloid cell–specific Atg5 knockout) point to autophagy as not only enhancing ehrlichial infection but also being required for replication.Citation11 Thus, E. chaffeensis does not simply escape innate immune clearance via cellular autophagy but rather takes advantage of autophagy for its proliferation.
Autophagy is induced by bacterial type IV secretion effectors
The best-known mechanisms of autophagy induction are those induced by amino-acid starvation initiated by the activation of ULK, which is mainly regulated by the MTOR kinase complex or by PRKA/AMPK (protein kinase, AMP-activated) independently of MTOR activity.Citation26,Citation27 However, E. chaffeensis induces autophagy independently of MTOR, ULK1, and PRKA/AMPK.Citation11 Upon their ubiquitination, intracellular aggregated proteins, overabundant proteins, and certain microbial proteins are selectively targeted to autophagosomes for degradation through the ubiquitin- and LC3-binding protein SQSTM1/p62 (sequestosome 1).Citation28,Citation29 Certain proteins of intracellular vacuoles that contain Salmonella, Streptococcus, or Legionella are ubiquitinated, and subsequent binding of p62 delivers the ubiquitin-marked vacuoles to autolysosomes for degradation.Citation30-Citation32 However, proteins of inclusions containing E. chaffeensis are not ubiquitinated as monoclonal antibody FK2 that recognizes both mono- and poly-ubiquitinated proteins did not label the inclusions, and the extent of overall (cellular/bacterial) protein ubiquitination is not increased in infected cells by western blot analysis using FK2 and monoclonal antibody FK1 that recognizes poly-ubiquitinated proteins.Citation11 How, then, does Ehrlichia induce autophagy?
E. chaffeensis has a functional type IV secretion systemCitation33 that mediates the transport of bacterial molecules, referred to as effectors/substrates, across the bacterial and host-cell membranes into the cytoplasm of host cells using bacterial-derived energy.Citation34 One of the type IV secretion system effectors, Etf-1 ( Ehrlichia translocated factor-1), interacts with BECN1, VPS34, and RAB511 and activates class III PtdIns3K, which induces autophagy independently of amino-acid or ATP depletion.Citation11
RAB5-regulated autophagy
RAB5 regulates early endosome maturation to late endosomes, thereby coordinating degradation of extracellular components. RAB5 also regulates degradation of intracellular components by coordinating the fusion of LC3-decorated autophagosomes with late endosomes to form intermediary compartments before fusion with lysosomes.Citation35 Furthermore, in an increasing number of cellular processes, RAB5 has been found to regulate autophagy upstream of LC3 conjugation,Citation36-Citation38 which is called as “RAB5-regulated autophagy.”Citation38 The exact factors that induce this type of autophagy vary widely depending on the cellular circumstances, but they are conceptually similar: constitutively active RAB5 induces autophagy by binding to VPS34, which binds to BECN1, an essential component and master regulator of class III PtdIns3K complex. On the other hand RAB5-DN cannot bind VPS34, thus it not only cannot induce autophagy, but also inhibits autophagy.
Etf-1 induces RAB5-regulated autophagy. VPS34 and BECN1 interact with Etf-1 only in the presence of GTP-bound (active) RAB5, but not GDP-bound RAB5.Citation11 Ectopic expression of Etf-1 in mammalian cells activates class III PtdIns3K and induces the biogenesis of numerous ATG5-containing structures (autophagosome precursors) and LC3-containing autophagosomes.Citation11 Deconvolution fluorescence microscopy has revealed that Etf-1–containing puncta line the inner side of the enlarged GFP-tagged constitutively active RAB5 endosomes when these 2 molcules are co-expressed in mammalian cells.Citation11
The details of RAB5-regulated autophagy have not been fully elucidated because this type of autophagy is not easily followed in the well-studied mammalian-cell systems in which autophagy is induced by amino-acid or ATP starvation. However, RAB5-regulated autophagy has been reported in several systems in which autophagy is induced independently of the activation of MTOR signaling systems, noted as follows.Citation11,Citation36,Citation38 First, ectopic expression of RAB5A-DN or treatment of cultured cells with 3-methyladenine blocks the progression of early ATG5-positive phagophores to form LC3-positive autophagosomes.Citation38 Second, upon growth-factor restriction, the class 1A PtdIns3K catalytic subunit β (PIK3CB/p110β) dissociates from growth factor receptor and binds RAB5.Citation36 This interaction blocks the binding of the RAB5 GAP, PIK3R1/p85α (class 1A PtdIns3K regulatory subunit 1) to RAB5, which consequently increases the amount of GTP-bound RAB5 and enhances the RAB5-VPS34 interaction to promote autophagy.Citation36 Third, the hepatitis C virus protein NS4B forms a complex with RAB5 and VPS34 and promotes RAB5-GTP–induced autophagy, which is required for virus replication.Citation37 Finally, eukaryotic proteasomes can only very poorly cleave (if at all) polyglutamine sequences such as polyglutamine tracts in huntingtin protein.Citation39 The expansion of polyglutamine tracts in huntingtin causes aggregation of polyglutamine-containing peptides, leading to the neurodegenerative genetic disorder Huntington's disease.Citation40 The protein huntingtin binds RAB5 via the RAB5 effector F8A1/HAP40 (coagulation factor VIII–associated 1) and is degraded during autophagy.Citation41 Interestingly, Etf-1–induced autophagy also clears an aggregation-prone mutant huntingtin protein in a class III PtdIns3K–dependent manner,Citation11 although whether the HAP40 adaptor is involved in this process is unknown.
In addition to these examples, the endoplasmic reticulum–localized transmembrane protein EMC6 (endoplasmic reticulum membrane protein complex subunit 6) interacts with both RAB5A and BECN1 to induce autophagosomes;Citation42 notably, EMC6 colocalizes with DFCP1, a marker of the omegasome (amino-acid starvation–induced precursor of isolation membrane). Ypt53, an isoform of RAB5 in yeast, is upregulated significantly under nutrient stress to maintain both Golgi–vacuole trafficking and vacuolar hydrolase activity.Citation43 Overall, published data suggest a broad involvement of RAB5 in autophagy, including amino-acid starvation–induced autophagy. In addition to RAB5, other RABs may be involved in autophagy regulation; several other RABs as well as a group of TBC (tre2-bub2-cdc16) domain–containing RAB GAPs and non-TBC RAB GAPs have been shown to regulate autophagosome formation (for review see refs. Citation44, Citation45).
RAB5-regulated autophagy as a bacterial nutrient acquisition mechanism
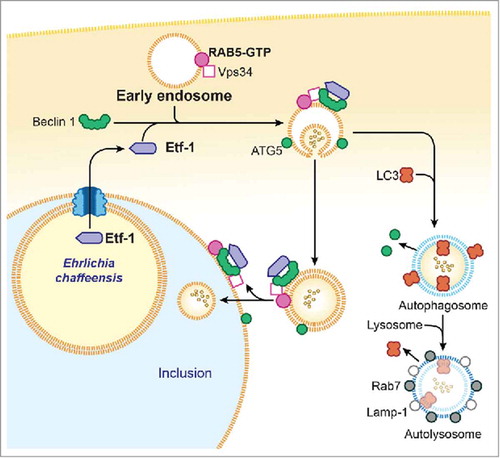
Etf-1–induced autophagy releases host-cell small-molecule catabolites into the host cytoplasm without actually starving host cells. An additional advantage of bacterial effector–induced autophagy is that it creates more host cytoplasmic space for bacterial growth without initially harming host cells. During the exponential growth stage of E. chaffeensis, the concentrations of free/cytoplasmic l-glutamine and l-glutamate in infected human monocytes are notably increased, making them available for ehrlichial growth.Citation11 Indeed, host cell–preincorporated radioactive amino acids can be readily taken up by E. chaffeensis in an autophagy-dependent manner, and the cytoplasmic autophagy cargo protein human GAPDHCitation46 can be delivered to E. chaffeensis inclusions.Citation11 However, how do inclusion-confined E. chaffeensis acquire host cytoplasmic molecules? Although the classical view has been that the autophagic and endocytic pathways converge at the phagolysosomal and lysosomal levels, autophagosomes have also been found to undergo fusion with components of the early endocytic pathway.Citation35,Citation47,Citation48 RAB5, VPS34, and Etf-1–containing vesicles are targeted to already-established E. chaffeensis inclusions, suggesting the recruitment of RAB5 endosomes during E. chaffeensis replication and expansion of the inclusion. Thus, E. chaffeensis inclusions can be considered as large amphisomes formed by fusion of early endosomes and early autophagosomes because several early-endosome markers, Etf-1, and the early autophagosome marker ATG5 (but not LC3) are present on the membrane of E. chaffeensis inclusions.Citation11 Because Etf-1–induced autophagosomes can capture host-cell cytoplasmic nutrients, if newly nucleated autophagosomes fuse with ehrlichial inclusions before their maturation into LC3-positive autophagosomes or autolysosomes, these autophagosomes can deliver the nutrients to the inclusions. Another possibility is that Etf-1–induced autophagosomes (vesicles) in infected cells are already amphisomes formed by fusion of early endosomes because these vesicles that are docked to inclusions also contain RAB5, VPS34, ATG5, and Etf-1 but not LC3, as does the inclusion membrane itself. Once these small amphisomes fuse with ehrlichial inclusions, they deliver host cytoplasmic nutrients into the inclusion lumen. Endosomes, themselves may also facilitate cytosolic nutrient delivery to ehrlichial inclusions. Multivesicular bodies (MVBs), which are morphologically distinct endosomes internally accumulate small membrane vesicles (60 to 80 nm) that contain cytoplasmic cargo molecules.Citation35,Citation49 Morphological evidence suggests that MVBs are the main endocytic fusion partner of autophagosomes, forming the amphisome.Citation35,Citation50 class III PtdIns3K and PtdIns3P are essential for the generation of the internal vesicles of MVBs, and RAB5 promotes the biosynthesis of PtdIns3P on endosomesCitation51 and phagosomes.Citation52 PtdIns3P accumulates within MVBs.Citation53 Thus, most labeling of 2 × FYVE-GFP that binds to PtdIns3P is associated with the internal membranes of MVBs, which contain only small amounts of the markers for late endosomes and lysosomes.Citation53 Results with 2 × FYVE-GFP–transfected cells has revealed large amounts of intraluminal PtdIns3P in large vesicles docked to E. chaffeensis inclusions, suggesting that these vesicles are MVBs.Citation11 If MVBs fuse with ehrlichial inclusions, then host cytoplasmic nutrients can be effectively delivered to inclusions. presents our proposed model of RAB5-regulated autophagosome nucleation and subversion by Etf-1 in E. chaffeensis–infected cells.
Figure 1. Proposed model for Etf-1-mediated autophagy fueling E. chaffeensis growth. Secreted Etf-1 interacts with RAB5, VPS34, and Beclin 1 to induce complex formation and localize to the ATG5-positive “precursor of preautophagosomes.” If not fused with E. chaffeensis inclusions, Etf-1 autophagosomes mature to autolysosomes to generate cytosolic nutrients (∘∘) (right side). When nascent Etf-1 preautophagosomes fuse with E. chaffeensis inclusions, they deliver captured cytosolic nutrients to the inclusions where lysosomal fusion is blocked (left side).
Conclusions
Intracellular pathogens such as bacteria and virus can utilize a recently defined pathway, namely RAB5-regulated autophagy, to their advantage. The possibility exists that RAB5-regulated autophagy may be induced by other unidentified eukaryotic, bacterial, and viral proteins that can interact with RAB5 or RAB5 regulatory proteins. Perhaps the most salient feature of RAB5-regulated autophagy in ehrlichial infection is that preexisting host-cell components and signaling pathways are repurposed in a way that is independent of other signaling pathways by using a bacterial molecule. The bacterial molecule can be used to predictably induce autophagy-related signaling to clear the abnormal intracellular accumulation of toxic molecules, damaged organelles, and pathogenic agents. Currently the only therapy for human monocytic ehrlichiosis is the broad-spectrum antibiotic doxycycline, which is effective only if initiated early, and no vaccine is available. Mechanistic understanding of nutritional virulence of E. chaffeensis will provide important scientific basis for novel anti-E. chaffeensis therapy and preventive measures.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Pfeffer SR. Rab GTPases: Master regulators that establish the secretory and endocytic pathways. Mol Biol Cell 2017; 28:712-5; PMID:28292916; https://doi.org/10.1091/mbc.E16-10-0737
- Zhen Y, Stenmark H. Cellular functions of Rab GTPases at a glance. J Cell Sci 2015; 128:3171-6; PMID:26272922; https://doi.org/10.1242/jcs.166074
- Fratti RA, Chua J, Vergne I, Deretic V. Mycobacterium tuberculosis glycosylated phosphatidylinositol causes phagosome maturation arrest. Proc Natl Acad Sci U S A 2003; 100:5437-42; PMID:12702770; https://doi.org/10.1073/pnas.0737613100
- Puri RV, Reddy PV, Tyagi AK. Secreted acid phosphatase (SapM) of Mycobacterium tuberculosis is indispensable for arresting phagosomal maturation and growth of the pathogen in guinea pig tissues. PloS One 2013; 8:e70514; PMID:23923000; https://doi.org/10.1371/journal.pone.0070514
- Alvarez-Dominguez C, Madrazo-Toca F, Fernandez-Prieto L, Vandekerckhove J, Pareja E, Tobes R, Gomez-Lopez MT, Del Cerro-Vadillo E, Fresno M, Leyva-Cobián F, et al. Characterization of a Listeria monocytogenes protein interfering with Rab5a. Traffic 2008; 9:325-37; PMID:18088303; https://doi.org/10.1111/j.1600-0854.2007.00683.x
- Mukherjee K, Parashuraman S, Raje M, Mukhopadhyay A. SopE acts as an Rab5-specific nucleotide exchange factor and recruits non-prenylated Rab5 on Salmonella-containing phagosomes to promote fusion with early endosomes. J Biol Chem 2001; 276:23607-15; PMID:11316807; https://doi.org/10.1074/jbc.M101034200
- Mallo GV, Espina M, Smith AC, Terebiznik MR, Aleman A, Finlay BB, Rameh LE, Grinstein S, Brumell JH. SopB promotes phosphatidylinositol 3-phosphate formation on salmonella vacuoles by recruiting Rab5 and Vps34. J Cell Biol 2008; 182:741-52; PMID:18725540; https://doi.org/10.1083/jcb.200804131
- Rikihisa Y. Molecular pathogenesis of Ehrlichia chaffeensis infection. Annu Rev Microbiol 2015; 69:283-304; PMID:26488275; https://doi.org/10.1146/annurev-micro-091014-104411
- Mohan Kumar D, Yamaguchi M, Miura K, Lin M, Los M, Coy JF, Rikihisa Y. Ehrlichia chaffeensis uses its surface protein EtpE to bind GPI-anchored protein DNase X and trigger entry into mammalian cells. PLoS Pathog 2013; 9:e1003666; PMID:24098122; https://doi.org/10.1371/journal.ppat.1003666
- Barnewall RE, Rikihisa Y, Lee EH. Ehrlichia chaffeensis inclusions are early endosomes which selectively accumulate transferrin receptor. Infect Immun 1997; 65:1455-61; PMID:9119487
- Lin M, Liu H, Xiong Q, Niu H, Cheng Z, Yamamoto A, Rikihisa Y. Ehrlichia secretes Etf-1 to induce autophagy and capture nutrients for its growth through RAB5 and class III phosphatidylinositol 3-kinase. Autophagy 2016; 12:2145-66; PMID:27541856; https://doi.org/10.1080/15548627.2016.1217369
- Barr F, Lambright DG. Rab GEFs and GAPs. Curr Opin Cell Biol 2010; 22:461-70; PMID:20466531; https://doi.org/10.1016/j.ceb.2010.04.007
- Abu Kwaik Y, Bumann D. Microbial quest for food in vivo: ‘Nutritional virulence’ as an emerging paradigm. Cell Microbiol 2013; 15:882-90; PMID:23490329; https://doi.org/10.1111/cmi.12138
- Niu H, Yamaguchi M, Rikihisa Y. Subversion of cellular autophagy by Anaplasma phagocytophilum. Cell Microbiol 2008; 10:593-605; PMID:17979984; https://doi.org/10.1111/j.1462-5822.2007.01068.x
- Niu H, Rikihisa Y. Ats-1: A novel bacterial molecule that links autophagy to bacterial nutrition. Autophagy 2013; 9:787-8; PMID:23388398; https://doi.org/10.4161/auto.23693
- Niu H, Xiong Q, Yamamoto A, Hayashi-Nishino M, Rikihisa Y. Autophagosomes induced by a bacterial Beclin 1 binding protein facilitate obligatory intracellular infection. Proc Natl Acad Sci U S A 2012; 109:20800-7; PMID:23197835; https://doi.org/10.1073/pnas.1218674109
- Yang Z, Klionsky DJ. Eaten alive: A history of macroautophagy. Nat Cell Biol 2010; 12:814-22; PMID:20811353; https://doi.org/10.1038/ncb0910-814
- Mizushima N, Komatsu M. Autophagy: Renovation of cells and tissues. Cell 2011; 147:728-41; PMID:22078875; https://doi.org/10.1016/j.cell.2011.10.026
- Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol 2009; 10:1215-21; PMID:19820708; https://doi.org/10.1038/ni.1800
- Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol 2009; 11:1233-40; PMID:19749745; https://doi.org/10.1038/ncb1967
- Dupont N, Lacas-Gervais S, Bertout J, Paz I, Freche B, Van Nhieu GT, van der Goot FG, Sansonetti PJ, Lafont F. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe 2009; 6:137-49; PMID:19683680; https://doi.org/10.1016/j.chom.2009.07.005
- Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol 2009; 183:5909-16; PMID:19812211; https://doi.org/10.4049/jimmunol.0900441
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 2004; 119:753-66; PMID:15607973; https://doi.org/10.1016/j.cell.2004.11.038
- Homer CR, Richmond AL, Rebert NA, Achkar JP, McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn's disease pathogenesis. Gastroenterology 2010; 139:1630-41 . 41 e1-2; PMID:20637199; https://doi.org/10.1053/j.gastro.2010.07.006
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 2008; 19:5360-72; PMID:18843052; https://doi.org/10.1091/mbc.E08-01-0080
- Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 2013; 15:741-50; PMID:23685627; https://doi.org/10.1038/ncb2757
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011; 13:132-41; PMID:21258367; https://doi.org/10.1038/ncb2152
- Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A 2008; 105:20567-74; PMID:19074260; https://doi.org/10.1073/pnas.0810611105
- Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, Kominami E, Yamane T, Tanaka K, Komatsu M. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem 2008; 283:22847-57; PMID:18524774; https://doi.org/10.1074/jbc.M802182200
- Fujita N, Morita E, Itoh T, Tanaka A, Nakaoka M, Osada Y, Umemoto T, Saitoh T, Nakatogawa H, Kobayashi S, et al. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J Cell Biol 2013; 203:115-28; PMID:24100292; https://doi.org/10.1083/jcb.201304188
- Barnett TC, Liebl D, Seymour LM, Gillen CM, Lim JY, Larock CN, Davies MR, Schulz BL, Nizet V, Teasdale RD, et al. The globally disseminated M1T1 clone of group A Streptococcus evades autophagy for intracellular replication. Cell Host Microbe 2013; 14:675-82; PMID:24331465; https://doi.org/10.1016/j.chom.2013.11.003
- Khweek AA, Caution K, Akhter A, Abdulrahman BA, Tazi M, Hassan H, Majumdar N, Doran A, Guirado E, Schlesinger LS, et al. A bacterial protein promotes the recognition of the Legionella pneumophila vacuole by autophagy. Eur J Immunol 2013; 43:1333-44; PMID:23420491; https://doi.org/10.1002/eji.201242835
- Rikihisa Y, Lin M. Anaplasma phagocytophilum and Ehrlichia chaffeensis type IV secretion and Ank proteins. Curr Opin Microbiol 2010; 13:59-66; PMID:20053580; https://doi.org/10.1016/j.mib.2009.12.008
- Backert S, Meyer TF. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol 2006; 9:207-17; PMID:16529981; https://doi.org/10.1016/j.mib.2006.02.008
- Berg TO, Fengsrud M, Stromhaug PE, Berg T, Seglen PO. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem 1998; 273:21883-92; PMID:9705327; https://doi.org/10.1074/jbc.273.34.21883
- Dou Z, Pan JA, Dbouk HA, Ballou LM, Deleon JL, Fan Y, Chen JS, Liang Z, Li G, Backer JM, et al. Class IA PI3K p110beta subunit promotes autophagy through Rab5 small GTPase in response to growth factor limitation. Mol Cell 2013; 50:29-42; PMID:23434372; https://doi.org/10.1016/j.molcel.2013.01.022
- Su WC, Chao TC, Huang YL, Weng SC, Jeng KS, Lai MM. Rab5 and class III phosphoinositide 3-kinase Vps34 are involved in hepatitis C virus NS4B-induced autophagy. J Virol 2011; 85:10561-71; PMID:21835792; https://doi.org/10.1128/JVI.00173-11
- Ravikumar B, Imarisio S, Sarkar S, O'Kane CJ, Rubinsztein DC. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci 2008; 121:1649-60; PMID:18430781; https://doi.org/10.1242/jcs.025726
- Venkatraman P, Wetzel R, Tanaka M, Nukina N, Goldberg AL. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol Cell 2004; 14:95-104; PMID:15068806; https://doi.org/10.1016/S1097-2765(04)00151-0
- Raspe M, Gillis J, Krol H, Krom S, Bosch K, van Veen H, Reits E. Mimicking proteasomal release of polyglutamine peptides initiates aggregation and toxicity. J Cell Sci 2009; 122:3262-71; PMID:19690053; https://doi.org/10.1242/jcs.045567
- Pal A, Severin F, Lommer B, Shevchenko A, Zerial M. Huntingtin-HAP40 complex is a novel Rab5 effector that regulates early endosome motility and is up-regulated in Huntington's disease. J Cell Biol 2006; 172:605-18; PMID:16476778; https://doi.org/10.1083/jcb.200509091
- Li Y, Zhao Y, Hu J, Xiao J, Qu L, Wang Z, Ma D, Chen Y. A novel ER-localized transmembrane protein, EMC6, interacts with RAB5A and regulates cell autophagy. Autophagy 2013; 9:150-63; PMID:23182941; https://doi.org/10.4161/auto.22742
- Nakatsukasa K, Kanada A, Matsuzaki M, Byrne SD, Okumura F, Kamura T. The nutrient stress-induced small GTPase Rab5 contributes to the activation of vesicle trafficking and vacuolar activity. J Biol Chem 2014; 289:20970-8; PMID:24923442; https://doi.org/10.1074/jbc.M114.548297
- Kern A, Dikic I, Behl C. The integration of autophagy and cellular trafficking pathways via RAB GAPs. Autophagy 2015; 11:2393-7; PMID:26565612; https://doi.org/10.1080/15548627.2015.1110668
- Lamb CA, Longatti A, Tooze SA. Rabs and GAPs in starvation-induced autophagy. Small GTPases 2016; 7:265-9; PMID:27669114; https://doi.org/10.1080/21541248.2016.1220779
- Sneve ML, Overbye A, Fengsrud M, Seglen PO. Comigration of two autophagosome-associated dehydrogenases on two-dimensional polyacrylamide gels. Autophagy 2005; 1:157-62; PMID:16874067; https://doi.org/10.4161/auto.1.3.2037
- Morvan J, Kochl R, Watson R, Collinson LM, Jefferies HB, Tooze SA. In vitro reconstitution of fusion between immature autophagosomes and endosomes. Autophagy 2009; 5:676-89; PMID:19337031; https://doi.org/10.4161/auto.5.5.8378
- Tooze J, Hollinshead M, Ludwig T, Howell K, Hoflack B, Kern H. In exocrine pancreas, the basolateral endocytic pathway converges with the autophagic pathway immediately after the early endosome. J Cell Biol 1990; 111:329-45; PMID:2166050; https://doi.org/10.1083/jcb.111.2.329
- Eskelinen EL. Maturation of autophagic vacuoles in mammalian cells. Autophagy 2005; 1:1-10; PMID:16874026; https://doi.org/10.4161/auto.1.1.1270
- Fader CM, Colombo MI. Autophagy and multivesicular bodies: Two closely related partners. Cell Death Differ 2009; 16:70-8; PMID:19008921; https://doi.org/10.1038/cdd.2008.168
- Shin HW, Hayashi M, Christoforidis S, Lacas-Gervais S, Hoepfner S, Wenk MR, Modregger J, Uttenweiler-Joseph S, Wilm M, Nystuen A, et al. An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J Cell Biol 2005; 170:607-18; PMID:16103228; https://doi.org/10.1083/jcb.200505128
- Vieira OV, Botelho RJ, Rameh L, Brachmann SM, Matsuo T, Davidson HW, Schreiber A, Backer JM, Cantley LC, Grinstein S. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J Cell Biol 2001; 155:19-25; PMID:11581283; https://doi.org/10.1083/jcb.200107069
- Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J 2000; 19:4577-88; PMID:10970851; https://doi.org/10.1093/emboj/19.17.4577
