ABSTRACT
Macrophages are important immune sentinels that detect and clear pathogens and initiate inflammatory responses through the activation of surface receptors, including Toll-like receptors (TLRs). Activated TLRs employ complex cellular trafficking and signalling pathways to initiate transcription for inflammatory cytokine programs. We have previously shown that Rab8a is activated by multiple TLRs and regulates downstream Akt/mTOR signalling by recruiting the effector PI3Kγ, but the guanine nucleotide exchange factors (GEF) canonically required for Rab8a activation in TLR pathways is not known. Using GST affinity pull-downs and mass spectrometry analysis, we identified a Rab8 specific GEF, GRAB, as a Rab8a binding partner in LPS-activated macrophages. Co-immunoprecipitation and fluorescence microscopy showed that both GRAB and a structurally similar GEF, Rabin8, undergo LPS-inducible binding to Rab8a and are localised on cell surface ruffles and macropinosomes where they coincide with sites of Rab8a mediated signalling. Rab nucleotide activation assays with CRISPR-Cas9 mediated knock-out (KO) cell lines of GRAB, Rabin8 and double KOs showed that both GEFs contribute to TLR4 induced Rab8a GTP loading, but not membrane recruitment. In addition, measurement of signalling profiles and live cell imaging with the double KOs revealed that either GEF is individually sufficient to mediate PI3Kγ-dependent Akt/mTOR signalling at macropinosomes during TLR4-driven inflammation, suggesting a redundant relationship between these proteins. Thus, both GRAB and Rabin8 are revealed as key positive regulators of Rab8a nucleotide exchange for TLR signalling and inflammatory programs. These GEFs may be useful as potential targets for manipulating inflammation.
Abbreviations: TLR: Toll-like Receptor; OCRL: oculocerebrorenal syndrome of Lowe protein; PI3Kγ: phosphoinositol-3-kinase gamma; LPS: lipopolysaccharide; GEF: guanine nucleotide exchange factor; GST: glutathione S-transferases; BMMs: bone marrow derived macrophages; PH: pleckstrin homology; GAP: GTPase activating protein; ABCA1: ATP binding cassette subfamily A member 1; GDI: GDP dissociation inhibitor; LRP1: low density lipoprotein receptor-related protein 1
Introduction
Rab8 is a multifunctional member of the small GTPase family of Rab molecular switches that commonly regulate membrane trafficking, cell shape, movement and receptor signalling [Citation1,Citation2]. There are two isoforms of Rab8: Rab8a and 8b, which share 83% peptide sequence homology [Citation3]. The expression levels of Rab8a and 8b vary in some cell types and in many instances the functional overlap or differences are unclear [Citation3,Citation4]. Rab8 functions in several guises as a regulator of cellular trafficking, including apical membrane morphogenesis and ciliogenesis in epithelial cells to facilitate processes such as migration, cell polarization and signalling [Citation3,Citation5,Citation6]. Rab8 can interact with a variety of effector molecules, including the Oculocerebrorenal syndrome of Lowe protein (OCRL), Optineurin, MICAL-L1 and L2 and myosins V and VI [Citation7–10]. Much less is known about the function of Rab8 in cells of the immune system. In macrophages, Rab8 is involved in controlling the turnover and recycling of cell surface proteins, such as the metalloproteinase MT1-MMP [Citation11] and the ATP-cassette transporter ABCA1 [Citation12] and we have shown that Rab8a has a role in TLR signalling in these cells [Citation12,Citation13].
Macrophages employ the pattern recognition receptors of the Toll-like receptor (TLR) family to detect molecular signals from different classes of pathogens, including bacteria and viruses [Citation13]. TLR4, which is activated by the lipopolysaccharide (LPS) of Gram-negative bacteria, is one of best-characterised innate immune receptors. LPS activation of TLR4 initiates complex signalling pathways mediated through cell surface and endosomal adaptors, that drive transcriptional programs for the synthesis and secretion of inflammatory cytokines and chemokines. We showed that Rab8a is activated (GTP-bound) by LPS, downstream of TLR4, and also in response to agonist activation of other TLRs [Citation14]. Rab8a in macrophages is enriched dynamically on cell surface ruffles and the early macropinosomes they give rise to, eventually being depleted from the macropinosome as it matures to be replaced by Rab5. Studies using a Rab8a biosensor demonstrated that Rab8a is active on early macropinosomes in macrophages as the likely site for LPS-induced signalling [Citation15]. Most recently we have shown that TLR crosstalk activation of the endocytic receptors low density lipoprotein receptor-related protein 1 (LRP1) recruits Rab8a for signalling downstream of TLRs [Citation16]. In this pathway, TLR-activated Rab8a recruits the class 1B PI3-kinase PI3Kγ, as its effector to enhance Akt and mTOR signalling [Citation17]. The serine/threonine kinase Akt, is recruited and activated upon recognition of the PI3Kγ product phosphatidylinositol (3,4,5)-trisphosphate [PtdIns(3,4,5)P3] [Citation18], triggering signalling cascades, such as mTORC1, that help to bias cytokine output [Citation19]. Under the influence of Rab8a with PI3Kγ, macrophages are reprogrammed to an anti-inflammatory or M2-like status that constrains inflammation [Citation14]. Thus, Rab8a has a key role in directing macrophage responses and in controlling TLR-induced inflammation. The molecules responsible for the TLR-induced activation of Rab8a are yet to be identified and will be the focus for this study.
The activation of Rab GTPases is typically dependent on guanine nucleotide exchange factors (GEFs) that promote nucleotide exchange and GTP binding [Citation20–23]. There are two well-known, structurally-similar and potent Rab8 specific GEFs: GRAB and Rabin8 [Citation24–26]. Rabin8 is arguably the better known of the two, having been first identified in a yeast-2 hybrid screen using a dominant negative form of the Rab8b isoform [Citation5]. Rabin8 is a mammalian homolog of the yeast GEF protein Sec2p and it has since been implicated in regulating Rab8 activation in multiple membrane trafficking and vesicular transport pathways, most notably the targeting of cellular components for ciliogenesis [Citation27,Citation28]. GRAB was first identified as a neuronal specific Rab3a GEF, facilitating the release of neurotransmitters via synaptic vesicle transport [Citation29] and then later shown to have a greater binding affinity for Rab8 compared to Rab3 [Citation26]. GRAB shares structural similarities with Rabin8, including a common Sec2 coiled-coil GEF domain and a C-terminal Rab11 binding site [Citation24]. GRAB and Rabin8 also demonstrate GEF activity in the same pathways, such as in neurite outgrowth [Citation30,Citation31]. However, despite their similarities, the roles of Rabin8 and GRAB have not been systematically studied side-by-side in a specific cellular function or pathway.
Our studies herein set out to test and directly compare the functions of GRAB and Rabin8 as candidate GEFs for regulating Rab8a recruitment of PI3Kγ and Akt signalling downstream of TLR4. Utilising biochemical and imaging approaches we report the interaction of GRAB and Rabin8 with Rab8a and assay changes in TLR signalling after single and double CRISPR-Cas9 deletion of both GEFs in macrophages. The findings reported here reveal that in macrophages, GRAB and Rabin8 both contribute to Rab8a activation in response to LPS to facilitate downstream TLR signalling. This identifies these proteins as new regulators of TLR signalling, specifically with the macrophage immune regulatory complex LRP1-Rab8a-PI3Kγ.
Materials and methods
Plasmid constructs
For the Rab8 activation assay, GST-OCRL-RBD was generated by sub-cloning human OCRL-RBD (residues 539–901) into pGEX-6p-1 and expressed in E. coli BL21(DE3) bound to GSH-Sepharose beads as used in previous studies [Citation14]. Full-length mouse GRAB and Rabin8 were PCR amplified from mouse RAW 264.7 cell cDNA and inserted into a pEF6-GFP-C1 backbone using BamHI/NotI and SpeI/NotI respectively. The following primers were used to generate the constructs: Rabin8 forward 5ʹ-AATTACTAGTATGGCTAACGACCCCTTG-3ʹ, Rabin8 reverse 5ʹ-AATTGCGGCCGCCGAGTTCCTCTTTGAAATA-3ʹ, GRAB forward 5ʹ- AATTGGATCCATGGAGATCCGAGAGAAG-3ʹ and GRAB reverse 5ʹ- AATTGCGGCCGCCGGCCTCCTGGGGGAAGAA-3ʹ. pEGFP-C1 vector (Clonetech) with full-length mouse Rab8a was a gift from M. Fukuda (Tohoku University), which was later sub-cloned into ptd-Tomato-C1 as described in previous work [Citation14,Citation17]. Full-length TRAM was cloned from RAW 264.7 cDNA into the Clontech vector pEGFP-N1 using BamHI/HindIII [Citation32]. The TagRFP-T-Akt1 construct was generously provided by James Burchfield (University of Sydney) [Citation33]. CRISPR selection resistance plasmid pFloxNeo-DTA was designed by Adam Wall (IMB, University of Queensland) [Citation14].
Cell culture and transfection
The mouse macrophage cell line RAW 264.7 was obtained from ATCC. Cells were maintained in complete RPMI medium (Lonza, Australia) supplemented with 10% heat-inactivated Fetal calf serum (Thermo Trace, Australia) and 2 mM L-glutamine (Invitrogen) at 37ºC in humidified 5% CO2. Primary mouse bone marrow-derived macrophages (BMMs) were differentiated from femur bone marrow cells for 7 days in RPMI medium supplemented with 10% heat-inactivated Fetal calf serum (Thermo Trace, Australia), 2 mM L-glutamine (Invitrogen), 20 unit/ml penicillin, 20 µg/ml streptomycin 100 ng/ml macrophage colony-stimulating factor-1 (CSF-1). For experimental procedures, all cells were treated with 10 ng/ml LPS unless stated otherwise. BMMs from TLR4 mice were kindly provided by Matt Sweet (IMB, The University of Queensland) [Citation34].
RAW 264.7 macrophages were transfected using Lipofectamine 2000TM (ThermoFisher Scientific, San Diego, CA) as per manufacturer’s instruction. Cells were seeded either on 25mm coverslips or Mattek 35mm glass bottom dishes (Mattek Cooporation) at a density of 0.1 × 106 cells/ml and incubated with transfection complexes for 2–4 hr before changing the medium, finally, cells were incubated overnight and then used for experiments.
Rab8 activation assay and immunoprecipitation
The Rab8 activation assay, based on capture and pull-down of GTP-Rab8a with GST-OCRL-RBD bound GSH-Sepharose resin, has been described previously [Citation14]. Briefly, cells ± LPS (100 ng/ml) were lysed in ice-cold lysis buffer A [25 mM TRIS pH 7.4, 150 mM NaCl, 5mM MgCl2, 1% NP-40, PhosSTOPTM (Roche Applied Science, Switzerland), EDTA-free cOmplete Mini protease inhibitor (Sigma-Aldrich, Australia) and 5% glycerol], and centrifuged at 14,000 × g for 15 min. Lysates were applied to GSH-Sepharose resin with bound GST-OCRL-RBD to capture active, GTP-loaded Rab8. Binding was performed in microspin columns (GE Healthcare) for 1 hr at 4ºC with constant agitation. For the loading calibration assay, cell lysates were applied to GSH-Sepharose resin and treated with lysis buffer containing either 1, 10, or 100 µM of GTP or 1 mM GDP for 15 min at room temperature with constant agitation. The nucleotide exchange reaction was terminated by placing the sample on ice and adding MgCl2 to a final concentration of 60 mM. The resin was later washed multiple times with ice-cold lysis buffer and bound proteins were eluted by boiling at 95ºC for 5 min in 2X SDS-PAGE sample buffer (1M Tris pH7.4, 20% glycerol, 6mM EGTA, 2.5% SDS, 6% β-Mercaptoethanol). Eluted samples were analysed by immunoblotting.
For immunoprecipitations, LPS treated cells were lysed by passage through 27-gauge needles in ice-cold lysis buffer B [25 mM TRIS pH 7.4, 150 mM NaCl, 5mM MgCl2, 1% NP-40, 1% Triton X-100 (Sigma-Aldrich), PhosSTOPTM (Roche Applied Science, Switzerland), EDTA-free cOmplete Mini protease inhibitor (Sigma-Aldrich, Australia) and 5% glycerol], and centrifuged at 14,000 × g for 15 min. A small sample of cleared cell lysate was saved as input while the rest was mixed with antibody bound protein G beads for 1 hr at 4◦C with constant agitation. The bound proteins were eluted from beads as described above, separated on 10% SDS-PAGE gels and analysed by immunoblotting. Pierce BCA Protein Assay Kits (#23,225) were used to quantify total protein in cell lysates, according to the manufacturer’s instructions.
Mass spectrometry
LPS treated cells were lysed as above (Immunoprecipitation, lysis buffer B) and the lysates applied to GSH-Sepharose resin with bound GST-Rab8a and the pulled-down samples were analysed by mass spectrometry using a LC MS/MS with a Shimadzu Prominence Nano HPLC (Japan) coupled to a Triple TOF 5600 mass spectrometer (ABSCIEX, Canada) equipped with a nano electrospray ion source at IMB, The University of Queensland as described previously [Citation35]. Protein identification was performed via database searching using ProteinPilot v4.5 (ABSCIEX, Canada) against the UniProt_Sprot_20130205 database (B106,000 entries of all species searched, FDR of 1%).
Immunoblotting
Cell lysates were fractionated using 10% SDS-PAGE gels and transferred onto to polyvinylidene difluoride (PVDF) membranes (Merck Immobilon-P®) using wet transfer apparatus (Bio-Rad). The membranes were blotted with antibodies and visualized with an ECL kit (SuperSignal™ West Pico) and X-ray films (FujiFilm Super RX). Antibodies used include Rab8 (1:1000, #610,845, BD Biosciences USA), Rabin8 (1:1000, #12,321–1-AP, ProteinTech, USA), GRAB (1:1000, #17,827–1-AP, ProteinTech, USA), GFP (1:2000, #A6455, ThermoFisher Scientific, Australia), Flotillin-1 (1/500, #610,820, BD Biosciences), β-actin (1:1000, #A1978, Sigma Aldrich) and from Cell Signaling Technology, USA: GAPDH (1:4000, #14C10), pAKT (1:1000, #9271, Ser473). pERK1/2 (1:3000, #9102, Thr202/Tyr204) and Myc (1:3000, #9B11). Secondary anti-rabbit (1:10 000, #G-21,234, ThermoFisher Scientific, Australia) or anti-mouse (1:10 000, #G-20,140, ThermoFisher Scientific, Australia) conjugated to horseradish peroxidase were used.
CRISPR/Cas9-knockout (KO) of GRAB and Rabin8
CRISPR targeting guideRNAs were generated using published genome sequences (NCBI). The third exon of Rabin8 and the second exon of GRAB, which contained the start site and downstream coding regions were chosen to create a specific double-stranded break which would result in non-homologous end joining leading to a frameshift mutation, effectively creating a KO. The following primer pairs were ordered from IDT: GRAB Forward 5ʹ-TAATACGACTCACTATAGGCAGGCGTGACACATCCAG-3ʹ; GRAB Reverse 5ʹ-TTCTAGCTCTAAAACCTGGATGTGTCACGCCTGC-3ʹ; Rabin8 Forward 5ʹ-TAATACGACTCACTATAGAGAGAGAAGGGCTACGAA-3ʹ; Rabin8 Reverse 5ʹ-TTCTAGCTCTAAAACTTCGTAGCCCTTCTCTCT-3ʹ. These primers were used to generate guideRNA sequences as per manufacturer’s instructions (GeneArt™ Precision gRNA Synthesis Kit, ThermoFisher Scientific) and combined with purified S.pyogenes Cas9 nuclease protein (IDT) to form the genome targeting complex. The complex was co-transfected with a pFloxNeo resistance plasmid into RAW 264.7 cells using the LipofectamineTM CRISPRMAXTM transfection reagent (ThermoFisher Scientific) and transfected cells were selected in RPMI medium with 1 mg/ml G418 (Geneticin, Invitrogen) and clonal lines were screened for loss of GRAB or Rabin8 protein expression by immunoblotting (Supplementary Figures S1A-C).
Fluorescence microscopy
RAW 264.7 cells transfected with GFP-GRAB or GFP-Rabin8 and td-Tomato-Rab8a ± LPS (30 min), were fixed in 4% PFA, washed with PBS and co-stained with Alexa350-phalloidin (1:500 for 30 min). Fixed cells were imaged using an upright Zeiss Axiolmager equipped with Apotome2 and Axiocam 506 camera with a mercury light source. Images were captured using a 63X plan apochromat objective oil immersion lens. For live cell imaging, cells expressing either GFP-Rab8a or soluble-GFP and TagRFP-T-Akt1 were incubated in Leibovitz L-15 medium (Invitrogen) supplemented with 10% heat-inactivated fetal calf serum (Thermo Trace, Australia) and 2 mM L-glutamine. The cells were treated with LPS and imaged using a Zeiss Axiovert 200 Inverted Microscope with CSU-X1 scanhead. GFP and RFP were sequentially imaged over 15–10 min at 5 sec intervals using a 63X LCI PlanN water immersion lens.
Membrane fractionation
RAW 264.7 cells treated with LPS (15 min) were lysed by passage through a 27-gauge needle in 500µl of fractionation buffer [20 mM HEPES pH 7.4, 10 mM KCl, 2mM MgCl2, 1 mM EDTA, 1mM EGTA, 1mM DTT and EDTA-free cOmplete Mini protease inhibitor (Sigma-Aldrich, Australia)]. The extract was processed through centrifugation steps to clarify the suspension as follows: 720 x g for 5 min (nuclei pellet), 10, 000 x g for 5 min (mitochondria pellet), 100, 000 x g for 1 hr (membrane pellet) leaving a cytoplasmic supernatant. The supernatant (cytosolic fraction) was retained, while the membrane pellet was washed in 400µl of fractionation buffer by pipetting and recentrifuged at 100, 000 x g for 1 hr. Finally, the membrane pellet is resuspended in 0.1% SDS in TBS and both cytosolic and membrane fractions were boiled in SDS-PAGE sample buffer and the samples were used for immunoblot analysis.
Image analysis software
Analysis of immunoblots and fluorescence imaging was performed using ImageJ software (version 2.0.0; NIH, USA). Adobe Photoshop CS6 was also used to crop regions of interest on images.
Statistics
Data are shown as arithmetic means ± s.e.m., unless otherwise stated. Data sets with normal distribution were analysed with Student’s t-test (assessed by Shapiro-Wilk test) to directly compare one experimental variation, while a Two-way ANOVA was used for multiple comparisons. Sidak’s method was used for analysing variants of two-way ANOVA during multiple comparisons. All analysed experiments used technical replicates to compute statistical significance. In all statistical analysis, a P value < 0.05 was considered statistically significant and calculated using GraphPad Prism version 7.0 (GraphPad Software, San Diego, CA).
Results
LPS-activated Rab8a is on ruffles and early macropinosomes in macrophages
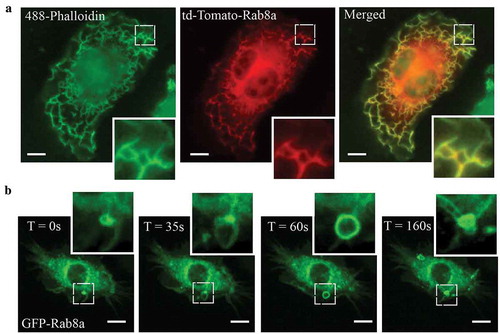
In LPS activated macrophages, Rab8a is recruited to and enriched on specific membrane domains at the dorsal ruffles and macropinosomes where it facilitates TLR-signalling functions [Citation14,Citation17]. Imaging on fixed and live transfected, LPS-treated RAW 264.7 macrophages demonstrates this membrane recruitment of Rab8a in response to LPS. shows tdTomato-Rab8a recruited abundantly to actin-rich dorsal ruffles in fixed cells. Live cell imaging of GFP-Rab8a shows formation of Rab8a positive macropinosomes which originate from ruffles, and Rab8a positive tubules that emerge from these macropinosomes during maturation (). The ruffles and early macropinosomes in particular, define the sites where LPS-activated Rab8a participates in signalling downstream of TLR4 in macrophages [Citation14,Citation15]. Thus, these membrane domains also define the sites where Rab-GEF activity is necessary for Rab8a activation to elicit TLR signalling responses.
Figure 1. Rab8a localises to Ruffles and Macropinosomes in LPS stimulated Mouse Macrophages. (a) Fluorescence microscopy images of fixed LPS-treated (30 mins) RAW 264.7 cells transiently overexpressing td-Tomato-Rab8a and stained with Alexa488-phalloidin. (b) Live cell confocal spinning disc images of LPS-treated RAW 264.7 cells transiently transfected with GFP-Rab8a showing enrichment of Rab8a on macropinosomes and subsequent internalisation via tubules. Movies were taken over 15 mins at 5 sec intervals. Scale bars, 10 µm
GRAB is a binding partner for Rab8a in macrophages
To identify Rab8a GEFs in RAW 264.7 macrophages, cells were activated with LPS (30 mins) and the cell lysate was screened for Rab8a binding proteins using GST-Rab8a pull-downs with enhanced enrichment strategies [Citation36]. The samples were separated on a gradient gel () and analysed by mass spectrometry. The results identified a number of known Rab8a binding partners, notably the effector PI3Kγ [Citation14], GDP dissociation inhibitor (GDI) alpha/beta [Citation37,Citation38], MSS4 [Citation17,Citation39–41] and, in addition, a known Rab8 regulatory GEF, GRAB () [Citation24,Citation31,Citation39]. This is the first time GRAB has been identified as a binding partner for Rab8a on these macrophage membranes, prompting interest in GRAB as a potential GEF during TLR-mediated activation of Rab8a.
Figure 2. GRAB interacts with Rab8a in an LPS dependent manner for its GEF activity. GSH-Sepharose beads with bound GST-Rab8a were used to pull-down proteins from LPS-treated (30 mins) RAW 264.7 cell lysate and eluted with proteases. The elutes were separated on a 7–15% SDS-PAGE gradient gel. (a) A faint band between 150 kDa and 100 kDa indicating the presence of previously identified effector PI3Kγ and another band above 37 kDa was identified as the Rab8a GEF GRAB (~43kDa), both of which were absent in the GST control sample. Mass spectrometry analysis identified two trypsin-digested peptides from GRAB with 95% confidence. (b) Schematic of peptide sequences of GRAB identified from the mass spectrometry. (c) Example list of identified known Rab8a interacting proteins from the mass-spectrometry analysis. (d) Immunoprecipitation of GRAB using anti-myc antibodies from cell lysates of Rab8a KO cells ± LPS (30 min) reconstituted with myc-Rab8a. Fluorescence microscopy images of fixed LPS-treated RAW 264.7 cells transiently overexpressing either (e) GFP-GRAB or (f) co-expressing td-Tomato-Rab8a and GFP-GRAB and stained with Alexa350-phalloidin. Scale bars, 10 µm. (g) Immunoblotting for levels of GTP-Rab8 pulled-down from the lysate of control and GRAB KO cells ± LPS (100 ng/ml, 15 min) using GSH-Sepharose beads with GST-OCRL-RBD. Levels of active Rab8 were quantified by using the densitometric ratio between the band intensities of captured Rab8 and GAPDH. Significance was measured via two-way analysis of variance (ANOVA) (*P < 0.05, ***P < 0.001, n = 3)
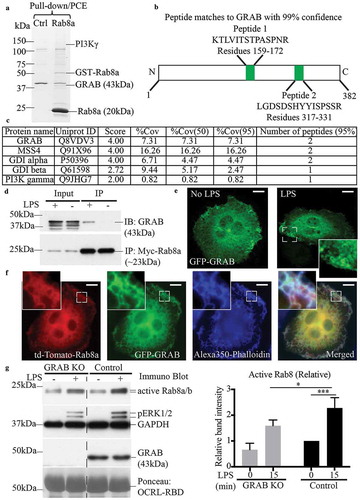
To verify GRAB’s interaction with Rab8a, we performed co-immunoprecipitations. As the commercially available Rab8a antibody was not a strong reagent for immunoprecipitations, we stably reconstituted myc-Rab8a by transfection in a Rab8a KO cell line previously generated using CRISPR/Cas9 editing in RAW 264.7 cells [Citation14]. The cells ± LPS (30 min) were lysed and immunoprecipitation experiments were performed using myc IgG antibodies on the cell lysates. The results confirmed that GRAB is expressed in these mouse macrophages and its protein levels are unchanged after acute LPS treatment; moreover, GRAB co-immunoprecipitated with myc-Rab8a but only in LPS-activated cells (). We next examined the localisation of GRAB in cells to determine membrane locations where GRAB and Rab8a might interact. RAW 264.7 macrophages transfected with GFP-GRAB were fixed with or without prior LPS (30 min) treatment. GFP-GRAB labelling was diffuse but with some localisation on membranes, notably including membranes at surface dorsal ruffles and on some macropinosomes that became more pronounced after LPS treatment (). Imaging of LPS treated RAW 264.7 cells transiently co-transfected with both GFP-GRAB and td-Tomato-Rab8a showed that both proteins are present together on ruffles and on other membranes throughout the cell (). Thus, we conclude that GRAB is available to interact with Rab8a on dorsal ruffles and macropinosome membranes. Taken together the results suggest that GRAB and Rab8a interact in LPS activated cells on these membranes for conjoint roles in LPS-induced responses.
GRAB functions investigated in GRAB-deleted macrophages
To assess if GRAB is responsible for TLR4-driven activation of Rab8, GRAB was genetically deleted from RAW 264.7 macrophages by CRISPR-Cas9 gene editing. Homozygous KO clones of GRAB KO cells showed a complete loss of GRAB protein expression (Supplementary Figure S1A) and these cells were used for subsequent studies. As a GEF, GRAB functions to facilitate GTP-loading of Rab8 for subsequent effector recruitment [Citation24]. To assay nucleotide loading on Rab8a, we employed a previously-described GTPase activation assay which uses an effector binding domain [GST-OCRL Ras binding domain (RBD)] for bead capture of GTP-loaded Rab8 (both Rab8a and Rab8b isoforms) (Supplementary Figure S2A) [Citation14]. First, we applied GDP- and GTP-loaded macrophage cell lysates to the beads to demonstrate their ability to quantitatively detect levels of GTP-Rab8 (Supplementary Figure S2B). Next, lysates from ± LPS (100 ng/ml, 15 min) control cells and GRAB KO cells were applied to this capture assay. Results showed that the control macrophages have a low basal level of GTP-bound Rab8, which increases significantly in the presence of LPS, and this activation of Rab8 is markedly impaired but not completely wiped out in the GRAB KO cells (). While activation of Rab8 was affected, the phosphorylation of the MAP kinase ERK1/2 remained unchanged between the wild-type and KO cells, suggesting that other unrelated pathways were unaffected by the deletion of GRAB.
To verify that this increase in GTP-Rab8a is LPS/TLR4 dependent, we repeated the GTP-Rab8 capture experiment on bone marrow derived macrophages (BMMs) from TLR4 deficient and wild-type mice. The results showed that LPS induces activation of Rab8 in primary macrophages and the deletion of TLR4 decreases GTP-loading of Rab8 (Supplementary Figure S2C). In addition, live cell confocal imaging of LPS-treated wild-type RAW 264.7 cells co-expressing td-Tomato-Rab8a and the TLR4 adaptor GFP-TRAM showed that both proteins are enriched on the same macropinosomal membranes (Supplementary Figure S2D). Thus, LPS associated activation of Rab8 is dependent on TLR4 receptor activation, and this likely occurs on macropinosomes. Taken together, these results demonstrate that GRAB activates Rab8 in an LPS-TLR4 dependent manner in mouse macrophages.
Conventionally, GEFs control the spatio-temporal activation of their GTPases by helping dictate their GTP/GDP loaded states, affecting subsequent recruitment to and retention on specific, relevant membrane domains [Citation42,Citation43]. We examined whether GRAB depletion affects the membrane recruitment and localisation of Rab8a. In live cell imaging of LPS-treated transfected control RAW 264.7 cells, GFP-Rab8a can be seen on membrane ruffles that form macropinosomes and is retained on tubules that emerge from these macropinosomes as they undergo sorting and maturation (). Interestingly, this membrane association and the behaviour of Rab8a remained unchanged in the GRAB KO cells reflecting similarly labelled ruffles, macropinosomes, tubules and membrane retention times (). From this, we conclude that while GRAB itself does play a role in activating Rab8, it is not necessary for Rab8a membrane attachment or its localisation on specific membrane domains where it functions in LPS/TLR4 signalling.
Figure 3. Absence of GRAB does not affect Rab8a recruitment or downstream TLR signalling. (a) Live cell confocal spinning disc imaging of LPS-treated control and GRAB KO cells transiently overexpressing GFP-Rab8a showing recruitment to macropinosomes and tubules. Quantification of Rab8a retention on macropinosomes was measured by the total number of timeframes Rab8a is spent enriched on each macropinosome, and 5 cells of each cell line was used for quantification (n = 5). Scale bars, 10 µm. (b) Immunoblotting for levels of Akt phosphorylation using phospho specific antibodies on the lysates of control and GRAB KO cells treated with LPS over a 60 min time course. The immunoblot is representative and levels of Akt phosphorylation was quantified using the densitometry ratio between the band intensities of GAPDH and phospho-Akt levels. Significance was measured via two-way analysis of variance (ANOVA) (n = 3)
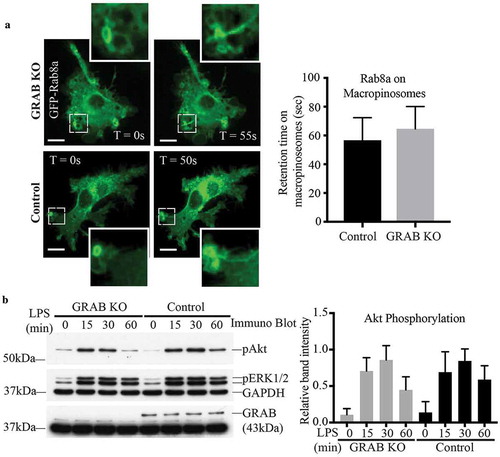
Despite GRAB not affecting Rab8a recruitment to these surface domains, absence of GRAB in the KO cells diminished the levels of LPS-induced Rab8 activation (). Therefore, we next examined if downstream TLR signalling in the GRAB deleted cells is likewise impaired. Control and GRAB KO cell lysates were collected over an LPS time course and probed by immunoblotting to detect the phosphorylation of the key signalling kinase, Akt [Citation14]. We observed an LPS induced temporal increase in phospho-Akt over 60 mins in the control cells and surprisingly, this signalling remained unchanged in the GRAB-depleted cells (). As another approach, Akt signalling was also examined in RAW 264.7 cells transiently overexpressing GFP-GRAB and treated to an LPS time course (Supplementary Figure S3A). As before, Akt phosphorylation was likewise not altered in these cells. Both these findings imply that loss or overexpression of GRAB itself does not affect Rab8a/PI3Kγ mediated signalling downstream of TLR4. This led us to investigate whether another Rab8 GEF may also be involved in Rab8a activation in this context.
Rabin8 is also a GEF for Rab8 in macrophages
Rabin8 is an alternative Rab8 GEF identified in other cell types, where it is known as a key regulator of Rab8 function in membrane trafficking [Citation24,Citation26]. We thus replicated our analysis of Rab8a activation and function in macrophages using Rabin8 as the focus. Like GRAB, Rabin8 is expressed in RAW 264.7 macrophages and it also co-immunoprecipitates with myc-Rab8a in an LPS-inducible manner (). Transient expression of GFP-Rabin8 in fixed RAW 264.7 cells showed localization on cell surface membranes in the presence of LPS () and in co-transfected LPS-treated cells, Rabin8 appeared to colocalize with td-Tomato-Rab8a on cell surface ruffles (). Following this, we generated a Rabin8 homozygous KO clonal cell line in RAW 264.7 cells using the CRISPR-Cas9 gene editing system (Supplementary Figure S1B) which had no expression of endogenous Rabin8, and these cells were then used for functional assays. In the Rab8 activation assay, the absence of Rabin8 in the KO cells did not affect the baseline levels of GTP-Rab8 but did reduce the amount of LPS-induced GTP-Rab8 recovered (). Despite this, live cell imaging of LPS-treated control and Rabin8 KO cells transiently expressing GFP-Rab8a revealed no change in Rab8a recruitment, localisation or retention on membrane ruffles and macropinosomes (). Thus, Rabin8 is similar in membrane distribution to GRAB in these cells but it is also dispensable for Rab8a membrane enrichment at these sites, despite having the ability to activate the GTPase.
Figure 4. Rab8 GEF Rabin8 also interacts with and activates Rab8 in LPS stimulated Macrophages. (a) Immunoprecipitation of Rabin8 using anti-myc antibodies from cell lysates of ± LPS (30 min) RAW 264.7 Rab8a KO cells reconstituted with myc-Rab8a. Fluorescence microscopy images of fixed (b) ± LPS (30 min) RAW 264.7 cells transiently overexpressing GFP-Rabin8 or (c) LPS-treated cells co-expressing td-Tomato-Rab8a and GFP-Rabin8 and stained with Alexa350-phalloidin. Scale bars, 10 µm. (d) Immunoblotting for levels of GTP-Rab8 pulled-down from the lysate of control and Rabin8 KO cells ± LPS (100 ng/ml, 15 min) using GSH-Sepharoase beads with GST-OCRL-RBD. Levels of active Rab8 was quantified by using the densitometric ratio between the band intensities of captured Rab8 and GAPDH. Significance was measured via two-way analysis of variance (ANOVA) (*P < 0.05, n = 3)
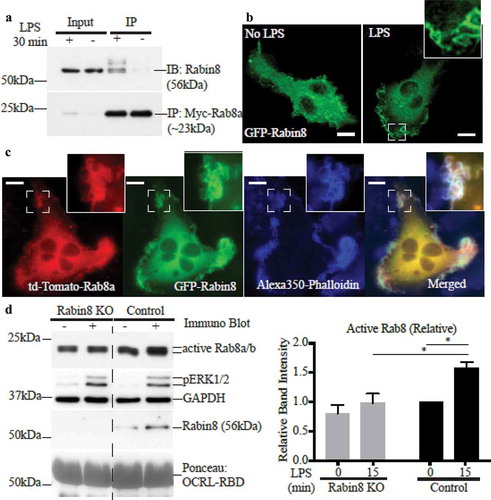
Figure 5. Similar to GRAB, absence of Rabin8 does not affect Rab8a localisation and TLR signalling. (a) Live cell confocal spinning disc imaging of LPS-treated control and Rabin8 KO cells transiently overexpressing GFP-Rab8a showing recruitment to macropinosomes and tubules. Quantification of Rab8a retention on macropinosomes was measured by the total number of timeframes Rab8a is spent enriched on each macropinosome, and 5 cells of each cell line was used for quantification (n = 5). Scale bars, 10 µm. (b) Immunoblotting for levels of phosphorylated Akt in control, Rabin8 KO and GRAB KO cells treated with LPS over a 60 min time course. Quantification of Akt phosphorylation was performed by using the densitometric ratio between the band intensities of phosphorylated Akt and GAPDH. The immunoblot is representative and significance was measured via two-way analysis of variance (ANOVA) (n = 3)
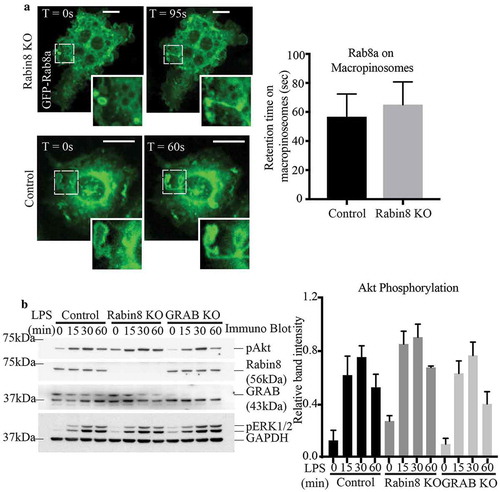
When signalling downstream of LPS-activated TLR4 was assayed, we found no change in Akt phosphorylation resulting from loss of Rabin8 in the KO cell line when compared to control cells (). Indeed, a direct comparison of Rabin8 KO to GRAB KO reveals that neither condition affects Akt signalling; moreover, Rabin8 overexpression cells also had no effect on Akt phosphorylation (Supplementary Figure S3B). Therefore, a change in expression of either GRAB or Rabin8 alone is not sufficient to affect TLR signalling driven by GTP-Rab8a (). However, given the striking similarities between GRAB and Rabin8 in both location and GEF activity in macrophages, we hypothesized that the lack of a functional outcome might be due to a compensatory relationship between these proteins.
Double knockouts reveal additive functions for GRAB and Rabin8
To further investigate the roles of both GEFs, a double KO of Rabin8 and GRAB together (double KO) was generated by performing a CRISPR-Cas9 KO of Rabin8 in the original GRAB KO cell line, leaving no detectable expression of either protein (Supplementary Figure S1C). A Rab8 activation assay was performed to compare the double KO and the original GRAB KO with control cells over a full 60 min LPS time course. In the double KO line, the level of LPS-induced GTP-loaded Rab8 was markedly lower than that in the single GRAB KO cells (). Notably, the basal level of GTP-Rab8 in the untreated cells was also decreased compared to both the GRAB KO and control cells. This demonstrates that GRAB and Rabin8 contribute additively to Rab8 activation in response to LPS, as well as co-operating to maintain a level of constitutively active Rab8 in these cells.
Figure 6. Both GRAB and Rabin8 contribute additively to LPS induced Rab8 activation but not Rab8 recruitment. (a) Immunoblotting for levels of GTP-Rab8 pulled-down using GSH-Sepharoase beads with GST-OCRL-RBD from the lysate of double KO, GRAB KO and control cells treated with LPS over a 60 min time course. Repeat activation assays of 15 min LPS treatments was used for quantification. Levels of active Rab8 was quantified by using the densitometric ratio between the band intensities of captured Rab8 and GAPDH. Significance was measured using via two-way analysis of variance (ANOVA) (*P < 0.05, **P < 0.01, n = 3). (b) Live cell confocal spinning disc imaging of LPS-treated double KO and control cells overexpressing GFP-Rab8a showing recruitment to macropinosomes and tubules. Quantification of Rab8a retention on macropinosomes was measured by the number of timeframes Rab8a is spent enriched on each macropinosome, and 5 cells of each cell line was used for quantification (n = 5). Scale bars, 10 µm
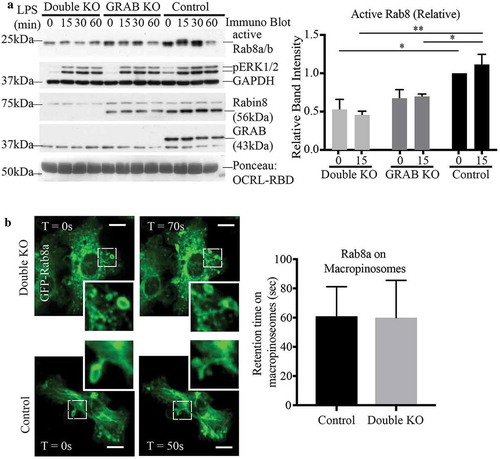
Unexpectedly, even with both GEFs absent, live cell imaging of GFP-Rab8a in double KO cells showed that Rab8a recruitment to, and retention on, surface ruffles, macropinosomes and tubules was unaffected (). To examine endogenous Rab8, we performed membrane fractionation experiments to compare Rab8 membrane association on wild-type and double KO RAW 264.7 cells treated with LPS (15 min) (Supplementary Figure S4). In these LPS activated cells, Rab8 exists mostly bound to membranes and this level was unchanged in the absence of both GEFs. Therefore, Rab8 appears to remain associated with cell membranes independent of its activation.
GRAB and Rabin8 both contribute to GTP-Rab8a-mediated Akt signalling
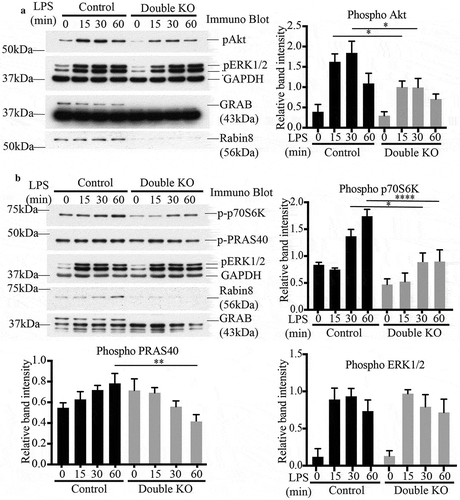
After establishing the additive roles of GRAB and Rabin8 in activating Rab8a, we next assayed TLR signalling in the double KO cells. Over an LPS time course, these cells, deleted of both GRAB and Rabin8, had a significant decrease in phosphorylated Akt levels compared to control cells (), mimicking what was observed in Rab8a KO cells [Citation14]. Thus, while single KO of either GRAB or Rabin8 did not seem to affect the phosphorylation of Akt, loss of both GEFs impaired the function of Rab8a in modulating TLR4 signalling. As a further test of disruption in this signalling pathway, we also measured the phosphorylation of signalling kinases downstream of Akt, including the p70S6 kinase and PRAS40 as an mTOR substrate () [Citation17]. Both kinases showed reduced levels of phosphorylation as a result of the double GEF KO. To ensure this effect is specifically due to the loss of function of these GEFs, we reintroduced both Rabin8 and GRAB via transiently co-transfecting GFP-tagged versions of these proteins into the double KO cells to measure Akt activation (Supplementary Figure S5). As expected, the double rescue cells showed an increase in Akt phosphorylation in response to LPS compared to the double KO cells. This confirms that the TLR signalling function of Rab8a is facilitated by the additive contribution of GRAB and Rabin8, and the depletion of both proteins was needed to sufficiently disrupt Rab8a-associated signalling. Having established the regulatory GEFs for macrophage Rab8a-TLR signalling, we turned our attention to identifying where this LPS associated Akt signalling is occurring.
Figure 7. Loss of both GRAB and Rabin8 perturbs Rab8a-associated TLR signalling. Control and double KO cells were treated with LPS over a 60 min time course and the samples were analysed for levels of phosphorylation of key signalling substrates. Representative immunoblots of the phosphorylation levels of (a) Akt and (b) p70S6K and PRAS40. Quantification of immunoblots was performed using the densitometric ratio between the band intensities of phosphorylated proteins and GAPDH. The gels are representative and significance was measured via two-way analysis of variance (ANOVA), (*P < 0.05, **P < 0.01, ****P < 0.0001, n = 3 each)
Absence of Rab8 GEFs affects the recruitment of Akt to signalling macropinosomes
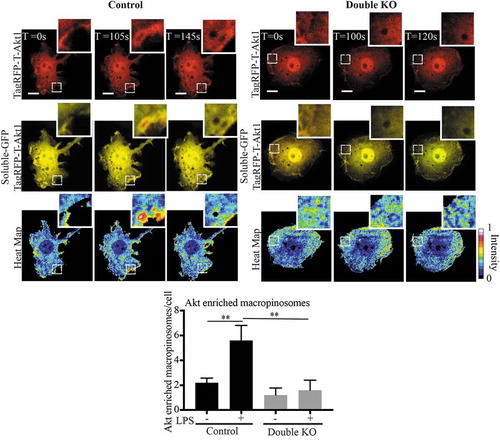
Using a reporter assay we previously pinpointed the site for activation of Rab8a and its PI3Kγ-associated signalling on early macropinosomes [Citation14]. Now, to directly visualise Rab8a/PI3Kγ associated Akt signalling, we made use of a fluorescent Akt reporter, TagRFP-T-Akt1 [Citation33]. Live cell imaging was performed on transfected double KO and control cells transiently co-expressing GFP alone as a soluble background label for the cells and the TagRFP-T-hAkt1 reporter. We constructed a ratio between the TagRFP-Akt and the soluble GFP to generate a ‘heat-map’ to identify membrane domains with Akt enrichment (), a method similarly used in a previous study to observe enrichment of YFP-tagged Pak1-binding domain on macropinosomes in macrophages [Citation44]. In LPS treated wild-type RAW 264.7 cells, we observed short bursts of enriched Akt reporter labelling on macropinosomal membranes denoting these as possible sites for TLR-generated Akt signalling. Using this approach, we quantified the number of macropinosomes that were enriched with Akt in LPS treated and untreated cells. As predicted from our biochemical signalling assays, the wild-type cells showed an increased number of Akt enriched macropinosomes in response to LPS, and this increase is abolished in the double KO cells (). This visually demonstrates that Rab8a-associated Akt signalling is occurring on macrophage macropinosomes and the decrease in Akt recruitment in the absence of both Rab8 GEFs GRAB and Rabin8, is consistent with reduced Rab8a activation and signalling downstream of LPS/TLR4.
Figure 8. Rabin8 and GRAB double KO reduces Akt enrichment on LPS induced Macropinosomes. Control and double KO cells were transiently co-transfected with C1-GFP and TagRFP-T-Akt1 and live cell spinning disc confocal microscopy was performed. A ratiometric ‘heat map’ of Akt intensity was generated by using the ratio of TagRFP-Akt1 signal intensity to the soluble GFP to identify regions of Akt membrane enrichment. The cells were imaged with and without LPS. Macropinosomes with Akt enrichment were counted as number of events per cell, expanded panels show examples of macropinosomes observed with and without Akt enrichment in the control and double KO cells respectively. Movies were taken over 15 min with 5 sec intervals and quantification was performed by counting the number of Akt enriched macropinosomes observed per cell over a sample of 5 cells for each cell line. Significance was measured via Student’s t-test. (*P < 0.05, **P < 0.01 and n = 5 cells). Scale bars, 10 µm
Discussion
In this study, we describe an expansion of the Rab8a complex that participates in TLR signalling in macrophages by identifying two GEFs responsible for activating Rab8a in this context. In its GTP-bound form, Rab8a recruits PI3Kγ for Akt/mTOR signalling to modulate the cytokine programs elicited through TLR pathways in infection and inflammation. We demonstrate that the Rab8 GEFs, GRAB and Rabin8, interact with Rab8a in macrophages in an LPS/TLR4 inducible fashion but gene deletion in vitro showed that neither GEF is required for Rab8a membrane localisation on macropinosome membranes. The absence of either GRAB or Rabin8 affects LPS induced Rab8 GTP-loading, and deletion of both GEFs together impairs the phosphorylation of key kinases and signalling molecules in the LPS-induced Akt/mTOR pathway. Finally, using a fluorescent Akt probe, we visually confirmed that Akt enrichment occurs on macropinosomes in response to LPS and this recruitment of Akt is diminished in the absence of both GEFs. Therefore, we define a previously unknown function of GRAB and Rabin8 in regulating macrophage TLR signalling for the control of inflammatory responses.
As depicted here and in a previous study [Citation14], Rab8a accumulates on membrane surface ruffles and on resulting early macropinosomes, before it is sorted onto tubulating membranes for redeployment. Here, we show that in response to LPS, the regulatory Rab8 GEFs, GRAB and Rabin8 are recruited to the same membrane domains as Rab8a, where they can interact with and activate this GTPase. The formation of ruffles, macropinosomes or tubules was found not to be affected by loss of either or both of the Rab8 GEFs, GRAB and Rabin8. This is consistent with our previous findings suggesting that Rab8a itself is not mechanistically responsible for these trafficking events, but rather a passenger on these membranes [Citation14,Citation45]. Similarly, deletion of the Rab8 GEFs did not affect Rab8a association with these membranes, even though depletion of either or both GRAB and Rabin8 reduced GTP-loading of Rab8a. This suggests that Rab8a remains bound to these membranes, independently of these GEFs or its activation state. Fractionation studies revealed that Rab8a is predominantly on membranes, rather than being in the cytosol, and this is independent of its activation or these GEFs. This implies that Rab8 remains stably associated in the membrane environment in these cells. In many instances, GEFs are important for both activating and targeting Rabs to relevant membranes for their function. An example being the depletion or mutation of Rabex-5 or Rabin8 causing mistargeting of Rab5a and Rab8a respectively in Cos-7 cells (34). This regulation can also take place as part of a Rab cascade in which upstream Rabs recruit the GEF of the next Rab, leading to membrane tethering, retainment and follow-up effector recruitment [Citation43]. This cascade is a likely scenario in macrophages, as during phagocytosis, we previously demonstrated a tight sequence of Rab GTPases recruited to phagosomes [Citation46]. However, GEFs and Rab activation are not always required for membrane attachment of the GTPases. For instance, Rab13 remains stably associated with endosomal or vesicular membranes, even in its GDP-loaded state [Citation47,Citation48]. Additionally, other factors can affect Rab8a membrane association, such as the prenylation of the Rab8 C-terminal CAAX motif that facilitates lipid binding [Citation49] or potential co-regulation of Rab8a escort proteins, GAPs and GDIs, all of which can modulate Rab membrane association [Citation50,Citation51]. Our most recent studies highlight important protein-protein interactions that contribute to the membrane recruitment of Rab8a to macrophage macropinosomes, where Rab8a is tethered to the endocytic coreceptor LRP1, which is crosstalk activated by LPS/TLR4 [Citation16]. Taken together, there are multiple mechanisms at play for recruiting and retaining Rab8a on macropinosome membranes that complement or transcend the role of Rabin8 and GRAB.
It is clear from our results that both GRAB and Rabin8 share many similarities in function in macrophages, these findings have emerged from a side-by-side comparison of both GEFs in this biological process. It is not apparent that GEF expression levels are altered significantly in these cells or processes. According to earlier data GRAB is relatively more abundant in RAW 264.7 cells compared to Rabin8 [Citation52] (BioGPS: http//biogps.org). In our study, the protein levels of GRAB and Rabin8 showed no compensatory change after KO of the opposite GEF. Furthermore, high-resolution crystal structures have shown that both GRAB and Rabin8 bind in identical fashion to Rab8, indirectly suggesting the potential for these proteins to have overlapping functions [Citation24,Citation53]. Following this, separate functional studies performed individually on GRAB and Rabin8 showed that both operate in some of the same complexes. For instance both GEFs are known to form a complex with GTP-loaded Rab11 to facilitate Rab8 recruitment for the subsequent targeting of exocytic membrane vesicles [Citation54,Citation55]. Both GEFs also function in the same pathways, such as regulating the transport of membrane vesicles for neurite outgrowth [Citation30,Citation31]. However, many of these studies showed obvious phenotypes obtained by single KO, knock-down (KD) or expression of GEF deficient mutants for either GRAB or Rabin8 in these pathways. In the present study we found that GRAB and Rabin8 have additive effects on Rab8a activation but both GEFs show overlap and redundancy in facilitating Rab8a-mediated TLR signalling. This may reflect the need for only a transient enrichment or a modest amount of active Rab8a required on signalling membranes to trigger activation of PI3Kγ and downstream signalling kinases. As compared to the higher thresholds of active Rab8a likely necessary to sustain trafficking functions.
In the context of a complex tethered by TLR-activated LRP1, GTP-Rab8a recruits PI3Kγ, which in turn recruits and activates Akt and mTOR kinases and our findings now show that deletion of both GRAB and Rabin8 affect this signalling. By utilising in-cell labelling of Akt we could detect its membrane enrichment on macropinosome membranes induced by LPS. Moreover, the effects of double GEF depletion on Akt membrane labelling provide key functional evidence that the macropinosomes are the site of LPS-induced Akt signalling. PI3Kγ [Citation56] and Akt signalling [Citation57] play an important role in determining the functional programming of macrophages and their polarization into M1 or M2 states. In turn, these macrophage programs underpin normal, physiological activation and resolution of inflammation, that if derailed, can contribute to inflammatory disease. Thus, we now reveal important roles for the Rab8 GEFs, GRAB and Rabin8, in TLR-induced inflammatory pathways. These are the first Rab GEFs implicated in signalling roles in these pathways and they highlight the mechanistic importance of Rab regulation in modulating macrophage responses in infection and inflammation.
Supplemental Material
Download PDF (5.5 MB)Acknowledgments
J.L.S, L.L and S.J.T conceived the project, designed the experiments and wrote the manuscript. S.J.T performed the experiments, analysed the data and drafted the manuscript. L.L designed and optimized the OCRL-RBD Rab activation assay and assisted with mass spectrometry analysis. A.A.W assisted with experimental design, microscopy and the generation of the CRISPR constructs. Y.H helped with fixed cell fluorescence microscopy. The authors thank Tatiana Khromykh for expert technical assistance in molecular cloning and Alun Jones for assistance with mass spectrometry. We acknowledge colleagues where stated for providing cells and reagents, our thanks to James Burchfield (The University of Sydney) for provision of the TagRFP-T-Akt1 construct and helpful advice.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10(8):513–525.
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2(2):107–117.
- Peränen J. Rab8 GTPase as a regulator of cell shape. Cytoskeleton. 2011;68(10):527–539.
- Armstrong J, Thompson N, Squire JH, et al. Identification of a novel member of the Rab8 family from the rat basophilic leukaemia cell line, RBL. 2H3. J Cell Sci. 1996;109(6):1265–1274.
- Hattula K, Furuhjelm J, Arffman A, et al. A Rab8-specific GDP/GTP exchange factor is involved in actin remodeling and polarized membrane transport. Mol Biol Cell. 2002;13(9):3268–3280.
- Hattula K, Furuhjelm J, Tikkanen J, et al. Characterization of the Rab8-specific membrane traffic route linked to protrusion formation. J Cell Sci. 2006;119(23):4866–4877.
- Luo N, West CC, Murga-Zamalloa CA, et al. OCRL localizes to the primary cilium: a new role for cilia in Lowe syndrome. Hum Mol Genet. 2012;21(15):3333–3344.
- Chibalina MV, Roberts RC, Arden SD, et al. Rab8‐optineurin‐myosin VI: analysis of interactions and functions in the secretory pathway. Methods Enzymol. 2008;438:11–24.
- Yamamura R, Nishimura N, Nakatsuji H, et al. The interaction of JRAB/MICAL-L2 with Rab8 and Rab13 coordinates the assembly of tight junctions and adherens junctions. Mol Biol Cell. 2008;19(3):971–983.
- Sun Y, Chiu TT, Foley KP, et al. Myosin Va mediates Rab8A-regulated GLUT4 vesicle exocytosis in insulin-stimulated muscle cells. Mol Biol Cell. 2014;25(7):1159–1170.
- Wiesner C, El Azzouzi K, Linder S. A specific subset of RabGTPases controls cell surface exposure of MT1-MMP, extracellular matrix degradation and 3D invasion of macrophages. J Cell Sci 2013;126(13), 2820–2833. jcs. 122358.
- Linder MD, Mäyränpää MI, Peränen J, et al. Rab8 regulates ABCA1 cell surface expression and facilitates cholesterol efflux in primary human macrophages. Arterioscler Thromb Vasc Biol. 2009;29(6):883–888.
- Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2(8):675–680.
- Wall AA, Luo L, Hung Y, et al. Rab8a recruited PI3Kγ regulates signaling and cytokine outputs from endosomal Toll-like receptors. J Biol Chem. 2017: jbc. M116. 766337.
- Wall AA, Condon ND, Luo L, et al. Rab8a localisation and activation by Toll-like receptors on macrophage macropinosomes. Philos Trans R Soc London Ser B Biol Sci. 2018;374(1765). DOI:10.1098/rstb.2018.0151.
- Luo L, Wall AA, Tong SJ, et al. TLR Crosstalk activates LRP1 to Recruit Rab8a and PI3Kγ for suppression of inflammatory responses. Cell Rep. 2018;24(11):3033–3044.
- Luo L, Wall AA, Yeo JC, et al. Rab8a interacts directly with PI3Kgamma to modulate TLR4-driven PI3K and mTOR signalling. Nat Commun. 2014;5:4407. PubMed PMID: 25022365.
- Bellacosa A, Chan TO, Ahmed NN, et al. Akt activation by growth factors is a multiple-step process: the role of the PH domain. Oncogene. 1998;17(3):313.
- Delgoffe GM, Pollizzi KN, Waickman AT, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12(4):295.
- Das A, Guo W. Rabs and the exocyst in ciliogenesis, tubulogenesis and beyond. Trends Cell Biol. 2011;21(7):383–386.
- Feng S, Knödler A, Ren J, et al. A Rab8 guanine nucleotide exchange factor-effector interaction network regulates primary ciliogenesis. J Biol Chem. 2012;287(19):15602–15609.
- Gutierrez MG. Functional role (s) of phagosomal Rab GTPases. Small GTPases. 2013;4(3):148–158.
- Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Nat Acad Sci. 2006;103(32):11821–11827.
- Guo Z, Hou X, Goody RS, et al. Intermediates in the guanine nucleotide exchange reaction of Rab8 protein catalyzed by guanine nucleotide exchange factors Rabin8 and GRAB. J Biol Chem. 2013;288(45):32466–32474.
- Wang J, Ren J, Wu B, et al. Activation of Rab8 guanine nucleotide exchange factor Rabin8 by ERK1/2 in response to EGF signaling. Proc Nat Acad Sci. 2015;112(1):148–153.
- Yoshimura SI, Gerondopoulos A, Linford A, et al. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J Cell Biol. 2010;191(2):367-381. jcb:201008051.
- Nachury MV, Loktev AV, Zhang Q, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129(6):1201–1213.
- Lim YS, Chua CEL, Tang BL. Rabs and other small GTPases in ciliary transport. Biol Cell. 2011;103(5):209–221.
- Luo HR, Saiardi A, Nagata E, et al. GRAB: a physiologic guanine nucleotide exchange factor for Rab3A, which interacts with inositol hexakisphosphate kinase. Neuron. 2001;31(3):439–451.
- Homma Y, Fukuda M. Rabin8 regulates neurite outgrowth in both GEF activity-dependent and -independent manners. Mol Biol Cell. 2016;27(13):2107–2118. PubMed PMID: 27170183; PubMed Central PMCID: PMCPMC4927283. .
- Furusawa K, Asada A, Urrutia P, et al. Cdk5 regulation of the GRAB-mediated Rab8-Rab11 cascade in axon outgrowth. J Neurosci. 2016;37(4):2197–2216.
- Wall AA, Luo L, Hung Y, et al. Small GTPase Rab8a-recruited phosphatidylinositol 3-Kinase γ regulates signaling and cytokine outputs from endosomal Toll-like receptors. J Biol Chem. 2017;292(11):4411–4422.
- Norris DM, Yang P, Krycer JR, et al. An improved Akt reporter reveals intra-and inter-cellular heterogeneity and oscillations in signal transduction. J Cell Sci. 2017;130(16):2757–2766.
- Hoshino K, Takeuchi O, Kawai T, et al. Cutting edge: toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol. 1999;162(7):3749–3752.
- Luo L, Bokil NJ, Wall AA, et al. SCIMP is a transmembrane non-TIR TLR adaptor that promotes proinflammatory cytokine production from macrophages. Nat Commun. 2017;18;8:14133. PubMed PMID: 28098138. .
- Luo L, King NP, Yeo JC, et al. Single‐step protease cleavage elution for identification of protein–protein interactions from GST pull‐down and mass spectrometry. Proteomics. 2014;14(1):19–23.
- Schalk I, Zeng K, Wu S-K, et al. Structure and mutational analysis of Rab GDP-dissociation inhibitor. Nature. 1996;381(6577):42.
- Ullrich O, Stenmark H, Alexandrov K, et al. Rab GDP dissociation inhibitor as a general regulator for the membrane association of rab proteins. J Biol Chem. 1993;268(24):18143–18150.
- Itzen A, Pylypenko O, Goody RS, et al. Nucleotide exchange via local protein unfolding—structure of Rab8 in complex with MSS4. Embo J. 2006;25(7):1445–1455.
- Raffaniello R, Fedorova D, Ip D, et al. Hsp90 Co-localizes with Rab-GDI-1 and regulates agonist-induced amylase release in AR42J cells. Cell Physiol Biochem. 2009;24(5–6):369–378.
- Zhang B, Zhang T, Wang G, et al. GSK3β-Dzip1-Rab8 cascade regulates ciliogenesis after mitosis. PLoS Biol. 2015;13(4):e1002129.
- Pfeffer SR. Rab GTPase regulation of membrane identity. Curr Opin Cell Biol. 2013;25(4):414–419.
- Blümer J, Rey J, Dehmelt L, et al. RabGEFs are a major determinant for specific Rab membrane targeting. J Cell Biol. 2013;200(3):287–300.
- Yoshida S, Hoppe AD, Araki N, et al. Sequential signaling in plasma-membrane domains during macropinosome formation in macrophages. J Cell Sci. 2009;122(18):3250–3261.
- Condon ND, Heddleston JM, Chew T-L, et al. Macropinosome formation by tent pole ruffling in macrophages. J Cell Biol. 2018;217(11):3873–3885. jcb. 201804137.
- Yeo JC, Wall AA, Luo L, et al. Sequential recruitment of Rab GTPases during early stages of phagocytosis. Cell Logist. 2016;6(1):e1140615. PubMed PMID: 27217977; PubMed Central PMCID: PMCPMC4861590.
- Ioannou MS, McPherson PS. Rab13 and the regulation of cancer cell behavior. J Biol Chem 2016;291(19):9929–9937. jbc. R116. 715193.
- Ioannou MS, Girard M, McPherson PS. Rab13 traffics on vesicles independent of prenylation. J Biol Chem 2016;291(20):10726–10735. jbc. M116. 722298.
- Pereira-Leal JB, Hume AN, Seabra MC. Prenylation of Rab GTPases: molecular mechanisms and involvement in genetic disease. FEBS Lett. 2001;498(2–3):197–200.
- Pylypenko O, Hammich H, Yu I-M, et al. Rab GTPases and their interacting protein partners: structural insights into Rab functional diversity. Small GTPases. 2018;9(1–2):22–48.
- Müller MP, Goody RS. Molecular control of Rab activity by GEFs, GAPs and GDI. Small GTPases. 2018;9(1–2):5–21.
- Hammer KD, Yum M-Y, Dixon PM, et al. Identification of JAK–STAT pathways as important for the anti-inflammatory activity of a Hypericum perforatum fraction and bioactive constituents in RAW 264.7 mouse macrophages. Phytochemistry. 2010;71(7):716–725.
- Sato Y, Shirakawa R, Horiuchi H, et al. Asymmetric coiled-coil structure with Guanine nucleotide exchange activity. Structure. 2007;15(2):245–252.
- Horgan CP, Hanscom SR, McCaffrey MW. GRAB is a binding partner for the Rab11a and Rab11b GTPases. Biochem Biophys Res Commun. 2013;441(1):214–219.
- Wang J, Deretic D. The Arf and Rab11 effector FIP3 acts synergistically with ASAP1 to direct Rabin8 in ciliary receptor targeting. J Cell Sci. 2015;128(7):1375–1385.
- De Henau O, Rausch M, Winkler D, et al. Overcoming resistance to checkpoint blockade therapy by targeting PI3Kγ in myeloid cells. Nature. 2016;539(7629):443.
- Vergadi E, Ieronymaki E, Lyroni K, et al. Akt signaling pathway in macrophage activation and M1/M2 polarization. J Immunol. 2017;198(3):1006–1014.
