Abstract
Colorectal cancers (CRC) develop in the face of an important immune system associated with the intestinal mucosal tissue. The immune response against the tumor has been proposed to affect the prognosis of patients undergoing treatment for CRC. In this study T cells infiltrating the tumor were compared with T cells populating the unaffected neighboring mucosal tissue and cells from the peripheral blood. We observed that T cells from the tumor harbor an activated phenotype, with engagement of the NKG2D pathway in CD8 T cells. We show that mucosal and tumor-infiltrating T cells are enriched in NKG2D CD4 T cells, which exhibit cytotoxic functions. Finally, T cell populations in the tumor were modified according to its oncogenetic status, with higher percentages of CD8 T cells isolated from patients with microsatellite instable tumor status.
Abbreviations:
- CRC, Colo Rectal Cancer
- LPL, Lamina propria lymphocytes
- MHC, Major Histocompatibility Complex
- MIC , MHC-I chain related
- MSI, Microsatellite instability
- MSS, Microsatellite stability
- NK, Natural Killer
- PBL, Peripheral blood lymphocytes
- PI3K, Phosphoinositide 3-kinase
- RAF, Rapidly accelerated fibrosarcoma
- RAS, Rat sarcoma
- TIL, Tumor infiltrating lymphocytes
- TNM, Tumor node metastasis
- ULBP, UL16 binding protein
Introduction
Recent advances in tumor immunology have highlighted the role of the immune response in the development, evolution, and outcome of cancers. The immune system is thought to actively edit out precancerous cells in tissues as they appear.Citation1 In certain circumstances, the tumor cells develop biological processes that enable escape from this immune surveillance.Citation1 The quality of the immune response against the tumor is an important prognostic factor in patients with colorectal cancer (CRC).Citation2
CRC is the third most common cancer and the fourth most common cancer cause of death globally, accounting for approximately 1 to 2 million new cases and 600,000 deaths per year.Citation3 The incidence of CRC strongly increases with age, and median age at diagnosis is approximately 65 years. A significant proportion of patients presenting with Stage I, II, or III disease (75% of patients) can be cured by surgical intervention. In the absence of adjuvant therapy, approximately 50% of colon cancer patients with resectable disease are cured by surgery alone whereas 50% relapse. Use of adjuvant chemotherapy after surgery rescues approximately 15% of patients from relapse.Citation3
The majority of CRCs are sporadic environmentally driven tumors (rather than familial heritable disease). Several studies have highlighted different types of mutation and developmental processes involved in CRC.Citation4–7 Some of these mutations are associated with better prognosis (e.g., microsatellite instability [MSI])Citation8 and others with poor outcome (e.g., BRAF mutations).Citation9,10 Some studies suggest that these differential outcomes are in part driven by the capacity of the tumor to induce a strong immune response.Citation3 Several other predictive biomarkers have been described, including mutations of the KRAS, NRAS, PIK3CA, and TP53 genes, but their prognostic role remains uncertain.Citation11
To fend off tumorigenesis, the immune system includes several layers of active immune cells capable of recognizing altered cells.Citation12 Natural killer (NK) cells detect the presence or absence of histocompatibility molecules at the surface of cells and are able to integrate these signals to either lyse the target cells, or not. Tumor-associated modified self-antigens can also be presented to T cells, enabling an adaptive immune response to the tumor that consists of helper CD4 T cells and drives the expansion and differentiation of cytotoxic CD8 T cells.Citation1,12 During this process the tumor microenvironment can influence the quality of the T-cell response and the generation of T CD4 regulatory cells that favor immunologic escape of the tumor.
The intestinal mucosa is a dynamic tissue that has to intensively renew in the face of a microbial community that induces important stress in the epithelial layer. A large number of immune cells are present in the epithelium to monitor this host–microbe interaction and tissue renewal, suggesting that the immune system could play a major role in CRC. Here, we show that T cells from colon tumors (tumor infiltrating lymphocytes [TIL]) displayed important differences from the lymphocytes isolated from peripheral blood (peripheral blood lymphocytes [PBL]) and the neighboring healthy intestinal lamina propria (lamina propria lymphocytes [LPL]). Phenotypically, TIL were closer to LPL than to PBL. We show that the surface receptor NKG2D was differentially expressed in LPL and TIL compared to PBL. Expression of NKG2D on CD4 T cells was associated with increased effector cytotoxic functions. NKG2D was downregulated on CD8 T cells from the tumor, suggesting that this pathway could play an important role in the antitumor immune response. Finally, oncogenic status of the cancer appears to influence the immune response within the tumor as T cell populations differed between microsatellite instable (MSI) and microsatellite stable (MSS) tumors and between KRAS/NRAS mutated tumors and their wild-type counterparts.
Results
Phenotypic study of mucosal T cells shows recruitment of CD4 T cells to colon tumor compared to neighboring tissue
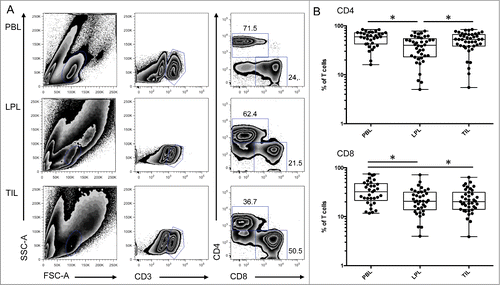
The intestine contains a large number of immune cells, including T cells, that participate in the response against microbial infections and in homeostasis of the mucosal tissue and possibly affect the development of colon cancer. We studied T lymphocytes present in colonic tumors and compared the phenotype of these cells to T cells present in peripheral blood and in neighboring unaffected mucosa (). Interestingly, the percentage of CD4 T cells was higher in the tumor (TIL) compared to LPL isolated at a distance from the tumor (). In accordance with previous studies,Citation13 the percentage of CD4 T cells was reduced in the colon lamina propria (LPL) compared to peripheral blood lymphocytes (PBL) (). In contrast, the percentage of CD8 T cells present in TIL and LPL was equivalent (). These results suggest the recruitment and/or expansion of CD4 T cells within the tumor compared to the normal tissue.
Figure 1. Differential representation of CD4 and CD8 T cells in peripheral blood, colon lamina propria, and corresponding tumor. (A) Representative FACS analysis of CD4 and CD8 T cell populations in the indicated compartment. (B) Compiled analysis of CD4 and CD8 T cell proportions among total CD3-positive cells in the different compartments (n = 42). The Wilcoxon paired non-parametric t-test was used for statistical analyses (*: P < 0.05). LPL, lamina propria lymphocytes; PBL, peripheral blood lymphocytes; TIL, tumor-infiltrating lymphocytes.
Decreased expression of NKG2D on CD8 T cells isolated from the tumor
To study the functionality of the T cells found in the tumor we performed specific phenotyping of TIL compared with LPL obtained from neighboring non-tumoral tissue. The NKG2D receptor has an important function in both mucosal immunity and antitumor responses. An increased proportion of CD4 T cells from LPL and TIL showed expression of NKG2D at their surface compared to cells from PBL (). However, there was no difference in the expression of NKG2D on CD4 T cells isolated from the tumor and those isolated from the mucosa.
Figure 2 (See previous page). Activation markers and NKG2D expression on mucosal CD4 T cells and tumor infiltrating CD8 T cells. (A) Representative FACS analysis of the indicated compartment for the expression of NKG2D on T cells according to their expression of CD4 and CD8. (B) Compiled analysis of NKG2D expression on CD4 and CD8 T cell populations defined as in 1A (n = 38). The Wilcoxon paired non-parametric t-test was used for statistical analyses (*: P < 0.05). (C) Intensity of NKG2D expression on CD4 and CD8 T cells in the 3 compartments (histograms: gray solid, PBL; thin black line, LPL; heavy black line, TIL). (D) Expression of NKG2D on cells from LPL (thin black line) and TIL (heavy black line) immediately after isolation from the tissues (solid lines) or after overnight culture (dotted lines). (E) Representative FACS analysis of CD4 and CD8 T cells defined as in 1A from PBL, LPL, and TIL for expression of CD103 and HLA-DR. (F) Compiled analysis of CD103 and HLA-DR expression on CD4 and CD8 T cells from the 3 compartments (PBL, LPL, and TIL) (n = 16 and n = 9, respectively). The Wilcoxon paired non-parametric t-test was used for statistical analyses (*: P < 0.05). LPL, lamina propria lymphocytes; PBL, peripheral blood lymphocytes; TIL, tumor-infiltrating lymphocytes.
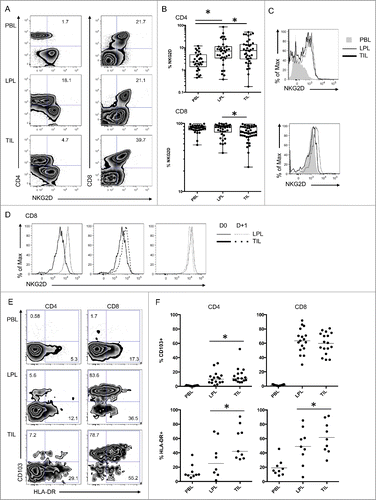
All CD8 T cells express high levels of NKG2D; however, this expression was significantly reduced in CD8 T cells from TIL compared to those from LPL (). Additionally, as indicated by fluorescence intensity on cells from these tissues extracted from the same donor, NKG2D levels on the surface of CD8 T cells from TIL were lower than those on CD8 T cells from LPL (). After overnight culture, the surface expression of NKG2D on CD8 T cells from TIL increased, whereas CD8 T cells from LPL showed no difference (). Taken together, these results suggest that the NKG2D molecule is differentially regulated in mucosal tissues and that the NKG2D pathway is probably engaged within the colon tumor.
T cells from the tumor show increased activation and epithelium interaction
The decreased expression of NKG2D at the surface of CD8 T cells in TIL suggests that the NKG2D pathway is engaged in colon cancer. We analyzed several markers that might indicate that these cells are activated in the tumor. Interestingly, tumor T cells harbored increased expression of the MHC class II molecule HLA-DR and the integrin CD103 (), suggesting an enhanced activation status and increased interaction with epithelial/tumor cells.
Additionally, we observed an increased percentage of cells with intracellular perforin in CD8 T cells infiltrating the tumor compared to neighboring LPL. Similarly, the percentage of CD4 T cells expressing perforin was increased in TIL compared to LPL (). Conversely, levels of granzyme A, a preformed cytotoxic molecule present in CD8 T cells, were reduced in TIL (), suggesting that this molecule was secreted into tumor tissues. The expression patterns of these molecules associated with cytotoxic functions strongly suggest that CD8 T cells infiltrating the tumor are engaged in a cytotoxic process.
Figure 3 (See previous page). Functions of tumor infiltrating lymphocytes compared with T cells from neighboring tissues. (A) Representative FACS analysis of CD4 and CD8 T cells in the indicated compartment (LPL and TIL) for expression of NKG2D and intracellular granzyme A and perforin. (B) Compiled analysis of granzyme A and perforin in mucosal T cells (LPL and TIL) (n = 9). The Wilcoxon paired non-parametric t-test was used for statistical analyses (*: P < 0.05). (C) Representative intracellular FACS analysis of CD4 and CD8 T cells from the indicated compartment (LPL and TIL) for expression of IFNγ and TNFα with or without PMA-ionomycin restimulation. (D) Compiled analysis of IFNγ and TNFα in mucosal T cells with or without PMA-ionomycin restimulation (n = 7). The Wilcoxon paired non-parametric t-test was used for statistical analyses (*: P < 0.05). IFNγ, ιντϵρφϵρµν γαμμα; LPL, lamina propria lymphocytes; PBL, peripheral blood lymphocytes; TIL, tumor-infiltrating lymphocytes; TNFα, tumor necrosis factor.
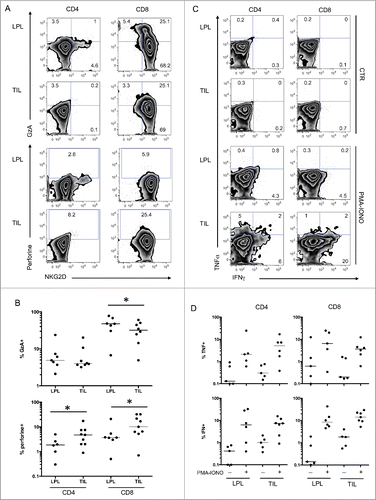
We also tested the capacity of T cells from TIL and LPL to produce effector molecules normally associated with T-cell functions, such as TNFα and IFNγ. The number of T cells able to produce TNFα and IFNγ after ex vivo restimulation by PMA and ionomycin was equivalent in TIL and LPL (). These results suggest that tumor T cells have normal responsiveness to classic triggers.
NKG2D expressing CD4 T cells harbor increased cytotoxic features
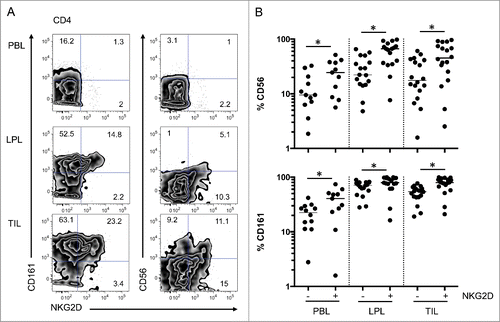
NKG2D-expressing CD4 T cells harboring increased effector functions are implicated in inflammatory bowel diseasesCitation13,14 and may also play a role in antitumor immunity.Citation15,16 We analyzed the phenotype and function of these cells in the mucosa and tumor of colon cancer patients (). We observed that CD4 T cells expressing NKG2D at their surface more frequently expressed other NK-like receptors (CD56 and CD161) in all compartments (PBL, LPL, and TIL) (). Accordingly, intracellular staining for molecules associated with cytotoxic functions (perforin, Granzyme A) were more frequent in cells expressing NKG2D compared to conventional CD4 T cells (). These results are consistent with our previous observations in another modelCitation13,14 and indicate that NKG2D-positive CD4 T cells might have cytotoxic functions.
Figure 4. Increased expression of CD161 and CD56 on NKG2D-positive CD4 T cells. (A) Representative FACS analysis of CD4 T cells from PBL, LPL, and TIL for co-expression of CD161 and CD56 with NKG2D. (B) Compiled analysis of CD161 and CD56 expression on NKG2D-positive or -negative CD4 T cells from the 3 compartments (n = 18). The Wilcoxon paired non-parametric t-test was used for statistical analyses (*: P < 0.05). LPL, lamina propria lymphocytes; PBL, peripheral blood lymphocytes; TIL, tumor-infiltrating lymphocytes.
Activation and cytotoxic functions of T cells induced by NKG2D
To test the cytotoxic potential of cells isolated from the different compartments, we performed a redirected cytotoxicity assay using the mastocytoma-derived cell line P815 and P815 cells stably transfected with the NKG2D ligand ULBP1 as target cells.Citation13,14 After isolation from the tumor, mucosal tissue, or blood, T cells were activated overnight with CD3-coated target cells. CD4 and CD8 T cells from PBL were activated, as demonstrated by upregulation of the activation markers CD69 and CD25 at their surface (). T cells from the tissue constitutively expressed CD69 at their surface (data not shown); however, they expressed CD69 and CD25 only when stimulated through CD3 (). Interestingly, increased activation was achieved when CD8 T cells were co-stimulated through CD3 and NKG2D (). More surprisingly, CD4 T cells were also responsive to ULBP1 stimulation (). These results show that T cells from TIL are activated in the same manner as cells from LPL or PBL, and that CD4 T cells could be co-stimulated through NKG2D similar to CD8 T cells.
Figure 5. Co-stimulation of CD4 and CD8 T cells through the NKG2D ligand ULBP1. (A) Representative FACS analysis of CD4 and CD8 T cells from PBL co-cultured with P815 cells in control conditions (CTR), P815 cells incubated with anti-CD3 antibodies (CD3), P815 cells expressing ULBP1 (ULBP1), or P815 cells expressing ULBP1 in the presence of anti-CD3 antibody (CD3 + ULBP1). Activation was assessed by the expression of CD69 and CD25 at the cell surface of T cells. (B) Compiled analysis of CD4 and CD8 T-cell activation from the 3 compartments (PBL, LPL, and TIL) (n = 3). Mean and SEM are represented. (C) Representative FACS analysis of CD8 T cells from PBL co-cultured with P815 cells as in (A). NKG2D and CD107a expression was measured to assess degranulation. (D) Compiled analysis of CD8 T-cell activation from the 3 compartments (PBL, LPL, and TIL) (n = 3). Mean and SEM are represented. LPL, lamina propria lymphocytes; PBL, peripheral blood lymphocytes; TIL, tumor-infiltrating lymphocytes.
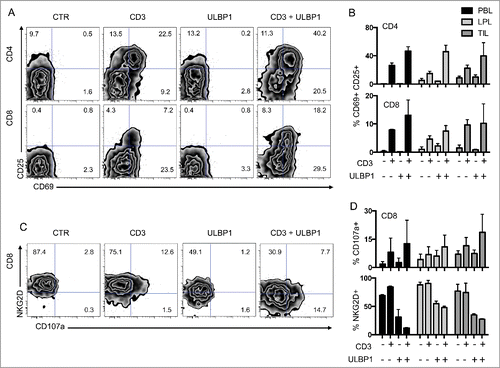
We then assessed the degranulation of CD8 T cells by the surface expression of CD107a as a marker of cytotoxic functions. CD107a was synergistically expressed at the surface of these cells when they were co-stimulated through CD3 and NKG2D, compared to CD3 alone (). Additionally, NKG2D surface expression decreased dramatically when these cells were stimulated in the presence of ULBP1, strongly suggesting that this trigger induced downregulation of NKG2D from the surface of T cells.
Expression of NKG2D ligands on epithelial and tumor cells
To analyze the engagement of NKG2D on T cells within the tumor, we quantified the expression of its ligands on tumor cells and epithelial cells from neighboring uninvolved tissue. Expression of the NKG2D ligands MIC-A, MIC-B, ULBP2, and ULP2 was detectable on epithelial cells, which displayed variable levels of expression from patient to patient. The fluorescence intensity of the staining of each ligand on the tumor cells was compared to their expression on epithelial cells from the same patient (Figure S1A). Increased expression of at least one NKG2D ligand was observed in 4 of 9 patients. However, there was no consistent difference in ligand expression among the cohort (Figure S1B). No correlation was observed between expression of the ligands and the level of expression of NKG2D on CD8 and CD4 T cells.
Microsatellite instability and KRAS/NRAS mutations correlate with an increased percentage of CD8 T cells in the tumor
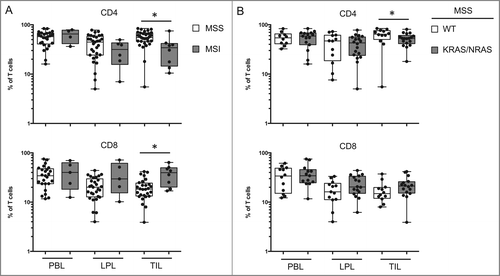
Among the genetic events involved in the development of colon cancer, some are thought to generate stronger immune responses than others.Citation6 We analyzed T cells from the tumor with regard to the genetic features of the tumors. Microstatellite instability (MSI) is associated with cancers that enable a good immune response and have a better prognosis. We observed a significantly lower percentage of CD4 T cells and a significantly higher percentage of CD8 T cells among TILs isolated from patients with MSI disease compared to those with microsatellite stable (MSS) disease (). The same was observed in the population of MSS patients with mutations in the KRAS or NRAS genes (). No significant differences were observed among T-cell populations with regard to the characteristics of the tumors (TNM grade, degree of differentiation of the cancerous cells) or the genetic status of the tumor (BRAF or TP53 mutations) (data not shown). The low rates of PIK3CA gene mutation precluded statistical analysis.
Figure 6. T-cell populations vary in function of the type of oncogenic status of the tumor. (A) Comparison of the percentage of CD4 and CD8 T cells in PBL, LPL, and TIL from patients with microsatellite instability (MSI) or stability (MSS). (B) Comparison of the percentage of CD4 and CD8 T cells in PBL, LPL, and TIL from MSS patients with or without identified KRAS or NRAS mutations. LPL, lamina propria lymphocytes; MSI, microsatellite instability; MSS, microsatellite stable; PBL, peripheral blood lymphocytes; TIL, tumor-infiltrating lymphocytes.
As anticipated, survival of patients with unfavorable tumor characteristics, as established by clinical and histological evidence, was decreased compared to that of patients with favorable clinical and pathological characteristics (Figure S2A, B). In accordance with recent studies,Citation9,10 patients with BRAF mutations had a poor prognosis (data not shown). Interestingly, patients with low expression of NKG2D on their CD8 T cells displayed a trend toward better survival, suggesting that the NKG2D pathway on these effector cells could have an impact on the immune response to the tumor (Figure S2C).
Discussion
A large body of evidence supports the influence of immunity on the outcome of patients with colorectal cancer.Citation2,17,18 High densities of Th1 and memory T cells in both the core and the invasive margin of a tumor were shown to have better prognostic value in colorectal cancer than the classic tumor node metastasis (TNM) classification.Citation19,20 Most reports focus exclusively on TILs and information on immune responses in the non-tumoral colonic mucosa is scarce. Our study design included concomitant evaluation of T-cell phenotype in the tumor, the non-tumoral mucosa, and the blood for all patients.
An interesting finding in the present study was that although phenotypically TILs displayed important differences from PBL T cells, they were rather similar to LPL T cells. The higher percentage of CD4 T cells among TIL compared to LPL suggests that the tumor induces specific modifications of T-cell populations present in healthy mucosa. Interestingly, there was no difference in the percentage of CD8 T-cell infiltrates between these 2 compartments although there was a significant decrease in each compared with the blood compartment. Taken together, these findings suggest that focusing on both compartments rather than on TIL alone might improve our comprehension of the immune mechanisms involved in colorectal carcinogenesis. We did not show any influence of TIL composition or phenotype on patient survival; however, this can probably be explained by the small number of patients and the short follow-up period.
Next, we explored involvement of the NKG2D pathway in CRC. We showed that T cells from the tumor were in an activated state compared to those in neighboring tissues, as reflected by increased expression of HLA-DR and CD103, high levels of intracellular perforin, and low levels of granzyme A, which is preformed in cytotoxic T cells. Moreover, decreased levels of NKG2D on the surface of CD8 TILs compared to blood and LPL, and its re-expression after overnight culture in the absence of cancer cells, strongly suggest engagement of the NKG2D pathway within the tumor. These results are in accordance with data from previous studies exploring the role of NKG2D-mediated antitumor activity in colorectal cancer.Citation16,20 In a retrospective review of 462 colon tumors, McGilvray et al.Citation22 observed that expression of NKG2D on colorectal tumors was heterogeneous and correlated with patient survival. Maccali et al.Citation21 studied a panel of human CRC lines and showed that triggering of NKG2D alone can lead to an antitumor immune response in CRC. Cytokine secretion was lower when NKG2D was the only engaged receptor than after co-stimulation of the TCR, suggesting that more efficient antitumor reactivity could be achieved when TCR is engaged and its signal is strengthened by co-engagement of NKG2D.
We performed co-stimulation assays on cells isolated from LPL and TIL and were able to show that this co-stimulation was very effective on CD8 T cells, leading to increased activation and degranulation as well as downregulation of NKG2D from the cell surface. More surprisingly, co-stimulation could be observed on CD4 T cells that express low levels of NKG2D. This apparent discrepancy might be explained by different regulation of this NK receptor at the surface of CD4 T cells.
Maccali et al. identified a subpopulation of CD4 T cells expressing NKG2D that is involved in CRC but their study was not designed to study it in detail.Citation21 In the present study we found significant tissue (tumor and mucosa) accumulation of NKG2D CD4 T cells compared to the blood. We then showed that NKG2D CD4 T cells were more likely to express other NK-like receptors (CD56, CD161) and intracellular molecules associated with cytotoxic functions (perforin, granzyme-A). Taken together, these results suggest that NKG2D CD4 T cells have cytotoxic functions in CRC, similar to our findings in Crohn disease.Citation13,14 Our results are in contrast with those published by Groh et al.Citation15 who explored NKG2D CD4 T cells in 26 cancer patients (6 with breast carcinoma, 8 with lung carcinoma, 4 with ovarian carcinoma, 4 with colon carcinoma, and 4 with melanoma). The authors assigned a suppressive function to NKG2D CD4 T cells, showing that NKG2D CD4 T cells (a) were associated with MIC-expressing epithelial tumors, (b) inhibited proliferation of other T-cell subsets, (c) had an unusual T regulatory type 1-like cytokine profile and induced FasL production, and (d) exerted suppressive activity that was NKG2D ligand-dependent and FasL-mediated.Citation15 One explanation for this discordance is that different cell subtypes may have different roles in different types of cancer. For example, a high density of tumor-infiltrating FOXP3 Tregs improves survival in colorectal cancer,Citation23 which is in contrast to several other solid cancer types.Citation24 Our analysis was restricted to colorectal cancer, which may account for the discordance with the study by Groh et al.Citation15
Nonetheless, the development of tumors in the face of the immune response suggests that cancer cells circumvented NKG2D-dependent recognition by T cells. The detection of very variable levels of NKG2D ligand at the surface of tumor cells in different patients might implicate the existence of several mechanisms that remain to be explored.
In their pivotal reviews, Hanahan and WeinbergCitation25,26 propose 8 hallmarks of cancer, which include immunity. In addition to the prototypical growth signaling circuit centered on Ras, other component circuits transmit antigrowth and differentiation signals or mediate commands to live or die by apoptosis; all of these circuits are coupled together and to a spectrum of extracellular cues. Functional assessment of these intracellular pathways is difficult in clinical practice and although several prognostic/predictive biomarkers have recently emerged their precise clinical significance in colorectal cancer is still uncertain. MSI statusCitation6,8 and BRAF mutationsCitation9,10 have been shown to be of prognostic utility, whereas KRAS/NRAS and PIK3CA mutations are predictive of outcome with EGFR-targeted agentsCitation27 and aspirin,Citation28 respectively. Although TP53 mutations resulting in p53 protein overexpression are frequent in colon cancer, the precise prognostic/predictive impact of such alterations is not clear.Citation11 Systematic analysis of these biomarkers in our center did not yield significant results. In accordance with other reports in the literatureCitation9,10 we found significant survival impairment in patients with mutated BRAF.
The immune system, which acts as an extrinsic tumor suppressor, manifests its effects only after transformed cells have circumvented their intrinsic tumor suppressor mechanisms coordinated by the above mentioned signals.Citation1 Hence, we tried to evaluate correlations between biomarkers reflecting modifications of intracellular signaling pathways and phenotypic characteristics of TILs. We observed that TILs from MSI-high patients have higher percentages of CD8 T cells compared to their MSS counterparts. In the literature, MSI tumors have been reported to be associated with a high density of TILsCitation3 and have a better prognosis than MSS tumors.Citation11 MSI induces frameshift somatic mutations within genes harboring repeated sequences in their coding frame including TGFβR2, which is mutated in 90% of cases.Citation8 These mutations lead not only to the inactivation of these genes but also to the appearance of potentially immunogenic neoantigens, suggesting an important role of the immune response to specific neoantigens in CRC with MSI and its potential involvement in the better prognosis of these tumors.Citation8 We found no other significant correlations between characteristics of TILs (surface expression of CD4, CD8, NKG2D, CD56, CD161; production of perforin, Granzyme A, IFNγ, TNFα) and tumor status regarding MSI, KRAS, NRAS, BRAF, PIK3CA, and p53. Failure to establish more consistent relationships between immunity and oncogenetics in colorectal cancer may be due to several factors. First, as pointed out by Hanahan and Weinberg, our understanding of the cell circuitry is still rudimentary; interplays between immunity and intrinsic oncogenesis may rely on pathways that are still unknown.Citation25 Second, the immune peritumoral infiltrate is heterogeneous and cells other than TILs may have a primary role in the abovementioned interplay.
In conclusion, we confirmed higher CD8 T-cell infiltration in patients with MSI tumors but were unable to establish direct correlations between MSI status and involvement of the NKG2D pathway in colon cancer. We also showed downregulation of NKG2D expression on the CD8 T cells infiltrating colon cancer, which suggests active engagement of the NKG2D pathway in colon cancer and was associated with a trend in improved survival.
Patients and Methods
Patients
Between November 2011 and December 2013, 42 patients who underwent resection of colon cancer at the Saint Louis Hospital in Paris were prospectively included in this study (). These patients are part of a cohort of 210 patients who underwent colonic resection during this period. Patients who underwent emergency surgery (n = 120) and patients who received preoperative chemotherapy (n = 35) were excluded. Thirteen patients were excluded because sample analysis was incomplete. Indication for surgery was decided following multidisciplinary consultation. Upfront colonic resection was offered to fit patients in the absence of distant metastasis and to patients with metastatic disease with life-threatening tumor-related complications (bleeding, perforation).
Table 1. Characteristics of the 42 patients with colorectal cancer
This study was approved by the ethical committee of Hospital Saint-Louis, and all subjects gave written informed consent.
Isolation of intestinal epithelial cells and lymphocytes
Isolation from surgical specimens was performed as described previously.Citation13,14 Mucosa samples were harvested from macroscopically healthy tissue at a distance of ≥5 cm from the tumor. Briefly, surgical specimens were washed extensively with PBS. The mucosa was stripped off from the submucosa, minced into small pieces, and placed in 1 mmol/L dithiothreitol for 10 min at room temperature. The pieces were washed in PBS and incubated in medium (RPMI 1640) containing 1.5 mmol/L MgCl2 and 1 mmol/L EDTA for 30 min at 37°C, with vortexing every 5 min. The supernatant, containing intestinal epithelial cells (IECs), was passed through a nylon filter (Falcon 2360; Becton Dickinson, BD Biosciences, Le Pont de Claix, France). Cells were washed twice in PBS and resuspended in RPMI 1640.
Lamina propria lymphocytes (LPL) were isolated from mucosal tissue incubated for 1 h at 37°C in medium containing 1 mg/mL collagenase (clostridiopeptidase A). The cell suspension was collected, centrifuged, washed, and resuspended in PBS.
Heparinized venous blood was collected from all patients, diluted 1:3 with PBS, layered on a Ficoll-Hypaque density gradient, and centrifuged for 30 min at 900 g. Peripheral blood mononuclear cells (PBMCs) were collected from the interface, washed 3 times with PBS, and resuspended in RPMI 1640.
Isolation of intratumoral lymphocytes
For the isolation of tumor infiltrating lymphocytes (TILs) from surgical specimens, tumor samples were washed with PBS and cut into pieces of 2 to 3 mm. The tissue fragments were incubated for 1 h in RPMI 1640 medium containing collagenase. The supernatant was then filtered and the recovered cells were resuspended in RPMI 1640 for isolation of intratumoral lymphocytes.
Redirected cytotoxicity assay
The redirected cytotoxicity assay was performed as previously described.Citation14 Briefly, murine mastocytome cells (P815 cells and P815 stably transfected with the NKG2D ligand ULBP1Citation14) were incubated with mouse anti-human CD3 antibodies bound by the Fc receptor. After washing, these cells were used as target cells for isolated T cells from the different compartments with a 4:1 ratio of effector:target cells. After overnight culture, activation and degranulation was assessed by flow cytometry.
Analysis by flow cytometry
Lymphocytes (TIL, LPL, PBL) were resuspended in PBS and incubated for 15 min in the presence of antibodies. The antibodies used for phenotypic analysis were directed against CD3, CD4, CD8, NKG2D, CD161, CD103, HLA-DR, and CD56 (BD, eBiosciences, Miltenyi Biotech). Lymphocytes were stimulated for 4 h with phorbol myristate acetate (PMA) and ionomycin before intracellular staining. Cells were incubated for 15 min with surface markers (CD8, CD3, CD4, NKG2D combined with amcyan, Pacific Blue, Allophycocyanin-H7, Phycoerythrin) followed by 10-min fixation in 3.5% formaldehyde solution. The cells were then incubated with FACS permeabilizing solution (BD Biosciences) for 10 min and with antibodies directed against interferon (IFN-γ), tumor necrosis factor (TNF-α), and isotype controls. Intracellular staining for perforin and Granzyme A was performed using 0.2% saponin. For activation and cytotoxicity assay, T cells were incubated with antibodies specific to CD3, CD4, CD8, NKG2D, CD69, CD25, and CD107a (BD, eBiosciences, Miltenyi Biotech).
IECs obtained from the colon or the tumor were resuspended in PBS and stained for analysis with anti-EpCAM and anti-CD45 antibodies (BD) to differentiate epithelial from hematopoietic cells. Antibodies specific for MIC-A, MIC-B, ULBP-1 and ULBP-2 (R&D Systems) were used against isotype controls to assess surface expression of these NKG2D ligands on IEC and tumor cells. Analyses were performed with a BD FACS CantoII-8 color scan.
Molecular Analyses
Extraction of nucleic acids from paraffin-embedded fixed tissues and frozen tissues
Tumor tissue sections were fixed and embedded in paraffin after overnight digestion by proteinase K. DNA was extracted using the QIAamp DNA mini-kit (Qiagen) as recommended by the manufacturer. When frozen tissue was available, DNA and RNA were manually extracted from frozen sections using the phenol-chloroform method. Tissue sections were examined by the pathologist, who selected tumor areas with a minimum percentage of tumor cells of 20%.
Detection of mutations in KRAS and NRAS
The top 7 KRAS mutations, which are located in exon 2 (codons 12 and 13), were analyzed by allelic discrimination: probes specific for each allele (mutated and wild type) were labeled with fluorescent reporter dyes at their 5´ end and analyzed by real-time PCR on a LC480 Lightcycler (Roche).Citation29 Other rare alterations of exon 2 were investigated using technical high resolution melting (HRM); any abnormal profile was analyzed by direct sequencing to identify the mutation.
Mutations in KRAS exons 3 and 4 and NRAS exons 2 and 3 were assessed by HRM using specific primers for each identified exon and by direct sequencing in the case of an abnormal HRM profile. In all cases, the final result was checked by a second independent analysis.
Detection of V600E BRAF mutations
The presence of the V600E BRAF mutation was determined by allelic discrimination on a LC480 Lightcycler.Citation30 We also conducted HRM analysis followed by direct sequencing of BRAF exon 15 to search for other modifications.
Detection of PIK3CA gene mutations in exons 9 and 20
The presence of 3 mutation hotspots, E542K, E545K, and H1047R, on the PIK3CA gene was evaluated by allelic discrimination. Less common exon 9 and 20 mutations were also assessed by HRM analysis followed by direct sequencing.
Determination of microsatellite instability status
To evaluate microsatellite instability (MSI), a pentaplex PCR comprising 5 quasi-monomorphic mononucleotide repeats (BAT-25, BAT-26, NR-21, NR-24, and NR-27) was used. Tumors with instability at ≥3 markers were defined as high microsatellite instability (MSI-H), whereas those with instability at <3 markers were considered microsatellite stable (MSS).Citation31
p53 functional status by FASAY
When frozen tumor samples were available, frozen tissue sections were used to determine p53 gene functional status using a highly efficient yeast functional assay (Functional Analysis of Separated Alleles in Yeast or FASAY), which evaluates the transactivation activity of p53 on a p53-responsive promoter stably integrated into the yeast genome. Citation32
RNA was extracted by the phenol-chloroform method, reverse transcribed, and p53 transcripts were amplified by PCR and transfected into yeast. Yeast colonies transformed with wild-type or mutated TP53 sequences appear white and large, or red and small, respectively. TP53 status was considered mutated when more than 10% of the yeast colonies were red and analysis using the split versions of the test could identify the defect in the 5´ or 3´ part of the gene.
Statistical Analysis
Results are reported as mean (±SD) or as counts (proportion). Quantitative variables were compared by the paired Wilcoxon rank-sum and Mann-Whitney tests as appropriate and qualitative variables by Fisher's exact test. Survival curves were estimated using the Kaplan Meier estimator and were compared by the log-rank test. A test was considered significant if P < 0.05 and all reported P values are 2-sided.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
KONI_A_1016698_SUP_FIG_2.pdf
Download PDF (48 KB)KONI_A_1016698_SUP_FIG_1.pdf
Download PDF (126.7 KB)Acknowledgments
The authors thank Ms. Laurence Françoise and Ms. Claire Bocquet for help in genetic marker assessment. The authors also thank Alexander Steinle from the Department of Immunology, Eberhard Karls University of Tuebingen, Germany for providing P815 NKG2D ligand transfectants.
Funding
This work was supported by grants from INSERM.
References
- Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol 2004; 22: 329-60; PMID:15032581; http://dx.doi.org/10.1146/annurev.immunol.22.012703.104803
- Pages F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, Mlecnik B, Kirilovsky A, Nilsson M, Damotte D, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med 2005; 353: 2654-66; PMID:16371631; http://dx.doi.org/10.1056/NEJMoa051424
- Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet 2014; 383: 1490-502; PMID:24225001; http://dx.doi.org/10.1016/S0140-6736(13)61649-9
- Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology 2010; 138: 2059-72; PMID:20420946; http://dx.doi.org/10.1053/j.gastro.2009.12.065
- Manne U, Shanmugam C, Katkoori VR, Bumpers HL, Grizzle WE. Development and progression of colorectal neoplasia. Cancer Biomark 2010; 9: 235-65; PMID:22112479
- Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med 2009; 361: 2449-60; PMID:20018966; http://dx.doi.org/10.1056/NEJMra0804588
- Al-Sohaily S, Biankin A, Leong R, Kohonen-Corish M, Warusavitarne J. Molecular pathways in colorectal cancer. J Gastroenterol Hepatol 2012; 27: 1423-31; PMID:22694276; http://dx.doi.org/10.1111/j.1440-1746.2012.07200.x
- Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology 2010; 138: 2073-87 e3; PMID:20420947; http://dx.doi.org/10.1053/j.gastro.2009.12.064
- Tol J, Nagtegaal ID, Punt CJ. BRAF mutation in metastatic colorectal cancer. N Engl J Med 2009; 361: 98-9; PMID:19571295; http://dx.doi.org/10.1056/NEJMc0904160
- Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 2013; 369: 1023-34; PMID:24024839; http://dx.doi.org/10.1056/NEJMoa1305275
- Tejpar S, Bertagnolli M, Bosman F, Lenz HJ, Garraway L, Waldman F, Warren R, Bild A, Collins-Brennan D, Hahn H, et al. Prognostic and predictive biomarkers in resected colon cancer: current status and future perspectives for integrating genomics into biomarker discovery. Oncologist 2010; 15: 390-404; PMID:20350999; http://dx.doi.org/10.1634/theoncologist.2009-0233
- Tougeron D, Fauquembergue E, Latouche JB. [Immune response and colorectal cancer]. Bull Cancer 2013; 100: 283-94; PMID:23501583
- Allez M, Tieng V, Nakazawa A, Treton X, Pacault V, Dulphy N, Caillat-Zucman S, Paul P, Gornet JM, Douay C, et al. CD4+NKG2D+ T cells in Crohn's disease mediate inflammatory and cytotoxic responses through MICA interactions. Gastroenterology 2007; 132: 2346-58; PMID:17570210; http://dx.doi.org/10.1053/j.gastro.2007.03.025
- Pariente B, Mocan I, Camus M, Dutertre CA, Ettersperger J, Cattan P, Gornet JM, Dulphy N, Charron D, Lémann M, et al. Activation of the receptor NKG2D leads to production of Th17 cytokines in CD4+ T cells of patients with Crohn's disease. Gastroenterology 2011; 141: 217-26, 26 e1-2; PMID:21600899; http://dx.doi.org/10.1053/j.gastro.2011.03.061
- Groh V, Smythe K, Dai Z, Spies T. Fas-ligand-mediated paracrine T cell regulation by the receptor NKG2D in tumor immunity. Nat Immunol 2006; 7: 755-62; PMID:16732291; http://dx.doi.org/10.1038/ni1350
- Nausch N, Cerwenka A. NKG2D ligands in tumor immunity. Oncogene 2008; 27: 5944-58; PMID:18836475; http://dx.doi.org/10.1038/onc.2008.272
- Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012; 12: 298-306; PMID:22419253; http://dx.doi.org/10.1038/nrc3245
- Pernot S, Terme M, Voron T, Colussi O, Marcheteau E, Tartour E, Taieb J. Colorectal cancer and immunity: what we know and perspectives. World J Gastroenterol 2014; 20: 3738-50; PMID:24833840; http://dx.doi.org/10.3748/wjg.v20.i14.3738
- Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, Tosolini M, Camus M, Berger A, Wind P, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006; 313: 1960-4; PMID:17008531; http://dx.doi.org/10.1126/science.1129139
- Mlecnik B, Tosolini M, Kirilovsky A, Berger A, Bindea G, Meatchi T, Bruneval P, Trajanoski Z, Fridman WH, Pagès F, et al. Histopathologic-based prognostic factors of colorectal cancers are associated with the state of the local immune reaction. J Clin Oncol 2011; 29: 610-8; PMID:21245428; http://dx.doi.org/10.1200/JCO.2010.30.5425
- Maccalli C, Pende D, Castelli C, Mingari MC, Robbins PF, Parmiani G. NKG2D engagement of colorectal cancer-specific T cells strengthens TCR-mediated antigen stimulation and elicits TCR independent anti-tumor activity. Eur J Immunol 2003; 33: 2033-43; PMID:12884870; http://dx.doi.org/10.1002/eji.200323909
- McGilvray RW, Eagle RA, Watson NF, Al-Attar A, Ball G, Jafferji I, Trowsdale J, Durrant LG. NKG2D ligand expression in human colorectal cancer reveals associations with prognosis and evidence for immunoediting. Clin Cancer Res 2009; 15: 6993-7002; PMID:19861434; http://dx.doi.org/10.1158/1078-0432.CCR-09-0991
- Salama P, Phillips M, Grieu F, Morris M, Zeps N, Joseph D, Platell C, Iacopetta B. Tumor-infiltrating FOXP3+ T regulatory cells show strong prognostic significance in colorectal cancer. J Clin Oncol 2009; 27: 186-92; PMID:19064967; http://dx.doi.org/10.1200/JCO.2008.18.7229
- Whiteside TL. Regulatory T cell subsets in human cancer: are they regulating for or against tumor progression? Cancer Immunol Immunother 2014; 63: 67-72; PMID:24213679; http://dx.doi.org/10.1007/s00262-013-1490-y
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000; 100: 57-70; PMID:10647931; http://dx.doi.org/10.1016/S0092-8674(00)81683-9
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646-74; PMID:21376230; http://dx.doi.org/10.1016/j.cell.2011.02.013
- Van Cutsem E, Kohne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D'Haens G, Pintér T, Lim R, Bodoky G, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med 2009; 360: 1408-17; PMID:19339720; http://dx.doi.org/10.1056/NEJMoa0805019
- Liao X, Lochhead P, Nishihara R, Morikawa T, Kuchiba A, Yamauchi M, Imamura Y, Qian ZR, Baba Y, Shima K, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med 2012; 367: 1596-606; PMID:23094721; http://dx.doi.org/10.1056/NEJMoa1207756
- Lievre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, Ychou M, Bouché O, Landi B, Louvet C, André T, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol 2008; 26: 374-9; PMID:18202412; http://dx.doi.org/10.1200/JCO.2007.12.5906
- Benlloch S, Paya A, Alenda C, Bessa X, Andreu M, Jover R, Castells A, Llor X, Aranda FI, Massutí B. Detection of BRAF V600E mutation in colorectal cancer: comparison of automatic sequencing and real-time chemistry methodology. J Mol Diagn 2006; 8: 540-3; PMID:17065421; http://dx.doi.org/10.2353/jmoldx.2006.060070
- Pawlik TM, Raut CP, Rodriguez-Bigas MA. Colorectal carcinogenesis: MSI-H versus MSI-L. Dis Markers 2004; 20: 199-206; PMID:15528785; http://dx.doi.org/10.1155/2004/368680
- Flaman JM, Frebourg T, Moreau V, Charbonnier F, Martin C, Chappuis P, Sappino AP, Limacher IM, Bron L, Benhattar J, et al. A simple p53 functional assay for screening cell lines, blood, and tumors. Proc Natl Acad Sci U S A 1995; 92: 3963-7; PMID:7732013; http://dx.doi.org/10.1073/pnas.92.9.3963
