ABSTRACT
Rnd proteins constitute a subfamily of Rho GTPases represented in mammals by Rnd1, Rnd2 and Rnd3. Despite their GTPase structure, their specific feature is the inability to hydrolyse GTP-bound nucleotide. This aspect makes them atypical among Rho GTPases. Rnds are regulated for their expression at the transcriptional or post-transcriptional levels and they are activated through post-translational modifications and interactions with other proteins. Rnd proteins are mainly involved in the regulation of the actin cytoskeleton and cell proliferation. Whereas Rnd3 is ubiquitously expressed, Rnd1 and 2 are tissue-specific. Increasing data has described their important role during development and diseases. Herein, we describe their involvement in physiological and pathological conditions with a focus on the neuronal and vascular systems, and summarize their implications in tumorigenesis.
1. Rnd, an atypical Rho GTPase family
1.1 The Rho GTPase family in human and emergence of Rnd during evolution
The Rho family of GTPases belongs to the Ras superfamily of small GTPases and contains, in humans, 20 members divided into 8 subfamilies according to their sequence homology: RhoA, Rac, Cdc42, RhoU/V, RhoD/F, RhoH, RhoBTB and Rnd. One important and common feature of Rho GTPase proteins is their ability to bind nucleotides and to cycle between GDP-bound (OFF) inactive state and GTP-bound (ON) active state. The switch between the two states occurs when the gamma phosphate group of the GTP is hydrolysed. The change in the structure of the GTPase on hydrolysis reduces binding to downstream effectors and turns off the protein. For most of Rho family members, three types of proteins regulate the transition between their ON and OFF states. Guanine nucleotide exchange factors (GEFs) play as activators allowing the replacement of GDP by GTP whereas GTPase-activating proteins (GAPs) inactivate them by promoting the GTP hydrolysis to GDP. Additional regulation is provided by Guanine nucleotide dissociation inhibitors (GDIs) which sequester GTPases away from their active subcellular compartments and prevent both nucleotide exchange and GTP hydrolysis. Within the RhoGTPase family, the Rnd subfamily is defined by three members sharing sequence identity. The Rnd subfamily consists of three proteins: Rnd1/Rho6/RhoS, Rnd2/Rho7/RhoN and Rnd3/Rho8/RhoE (). The human RND1 gene is located on chromosome 12 (q12-q13), while the RND2 and RND3 genes are located, respectively, on chromosome 17 (q21) and on chromosome 2 (q23). Rnd protein family emerged in the ancestral chordates, only expressed in vertebrates and shows a relative late evolution of history. This finding suggests that Rnd proteins are implicated in vertebrate-specific biological mechanisms [Citation1].
Table 1. Specific features of Rnd proteins
1.2 Specific features of Rnd proteins
Foster and collaborators identified the first member of the Rnd family, called RhoE by yeast two-hybrid screen with the GAP, p190RhoGAP-A (encoded by the ARHGAP35 gene), as bait [Citation2] (). They showed by sequence alignments that RhoE protein has similarity to different members of the Rho GTPase family and exhibits strong homology to the mammalian RhoA, RhoB and RhoC (54% identity). Nevertheless, RhoE was described as an unusual member of Rho family existing only in the GTP-bound form and lacking intrinsic GTPase activity. It is resistant to RhoGAP activity; incubation of GTP-loaded RhoE with purified preparation of p190RhoGAP or Cdc42GAP failed to promote any detectable GTP hydrolysis by RhoE [Citation2]. Two years later, the complete human cDNA for three members of the same family was obtained. These new members were nominated Rnd (‘round’) for their ability to induce cell rounding [Citation3]. Like RhoE, renamed Rnd3, Rnd1 and Rnd2 have altered sequences at residues critical for GTP hydrolysis (mainly serine instead of the conserved glutamine in classical GTPases) and are permanently in the GTP-bound form with no detectable GTPase activity (). As previously described for pseudokinases that lack kinase activity, Rnd proteins are considered pseudoGTPases. They indeed belong to a recently described pseudoGTPase subgroup (class ii) able to bind nucleotide but with no catalytic activity [Citation4]. Rnd proteins share 54–63% identity and display substantial differences compared to the classical RhoGTPase molecular switches. The molecular weights of Rnd proteins (~26-27 kD) are higher than Rho/Rac/Cdc42 (~22kD) due to the NH2-terminal extensions (5–21 residues) and COOH-terminal extensions (about 30 residues) for all three (). Rnd proteins are positively charged with basic isoelectric point (pls 8.1 to 8.7) whereas most Rho-family proteins have slightly acidic pls. Unlike most other Rho proteins which have a C-terminus ‘CAAX box’ with leucine and are predominantly geranyl-geranylated, Rnd proteins are farnesylated at the last methionine residue [Citation3,Citation5]. The recognition of the CAAX mediated by farnesyltransferase (FT) and/or genylgeranyltransferase type I (GGTase-I) is essential for all Rho family GTPases to undergo a series of posttranslational modifications promoting subcellular localization to the plasma membrane and/or endomembranes. Indeed, localization of all three Rnd is extremely sensitive to FT inhibitors (FTI) but not to GGTase-I inhibitors (GGTI) and resulted in the loss of plasma membrane localization, and increased cytoplasmic and nuclear accumulation of all three Rnd proteins. Thus, the subcellular localization of Rnd1 and Rnd3 proteins is predominantly plasma membrane-associated while Rnd2 lacks this association and appears to be in the cytosol and endosomes [Citation6], directly bound to vacuolar protein sorting 1-A (Vps4-A), the central protein regulating early endosome trafficking [Citation7] (). Interestingly, replacement of Rnd2 C-terminal domain with that of Rnd3 allowed Rnd2 to be localized at the plasma membrane and compensate for Rnd3 loss of function in cortical neurons [Citation8]. The phosphorylation of Rnd3 by the serine threonine kinases ROCK1, protein kinase C (PKCα) or TAO kinase (TAOK1, TAOK2) induces the protein translocation from the plasma membrane to the cytosol [Citation9–11]. Moreover, the interaction of the phosphorylated forms of Rnd1 and Rnd3 with 14-3-3 proteins also favours this translocation inhibiting Rnd-induced cell rounding [Citation12]. Rnd1 is concentrated at point of cell-cell contacts in confluent 3T3 fibroblasts and in epithelial cells with a peripheral staining of Rnd1 colocalizing with pan-cadherin [Citation3]. Similarly, Rnd3 was shown to localize to cell-cell contacts during tumour cell collective migration [Citation13] and to cell-cell collision zones upon the migration of human cancer-associated fibroblasts in extracellular matrices [Citation14]. Rnd3 protein was also found to localize with the Rho kinase ROCK1, one of its partner (), on the Golgi apparatus in COS-7 fibroblasts [Citation15]. Finally, Rnd1 and Rnd3 were found associated to lipid rafts thanks to a KERRA (Lys-Glu-Arg-Arg-Ala) sequence present in their N-terminus in various cell lines including COS-7, HeLa, HEK-293 and NIH-3T3 cells [Citation16].
Table 2. Rnd partners and associated functions
1.3 Rnd expression and functions
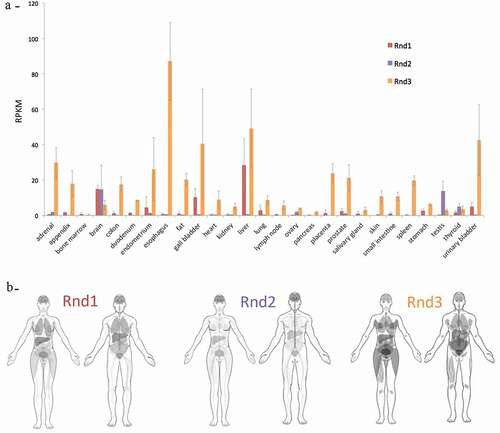
Early studies have demonstrated that all three Rnds display distinct patterns of expression [Citation2,Citation3]. Recent RNA sequencing performed on adult human tissues confirmed those data (). Rnd1 is expressed mostly in the liver and several brain tissues. Rnd2 is highly abundant in testis and in the brain. Rnd3 is ubiquitously expressed, showing expression in all tissues tested, with lower levels in the pancreatic tissue and bone marrow () [Citation18]. In foetal brain, lung, kidney and liver tissues, the patterns of Rnds expression are similar to that observed in adults [Citation3]. Thus, although the three Rnds show different pattern of expression, they are all expressed in the brain.
Figure 1. Expression of RND in healthy human tissues. A) Data extracted from the Gene database (https://www.ncbi.nlm.nih.gov/gene). RNA-seq was performed of tissue samples from 95 human individuals representing 27 different tissues in order to determine tissue-specificity of the three RND protein-coding genes (BioProject: PRJEB4337). The y axis shows RPKM (Reads Per Kilobase Million) reflecting expression level, whereas the x axis indicates human tissues. B) Anatomogram extracted from the human protein Atlas database, showing tissue mRNA expression for Rnd1 (https://www.proteinatlas.org/ENSG00000172602-RND1/tissue), Rnd2 (https://www.proteinatlas.org/ENSG00000108830-RND2/tissue), and Rnd3 (https://www.proteinatlas.org/ENSG00000115963-RND3/tissue). Image credit: Human Protein Atlas
As described above, Rnd proteins are not regulated by the typical GDP/GTP cycle; thus, biogenesis and decay of mRNA and proteins play an important role in their regulation. Besides intrinsic stability of each individual mRNA, transcriptional and post-transcriptional regulation of Rnds may change in response to a variety of extracellular stimuli. Indeed, it is now clear that all Rnd proteins are highly regulated and modifications of their expression in response to multiple different stimuli, such as growth factors, and conditions, such as hypoxia, are well documented [Citation19–23]. At the protein level, no specific guanine nucleotide exchange factors or GTPase activating proteins for Rnds are known. Instead, Rnds are activated through post-translational modifications (i.e. phosphorylation) or interaction with other proteins and inactivated through cytosolic sequestration or ubiquitinylation and destruction by the proteasome [Citation12,Citation24,Citation25]. For example, the interaction of Syx (also named PLEKHG5) via its ubiquitin-like ‘Rnd-binding domain’ (RndBD) with Rnd3 elevates the half-life of Rnd3, thus increasing Rnd3 protein stability (). The effect of Syx on Rnd3 is also observed on the other Rnd proteins, but the interaction of Syx with Rnd1 or Rnd2 is weaker than the one with Rnd3 [Citation24]. Syx is also considered as an Rnd effector as Rnd proteins potentially modulate RhoA activation through this RhoGEF [Citation26].
Over past years, members of the Rho GTPase family have been demonstrated as principal actors in many cellular processes including cytoskeletal organization, cell cycle, proliferation, cell shape and movement. Rnd proteins have also been implicated in these processes, as they are known to act mainly as antagonists of the RhoA/ROCK pathway, even if they also function through specific effectors. Rnd regulation of these key cellular events was mainly described in the neuronal and vascular systems, and during tumorigenesis. Thus, in this review, we focus on the pathophysiological functions of Rnds in these three contexts.
2. Rnd proteins in the neuronal system
2.1 In physiological conditions
As mentioned previously, the three members of the Rnd family are highly expressed in the brain and over the past several years, it has become clear that these RhoGTPases play a central role in various aspects of neuronal development (). The development of a neuron is a multistep process that begins with its generation from a neuronal progenitor cell residing in a germinal zone of the nascent brain. Then, after migration from its birthplace to its final location, the newborn neuron develops specialized processes, axon and dendrites, towards proper target regions and finally induces spine formation and synaptic connections with appropriate partners.
Table 3. Summary of Rnd protein functions in neuronal development
2.1.1 Rnd functions in neuronal progenitors
Cortical progenitors
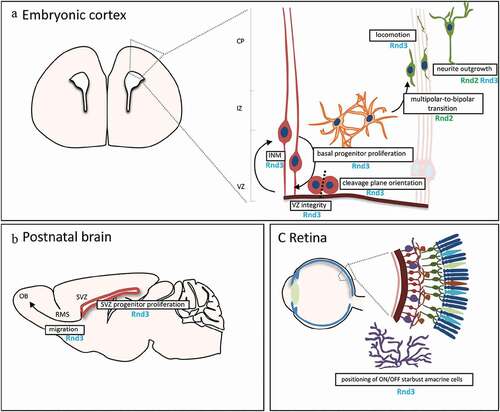
So far, only Rnd3 has been described to play a role in neuronal stem cells/progenitors. In the embryonic cortex, using in utero electroporation of Rnd3 shRNA, Rnd3 was shown, in radial glial cells (RGCs), to regulate the movement of the nuclei occurring during interkinetic nuclear migration (a process whereby nuclei change position along the apico–basal axis during the course of the cell cycle), to exert a control on the orientation of the cleavage plane and to be required for the maintenance of adherens junctions [Citation27]. Importantly, Rnd3 regulates these early steps of neurogenesis by inhibiting actin filament polymerization since the defects observed after Rnd3 knockdown are rescued when a constitutively active form of cofilin (cofilinS3A), which disrupts actin filaments, is co-electroporated [Citation27]. In addition, Rnd3 also limits the divisions of basal progenitors, but, interestingly, this involves the suppression of cyclin D1 translation. Rnd3 thus exerts pleiotropic functions in cortical progenitors but employs distinct mechanisms in RGCs and basal progenitors.
SVZ progenitors
Besides these functions in cortical progenitors, data obtained with Rnd3 mutant suggest that Rnd3 also controls progenitor behaviour in the subventricular zone (SVZ) [Citation28]. This zone along the lateral ventricle wall represents a reservoir of neural stem/progenitor cells in the postnatal and adult brain. There, neuroblasts are continuously produced by stem cells/progenitors and these neuroblasts then migrate through the rostral migratory stream (RMS) to reach the olfactory bulb (OB) where they differentiate in two principal cell types, granule cells and periglomerular cells [Citation29]. Using BrdU labelling, Ballester-Lurbe and colleague observed an excessive proliferation in the SVZ at postnatal day 9 (P9) in the Rnd3 mutants compared to control mice, but not at birth [Citation28]. This phenotype can be attributed to enhanced Notch signalling, which acts in coordination with proneural transcription factors to balance proliferation and differentiation of NSCs. Increased levels of the activated form of Notch (NICD: Notch intracellular domain) and of the Notch target Hes1 have been reported in the postnatal SVZ of Rnd3 knockouts and this increase is what drives the aberrant growth of NSCs, since Notch inhibition abolished the phenotype [Citation30]. So, like in cortical basal progenitors, Rnd3 limits proliferation of SVZ progenitors, but the mechanisms involved are different.
2.1.2 Rnd functions in neuronal migration
Migration of cortical neurons
In addition to defects in cortical progenitors, the in vivo knockdown of Rnd3 in the embryonic cortex perturbs the radial migration of projection neurons [Citation31]. Briefly, after detachment from the ventricular surface, the nascent cortical neurons become bipolar and move radially away to the SVZ/intermediate zone (IZ), where they acquire a multipolar shape. Then, neurons enter into the cortical plate (CP) and become bipolar, extending a leading process (future apical dendrite) towards the pial surface and a trailing process (future axon) in the opposite direction. Upon multi to bipolar transition, neurons establish dynamic contacts with RGC fibres and subsequently use them as a scaffold to migrate to the upper part of the CP. During this phase, neurons use a mode of migration called locomotion, which is characterized by repetitive migratory cycles of extension of the leading process, translocation of the nucleus, and retraction of the trailing process.
After Rnd3 silencing, neurons display an abnormal morphology and movement in the CP. Indeed, instead of being bipolar, Rnd3 deficient neurons exhibit an enlarged leading process with numerous branches and the coupling between the nucleus and centrosome is perturbed, which altogether disrupt the coordinated movement of the locomoting neurons [Citation8] (). Rnd2 has been shown to be also important for this process of radial migration but this Rnd protein, in contrast to Rnd3, regulates the multipolar to bipolar transition [Citation32–34] (). Indeed, Rnd2-deficient neurons display longer processes at the multipolar stage and fail to leave the IZ, which eventually leads to the accumulation of cells in the IZ of Rnd2-knocked down cortices and a concomitant reduction of neurons reaching the CP, in comparison to control cortices [Citation8,Citation33]. Despite these different functions, both Rnd2 and Rnd3 regulate neuronal migration by inhibiting RhoA, but only partially for Rnd2. The inhibitory effect of Rnd3 on RhoA activity depends on its interactions with p190RhoGAP, whereas Rnd2’s RhoA inhibitory activity does not. Moreover, it has been demonstrated that Plexin B2 competes with p190RhoGAP for binding to Rnd3, thus blocking the Rnd3-mediated inhibition of RhoA, which allows a cell-extrinsic control of RhoA activity in migrating neurons [Citation35]. In addition, although both Rnd2 and Rnd3 regulate actin dynamics in migrating neurons, only Rnd3 promotes neuronal migration by inhibiting RhoA-mediated actin polymerization and remodelling [Citation8]. Interestingly, the distinct subcellular localization of Rnd2 and Rnd3 and the resultant modulation of RhoA activity in different cell compartments underlie the difference in their effects. Rnd3 owes its distinct role in neuronal migration to its localization and interaction with RhoA at the plasma membrane. Rnd2 is expressed in early endosomes [Citation8] but the mechanisms by which Rnd2 promotes cortical neuron migration and inhibits RhoA remains unknown. Two important factors that could mediate Rnd2 activity in the embryonic cortex are the vacuolar protein sorting 1-A (Vps4-A) [Citation7] that regulates endosome recycling and the proteins Bacurd1/Kctd13 and Bacurd2/Tnfaip1 that could reduce RhoA levels through their BTB-domain containing adaptors for Cul3-mediated RhoA degradation (). In particular, Bacurd1/Kctd13 and Bacurd2/Tnfaip1 have been shown to control long-term positioning of cortical neurons within the postnatal mouse cerebral cortex and thus their interaction with both Rnd2 and Rnd3 might be relevant to ensure correct neuronal migration [Citation36].
Figure 2. In vivo roles of Rnd proteins in the nervous system. A) Schematic of the murine embryonic cortex, showing the cellular events regulated by Rnd proteins. Neural stem cells (NSCs, in red) are attached to one another in the ventricular zone (VZ) by apically located adherens junctions. NSCs coordinate nuclear movement with the cell cycle in a process known as INM (interkinetic nuclear migration). Once NSCs start differentiating, they generate neuronal committed basal progenitors (in orange) that divide in the intermediate zone (IZ), away from the VZ. Basal progenitors and recently born neurons in this basal region exhibit multipolar morphology and transit to a bipolar shape (in green) before entering the cortical plate (CP) and migrate through locomotion to the pial surface. Nascent neurons establish their polarity while they migrate, with a trailing and a leading process that will finally develop into axon and dendrites, respectively. B) Scheme of the postnatal brain in a sagittal section. After proliferation of NSCs in the subventricular zone (SVZ) along the lateral ventricles, newborn neurons migrate to the olfactory bulb (OB) through the rostral migratory stream (RMS). C) Scheme of the eye and the retina, showing the different neuronal layers in the retina. Position of the amacrine cells (in purple) is regulated by Rnd3
Importantly, different transcription factors control radial migration in the developing cerebral cortex via Rnd2 or Rnd3 [Citation31]. Indeed, the proneural transcription factor Neurogenin2 (Neurog2) has been shown to promote the migration of nascent cortical neurons by inducing the expression of Rnd2 while another proneural factor, Ascl1, promotes neuronal migration in the cortex through direct regulation of Rnd3 [Citation8,Citation33]. Neurog2 and Ascl1 directly bind to E-box DNA sequences within enhancers located 3ʹ to the Rnd2 and Rnd3 gene, respectively [Citation8,Citation33]. Other transcription factors such as RP58, Scratch2, and COUP-TFI promote cortical radial migration, but in contrast to Neurog2, by repressing Rnd2 expression [Citation37,Citation38,Citation41]. In particular, RP58 and Scratch2 regulate the same 3ʹ enhancer as Neurog2, suggesting that the three factors might compete to fine-tune appropriate levels of Rnd2 in the developing cortex [Citation39,Citation40,Citation41].
Migration of neuroblasts in the rostral migratory stream
In addition to an excessive proliferation in the SVZ, cell migration through the RMS is impaired in mice lacking Rnd3 expression [Citation28] (). In this RMS, newly generated neuroblasts migrate tangentially in chains surrounded by astrocytes in a well-organized manner. In Rnd3-deficient mice, the migrating cells do not form a compact structure but instead are scattered and their nucleus and cytoplasm are oriented in different directions [Citation28]. Moreover, after BrdU injection, most of the BrdU-labelled cells in the mutant remain in the caudal RMS, closer to the SVZ, and only a small percentage arrives to the OB [Citation28]. In vitro, neuroblasts from Rnd3-null SVZ-derived neurospheres migrate slower and cover lesser distances compared to control cells, which further suggests that Rnd3 is essential for the proper migration of neuroblasts in the RMS [Citation28].
Migration of retinal interneurons
Recently, Rnd3 has been shown to be also crucial for correct positioning of retinal interneurons called starburst amacrine cells (SACs) [Citation42] (). These neurons occur as pairs, with one member responding to increases (ON cells) and the other to decreases (OFF cells) in light intensity. SACs in the central mouse retina are born at the apical edge of the retinal neuroblast layer from around embryonic day (E) 10 to E14. They then migrate basally to reach the border of the outer and inner neuroblast layers (ONBL, INBL). After E16, about half continue basally to the INBL, where they become ON SACs; the others remain in the ONBL and become OFF SACs. SACs are fully integrated into direction-selective circuits by the time of eye-opening at P14. Interestingly, Rnd3 is selectively expressed by OFF SACs and regulates their migration but is repressed by Fezf1 in ON SACs, allowing their entry into the INBL [Citation42]. Rnd3 acts thus to enable distinct positioning of ON and OFF SACs in the developing retina () but the mode of action of Rnd3 in these cells remains to be elucidated.
2.1.3 Rnd functions in neuronal polarization
A mature neuron typically has several dendrites that receive inputs from presynaptic neurons and one axon that relays information to post-synaptic neurons. The development of these specialized structures is preceded by the formation of neurites, which are small protrusions sprouting from the cell body and lacking molecular and structural characteristics of mature axonal or dendritic processes, such as the expression of Tau or Microtubule-associated protein 2 (MAP2) respectively.
Neurite outgrowth
The three Rnd members have been implicated in the regulation of neurite outgrowth and this has been mainly demonstrated in vitro in PC12 cells. These cells, derived from a rat pheochromocytoma, have been used extensively over the years to study neuronal differentiation because they exhibit a typical phenotype of neuronal cells sending out neurites in response to nerve growth factor (NGF) [Citation43] or basic fibroblast growth factor (bFGF) [Citation44]. Forced expression of Rnd1 or Rnd3 in these cells, in the absence of growth factors, causes the formation of many neuritic processes [Citation45–47] while their knockdown suppresses the FGF or NGF-induced neurite outgrowth, respectively, [Citation47,Citation48]. Both Rnd1 and Rnd3 stimulate neurite outgrowth in PC12 cells through inhibition of RhoA signalling [Citation47,Citation48]. More precisely, in the resting state, Rnd1 interacts with FRS2β (), a docking protein of the fibroblast growth factor (FGF) receptor-1, and this prevents the interaction of Rnd1 with other molecules [Citation48]. In the presence of FGF, the receptor FGFR1 is activated and phosphorylates FRS2β. This phosphorylation releases Rnd1 and the liberated Rnd1 then inhibits RhoA activity [Citation48]. Rnd1 thus plays an important role in the FGF signalling during neurite development in PC12 cells and its activity is regulated by FGF receptor through FRS2β. Moreover, the Stress-Inducible Protein-1 (STI1), a co-chaperone that associates directly with Hsp70/Hsp90 heat shock proteins and modulates their chaperone activities has been shown to specifically bind to Rnd1 and regulates its activity in PC12 cells [Citation46] (). Regarding Rnd3, similar results have been obtained in cultured hippocampal neurons from mice lacking Rnd3 expression [Citation49]. Indeed, Rnd3-deficient neurons exhibit a decrease in neurite outgrowth and the RhoA/ROCK pathway, like in PC12 cells [Citation47], is involved in this neuronal alteration [Citation49].
In contrast to Rnd1 and Rnd3, Rnd2 induces inhibition of NGF-induced neurite outgrowth in PC12 cells and stimulates RhoA activity through Pragmin (), a catalytically inactive pseudo-kinase, although there is no direct biochemical connection between Pragmin and RhoA [Citation50]. Moreover, Rnd2 stimulates neurite branching in these cells through Rapostlin [Citation51], a molecule involved in the formation of endocytic vesicles [Citation52]. Rapostlin, also named FBP17, binds to Rnd2 through an HR1 domain, related to those identified in RhoA/Rac1 effectors such as PRK/PKN. It acts through the ability of its F-BAR domain to promote membrane invagination [Citation53]. Rnd2 may be thus involved in the regulation of different aspects of neuronal morphology development through different effectors, such as neurite branching with Rapostlin and neurite outgrowth inhibition with Pragmin.
Although the regulation of neurite outgrowth by Rnd proteins has been studied extensively in vitro, the control of the corresponding developmental process in vivo has been poorly addressed. Only very few studies have looked at this aspect in the developing cortex for Rnd2 and Rnd3. Rnd3-silenced neurons that reach the CP present multiple thin processes extending from the cell body and the leading process suggesting that Rnd3 knockdown in this system promotes neurite outgrowth, which is in contrast with the in vitro data previously described [Citation47]. Moreover, Rnd2 deficient neurons exhibit more and longer processes in the IZ [Citation8,Citation33] while its overexpression results in the disappearance of neurites and in the rounding of IZ cortical neurons [Citation33]. Rnd2 and Rnd3 are thus required for the proper initiation of polarization in nascent pyramidal cortical neurons, an essential step for their concomitant migration.
Axon and dendrite development
The development of the axon and dendrites has been studied extensively in rat primary hippocampal cultures and described as follows [Citation54,Citation55]. After 1.5–3 days in vitro, one of the neurites elongates rapidly without retraction to form the axon and several days later (days 4–7) the remaining neurites grow at a slower rate to form the dendrites. Lastly, the axon develops further and dendrites continue to branch. Using this model, it has been shown that Rnd1 knockdown suppresses axon formation whereas its overexpression induces the generation of multiple axons [Citation56]. Moreover, the loss of the microtubule-destabilizing protein Superior Cervical Ganglion10 (SCG10) blocks the formation of axons after Rnd1 overexpression, indicating that SCG10 acts downstream of Rnd1. Interestingly, Rnd1 interacts directly with SCG10 and enhances its activity (), which suggests that Rnd1 regulates axon extension by modulating microtubule dynamics through SCG10 [Citation56]. Besides this role, Rnd1 also controls growth cone collapse in the developing axons of hippocampal neurons, an essential step for correct targeting of growing axons [Citation57]. The growth cones, located at the tip of the growing axons, respond to attractive and repulsive cues in the local environment to guide the axons towards their specific targets. Among these cues, the semaphorins were originally identified as repulsive axonal guidance molecules and their function is mediated by plexins, which are classified into four subfamilies: Plexin-A1–4, PlexinB1–3, Plexin-C1, and Plexin-D1 [Citation58]. Interestingly, Rnd1 directly interacts with the cytoplasmic domain of Plexin-B1 [Citation59] () and the Plexin-B1-Rnd1 complex stimulates the GTPase activity of R-Ras, inducing growth cone collapse in hippocampal neurons in response to semaphorin 4D (Sema4D) and thus repulsion [Citation57]. Rnd1 also binds to Plexin-A1 [Citation60] and is required for the Sema3A-Plexin-A1–mediated repulsion but this has been demonstrated in COS-7 cells, so not in a neuronal context [Citation61].
In addition to Rnd1, Rnd2 and Rnd3 are also critical for axon development, especially for its outgrowth. Indeed, Rnd3-deficient hippocampal neurons display shorter axons compared to control neurons and this action seems to depend on the RhoA/ROCK signalling [Citation49]. Regarding Rnd2, its action involves the plexin-D1 receptor. Indeed, Rnd2 binds to Plexin-D1 in cortical neurons () and this interaction is required for Sema3E/Plexin-D1-induced inhibition of axon outgrowth in these neurons [Citation62].
Regarding dendritogenesis, although the role of the RhoGTPases and in particular the functions of RhoA, Rac1 and Cdc42 have been extensively characterized [Citation63], only one study has described an involvement of Rnd1 in the regulation of this process [Citation64]. In this study, overexpression of Rnd1 stimulates dendritic growth and branching in cultured hippocampal neurons. Inversely suppression of endogenous Rnd1 expression by RNA interference abolishes dendritic development promoted by KCl, an agent that mimics the depolarizing effects of persistent neuronal activity, or by brain-derived neurotrophic factor (BDNF) [Citation64]. However, in the absence of these treatments, the knockdown of Rnd1 does not affect significantly the dendrites of the cultured neurons suggesting that Rnd1 is involved in signalling pathways of neuronal activity-dependent dendritic development [Citation64]. Moreover, like for neurite outgrowth, the action of Rnd1 seems to be linked to an inhibition of the Rho/ROCK pathway [Citation64]. While a direct role for Rnd2 and Rnd3 in dendritogenesis is missing, it is important to mention that their interacting partners Bacurd1/Kctd13 and Bacurd2/Tnfaip1 play an essential role in dendritic maturation of cerebral cortical neurons [Citation36]. Moreover, the activity of Bacurd1/Kctd13 in synaptic transmission partially works via RhoA regulation [Citation65], thus reinforcing potential Rnd involvement in the pathway.
2.1.4 Rnd functions in spine formation
The final step in the development of a neuron is the formation of spines. These small dendritic protrusions contain receptors and other proteins necessary for synaptic transmission, and thus allow communication with appropriate targets into neural circuits.
The three members of the Rnd family have been shown to control spine morphogenesis, at least in vitro. In primary hippocampal neurons, Rnd1 is strongly localized in spines [Citation66]. Ectopical overexpression of Rnd1 promotes the elongation of dendritic spines, whereas its silencing causes an increase in the percentage of headless protrusions accompanied by a reduction in spine number [Citation66]. Knockdown of Rnd2 also reduces spine density in cultured hippocampal neurons [Citation53]. Rnd1 and Rnd2 are thus critical for normal spine formation but the downstream mechanisms remain to be understood. Similarly, suppression of Rnd3 by shRNA constructs reduces dendritic spine density and its overexpression has an opposite effect [Citation67]. Moreover, knockdown of Rnd3 completely blocks BDNF-stimulated spine formation in cultured hippocampal neurons indicating that Rnd3 is crucial for BDNF-dependent spinogenesis [Citation67]. Interestingly, these changes in the number of dendritic spines appear to reflect changes in the number of functional synapses, as the number of miniature excitatory post-synaptic currents (mEPSCs) correlates with changes in spine number. Indeed, expression of Rnd3 increases the frequency of mEPSCs approximately twofold over control, and targeted knockdown of Rnd3 modestly reduces mEPSC frequency, and significantly decreases the effect of BDNF on mEPSC frequency [Citation67]. Finally, Rnd3 appears to regulate spinogenesis via p190RhoGAP regulation of RhoA, as its effect on spines is blocked by expression of a constitutively active mutant of RhoA or suppression of p190RhoGAP [Citation67].
Taken together, these studies demonstrate that the three members of the Rnd family play multiple and crucial functions during neuronal development (). However, future studies are needed to better characterize these functions in vivo and address their roles in different brain regions at specific developmental stages.
2.2 In pathological conditions
As a consequence of their important physiological role in neuronal development, alterations in Rnd protein expression could contribute to the pathogenesis of many brain disorders. Perturbations in Rnd expression have been described in various forms of cortical malformations and other pathological events. However, so far, there is no direct evidence to support a role for these proteins in the aetiology and progression of these diseases.
Upregulation of Rnd2 expression has been found in brains of epileptic patients [Citation68] and in patients with focal cortical dysplasia (FCD), who also suffer from drug-resistance epileptic seizures [Citation69]. The brain of FCD patients exhibits various gross malformations, such as abnormal gyrification, thickened cortex and local areas of subcortical neurons, which can be attributed to altered neuronal generation and migration. Recent genetic studies of FCD patients revealed a decrease in certain microRNAs, one of which targets the proneural transcription factor Neurog2, which is known to control Rnd2 expression in the embryonic cortex [Citation33]. Thus, when the levels of this miRNA (mir-34a) decrease, Neurog2 and its transcriptional target Rnd2 are upregulated [Citation69]. Considering the crucial role that Rnd2 plays in neuronal migration in the cortex, it is possible that variations in Rnd2 levels observed in these patients contribute to the aetiology of cortical malformations.
Abnormal expression levels of Rnd3 are also thought to be involved in cortical malformations, although direct evidence is still missing. Rnd3 is one of the genes upregulated in human brain organoids derived from patients with periventricular heterotopia (PH), a malformation characterized by the accumulation of newborn neurons close to lateral ventricles [Citation70]. These results echo the block in migration observed upon Rnd3 overexpression in the embryonic murine cortex [Citation8]. However, how Rnd3 upregulation might be controlling neuronal stalling is not known. It is possible that Rnd3 regulation of RhoA activity, as shown in neurogenesis [Citation8,Citation35], might be involved. Indeed, Rnd3 acts via inhibiting RhoA in nascent cortical neurons and, since RhoA deletion in mice causes cortical malformations similar to human subcortical band heterotopia [Citation71,Citation72], it is possible that dysregulation of Rnd3 expression might lead to cortical malformations. Further studies are needed to directly assess this hypothesis.
Additional indirect evidence suggesting that Rnd3 might play important roles in human brain disorders comes from the analysis of Rnd3 knockout animals. Indeed, mice lacking Rnd3 die shortly after birth and exhibit neuromotor impairment and neuromuscular alterations, such as the loss of the peroneal nerve and reduction in the number of spinal motor neurons, which can be attributed to abnormal development and maturation of the nervous system [Citation73]. In another model of Rnd3 knockout animals, the genetic deletion of Rnd3 induces an excessive proliferation of the ependymal cells, which form the epithelial lining of the brain ventricles [Citation18]. This effect, which is due to an up-regulation of Notch signalling in these cells, blocks the cerebral aqueducts and leads to the development of hydrocephalus [Citation18]. Moreover, Rnd3 knockout animals exhibit an enlarged and disorganized postnatal SVZ, which is due to increased NSC proliferation [Citation28,Citation30]. Since the SVZ contains neural stem and progenitor cells that produce new neurons over the course of adult life, aberrant SVZ neurogenesis might lead to long-term effect on adult neurogenesis and eventually contribute to mental disorders and/or neurodegenerative neurological disorders.
A fundamental process contributing to neurodegeneration is neuroinflammation, which can lead to neuronal cell death in diseases such as Parkinson’s and Alzheimer’s. Importantly, Rnd3 has been shown to promote the expression of the pro-survival factor BDNF by microglia cells, the immune cell compartment of the nervous system, and to exert in this way a neuroprotective role [Citation74]. Moreover, the expression of another Rnd protein, Rnd1, has been found to be increased in the hippocampus of 2 out of 7 Alzheimer patients, in comparison to healthy controls [Citation75]. Altogether these works indicate a potential role for Rnd proteins in neurodegenerative disorders. However, whether Rnd1 and Rnd3 play an active role in neurodegeneration and contribute to the formation of the pathological features or whether Rnd expression in this context represents a neuroprotective response is currently unknown. Future work should focus on the role of Rnd proteins in neurodegeneration in order to provide more mechanistic insights into these degenerative disorders that affect millions of people worldwide.
3. Rnd proteins in the vascular system
Rnd proteins are expressed in various cells of the vascular bed, such as endothelial cells and smooth muscle cells. As contractility is a key mechanism for the performance of the vascular system, many studies addressed the involvement of the RhoA/ROCK pathway and Rnd proteins in this context. Rnd1 and Rnd3 were described to be involved in physiological conditions, whereas the majority of studies carried out to date on the role of Rnd protein family in vascular diseases mainly focused on Rnd3.
3.1 In physiological conditions
Rnd1 expression was found modulated upon various stimuli in endothelial cells, even if the pathways are not always deciphered. The vascular endothelial growth factor (VEGF) is a key regulator of endothelial cell migration, proliferation, and inflammation, which leads to activation of several signalling cascades. The expression of Rnd1 was found positively regulated by the NFATc1 (nuclear factor of activated T cells) transcription factor in VEGF-stimulated primary-cultured endothelial cells [Citation22]. Silencing of RND1 impaired vascular barrier function, caused RhoA hyperactivation, and further stimulated VEGF-mediated vascular outgrowth from aortic rings [Citation22]. Thus, Rnd1 is one of the downstream targets of VEGF involved in endothelial cell activation. An upregulation of Rnd1 expression was also observed in human microvascular endothelial cells (HMVEC) after interleukin IL-1β treatment [Citation76] and in mouse heart-isolated endothelial cells upon lipopolysaccharride (LPS) stimulation [Citation77] suggesting that Rnd1 may be involved in vascular inflammation.
Rnd1 was found specifically expressed in human saphenous vein smooth muscle [Citation78]. Vascular smooth muscle contractility is primarily regulated by the level of phosphorylation of myosin light chain. Beside RhoA/ROCK playing major roles in the Ca2+ sensitization through the phosphorylation and inhibition of the subunit of MLCP (Myosin light chain phosphatase), Rnd proteins were also implicated in muscle contraction by modification of their expression level. In smooth muscle, it has been showed that Rnd1 inhibits Ca2+ sensitization and interferes with the RhoA-dependent mechanism. Moreover, the sex hormone steroids such as oestradiol, known to induce the contractility of vascular smooth muscles increase the expression of Rnd1 [Citation79], Rnd2 and Rnd3 [Citation80] in muscles and decrease agonist-induced Ca2+ sensitization. Interestingly, large transcriptome studies using DNA array technology and RT-qPCR analyses found Rnd1 [Citation81], Rnd2 and Rnd3 [Citation82,Citation83] upregulated in animal myometrium contractility during pregnancy indicating that they may indeed play an important role via the regulation of RhoA/Ca2+ sensitization.
Recent emerging evidence shows the important physiological role of Rnd1 and Rnd3 in animal heart. Rnd1 was recently shown to participate in the cardiac remodelling in response to mechanical or humoral stress [Citation84]. Rnd1 was indeed discovered as one of the significantly upregulated genes in rat ventricular cardiomyocytes upon dynamic mechanical stretch or phenylephrine stimulation or in mice after pressure overload induced by transverse aortic constriction. Rnd1 overexpression induces cardiomyocyte proliferation and hypertrophy. Thus, Rnd1 is described as a sensor of biomechanical stress in the heart [Citation84]. Using loss-of-function approaches, Rnd3 gene was found indispensable for mouse development and for the intracellular calcium homoeostasis regulation in cardiomyocytes. Indeed, Rnd3-null mice died at the embryonic stage with foetal heart arrhythmias [Citation85]. This phenotype is due to destabilizing ryanodine receptor type 2 channels, contributing to abnormal intracellular Ca2+ release. Rnd3 deficiency mediates the activation of kinase PKA signalling leading to destabilizing ryanodine receptor type 2 by hyperphosphorylation. The same results were obtained with different cell types either after Rnd3 inhibition in neonatal rat cardiomycytes, HEK293T epithelial cells and C2C12 myoblasts or in fibroblasts isolated from Rnd3-/- mice. The regulatory role of Rnd3 is not only important in an embryonic heart, but also in adult. Indeed, the cardiomyocytes isolated from adult Rnd3± mouse hearts display significantly higher calcium transient frequency compared to the control cells, a phenotype that is attenuated by PKA inhibitor. Mechanistically, it has been shown that Rnd3 is a regulator of the beta2-adrenergic receptor (β2AR). Thus, the lack of Rnd3 attenuates the ubiquitinylation of β2AR, resulting in the accumulation of β2AR protein, which promotes the activation of PKA [Citation85].
3.2 In pathological conditions
Several studies suggest a link between Rnd3 and the progression of vascular diseases. Rnd3 can serve as a potential therapeutic tool to control the endothelial barrier integrity under certain inflammatory conditions. It is detected in both the rat mesenteric microcirculation and in cultured HUVEC endothelial cells. The ectopic expression of Rnd3 in endothelial cells challenged with an inflammatory stimulus such as thrombin promotes enhanced barrier function. This effect reduces the thrombin-induced changes in the actin cytoskeleton and prevents cellular retraction. Moreover, Rnd3 delivered in the mesenteric venules isolated from rats following haemorrhagic shock reduces the microvascular permeability [Citation86].
The involvement of Rnd3 in the control of endothelial barrier function adds to the growing list of Rnd3 functions, including Ca2+ homoeostasis in the heart and a potential role in cardioprotection against heart failure. Three studies from the Chang laboratory explored the involvement of Rnd3 in cardiomyopathies. They revealed a significant decrease of Rnd3 mRNA levels in human failing hearts [Citation87]. To investigate the biological function of Rnd3 in heart failure, they established a patient-relevant mouse model, Rnd3± haploinsufficient mice. The Rnd3± haploinsufficient mice are viable and survived into adulthood without detectable abnormalities in heart morphology compared to the wild-type animal under physiological conditions. However, these mice developed heart failure after haemodynamic stress by transverse aortic constriction challenge. This abnormality is associated with an elevated ROCK activity. Treatment with Fasudil, the only clinically approved ROCK inhibitor, attenuates the apoptotic cardiomyopathy observed in Rnd3± mice and improves cardiac functions [Citation88]. The same group further demonstrates that Rnd3 deficiency resulted in an angiogenesis defect in the mouse heart in response to pressure overload. Thus, they demonstrated that Rnd3 acts as a pro-angiogenic factor involved in the cardiac responsive angiogenesis [Citation87]. In a more recent study, Rnd3 was found to protect the heart from the inflammatory response during the cardiac healing process after myocardial infarction (MI) [Citation89]. Indeed, Rnd3 expression is rapidly increased during the acute inflammatory phase in animal heart after cardiac ischaemia. The use of cardiomyocyte-specific Rnd3 heterozygous mice revealed an excessive inflammatory response with deleterious cardiac function after MI. Mechanistically, Rnd3 negatively regulates the proinflammatory gene expression programme by directly blocking the key inflammatory regulator NF-κB nuclear translocation. Rnd3 is able to interact with the NF-κB components p50 and p65 in the cytosol, interrupting p65/p50 heterodimerization (). Manipulation of Rnd3 could serve as a potential therapeutic approach as the authors found that patients with a higher expression level of Rnd3 exhibit a better prognosis in cardiac function recovery [Citation89].
Primary studies showed links between Rnd3 and the progression of cardiovascular diseases. The stress hormone urocortin is known to confer significant protection against myocardial ischaemia/reperfusion (I/R) injury. Microarray analysis showed an upregulation of Rnd3 in rat infused with urocortin before the onset of reperfusion [Citation90]. However, the involvement of Rnd3 in urocortin-mediated cardioprotection remains to be determined. Interestingly, studies have evidenced that urocortin is also key at the end of the pregnancy period as low level of urocortin is required to provoke an increase in the contractility of the uterus [Citation91]. Whether a decrease of Rnd3 expression is also required in this context has not been explored. But, for the maintenance of successful pregnancy, fusion of placental villous cytotrophoblasts with the overlying syncytiotrophoblast is essential, and disturbances in this process have been implicated in pathological conditions such as pre-eclampsia (PE). Rnd3 expression was found to promote the fusion of BeWO choriocarcinoma cells, a model of villous cytotrophoblast fusion [Citation92]. Moreover, Johnson & collaborators defined the UTR-3 RND3 SNP (rs115015150) as a risk factor for the development of maternal cardiovascular disease and pre-eclampsia [Citation93]. PE is a pregnancy-specific syndrome, characterized by maternal new-onset hypertension and proteinuria by an abnormal placental function due to trophoblast invasion and failure of spiral artery remodelling. Interestingly, Rnd3 expression levels were found elevated in PE placental tissues [Citation94]. This increase of Rnd3 expression was attributed to the decrease of two Rnd3-targeting long non-coding (lnc) RNAs, TUG1 and HOXA11-AS, levels in PE [Citation94,Citation95]. In addition, Li and collaborators determined that the lncRNA HOTTIP expression is lower in PE placenta than in normal tissue, causing also the overexpression of Rnd3 [Citation96]. Functional in vitro studies further determined that an upregulation of Rnd3 inhibits trophoblast cell proliferation and migration [Citation94–96]. Thus, Rnd3 appears as a key protein involved in the progression of PE. The expression of Rnd3 in PE may also be regulated by miRNAs. However, in sharp contrast, Rnd3 expression was described as a target of miR-182-5p found highly expressed in placenta of PE patients [Citation97]. The overexpression of miR-182-5p induces the invasion and migration of trophoblasts by regulation of Rnd3 expression. Indeed, Rnd3 expression reversed the effects exerted by the overexpression of miR-182-5p [Citation97]. Overall, these results suggest that Rnd3 is an important factor in PE disease.
4. Rnd proteins in cancers
Deregulation of Rnd proteins is associated with cancer development and progression. The role of Rnd1 and Rnd3 in oncogenesis has been the subject of numerous studies, while Rnd2 function has been less explored. As the role of Rnd1 and Rnd3 in cancers has been extensively discussed elsewhere, we refer the reader to two recent reviews [Citation20,Citation98]. Here, we summarize the degree of altered expression and the roles of all three Rnd family members in human malignancies. We made use of the large number of cancer genome-sequencing data to compare Rnd1, Rnd2 and Rnd3 involvement in cancers.
4.1 Rnd genetic alteration in cancers
Although a few cancer genes, such as RAS, are mutated in a high proportion of tumours of a given type (>20%), most are mutated at intermediate frequencies (2–20%). RHOA and RAC1 are part of this latter category. Genetic alterations, including gene amplification, deletion or fusion, and missense or truncating mutations, for RND1, RND2 and RND3 genes are rather rare events in tumours. Indeed, data from the Cancer Genome Atlas (TCGA) PanCancer Atlas (querying 10,967 human samples in 32 studies) extracted from the CBioPortal database show genetic alteration in 0.9% of the samples for each RND1, RND2 or RND3 gene (). As most of these alterations are mutually exclusive, this raises to about 2.7% for all three genes (290 of queried samples). Gene fusions were found in only two samples in this cohort: DDX23-RND1 fusion in hepatocellular carcinoma and MRC2-RND2 fusion in glioblastoma multiforme. Locus deletion events were reported to be responsible for down-regulation of RND expression in few tumour types. The 12q12-q13 region, which comprises the RND1 gene, is deleted in several cancers, such as adenoid cystic carcinoma [Citation99], pancreas cancer [Citation100] and breast cancer [Citation21]. In most of the cases these deletions are heterozygous, requiring additional mechanisms to totally inactivate RND1 expression in tumours. Again, less is known about RND2. Of interest, RND2 human gene is the centromeric neighbour of the breast-ovarian cancer susceptibility gene BRCA1 on chromosome 17. One study describes the first case of a patient, with no strong family history of the disease, who developed early-onset bilateral breast cancer with a de novo complete BRCA1 gene deletion in the germinal line [Citation101]. Even if the large genomic rearrangement detected removed the RND2 gene, the involvement of Rnd2 in the onset of the disease remains to be studied. In addition, data from the TCGA PanCancer Atlas revealed that deep deletion of RND2 gene are also found in adrenocortical carcinomas, prostate adenocarcinoma, uveal melanoma and ovarian epithelial tumours, even if no study so far focused on their implication (). Even if deep deletions for the RND3 gene were also found in global approaches (), in prostate adenocarcinoma, so far, no study confirmed these events. Similarly, amplifications that may lead to up-regulation of RND expression are detected in few cancer types (). In non-small cell lung cancer, overexpression of RND3 expression was significantly associated with the patients’ smoking history and DNA copy number changes [Citation102].
Figure 3. Genetic alterations of RND genes in cancers. Data extracted from the CBioPortal database (https://www.cbioportal.org/). Data from TCGA PanCancer Altlas studies pooling 32 studies and 10,967 samples were analysed for RND1, RND2 and RND3 genes. (a) Shown is the OncoPrint. Note that only tumours with RND alterations (n = 290) are present on the schema. (b) Shown is the Cancer Types Summary. The y axis shows the frequency of alterations, whereas the x axis indicates tumour types
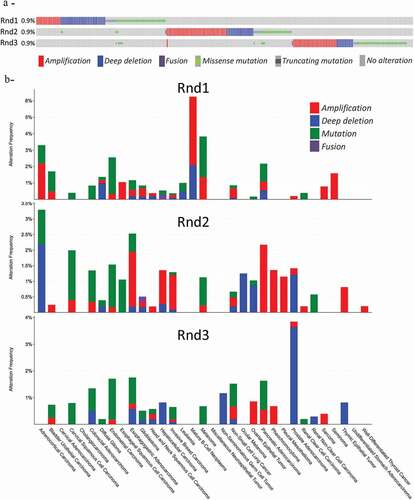
Somatic mutations, either missense or truncating mutations, are found at a frequency of 0.5%, 0.3% and 0.4% for respectively RND1, RND2 and RND3 genes in this dataset. Missense mutations (in green on ) were found all along the coding sequence, without hot spots. The impact of most of these mutations on their respective proteins is currently unknown. Available data concern only RND1; three missense mutations, which replaced evolutionarily conserved amino acids and found in a cohort of 96 breast cancers, were characterized [Citation21]. Unlike wild-type Rnd1, G70R, E98D, and F180C mutants expressed in MCF-10A cells are not able to inhibit tumour cell growth. Moreover, they are affected in their subcellular localization and in their ability to bind to plexin B1 to inhibit the downstream Raf-Erk cascade [Citation21]. Thus, these mutations participate to repress Rnd1 properties in mammary tumour cells. Even if RND1 point mutations are rare in tumours, this study suggests that in combination with gene deletion or epigenetic silencing they may inactivate RND1 in human breast cancer and favour breast tumour initiation and progression through activation of Ras-MAPK signalling.
4.2 Alteration of Rnd expression in cancers
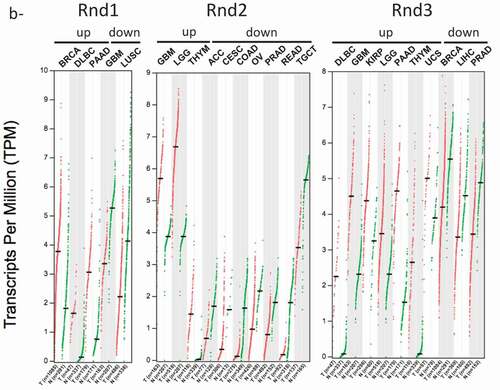
The most frequent alterations found in tumours remain deregulation of mRNA or protein expression. Thanks to the fact that Rnd is deficient in GTPase activity, compared to other Rho members, an alteration of their expression reflects more an alteration of their activity. We used the GEPIA (Gene Expression Profiling Interactive Analysis) web server, a valuable and highly cited resource, to analyse RND1, RND2 and RND3 gene expression in tumour and normal samples [Citation103]. This publicly accessible web interface (http://gepia2.cancer-pku.cn/) compiles RNA sequencing data from the TCGA and the GTEx projects. presents the expression of RND transcripts in 32 different types of cancer and shows that, depending on cancer types, RND expression is either down- or up-regulated.
Figure 4. Expression data of RND genes in cancers. Data extracted from the GEPIA2 (Gene expression Profiling Interative Analysis) database (http://gepia2.cancer-pku.cn/). Data from TCGA PanCancer Altlas studies pooling 32 studies and 10,967 samples were analysed for RND1, RND2 and RND3 genes. (a) Bar plot indicating the gene expression profile across all tumour samples (red bars) and paired normal tissues. The height of bar represents the median expression of certain tumour type or normal tissue (black bars). Tumours with a significant increase or decrease of Rnd expression are marked by an asteric (*) or a dièze (#), respectively. (b) Dot plot indicating the gene expression profile across indicated tumour samples (red dots) and paired normal tissues (green dots). Each dot represents expression of samples. Only tumour types with either up- or down-regulated Rnd gene expression are indicated. Tumour types: ACC, Adrenocortical carcinoma; BLCA, Bladder Urothelial Carcinoma; BRCA, Breast invasive carcinoma; CESC, Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, Cholangio carcinoma; COAD, Colon adenocarcinoma; DLBC, Lymphoid Neoplasm Diffuse, Large B-cell Lymphoma; ESCA, Oesophageal carcinoma; GBM, Glioblastoma multiforme; HNSC, Head and Neck squamous cell carcinoma; KICH, Kidney Chromophobe; KIRC, Kidney renal clear cell carcinoma; KIRP, Kidney renal papillary cell carcinoma; LAML, Acute Myeloid Leukaemia; LGG, Brain Lower Grade Glioma; LIHC, Liver hepatocellular carcinoma; LUAD, Lung adenocarcinoma; LUSC, Lung squamous cell carcinoma; MESO, Mesothelioma OV Ovarian serous cystadenocarcinoma; PAAD, Pancreatic adenocarcinoma; PCPG, Pheochromocytoma and Paraganglioma; PRAD, Prostate adenocarcinoma; READ, Rectum adenocarcinoma; SARC, Sarcoma; SKCM, Skin Cutaneous Melanoma; STAD, Stomach adenocarcinoma; TGCT, Testicular Germ Cell Tumours; THCA, Thyroid carcinoma; THYM, Thymoma; UCEC, Uterine Corpus Endometrial Carcinoma; UCS, Uterine Carcinosarcoma; UVM, Uveal Melanoma
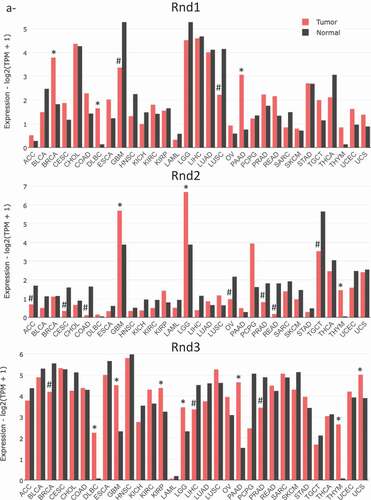
Using this global approach, RND1 is found significantly overexpressed in breast invasive carcinoma, diffuse large B-cell lymphoma and pancreatic adenocarcinoma. In addition, one study described the upregulation of RND1 transcripts in oesophageal squamous cell carcinoma, where Rnd1 expression promotes the growth and the migration of cancer cells [Citation104]. In another study on breast cancer, Rnd1 was found to behave as a tumour suppressor, in contrast with the TCGA data, highlighting the heterogeneity of breast carcinoma. Indeed, in mammary epithelium, RND1 loss was shown to activate Ras and the downstream Raf-Erk pathway, leading to uncontrolled proliferation and disruption of epithelial adhesion [Citation21]. In mice, inactivation of Rnd1 in mammary epithelial cells induced highly undifferentiated and invasive tumours. Thus, genetic and epigenetic inactivation of RND1 was proposed to underlie activation of the Ras-ERK pathway in a subset of breast cancers and participate in tumour initiation and progression [Citation21]. RND1 expression was also found significantly down-regulated in glioblastoma and lung squamous cell carcinoma (). Interestingly, RND1 downregulation was found to favour migration of glioblastoma stem-like cells derived from human samples [Citation105]. In patients, low levels of RND1 were shown to correlate with a decreased overall survival. Moreover, an RND1low signature of six genes was proposed as an independent prognostic factor in glioblastoma [Citation105]. Low expression of RND1 was also associated with a bad prognosis for hepatocellular carcinoma (HCC) patients. Indeed, two studies described RND1 as a tumour suppressor in liver cancers [Citation106,Citation107]. RND1 expression was found decreasing as HCC progressed, with a lower expression in advanced stages. In vitro, RND1 knockdown enhanced the malignant characteristics of HCC (Huh7 and PLC/PRF/5) cells, such as proliferation, invasion, and chemoresistance [Citation106]. Thus, RND1 was proposed as a novel biomarker for predicting prognosis of HCC patients.
Finally, recent data suggest that RND1 is involved in response to anticancer agents. RND1 expression was found rapidly induced upon treatment with the selective topoisomerase I inhibitor, camptothecin, in various cancer cells, including human osteosarcoma (U2OS), glioblastoma (U87), and colon carcinoma (HCT116) cells, and murine melanoma (B16F10) [Citation108]. As shown for osteosarcoma cells, RND1 expression protects them against drug-induced apoptosis, and thus, favours cellular resistance to topoisomerase I inhibitors [Citation108]. This response to cancer therapeutics is not restricted to camptothecin, as RND1 expression was also linked to cisplatin and sorafenib resistance in HCC patients [Citation106,Citation107]. Moreover, by analysing the correlation between RND1 expression and the cellular response to 20 anticancer agents with the CCLE database (https://portals.broadinstitute.org/ccle), Mouly and collaborators [Citation20] suggest a role for RND1 expression in the sensitivity of lung tumours to EGFR signalling pathway inhibitors.
RNA sequencing data revealed that RND2 expression is up-regulated in brain tumours, either low-grade (brain low-grade glioma) or high-grade (glioblastoma) and in thymoma (). On the contrary, it is down-regulated in adrenocortical carcinoma, and in colon and rectum adenocarcinomas. Moreover, in men, RND2 expression, which is normally high in normal testis tissue, is found strongly decreased in testicular germ cell tumours and in prostate adenocarcinoma. In women, it is also down-regulated in cervical and ovarian carcinomas. So far, no study reported the impact of these deregulations of RND2 on tumour development and progression. Thus, these data obtained from global approaches remain to be confirmed by further studies.
From GEPIA data, RND3 expression is significantly overexpressed in seven types of tumours including diffuse large B-cell lymphoma, low- and high-grade brain tumours (i.e. low-grade glioma and glioblastoma), kidney renal papillary cell carcinoma, pancreatic adenocarcinoma, thymoma and uterine carcinosarcoma. Most of these up-regulations remain to be confirmed by molecular and cellular studies. RND3 expression is down-regulated in breast invasive carcinoma, liver hepatocellular carcinoma and prostate adenocarcinoma. Indeed as we described earlier, depending on the tumour type, it may behave either as a tumour suppressor or as a tumour promoter [Citation98]. These data were confirmed for hepatocellular carcinoma [Citation109–111], and prostate adenocarcinoma [Citation112,Citation113], where RND3 down-regulation is associated with poor prognosis and favours malignant characteristics of tumour cells. Thus, RND3 loss was associated with liver and prostate cancer cells proliferation, invasion, and chemoresistance [Citation109,Citation110,Citation111,Citation113,Citation114]. Loss of Rnd3 was even found to favour epithelio-mesenchymal transition (EMT) by activating the Raf/MEK/ERK cascade and/or the ZEB2 EMT factor in hepatocellular carcinoma cells [Citation107,Citation109]. On the contrary, RND3 up-regulation was described in gastric adenocarcinoma [Citation23,Citation115,Citation116], non-small cell lung cancer [Citation102,Citation117,Citation118,Citation119] and melanoma [Citation120,Citation121,Citation122], by both cellular and clinical approaches. In these malignancies, high RND3 expression positively correlated with cancer cell aggressiveness and tumour progression. Concerning high-grade brain tumours, several studies reported RND3 status in human glioblastoma multiform (GBM). RND3 expression was described down-regulated by some [Citation123], but found up-regulated by others [Citation124,Citation125], probably reflecting the heterogeneity and the complexity of these tumours. Early work performed in U87 cells reported that RND3 up-regulation inhibited glioma cell proliferation and induced apoptosis [Citation122]. More recently, Liu et al. also described RND3 as a tumour suppressor in GBM. Its expression correlated positively with GBM patient survival, and is inversely associated with tumour size and tumour cell proliferation. They found that forced expression of Rnd3 inhibited GBM cell proliferation in vitro and tumour growth in mice by inhibiting Notch signalling activity [Citation123]. In addition, the same group found downregulation of Rnd3 transcript and protein in human GBM samples, and this decrease correlated positively with E-Cadherin expression and negatively with the EMT marker, SNAIL1. They, thus, reported that RND3 loss favours migration and invasion of GBM cells [Citation126]. In addition, by acting on the NF-κB signalling pathway, RND3 was recently involved in the apoptosis of GBM cells [Citation127]. In striking contrast, at least two other studies described RND3 as a tumour promoter in glial cells [Citation124,Citation125]. In these studies, high Rnd3 levels correlated with poor prognosis and an increase in GBM cell proliferation and invasion.
Altogether, such data revealed that Rnd proteins are often altered in tumours and can act as both tumour suppressors and tumour promoting factors depending on the context. Whether the same tumour may jointly exhibit alteration of more than one Rnd family members remains to be studied. From the literature, it is clear that both RND1 and RND3 expressions are down-regulated in hepatocellular carcinoma supporting the rationale that Rnd1 and Rnd3 may act as tumour suppressor genes in liver cancer. Similarly, both RND1 and RND3 expressions are altered in oesophageal squamous cell carcinoma. But, whereas Rnd1 is up-regulated in these tumours and promotes the growth and migration of cancer cells [Citation104], Rnd3 is low and exerts tumour-suppressing effects [Citation128,Citation129]. Finally, in glioblastoma, besides the controversy for RND3, it looks like all three members are deregulated in those high-grade brain tumours (). Further studies are required to better understand the specific and redundant roles of Rnd proteins in malignancies.
5. Conclusions and future directions
Rnd proteins form a distinct group of the Rho family of small GTP-binding proteins with unusual biochemical properties. It consists of three proteins that play redundant functions in the regulation of the actin cytoskeleton and cell adhesion, but also exhibit unique and non-redundant roles in different tissues. Indeed, alterations in one of the three Rnds generate different pathologies. They were first described as endogenous antagonists of the RhoA/ROCK pathway; hence, their ability to inhibit the formation of contractile actomyosin cables. Thus, Rnd proteins regulate the actin cytoskeleton organization in several tissues. Moreover, Rnd proteins are now shown to act through different pathways increasing their mode of action. In the brain, they control multiple steps of neuronal development from proliferation to integration whereas in smooth muscles, they modulate contractility. To provide a better understanding of how these proteins regulate these functions, it will be important to identify novel partners for Rnd proteins, deciphering new pathways for Rnd1, Rnd2 and/or Rnd3. Indeed, proteins in general do not act as single units but perform cellular functions as components of complexes. The establishment of Rnd3-null mice allowed researchers to better understand the role of this member in cardiovascular diseases and neuronal functions. However, future studies should focus on the role of the two others Rnds in particular via the analysis of Rnd1 or Rnd2 knockout mice and conditional mutants. As a consequence of their important role in different cellular functions such as proliferation, migration and invasion, aberrant Rnd expression has been frequently observed in different types of cancer. Additional investigations on the molecular mechanisms are necessary to clarify the involvement of Rnd proteins in tumours. Interestingly, Rnd1 and Rnd3 are implicated in the chemotherapeutic resistance of cancer cells; so new research that could target Rnds is needed in order to expand our knowledge of anticancer agents.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Boureux A, Vignal E, Faure S, et al. Evolution of the Rho family of ras-like GTPases in eukaryotes. Mol Biol Evol. 2007;24:203–216.
- Foster R, Hu KQ, Lu Y, et al. Identification of a novel human Rho protein with unusual properties: gTPase deficiency and in vivo farnesylation. Mol Cell Biol. 1996;16:2689–2699.
- Nobes CD, Lauritzen I, Mattei MG, et al. A new member of the Rho family, Rnd1, promotes disassembly of actin filament structures and loss of cell adhesion. J Cell Biol. 1998;141:187–197.
- Stiegler AL, Boggon TJ. p190RhoGAP proteins contain pseudoGTPase domains. Nat Commun. 2017;8:506.
- Guasch RM, Scambler P, Jones GE, et al. RhoE regulates actin cytoskeleton organization and cell migration. Mol Cell Bio. 1998;18:4761–4771.
- Roberts PJ, Mitin N, Keller PJ, et al. Rho Family GTPase modification and dependence on CAAX motif-signaled posttranslational modification. J Biol Chem. 2008;283:25150–25163.
- Tanaka H, Fujita H, Katoh H, et al. Vps4-A (vacuolar protein sorting 4-A) is a binding partner for a novel Rho family GTPase, Rnd2. Biochem J. 2002;365:349–353.
- Pacary E, Heng J, Azzarelli R, et al. Proneural transcription factors regulate different steps of cortical neuron migration through Rnd-mediated inhibition of RhoA signaling. Neuron. 2011;69:1069–1084.
- Garg R, Koo CY, Infante E, et al. Rnd3 interacts with TAO kinases and contributes to mitotic cell rounding and spindle positioning. J Cell Sci. 2020;133:jcs235895.
- Madigan JP, Bodemann BO, Brady DC, et al. Regulation of Rnd3 localization and function by protein kinase C alpha-mediated phosphorylation. Biochem J. 2009;424:153–161.
- Riento K, Totty N, Villalonga P, et al. RhoE function is regulated by ROCK I-mediated phosphorylation. Embo J. 2005;24:1170–1180.
- Riou P, Kjaer S, Garg R, et al. 14-3-3 proteins interact with a hybrid prenyl-phosphorylation motif to inhibit G proteins. Cell. 2013;153:640–653.
- Hidalgo-Carcedo C, Hooper S, Chaudhry SI, et al. Collective cell migration requires suppression of actomyosin at cell-cell contacts mediated by DDR1 and the cell polarity regulators Par3 and Par6. Nat Cell Biol. 2011;13:49–58.
- Park D, Wershof E, Boeing S, et al. Extracellular matrix anisotropy is determined by TFAP2C-dependent regulation of cell collisions. Nat Mater. 2020;19:227–238.
- Riento K, Guasch RM, Garg R, et al. RhoE binds to ROCK I and inhibits downstream signaling. Mol Cell Biol. 2003;23:4219–4229.
- Oinuma I, Kawada K, Tsukagoshi K, et al. Rnd1 and Rnd3 targeting to lipid raft is required for p190 RhoGAP activation. Mol Biol Cell. 2012;23:1593–1604.
- Vayssiere B, Zalcman G, Mahe Y, et al. Interaction of the Grb7 adapter protein with Rnd1, a new member of the Rho family. FEBS Lett. 2000;467: 91–96
- Lin X, Liu B, Yang X, et al. Genetic deletion of Rnd3 results in aqueductal stenosis leading to hydrocephalus through up-regulation of Notch signaling. Proc Natl Acad Sci USA. 2013;110:8236–8241.
- Jie W, Andrade KC, Lin X, et al. Pathophysiological Functions of Rnd3/RhoE. Compr Physiol. 2015;6:169–186.
- Mouly L, Gilhodes J, Lemarie A, et al. The RND1 Small GTPase: main functions and emerging role in oncogenesis. Int J Mol Sci. 2019';20:3612.
- Okada T, Sinha S, Esposito I, et al. The Rho GTPase Rnd1 suppresses mammary tumorigenesis and EMT by restraining Ras-MAPK signalling. Nat Cell Biol. 2015;17:81–94.
- Suehiro J, Kanki Y, Makihara C, et al. Genome-wide approaches reveal functional vascular endothelial growth factor (VEGF)-inducible nuclear factor of activated T cells (NFAT) c1 binding to angiogenesis-related genes in the endothelium. J Biol Chem. 2014;289:29044–29059.
- Zhou J, Li K, Gu Y, et al. Transcriptional up-regulation of RhoE by hypoxia-inducible factor (HIF)-1 promotes epithelial to mesenchymal transition of gastric cancer cells during hypoxia. Biochem Biophys Res Commun. 2011;415:348–354.
- Goh LL, Manser E. The GTPase-deficient Rnd proteins are stabilized by their effectors. J Biol Chem. 2012;287:31311–31320.
- Lonjedo M, Poch E, Mocholi E, et al. The Rho family member RhoE interacts with Skp2 and is degraded at the proteasome during cell cycle progression. J Biol Chem. 2013;288:30872–30882.
- Goh LL, Manser E. The RhoA GEF Syx is a target of Rnd3 and regulated via a Raf1-like ubiquitin-related domain. PloS One. 2010;5:e12409.
- Pacary E, Azzarelli R, Guillemot F. Rnd3 coordinates early steps of cortical neurogenesis through actin-dependent and -independent mechanisms. Nat Commun. 2013;4:1635.
- Ballester-Lurbe B, Gonzalez-Granero S, Mocholi E, et al. RhoE deficiency alters postnatal subventricular zone development and the number of calbindin-expressing neurons in the olfactory bulb of mouse. Brain Struct Funct. 2015;220:3113–3130.
- Lim DA, Alvarez-Buylla A. The adult ventricular-subventricular zone (V-SVZ) and olfactory bulb (OB) neurogenesis. Cold Spring Harbor Perspect Biol. 2016;8:a018820.
- Dong H, Lin X, Li Y, et al. Genetic deletion of Rnd3 in neural stem cells promotes proliferation via upregulation of notch signaling. Oncotarget. 2017;8:91112–91122.
- Azzarelli R, Guillemot F, Pacary E. Function and regulation of Rnd proteins in cortical projection neuron migration. Front Neurosci. 2015;9:19.
- Gladwyn-Ng IE, Li SS, Qu Z, et al. Bacurd2 is a novel interacting partner to Rnd2 which controls radial migration within the developing mammalian cerebral cortex. Neural Dev. 2015;10:9.
- Heng JI, Nguyen L, Castro DS, et al. Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature. 2008;455:114–118.
- Nakamura K, Yamashita Y, Tamamaki N, et al. In vivo function of Rnd2 in the development of neocortical pyramidal neurons. Neurosci Res. 2006;54:149–153.
- Azzarelli R, Pacary E, Garg R, et al. An antagonistic interaction between PlexinB2 and Rnd3 controls RhoA activity and cortical neuron migration. Nat Commun. 2014;5:3405.
- Gladwyn-Ng I, Huang L, Ngo L, et al. Bacurd1/Kctd13 and Bacurd2/Tnfaip1 are interacting partners to Rnd proteins which influence the long-term positioning and dendritic maturation of cerebral cortical neurons. Neural Dev. 2016;11:7.
- Alfano C, Viola L, Heng JI, et al. COUP-TFI promotes radial migration and proper morphology of callosal projection neurons by repressing Rnd2 expression. Development. 2011;138:4685–4697.
- Clement O, Hemming IA, Gladwyn-Ng IE, et al. Rp58 and p27(kip1) coordinate cell cycle exit and neuronal migration within the embryonic mouse cerebral cortex. Neural Dev. 2017;12:8.
- Heng JI, Qu Z, Ohtaka-Maruyama C, et al. The zinc finger transcription factor RP58 negatively regulates Rnd2 for the control of neuronal migration during cerebral cortical development. Cereb Cortex. 2015;25:806–816.
- Ohtaka-Maruyama C, Hirai S, Miwa A, et al. RP58 regulates the multipolar-bipolar transition of newborn neurons in the developing cerebral cortex. Cell Rep. 2013;3:458–471.
- Paul V, Tonchev AB, Henningfeld KA, et al. Scratch2 modulates neurogenesis and cell migration through antagonism of bHLH proteins in the developing neocortex. Cereb Cortex. 2014;24:754–772.
- Peng YR, James RE, Yan W, et al. Binary fate choice between closely related interneuronal types is determined by a Fezf1-dependent postmitotic transcriptional switch. Neuron. 2020;105(464–474):e466.
- Greene LA, Tischler AS. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci USA. 1976;73:2424–2428.
- Rydel RE, Greene LA. Acidic and basic fibroblast growth factors promote stable neurite outgrowth and neuronal differentiation in cultures of PC12 cells. J Neurosci: The Official Journal of the Society for Neuroscience. 1987;7:3639–3653.
- Aoki J, Katoh H, Mori K, et al. Rnd1, a novel rho family GTPase, induces the formation of neuritic processes in PC12 cells. Biochem Biophys Res Commun. 2000;278:604–608.
- de Souza LE, Moura Costa MD, Bilek ES, et al. STI1 antagonizes cytoskeleton collapse mediated by small GTPase Rnd1 and regulates neurite growth. Exp Cell Res. 2014;324:84–91.
- Talens-Visconti R, Peris B, Guerri C, et al. RhoE stimulates neurite-like outgrowth in PC12 cells through inhibition of the RhoA/ROCK-I signalling. J Neurochem. 2010;112:1074–1087.
- Harada A, Katoh H, Negishi M. Direct interaction of Rnd1 with FRS2 beta regulates Rnd1-induced down-regulation of RhoA activity and is involved in fibroblast growth factor-induced neurite outgrowth in PC12 cells. J Biol Chem. 2005;280:18418–18424.
- Peris B, Gonzalez-Granero S, Ballester-Lurbe B, et al. Neuronal polarization is impaired in mice lacking RhoE expression. J Neurochem. 2012;121:903–914.
- Tanaka H, Katoh H, Negishi M. Pragmin, a novel effector of Rnd2 GTPase, stimulates RhoA activity. J Biol Chem. 2006;281:10355–10364.
- Fujita H, Katoh H, Ishikawa Y, et al. Rapostlin is a novel effector of Rnd2 GTPase inducing neurite branching. J Biol Chem. 2002;277:45428–45434.
- Kamioka Y, Fukuhara S, Sawa H, et al. A novel dynamin-associating molecule, formin-binding protein 17, induces tubular membrane invaginations and participates in endocytosis. J Biol Chem. 2004;279:40091–40099.
- Wakita Y, Kakimoto T, Katoh H, et al. The F-BAR protein Rapostlin regulates dendritic spine formation in hippocampal neurons. J Biol Chem. 2011;286:32672–32683.
- Craig AM, Banker G. Neuronal polarity. Annu Rev Neurosci. 1994;17:267–310.
- Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. J Neurosci: The Official Journal of the Society for Neuroscience. 1988;8:1454–1468.
- Li YH, Ghavampur S, Bondallaz P, et al. Rnd1 regulates axon extension by enhancing the microtubule destabilizing activity of SCG10. J Biol Chem. 2009;284:363–371.
- Oinuma I, Ishikawa Y, Katoh H, et al. The Semaphorin 4D receptor Plexin-B1 is a GTPase activating protein for R-Ras. Science. 2004;305:862–865.
- Kruger RP, Aurandt J, Guan KL. Semaphorins command cells to move. Nat Rev Mol Cell Biol. 2005;6:789–800.
- Oinuma I, Katoh H, Harada A, et al. Direct interaction of Rnd1 with Plexin-B1 regulates PDZ-RhoGEF-mediated Rho activation by Plexin-B1 and induces cell contraction in COS-7 cells. J Biol Chem. 2003;278:25671–25677.
- Rohm B, Rahim B, Kleiber B, et al. The semaphorin 3A receptor may directly regulate the activity of small GTPases. FEBS Lett. 2000;486:68–72.
- Zanata SM, Hovatta I, Rohm B, et al. Antagonistic effects of Rnd1 and RhoD GTPases regulate receptor activity in Semaphorin 3A-induced cytoskeletal collapse. J Neurosci. 2002;22:471–477.
- Uesugi K, Oinuma I, Katoh H, et al. Different requirement for Rnd GTPases of R-Ras GAP activity of Plexin-C1 and Plexin-D1. J Biol Chem. 2009;284:6743–6751.
- Govek EE, Newey SE, Van Aelst L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19:1–49.
- Ishikawa Y, Katoh H, Negishi M. Small GTPase Rnd1 is involved in neuronal activity-dependent dendritic development in hippocampal neurons. Neurosci Lett. 2006;400:218–223.
- Escamilla CO, Filonova I, Walker AK, et al. Kctd13 deletion reduces synaptic transmission via increased RhoA. Nature. 2017;551:227–231.
- Ishikawa Y, Katoh H, Negishi M. A role of Rnd1 GTPase in dendritic spine formation in hippocampal neurons. Journal Neurosci: The Official Journal of the Society for Neuroscience. 2003;23:11065–11072.
- Lesiak A, Pelz C, Ando H, et al. A genome-wide screen of CREB occupancy identifies the RhoA inhibitors Par6C and Rnd3 as regulators of BDNF-induced synaptogenesis. PloS One. 2013;8:e64658.
- Yang X, Yang B, Li H, et al. Upregulation of Rho7 in the temporal lobe tissue of humans with intractable epilepsy. Mol Med Rep. 2017;16:9613–9619.
- Avansini SH, Torres FR, Vieira AS, et al. Dysregulation of NEUROG2 plays a key role in focal cortical dysplasia. Ann Neurol. 2018;83:623–635.
- Klaus J, Kanton S, Kyrousi C, et al. Altered neuronal migratory trajectories in human cerebral organoids derived from individuals with neuronal heterotopia. Nat Med. 2019;25:561–568.
- Cappello S. Small Rho-GTPases and cortical malformations: fine-tuning the cytoskeleton stability. Small GTPases. 2013;4:51–56.
- Cappello S, Bohringer CR, Bergami M, et al. A radial glia-specific role of RhoA in double cortex formation. Neuron. 2012;73:911–924.
- Mocholi E, Ballester-Lurbe B, Arque G, et al. RhoE deficiency produces postnatal lethality, profound motor deficits and neurodevelopmental delay in mice. PloS One. 2011;6:e19236.
- Neubrand VE, Forte-Lago I, Caro M, et al. The atypical RhoGTPase RhoE/Rnd3 is a key molecule to acquire a neuroprotective phenotype in microglia. J Neuroinflamm. 2018;15:343.
- Zhang C, Lu J, Liu B, et al. Primate-specific miR-603 is implicated in the risk and pathogenesis of Alzheimer’s disease. Aging (Albany NY). 2016;8:272–290.
- Warton K, Foster NC, Gold WA, et al. A novel gene family induced by acute inflammation in endothelial cells. Gene. 2004;342:85–95.
- Jambusaria A, Hong Z, Zhang L, et al. Endothelial heterogeneity across distinct vascular beds during homeostasis and inflammation. Elife. 2020;9:e51413.
- Cario-Toumaniantz C, Evellin S, Maury S, et al. Role of Rho kinase signalling in healthy and varicose human saphenous veins. Br J Pharmacol. 2002;137:205–212.
- Loirand G, Cario-Toumaniantz C, Chardin P, et al. The Rho-related protein Rnd1 inhibits Ca2+ sensitization of rat smooth muscle. J Physiol. 1999;516(Pt 3):825–834.
- Shimomura A, Ohama T, Hori M, et al. 17Beta-estradiol induces gastrointestinal motility disorder by decreasing CPI-17 phosphorylation via changes in rho-family G-protein Rnd expression in small intestine. J Vet Med Sci. 2009;71:1591–1597.
- Kim YS, Kim B, Karaki H, et al. Up-regulation of Rnd1 during pregnancy serves as a negative-feedback control for Ca2+ sensitization of contractile elements in rat myometrium. Biochem Biophys Res Commun. 2003;311:972–978.
- Cario-Toumaniantz C, Reillaudoux G, Sauzeau V, et al. Modulation of RhoA-Rho kinase-mediated Ca2+ sensitization of rabbit myometrium during pregnancy - role of Rnd3. J Physiol. 2003;552:403–413.
- Lartey J, Gampel A, Pawade J, et al. Expression of RND proteins in human myometrium. Biol Reprod. 2006;75:452–461.
- Kluge A, Rangrez AY, Kilian LS, et al. Rho-family GTPase 1 (Rnd1) is a biomechanical stress-sensitive activator of cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2019;129:130–143.
- Yang X, Wang T, Lin X, et al. Genetic deletion of rnd3/rhoe results in mouse heart calcium leakage through upregulation of protein Kinase A signaling. Circ Res. 2015;116:1.
- Breslin JW, Daines DA, Doggett TM, et al. Rnd3 as a Novel Target to Ameliorate Microvascular Leakage. J Am Heart Assoc. 2016;5:e003336.
- Yue X, Lin X, Yang T, et al. Rnd3/RhoE modulates hypoxia-inducible Factor 1alpha/vascular endothelial growth factor signaling by stabilizing hypoxia-inducible Factor 1alpha and regulates responsive cardiac angiogenesis. Hypertension. 2016;67:597–605.
- Yue X, Yang X, Lin X, et al. Rnd3 haploinsufficient mice are predisposed to hemodynamic stress and develop apoptotic cardiomyopathy with heart failure. Cell Death Dis. 2014;5:e1284.
- Dai Y, Song J, Li W, et al. RhoE Fine-Tunes Inflammatory Response in Myocardial Infarction. Circulation. 2019;139:1185–1198.
- Barry SP, Lawrence KM, McCormick J, et al. New targets of urocortin-mediated cardioprotection. J Mol Endocrinol. 2010;45:69–85.
- Vitale SG, Lagana AS, Rapisarda AM, et al. Role of urocortin in pregnancy: an update and future perspectives. World J Clin Cases. 2016;4:165–171.
- Collett GP, Goh XF, Linton EA, et al. RhoE is regulated by cyclic AMP and promotes fusion of human BeWo choriocarcinoma cells. PloS One. 2012;7:e30453.
- Johnson MP, Brennecke SP, East CE, et al. Genetic dissection of the pre-eclampsia susceptibility locus on chromosome 2q22 reveals shared novel risk factors for cardiovascular disease. Mol Hum Reprod. 2013;19:423–437.
- Xu Y, Ge Z, Zhang E, et al. The lncRNA TUG1 modulates proliferation in trophoblast cells via epigenetic suppression of RND3. Cell Death Dis. 2017;8:e3104.
- Xu Y, Wu D, Liu J, et al. Downregulated lncRNA HOXA11-AS affects trophoblast cell proliferation and migration by regulating RND3 and HOXA7 expression in PE. Mol Ther Nucleic Acids. 2018;12:195–206.
- Li L, Zhang X, Hong SL, et al. Long non-coding HOTTIP regulates preeclampsia by inhibiting RND3. Eur Rev Med Pharmacol Sci. 2018;22:3277–3285.
- Fang YN, Huang ZL, Li H, et al. Highly expressed miR-182-5p can promote preeclampsia progression by degrading RND3 and inhibiting HTR-8/SVneo cell invasion. Eur Rev Med Pharmacol Sci. 2018;22:6583–6590.
- Paysan L, Piquet L, Saltel F, et al. Rnd3 in cancer: a review of the evidence for tumor promoter or suppressor. Mol Cancer Res. 2016;14:1033–1044.
- Rutherford S, Hampton GM, Frierson HF, et al. Mapping of candidate tumor suppressor genes on chromosome 12 in adenoid cystic carcinoma. Lab Invest. 2005;85:1076–1085.
- Aguirre AJ, Brennan C, Bailey G, et al. High-resolution characterization of the pancreatic adenocarcinoma genome. Proc Natl Acad Sci USA. 2004;101:9067–9072.
- Garcia-Casado Z, Romero I, Fernandez-Serra A, et al. A de novo complete BRCA1 gene deletion identified in a Spanish woman with early bilateral breast cancer. BMC Med Genet. 2011;12:134.
- Cuiyan Z, Jie H, Fang Z, et al. Overexpression of RhoE in non-small cell lung cancer (NSCLC) is associated with smoking and correlates with DNA copy number changes. Cancer Biol Ther. 2007;6:335–342.
- Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560.
- Xiang G, Yi Y, Weiwei H, et al. RND1 is up-regulated in esophageal squamous cell carcinoma and promotes the growth and migration of cancer cells. Tumour Biol. 2016;37:773–779.
- Boyrie S, Delmas C, Lemarie A, et al. RND1 regulates migration of human glioblastoma stem-like cells according to their anatomical localization and defines a prognostic signature in glioblastoma. Oncotarget. 2018;9:33788–33803.
- Komatsu H, Iguchi T, Masuda T, et al. Attenuated RND1 expression confers malignant phenotype and predicts poor prognosis in hepatocellular carcinoma. Ann Surg Oncol. 2017;24:850–859.
- Qin CD, Ma DN, Zhang SZ, et al. The Rho GTPase Rnd1 inhibits epithelial-mesenchymal transition in hepatocellular carcinoma and is a favorable anti-metastasis target. Cell Death Dis. 2018;9:486.
- Mouly L, Mamouni K, Gence R, et al. PARP-1-dependent RND1 transcription induced by topoisomerase I cleavage complexes confers cellular resistance to camptothecin. Cell Death Dis. 2018;9:931.
- Grise F, Sena S, Bidaud-Meynard A, et al. Rnd3/RhoE is down-regulated in hepatocellular carcinoma and controls cellular invasion. Hepatology. 2012;55:1766–1775.
- Luo H, Dong Z, Zou J, et al. Down-regulation of RhoE is associated with progression and poor prognosis in hepatocellular carcinoma. J Surg Oncol. 2012;105:699–704.
- Ma W, Wong CC, Tung EK, et al. RhoE is frequently down-regulated in hepatocellular carcinoma (HCC) and suppresses HCC invasion through antagonizing the Rho/Rho-kinase/myosin phosphatase target pathway. Hepatology. 2013;57:152–161.
- Bektic J, Pfeil K, Berger AP, et al. Small G-protein RhoE is underexpressed in prostate cancer and induces cell cycle arrest and apoptosis. Prostate. 2005;64:332–340.
- Trojan L, Schaaf A, Steidler A, et al. Identification of metastasis-associated genes in prostate cancer by genetic profiling of human prostate cancer cell lines. Anticancer Res. 2005;25:183–191.
- Ma W, Sze KM, Chan LK, et al. RhoE/ROCK2 regulates chemoresistance through NF-kappaB/IL-6/STAT3 signaling in hepatocellular carcinoma. Oncotarget. 2016;7:41445–41459.
- Chang L, Guo F, Wang Y, et al. MicroRNA-200c regulates the sensitivity of chemotherapy of gastric cancer SGC7901/DDP cells by directly targeting RhoE. Pathol Oncol Res. 2014;20:93–98.
- Feng B, Li K, Zhong H, et al. RhoE promotes metastasis in gastric cancer through a mechanism dependent on enhanced expression of CXCR4. PloS One. 2013;8:e81709.
- Kratz JR, He J, Van Den Eeden SK, et al. A practical molecular assay to predict survival in resected non-squamous, non-small-cell lung cancer: development and international validation studies. Lancet. 2012;379:823–832.
- Raz DJ, Ray MR, Kim JY, et al. A multigene assay is prognostic of survival in patients with early-stage lung adenocarcinoma. Clin Cancer Res: An Official Journal of the American Association for Cancer Research. 2008;14:5565–5570.
- Zhang C, Zhou F, Li N, et al. Overexpression of RhoE has a prognostic value in non-small cell lung cancer. Ann Surg Oncol. 2007;14:2628–2635.
- Klein RM, Aplin AE. Rnd3 regulation of the actin cytoskeleton promotes melanoma migration and invasive outgrowth in three dimensions. Cancer Res. 2009;69:2224–2233.
- Klein RM, Higgins PJ. A switch in RND3-RHOA signaling is critical for melanoma cell invasion following mutant-BRAF inhibition. Mol Cancer. 2011;10:114.
- Poch E, Minambres R, Mocholi E, et al. RhoE interferes with Rb inactivation and regulates the proliferation and survival of the U87 human glioblastoma cell line. Exp Cell Res. 2007;313:719–731.
- Liu B, Lin X, Yang X, et al. Downregulation of RND3/RhoE in glioblastoma patients promotes tumorigenesis through augmentation of notch transcriptional complex activity. Cancer Med. 2015;4:1404–1416.
- Clarke K, Daubon T, Turan N, et al. Inference of low and high-grade Glioma gene regulatory networks delineates the role of Rnd3 in establishing multiple hallmarks of cancer. PLoS Genet. 2015;11:e1005325.
- Shang C, Hong Y, Guo Y, et al. miR-128 regulates the apoptosis and proliferation of glioma cells by targeting RhoE. Oncol Lett. 2016;11:904–908.
- Liu B, Dong H, Lin X, et al. RND3 promotes Snail 1 protein degradation and inhibits glioblastoma cell migration and invasion. Oncotarget. 2016;7:82411–82423.
- Sun Q, Dong H, Li Y, et al. Small GTPase RHOE/RND3, a new critical regulator of NF-kappaB signalling in glioblastoma multiforme? Cell Prolif. 2019;52:e12665.
- Wang H, Wang Y, Liang B, et al. The Rho GTPase RhoE exerts tumor-suppressing effects in human esophageal squamous cell carcinoma via negatively regulating epidermal growth factor receptor. J Cancer Res Ther. 2016;12:60–63.
- Zhao H, Yang J, Fan T, et al. RhoE functions as a tumor suppressor in esophageal squamous cell carcinoma and modulates the PTEN/PI3K/Akt signaling pathway. Tumour Biol. 2012;33:1363–1374.
- Ogata S, Morokuma J, Hayata T, et al. TGF-beta signaling-mediated morphogenesis: modulation of cell adhesion via cadherin endocytosis. Genes Dev. 2007;21:1817–1831.
- Karaulanov E, Bottcher RT, Stannek P, et al. Unc5B interacts with FLRT3 and Rnd1 to modulate cell adhesion in Xenopus embryos. PloS One. 2009;4:e5742.
- Naud N, Toure A, Liu J, et al. Rho family GTPase Rnd2 interacts and co-localizes with MgcRacGAP in male germ cells. Biochem J. 2003;372:105–112.
- McColl B, Garg R, Riou P, et al. Rnd3-induced cell rounding requires interaction with Plexin-B2. J Cell Sci. 2016;129:4046–4056.
- Katoh H, Harada A, Mori K, et al. Socius is a novel Rnd GTPase-interacting protein involved in disassembly of actin stress fibers. Mol Cell Biol. 2002;22:2952–2964.
- Wennerberg K, Forget MA, Ellerbroek SM, et al. Rnd proteins function as RhoA antagonists by activating p190 RhoGAP. Curr Biol. 2003;13, 1106–1115