Abstract
Non-Hodgkin's lymphomas (NHLs) are malignant neoplasms which are clinically and biologically diverse. Their incidence is constantly increasing and despite treatment advances, there is a need for novel targeted therapies. Here, we identified Lectin-like transcript 1 (LLT1) as a biomarker of germinal center (GC)-derived B-cell NHLs. LLT1 identifies GC B cells in reactive tonsils and lymph nodes and its expression is maintained in B-cell NHLs which derive from GC, including Burkitt lymphoma (BL), follicular lymphoma (FL), and GC-derived diffuse large B-cell lymphoma (DLBCL). We further show that LLT1 expression by tumors dampens natural killer (NK) cell functions following interaction with its receptor CD161, uncovering a potential immune escape mechanism. Our results pinpoint LLT1 as a novel biomarker of GC-derived B-cell NHLs and as a candidate target for innovative immunotherapies.
Abbreviations:
- BL, Burkitt Lymphoma
- CLL, chronic lymphocytic leukemia
- DC, dendritic cell
- DLBCL, diffuse large B-cell lymphoma
- FFPE, formalin-fixed paraffin-embedded
- FL, follicular lymphoma
- GC, germinal center
- ICPs, immune check points
- IF, Immunofluorescence
- IFZ, interfollicular zone
- IHC, immunohistochemistry
- mAb, monoclonal antibody
- MALT, mucosa associated lymphoid tissue
- MCL, mantle cell lymphoma
- NHL, non-Hodgkin's lymphoma
- NK, natural killer
- PBMCs, peripheral blood mononuclear cells
- SLL, small-cell lymphocytic lymphoma
- TFH, T follicular helper cells
- TSA, Tyramide signal amplification
Introduction
NHLs are heterogeneous lymphoproliferative malignancies which show a considerable heterogeneity in their morphologic and immunophenotypic characteristics as well as in their clinical behaviors, ranging from indolent to aggressive clinical courses.Citation1-3 The vast majority of NHLs derive from B cells, the most frequent subtypes being FL and DLBCL. Treatments of B-cell NHLs have been improved by the introduction of immunotherapies, mainly the use of anti-CD20 antibody.Citation4 However, a significant proportion of NHL patients still remains not curable to date and there is a need to identify new targets for therapy. Citation5-7
A majority of genes expressed by malignant cells are shared with genes expressed in their non-transformed cellular progenitor. Most B-cell NHLs in humans are derived from germinal center (GC) and post-GC B cells consequently to genetic abnormalities.Citation8,9 B-cell NHLs originating from GC B cells include FL, BL and a subtype of DLBCL identified by gene expression profiling which expresses GC genes (GC B-like or GC-DLBCL). A second subtype of DLBCL displays the molecular features of activated peripheral blood B cells (activated B-like DLBCL or ABC-DLBCL).Citation10 GC-DLBCL and ABC-DLBCL differ strongly by their prognosis.Citation11 Gene expression profiling has considerably helped the classification of B-cell NHLs and the identification of GC specific genes may direct toward novel biomarkers and therapeutic targets for GC-derived B-cell NHLs.
The last decades have provided overwhelming evidence for a control of tumor development by the immune system, the so-called “immunoediting.”Citation12 A better understanding of the interactions of tumors with their microenvironment is leading to novel therapeutic opportunities. Major clinical benefits have indeed recently been obtained with the use of agonists or blockers of immune check points (ICPs).Citation13 In NHL patients, our understanding of the role of the microenvironment in the maintenance of B-cell lymphomas is increasing steadily.Citation14-17 Host immunity is often suppressed or diverted by lymphoma cells. Several mechanisms have been identified which lead to inefficient antitumor responses. These include downregulation of MHC class I and II,Citation18,19 loss of CD58,Citation19 disruption of the T cell/malignant B cell synapse, Citation20 T cell exhaustionCitation21 or recruitment/amplification of regulatory T cells.Citation22-24 Expression of ligands of inhibitory co-receptors has been detected on lymphomas and can subvert NK and T cell immune functions. Among those relatively well described molecules are programmed death ligand 1 (PD-L1) interacting with PD-1, a negative regulator of T cell proliferation and function, and Herpesvirus entry mediator (HVEM) interacting with B and T lymphocyte attenuator (BTLA) on αβ and γδ T cells.Citation25-27 In addition, NK cells are active players of the antitumoral immune response. They largely contribute to the efficacy of anti-CD20 immunotherapy, mediating antibody-dependent cell-mediated cytotoxicity (ADCC) through their FcγRIIIA (CD16).Citation28,29
We previously identified LLT1 as an antigen expressed on activated B cells and interacting with its receptor CD161 (hNKR-P1A) on NK cells and subsets of T cells.Citation30-32 While LLT1/CD161 interaction inhibits NK cell functions, it costimulates T cells.Citation30,32 We herein further characterized LLT1 expression in reactive and neoplastic human lymphoid cells and tissues and found that LLT1 identifies GC B cells and GC-derived B-cell NHLs. We then assessed the role of LLT1 in the modulation of NK cell antitumoral activity. Our results suggest that LLT1 may be used as a biomarker and a therapeutic target for these subtypes of NHL.
Results
LLT1 identifies GC B cells in human tonsils and reactive lymph nodes
To study LLT1 expression in human tissues, we developed highly sensitive IHC and immunofluorescence (IF) staining protocols of formalin-fixed paraffin-embedded (FFPE) sections using anti-LLT1 mAb clone 2F1 we generated and characterized.Citation33 IHC and IF stainings with anti-LLT1 mAb revealed a predominant cytoplasmic staining pattern as well as a membranous staining, consistent with our previous data showing both intracellular and cell surface expression of LLT1Citation32,33 (). Importantly, the IHC staining of human tonsils (, top panel) and lymph nodes (, bottom panel) revealed a predominant expression of LLT1 in GCs (arrows). The mantle cells surrounding the GCs were negative, with scarce positive cells detected at high magnification (, filled stars). A positive signal was also observed in a few scattered cells in the interfollicular zone (IFZ) surrounding the GCs (, empty stars).
Figure 1 (See previous page). Predominant expression of LLT1 in B cells from germinal centers. (A) IHC staining of FFPE sections from human tonsils (top panel) and human reactive lymph nodes (bottom panel) with hematoxylin (HE), isotype control or anti-LLT1 (clone 2F1). (B) Double immunofluorescence staining of FFPE tonsil sections using anti-CD20 or anti-LLT1 (clone 2F1) and colocalization together with Hoechst staining of nucleus. Top panel shows GC, mantel cell, and interfollicular (IFZ) zones and bottom panel shows GC B cells at higher magnification (A–B) show GCs (arrows) surrounded by mantle cell zone (filled stars) and IFZ (empty stars). Data are representative of 4 to 12 independent experiments. (C) Frequency among B and T cells in PBMCs and tonsils from flow cytometry analysis. n = 10, ****, p < 0.0001 Mann–Whitney U-test. (D) LLT1 detection by protein gel blot using anti-LLT1 (clone 2F1) in whole cell lysates of the indicated cells, digested or not with Endo H. β-actin is used as an internal control. Two differentially glycosylated forms of LLT1 are visualized: Endo H resistant form (black arrow) and Endo H sensitive form yielding two bands (white arrows 1 and 2). Data are representative of 2 independent experiments. (E–I) Multiparameter flow cytometry analysis of LLT1 expression. (E) Representative flow cytometry staining using anti-LLT1 (clone 4F68). (F) Representative expression of LLT1 and (G) frequency on gated CD19+ B cells analyzed according to CD38 expression. n =7, ***, p< 0.001 Mann–Whitney U-test. (H) Distribution of centroblast (CB) and centrocyte (CC) populations within CD19+ CD38+ and LLT1+ CD19+CD38+ gated tonsil cells according to the expression of CXCR4 and CD83. Citation35,36 (I) Scheme of the differentiation stages of mature B-cell development adapted from. Citation34 Each stage of B cell differentiation is visualized in the rectangles in the dot-plots below. Cells (gray dots) were gated using the markers indicated above each dot-plot and displayed according to markers on the dot-plot axis. On these dot-plots, an overlay of the gated CD3−CD19+LLT1+ B cells is shown and LLT1+ cells are visualized as black dots. (E–F–H–I) Data are representative of 3–6 independent experiments.
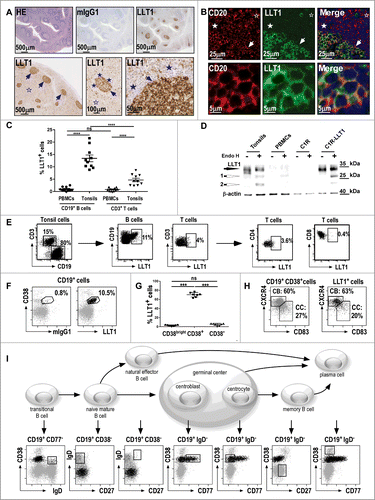
To further characterize the cells expressing LLT1, we performed double IF stainings of human tonsil FFPE sections ( and Fig. S1). LLT1 was found to co-localize with CD20-positive B cells within the GCs (). Naive B cells from the mantle zone and B cells from the T cell-rich IFZ did not stain for LLT1 ( and Fig. S1). Only a few CD3-positive T cells expressed LLT1, whereas LLT1 expression could not be detected on CD138+ plasma cells, CD303+ plasmacytoid dendritic cells (DCs), CD68+ macrophages, and CD83+CD11c+ DCs (Fig. S1).
The phenotype of LLT1+ cells in tonsils was further analyzed using multicolor flow cytometry and protein gel blot analyses (). As previously reported,Citation32 LLT1 cell surface expression could not be detected on PBMCs but LLT1 was expressed on a significant proportion of CD3−CD19+ B cells from tonsils (). This was consistent with the detection of Endo H (Endoglycosidase)-resistant and sensitive forms of LLT1 by protein gel blot on whole cell lysates from tonsils but not from PBMCs (). Cell surface expression of LLT1 correlated with acquisition of sugars that are resistant to Endo H digestion as found in tonsils and control C1R-LLT1 cells (). LLT1 could be detected on a small proportion of tonsil T cells, mostly CD4+ T cells but when compared to B cells, we consistently measured much lower level of LLT1 expression on T cells (). Interestingly, within tonsil B cells, LLT1 was specifically found in a subset expressing CD38 ( and 1G). Its detection on CD38+ but not on CD38bright and CD38− B cells suggested an expression limited to GC B cells. The differentiation stages of mature B cell development can be distinguished by their expression of specific cell surface markers. Based on the work of Van Zelm et alCitation34 using CD3, CD19, CD38, IgD, CD77 and CD27, we demonstrated a specific expression of LLT1 on the cell surface of centroblasts and centrocytes within GCs while no expression was found on transitional, naive mature, natural effector, memory B cells as well as plasma cells (). Indeed, gated CD3−CD19+LLT1+ cells solely overlaid with the GC B cells. Additional markers such as CD20, CD10, CD44, CD184 (CXCR4), CD83 and CD86 have also been used to better define centroblasts and centrocytes.Citation35,36 The combination of CXCR4 and CD83 defines particularly well the 2 subsets of GC B cells, allowing the detection of around 60% of centroblasts and 27% of centrocytes among CD3−CD19+CD38+ B cells ().Citation36 A similar proportion was found among LLT1+ B cells (). LLT1+ B cells were also found to be CD20+, CD10+, CD44−/low, IgM−, and IgG+ or − (Fig. S1).
Infiltration of CD161+ NK and T cells in the interfollicular zone surrounding GC
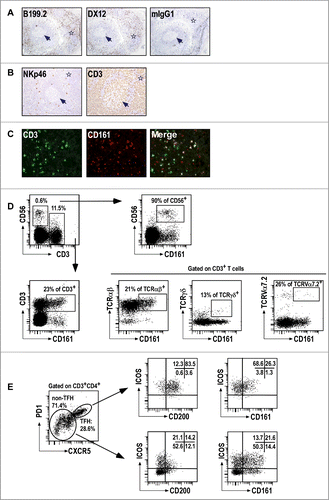
As LLT1 interacts with CD161 receptor expressed by NK and subsets of T cells, we then investigated whether such cells could be found within tonsils. We performed IHC stainings of human frozen tonsil sections using 2 independent anti-CD161 mAbs, clones B199.2 and DX12. CD161+ cells were mostly found in the IFZ surrounding GC (). Scattered NKp46+NK cells were also located in the IFZ while CD3+ T cells were predominantly found in the IFZ and also significantly within GC (). Double IF stainings for CD161 and CD3 identified both CD3− and CD3+ cells expressing CD161 in the IFZ, consistent with the presence of CD161+ NK and T cells, respectively (). Multicolor flow cytometry stainings were also performed which identified CD161+ CD56+ CD3− NK cells and CD161+CD3+ T cells in tonsils (). CD161 was expressed by the majority of NK cells and by a significant proportion of T cells which were mostly composed of TCRαβ+ T cells but also of small numbers of TCRγδ+ T cells and TCRVα7.2+mucosal-associated invariant T cells characterized by the expression of high level of CD161 (). Citation37 The combination of CD45RA and CCR7 is usually used to identify naïve (CD45RA+CCR7+), terminally differentiated effector (CD45RA+CCR7−), effector memory (CD45RA−CCR7−) and central memory (CD45RA−CCR7+) T cells. Citation38 In agreement with CD161 expression pattern on T cells from the periphery,Citation39 CD161 expression on tonsil T cells was primarily found on effector memory and central memory T cells, with CD161+ CD8+ T cells being mostly of effector memory phenotype while CD161+CD4+ T cells were mostly central memory T cells (Fig. S2). Follicular T helper (TFH) cells are found within GC and are characterized by a bright expression of PD1, CXCR5, ICOS and CD200.Citation24,40 We detected a significant proportion of TFH among CD3+CD4+ T cells and found similar proportion of CD161+ cells in TFH and non-TFH CD3+CD4+ T cells ( and data not shown). Interestingly, the level of CD161 expression on TFH was significantly lower, probably explaining that we did not detect CD161 staining within GC by IHC ( and data not shown).
Figure 2. CD161+ NK and T cells in tonsils. (A–B) IHC staining of frozen tissue sections from human tonsils with the indicated (A) anti-CD161 mAbs, and (B) anti-NKp46 and anti-CD3 mAbs. (C) Double IF staining of the IFZ on tonsil frozen tissue sections using anti-CD3 and anti-CD161. (D, E) Multiparameter flow cytometry analysis of CD161 expression. Representative expression of CD161 in CD56+CD3−NK cells and subsets of T cells (D), including TFH (E). Data are representative of 3 independent experiments.
Together, these results demonstrate that CD161-expressing NK and T cells are present within tonsils where they can interact with LLT1+ GC B cells.
LLT1 expression is maintained in GC-derived B-cell non-Hodgkin's lymphomas
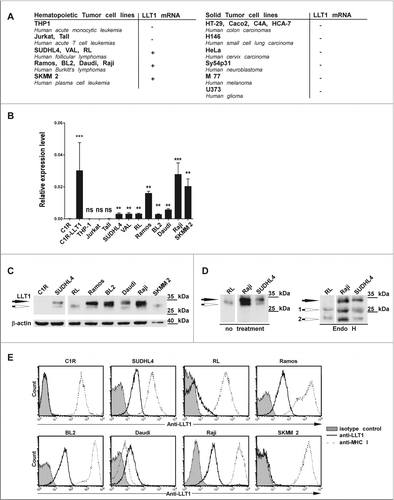
Since we found that LLT1 was brightly expressed by GC B cells, we investigated whether LLT1 expression could be maintained in subtypes of B-cell lymphomas. LLT1 expression was first assessed in a panel of cell lines derived from various types of cancers, both at the transcriptional () and at the protein levels (). CLEC2D transcript variant 1 encoding for LLT1 could not be detected in human cell lines derived from solid tumors, including colon, small cell lung and cervix carcinomas, neuroblastoma, melanoma and glioma (). Among the haematopoietic cancer cell lines tested, LLT1 mRNA was detected in tumors of B-cell but not of T cell or myeloid origin (). LLT1 protein was also detected in a variety of B lymphoma cell lines as shown by protein gel blot analysis of whole cell lysates () and by flow cytometry staining (). Interestingly, cell surface expression of LLT1 was correlated with acquisition of sugars and Endo H resistance as shown by the detection of individual bands by protein gel blot, the upper band being Endo H resistant (filled arrow) and the lower bands Endo H sensitive (empty arrows) (). Expression of LLT1 on the cell surface was restricted to cell lines derived from transformed FL and BL, with RL cell line expressing lower level. By contrast, LLT1 remained expressed intracellularly in SKMM2 cell line derived from plasma cell leukemia ().
Figure 3. LLT1 expression in B lymphoma cell lines. (A–B) CLEC2D transcript variant 1 coding for LLT1 quantified by real-time RT-PCR in the indicated cell lines, summarized in (A) and expressed relative to β-actin in (B). Statistical significance against LLT1− C1R cells was calculated n = 4 to 30, ***, p < 0.001, **, p< 0.01, Mann–Whitney U-test. (A–B) Data are representative of 3 independent experiments. (C–D) LLT1 expression detected by protein gel blot analysis of whole cell lysates digested or not with Endo H using anti-LLT1 mAb (clone 2F1). β-actin is used as an internal control. Two differentially glycosylated forms of LLT1 are visualized: Endo H resistant form (black arrow) and Endo H sensitive form (white arrows 1 and 2). Data are representative of 3 experiments. (E) LLT1 cell surface expression monitored by flow cytometry with anti-LLT1 mAb (clone 4F68) compared to isotype mIgG1 control and anti-MHC class I mAb (clone DX17). Data are representative of 8 independent experiments.
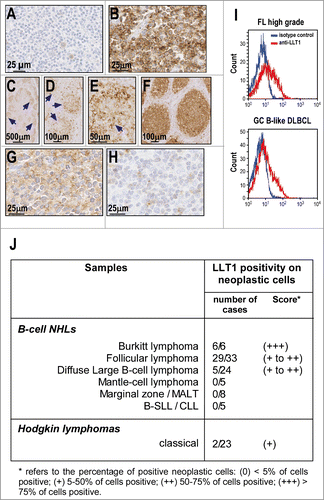
We then examined LLT1 expression in human primary NHL samples using IHC staining of FFPE sections. Among B-cell NHLs, LLT1 expression was restricted to BL, FL and some DLBCL and was not found in other B-cell NHLs (). Gastric marginal B-cell lymphoma (MALT type) is shown as a representative negative case (). The strongest positive signal for LLT1 was observed in BL samples, in which virtually 100% of malignant B cells stained at a high intensity in all cases analyzed (6/6, score +++) (). In FL samples, positive staining was observed in neoplastic B-cell follicles in the majority of cases (29/33, score + to ++) (). The positivity was stronger in large centroblastic cells than in centrocytic cells (). This resulted in a tendency of high grade FL () to exhibit a stronger expression than low grade FL () as the former displays a higher content of centroblasts.Citation1 Among DLBCL, positivity of neoplastic cells was observed in a minority of samples (5/24, score + to ++) (). The positive cases were all classified as GC-derived according to the profile of CD10, BCL6 and MUM1 expression, as previously described.Citation41 By contrast, other subtypes of B-cell NHLs including mantle cell lymphoma (MCL) (0/5), marginal zone/mucosa associated lymphoid tissue (MALT) lymphoma (0/8) and small-cell lymphocytic lymphoma/chronic lymphocytic leukemia (B-SLL/CLL) (0/5) exhibited no LLT1 positivity among the malignant cell population. Moreover, most Hodgkin's lymphoma samples examined displayed no LLT1 expression in neoplastic Reed–Sternberg cells, except in rare cases (2/23, score +) ( and data not shown).
Figure 4. LLT1 expression in primary B-cell NHLs. (A–H) Representative IHC staining with anti-LLT1 mAb (clone 2F1) of lymph node biopsies from patients: (A) gastric marginal B-cell lymphoma (MALT type); (B) Burkitt lymphoma; (C–E) low grade follicular lymphoma; (F) high grade follicular lymphoma; (G) Diffuse large B-cell lymphoma of GC type and (H) DLBCL of activated B-cell type. (C–E) Of note, the level of expression in low grade FL appears heterogenous within the neoplastic follicles (arrows) due to a stronger expression in centroblasts than in centrocytes. Accordingly, the expression appears higher in high grade FL (F) than in low grade FL (C–E). (I) LLT1 cell surface expression on the indicated primary NHLs monitored by flow cytometry with anti-LLT1 (clone 4F68). (J) Summary of immunohistochemical analysis of LLT1 expression in lymphoid neoplasms.
Consistent with the IHC staining results, flow cytometry analysis of isolated primary lymph node cells from NHL patients revealed that LLT1 was expressed at the cell surface of FL and DLBCL tumors ().
Altogether, our findings identify LLT1 as a novel biomarker predominantly associated with GC-derived B-cell lymphomas.
LLT1 expressed by GC-derived B-cell lymphomas is functional and inhibits NK cell functions
LLT1 interacts with CD161, an inhibitory receptor expressed by the majority of NK cells.Citation30,31 As NK cells are key actors of the antitumoral response and are also crucial effector cells in antibody therapy, we investigated whether LLT1 expression on B-cell NHLs controls NK cell activation, cytotoxicity and production of IFNγ. Blocking LLT1/CD161 interaction with anti-LLT1 mAb clone 4F68,Citation32 either as mIgG1, or as F(ab')2 fragment and chimeric 4F68-hIgG4 increased NK cell degranulation in the presence of LLT1-expressing lymphoma cell lines such as Raji, SUDHL4, Ramos and Daudi but had little effect in the presence of LLT1−/low RL cells ( and Fig. S3A). NK cell cytotoxicity toward LLT1+ SUDHL4 lymphoma was also significantly increased in the presence of blocking anti-LLT1 (). Similarly, the blocking of LLT1/CD161 interaction with anti-LLT1 mAb increased IFNγ production by NK cells in the presence of lymphoma cell lines ( and data not shown). Blocking the receptor CD161 with a specific anti-CD161 also increased NK cell degranulation and IFNγ production ( and C, and Fig. S3A, and data not shown). Interestingly, we observed an additive effect when LLT1/CD161 interaction was blocked together with MHC class I molecule interaction with inhibitory KIRs and CD94/NKG2A ( and Fig. S3A). These results therefore indicate that LLT1 expression by B-cell lymphomas is functional and contributes to protect them from NK cell attack. Importantly, LLT1 synergizes with MHC class I molecules to provide inhibitory signals to NK cells.
Figure 5 (See previous page). LLT1 expression by B-cell lymphoma inhibits NK cell functions. Polyclonal NK cells were incubated with the following lymphoma cell lines (A, C) Raji, (A,B) SUDHL4 in the presence or absence of blocking mAbs as indicated. (A) NK cell degranulation assessed by expression of CD107a on the cell surface. (B) NK cell cytotoxicity toward SUDHL4 in the presence or absence of indicated antibodies. n = 3 to 6, **, p < 0.01 Mann–Whitney U-test. (C) Intracellular secretion of IFNγ detected by flow cytometry staining. (A, C) Data are representative of 3 to 6 independent experiments. (D) Frequency of CD56+CD3−NK cells (left panel) and CD161+ cells among NK cells (right panel) in PBMCs, tonsils and NHL patients including 4 FL and 3 GC-DLBCL. n = 7, ***, p< 0.001, **, p< 0.01, Mann–Whitney U-test. (E) Representative expression of CD161 in CD56+CD3−NK cells and CD3+ T cells.
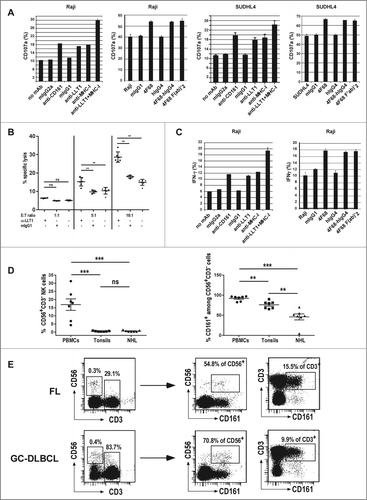
Within the tumor microenvironment, we detected the presence of NK cells as depicted in , showing respectively their frequency and representative flow cytometry stainings of FL and GC-DLBCL patients. NK cell frequency in the tumor microenvironment is not different from tonsils but is significantly lower than in blood (, left panel). Interestingly, we observed a modulation of the level of CD161 expression on NK cells with significant decrease in NHL patients as well as in tonsils, although to a lesser extend (, right panel). CD161 was also expressed on a proportion of T cells those phenotypes remain to be fully defined. Of note, TFH cells could be detected in FL patients but were absent in GC-DLBCL, consistent with previous reportsCitation16 (Fig. S3B). The level of CD161 expression was much lower on TFH compared to non-TFH CD4+ T cells, consistent with our findings in tonsils (Fig. S3B and data not shown).
In conclusion, we have shown that LLT1 expressed by B-cell lymphomas is functional and can interact with NK cells present within the tumor microenvironment, dampening their effector functions.
Discussion
B-cell NHLs form a group of heterogeneous lymphoproliferative malignancies, often clinically and histologically difficult to distinguish and of diverse prognosis. The cure of B-cell NHLs remains a crucial medical challenge that may be met through rational development of targeted therapies. In our study, we show for the first time that LLT1 identifies GC B cells and GC-derived B-cell NHLs. Through its interaction with CD161, LLT1 modulates immune responses and may contribute to tumor immune escape. LLT1 may therefore constitute a novel biomarker and a candidate target for GC-derived B-cell NHLs.
B-cell NHLs bear resemblance to particular stages of B cell differentiation and it has been postulated that for most B-cell NHL subtypes, this “normal B cell counterpart” corresponds to the cell of origin.Citation3,8 GC B cells are considered as the precursor cell of some B-cell NHLs, including FL, BL and the GC-DLBCL. Our finding that LLT1 is specifically detected on both normal GC B cells and GC-derived B-cell NHLs, but not on the other B-cell NHLs reinforce the GC origin of FL, BL and GC-DLBCL. LLT1 may thus serve as a marker for the diagnosis of GC-derived B-cell NHLs and may be particularly useful for the discrimination of DLBCLs which can be divided into two prognostically significant subgroups, GC-DLBCL and ABC-DLBCL, histologically difficult to distinguish.Citation10,42 Their definitive classification requires cDNA microarray gene profiling which is not widely available.Citation10,43 The anti-LLT1 mAb clone 2F1 we developed is suitable for IHC staining on paraffin sections and it therefore offers a potential accessible routine test that could facilitate and speed up diagnosis. While pioneer microarray studies used non-specific probes for CLEC2D-encoding LLT1,Citation10,44,45 a recent study using specific probes have classified CLEC2D within the top 5% genes significantly upregulated in GC-DLBCL compared to ABC-DLBCL,Citation46 thus in agreement with our findings. Further studies on large cohorts are warranted to conclude whether LLT1 can serve as a distinctive marker of DLBCL subsets. Similarly, LLT1 IHC staining may help the current diagnosis of BL which often requires the use of cytogenetics due to the border line cases displaying intermediate patterns between DLBCL and BL.Citation1 In addition, our finding of a predominant expression of LLT1 on centroblasts may prove useful for the grading of FL patients. Indeed, high grade FL biopsies display the highest frequency of centroblasts,Citation1 and as a result, they were strongly stained by anti-LLT1 mAb. Predominant expression of LLT1 on centroblasts might be correlated with their high capacity to proliferate as we previously reported that LLT1 expression was induced upon activation on proliferating cells.Citation32
Besides being a potential novel biomarker, LLT1 might constitute a relevant therapeutic target for GC-derived B-cell NHLs, either as an antigen to target, or as a modulator of antitumoral immunity. One advantage of targeting LLT1 is its increased specificity for cancer cells compared to CD20 more ubiquitously expressed. The development of anti-LLT1 mAbs with depleting properties may prove efficient as an alternative or in combination with anti-CD20 therapy and especially useful for patients refractory/resistant to anti-CD20 rituximab treatments.Citation5 Blocking or enhancing LLT1/CD161 interaction with anti-LLT1 mAb to boost antitumoral NK and T cell activities may also constitute an alternative approach in therapy. Recent findings have highlighted the benefit of combining blockers of ICPs, such as anti-PD1, with rituximab in patients with relapsed FLs.Citation47 The use of therapeutic agents targeting ICPs has been shown to enhance endogenous antitumor immune responses in several settings and has raised new hopes in cancer treatment.Citation13 The involvement of LLT1/CD161 interaction in the regulation of antitumor responses is still to demonstrate but current findings strongly suggest it can modulate NK and T cell responses.Citation30 Accumulating evidence show a predominant role of the microenvironment in the establishment and the maintenance of B-cell lymphomas.Citation14,16 Its composition varies between lymphoma subtypes.Citation16 CD56+ NK cells were detected among cells of the tumor microenvironment in FL and GC-DLBCL patients. The antitumoral role of NK cells is well documented in vitro, although it remains poorly appreciated in vivo.Citation14 Our finding that blocking LLT1/CD161 interaction increases NK cell secretion of IFNγ and NK cell killing of B-cell lymphoma cell lines suggests that LLT1 expression may constitute a novel immune escape mechanism. Counteracting this immune escape mechanism with blocking anti-LLT1 mAbs may not only improve the control of tumor growth by the immune system but also enhance the efficiency of anti-CD20 immunotherapies which rely in part on NK cells as effector cells triggering ADCC.Citation28,29 Besides NK cells, the tumor microenvironment is composed of multiple T cell subsets with diverse functions. Citation16 We have shown that CD161 is expressed by these different T cell subsets and thus may participate to their activation and/or exhaustion, as we and other demonstrated that CD161 is a costimulatory molecule on T cells.Citation32,37,48 Interestingly, we found that TFH which are in contact with LLT1-expressing GC B cells express lower level of CD161 compared to non-TFH cells. Whether this relates to downregulation of CD161 upon interaction with LLT1 as previously reported,Citation30,37 or to low level of expression on TFH remains to be addressed. A better understanding of the physiological role of LLT1/CD161 interaction within GC is necessary to predict the best way of modulating the interaction for therapeutic purposes.
Our study has shown for the first time that LLT1 is highly expressed by GC B cells. Signals that trigger LLT1 are currently unknown but may be linked to the GC microenvironment. BCL6, a master regulator of GC B cell differentiationCitation49-51 is one of the candidates as it matches LLT1 expression pattern and it facilitates B cell proliferation. GCs are the site of affinity maturation of antibody-dependent immune responses and are therefore at the center of the immune response against invading microorganisms.Citation52,53 Activation of GC precursor cells is antigen-dependent and relies on T-B cognate interactions involving cytokines such as IL-4 or adhesion and costimulatory molecules such as CD40/CD40L and the B7 family of ligands/receptors. Induction of LLT1 may result from these types of interactions since signals that can induce LLT1 on peripheral B cells include cross-linking of the BCR and CD40 or TLR signaling with a synergistic effect of IFNγ.Citation32 Once expressed, LLT1 may contribute to the GC reaction along with costimulatory receptors and TLRs. Interestingly, TLR agonists have been shown to promote GC development.Citation54,55 Whether LLT1 is also involved in this process needs to be investigated. Within GCs, survival and death which contribute to the selection of the clones secreting the mAbs with the highest affinity are determined by a complex interplay of positive and negative signals.Citation52,53 It also remains to establish whether LLT1/CD161 interaction contributes to this balance.
In conclusion, we report here the identification of LLT1 as a novel biomarker of B-cell NHLs and as a novel player in B cell biology. Furthermore, we have developed anti-LLT1 mAbs that could not only facilitate and speed up diagnosis but may be considered as innovative therapeutic tools for the treatment of GC-derived B-cell NHLs.
Materials and Methods
Cell lines and primary cells
B lymphoma cell lines, C1R and C1R-LLT1 cells were maintained as previously described.Citation32 Tonsil cells were obtained from discarded benign tonsils after informed consent from patients undergoing routine tonsillectomies at the Lenval Hospital, Nice. Cells were mechanically collected. Lymph node suspension cells were collected and processed as previously described Citation56 at diagnosis from FL and DLBCL patients. Recruitment was performed under institutional review board approval (University Hospital of Rennes) and informed consent process according to the Declaration of Helsinki. Diagnoses were based on histopathologic, immunophenotypic and molecular analyses according to the World Health Organization (WHO) classification of lymphoid tumors. Citation1 Classification of DLBCL into cell-of-origin subtypes was based on Hans algorithm.Citation57 Purified NK cells were generated from blood purchased from the Etablissement Français du Sang (Marseille). The purification was performed as previously described.Citation32 All NK cells used contained <5% CD3+CD56+ T cells.
Tissue sampling
A total of 104 lymphoma biopsy samples were analyzed from patients recruited under institutional review board approval and informed consent process according to the Declaration of Helsinki, from Paoli Calmettes Institute (Marseille, France) and University Hospital (Rennes, France). These samples included 23 cases of Hodgkin's Lymphomas and 81 specimens of NHLs, which were classified according to the WHO classification.Citation1 Diagnosis was based on morphological examination of FFPE material and IHC using monoclonal antibodies (mAb) recognizing B cells, T cells, or Reed-Sternberg cells. A control group of non-neoplastic lymphoid tissues from reactive lymphadenitis (n = 12) and tonsils (n = 4) was also tested.
Immunohistochemistry
IHC staining for LLT1 was performed with mouse (Mo) anti-LLT1 clone 2F1 mAb Citation33 on FFPE tissue sections (4 μm). FFPE sections were deparaffinized and rehydrated in successive baths of xylene, ethanol and water followed by heat-induced epitope retrieval in Tris/EGTA buffer pH 9. Endogenous peroxidase activity and biotin were blocked by H2O2 (0.5% v/v) and avidin–biotin blocking kit (DAKO), respectively. Unspecific protein-binding was blocked with 7% donkey serum (Do), 3% human serum, 3% bovine serum albumin, 3% skim milk in Tris Buffer Saline (TBS). Sections were washed in TBS with 0.05% Tween20 between steps. FFPE sections were incubated with Mo anti-LLT1 mAb (2F1) or isotype mIgG1 control (R&D Systems) at 0.1 µg/mL. Antibody binding was detected with biotinylated Do anti-Mo IgG secondary antibody (Jackson Immunoresearch) at 0.3 µg/mL and visualized using avidin–biotin-HRP complexes (Vectastain, Vector) in combination with the highly sensitive detection method Tyramide signal amplification (TSA), according to manufacturer's instructions (Perkin Elmer). Nuclei were counterstained with hematoxylin. Immunostained slides were analyzed by light microscopy using an AX70 microscope (Olympus). Pictures were captured with a DP71 camera and Cell A software (Olympus). Immunostaining in a given cell population was evaluated by the hematopathologist and marked (0) if less than 5% of the cells were positive; (+) if 5 to 50% of the cells were positive; (++) if 50 to 75% of the cells were positive and (+++) if greater than 75% of the cells were positive. The specificity of the staining was validated using another anti-LLT1 mAb, clone MAB3480 (R&D) which stained the same structures as the 2F1 clone, although with a much weaker signal (data not shown).
IHC stainings for CD161, NKp46, and CD3 were performed on frozen tissue sections (5 μm) fixed in acetone. Endogenous peroxidase activity and unspecific protein-binding were blocked as described above. Primary antibodies: Mo anti-CD161 (B199.2, AbD Serotech), Mo anti-NKp46 (9F2, BD Pharma), and Rb anti-CD3 (SP7, Neomarkers) were used at 0.1 μg/mL and Mo anti-CD161 (DX12, BD Biosciences) was used at 1 μg/mL. Antibody binding was detected with biotinylated anti-Mo or Rb IgG secondary antibodies (Jackson Immunoresearch) at 0.17 μg/mL and visualized as described above.
Immunofluorescence
FFPE sections were deparaffinized, pre-treated and incubated with Mo anti-LLT1 mAb (2F1)Citation33 or isotype mIgG1 control (R&D) at 1 µg/mL followed by secondary biotinylated Do anti-Mo IgG as described above. Antibody binding was fluorescently stained using Vectastain and TSA labeled with Alexa-488. For double-IF staining, pretreatment was repeated as described above followed by application of anti-CD20cy (clone L26, Dako) at 0.5 µg/mL to label B cells or isotype mIgG2a control (Dako). Antibody binding was detected by corresponding biotinylated Do anti-Mo IgG antibodies and visualized with Vectastain and TSA labeled with Alexa-594. Nuclei were counterstained with Hoechst and sections were mounted in Fluorescence Mounting Medium (FMM, Dako). Immunostained slides were analyzed by epifluorescence microscopy using an Axio Imager M2 microscope (Zeiss). Pictures were captured with an AxioCam MRm camera and ZEN software (Zeiss).
Double IF staining for CD161 and CD3 was performed on frozen tissue sections (5 μm) using rat anti-CD3 (CD3-12, Abd Serotech) at 0.3 μg/mL, Mo anti-CD161 (DX12, BD Biosciences) at 3 μg/mL, or the corresponding isotype controls followed by secondary Alexa-488-anti-rat IgG and biotinylated anti-Mo IgG at 0.3 μg/mL. Anti-CD161 binding was detected as described above with Alexa-594-Streptavidin.
Real-time RT-PCR
LLT1 mRNA was quantified by real-time RT-PCR as previously described.Citation33 Transcript levels were expressed relative to β-actin.
Biochemistry and protein gel blot
Whole cell lysates were prepared as previously describedCitation33 and digested or not with Endo H according to the manufacturer's instructions (New England Biolabs). Proteins were loaded on a 12% SDS-PAGE and transferred to nitrocellulose membranes. LLT1 or β-actin were detected as previously described.Citation33
Flow cytometry
Lymphoma cell lines were stained with purified Mo anti-LLT1 (4F68)Citation33 and anti-MHC class I (DX17, kindly provided by L. Lanier) mAbs followed by APC-conjugated goat anti-Mo Ig (BD Biosciences). Tonsil and primary lymphoma cells were treated as previously described.Citation32 Briefly, cells were incubated in PBS, 0.1% BSA, 10% normal mouse serum and 100 μg/mL human Ig for 30 min on ice prior to incubation with biotinylated anti-LLT1 (4F68) mAb or isotype mIgG1 control, followed by PE-CF594-conjugated streptavidin (BD Biosciences) and additional labeled antibodies to CD3 (UCHT1), CD4 (L200), CD8 (SK1), CD16 (3G8), CD19 (SJ25C1), CD27 (M-T271), CD38 (HIT2), CD56 (B159), CD77 (5B5), CD83 (HB15e), CD184-CXCR4 (12G5), CD200 (MRC OX-104), CD278-ICOS (DX29), CD279-PD1 (MIH4), IgD (IA6-2) (BD Biosciences) as well as to CD161 (191B8) (Miltenyi), to CXCR5 (M-T271) (e-Biosciences), to TCRVα7.2 (3C10) (Biolegend), to TCR-αβ (BMA 031) and to TCRγδ (Immu 510) (Beckman Coulter). Fluorescence was analyzed on a BD LSRFortessa™ cell analyzer with BD FACSDiva software (BD Biosciences).
NK cell assays
Polyclonal NK cells were stimulated for 4–5 h with B lymphoma cell lines at an E:T ratio of 1:3 in the presence or not of the indicated mAbs either as mIgG1, F(ab')2 fragments, or hIgG4 chimeric mAb (10 μg/mL). NK cell degranulation and IFNγ production were monitored as described previously.Citation32 NK cell cytototoxicity was measured using a CFSE-based assay adapted from.Citation58 Briefly, SUDHL4 cell line was labeled with 0.5 μM CFSE (Molecular Probes) for 10 min at 37°C. Reaction was blocked with cold FCS on ice for 5 min and cells were washed twice. 20000 CFSE-labeled target cells were co-cultured for 4 h in a humidified atmosphere of 5% CO2 at 37°C with polyclonal NK cells at the indicated effector to target (E:T) ratio, in the absence or presence of 20 μg/mL mIgG1 or anti-LLT1 (clone 4F68) blocking mAb. 0.1 μg/mL propidium iodide (PI) was added before analysis on a BD LSRFortessa™ cell analyzer. Spontaneous lysis was typically around 10%. The percentage of specific lysis was calculated using the percentage of PI-positive cells among CFSE-labeled targets.
Statistical analysis
The Mann–Whitney U-test was used to estimate the level of statistical significance of the difference between groups of data. A p value <0.05 was considered as evidence for statistical significance.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
supplementary_file.doc
Download MS Word (1.1 MB)Funding
This work was supported by Novo Nordisk S/A, Agence Nationale de Recherches sur le Syndrome de l'Immunodeficience-Acquise (SIDA), Ensemble contre le SIDA, Fondation ARC, Agence Nationale de la Recherche (ANR), Cancéropole PACA, the Centre National de la Recherche Scientifique and the French Government (National Research Agency, ANR) through the “Investments for the Future” LABEX SIGNALIFE: program reference #ANR-11-LABX-0028-01.
References
- Jaffe ES. The 2008 WHO classification of lymphomas: implications for clinical practice and translational research. Hematology Am Soc Hematol Educ Program 2009:523-31; PMID:20008237; http://dx.doi.org/10.1182/asheducation-2009.1.523
- Turner JJ, Morton LM, Linet MS, Clarke CA, Kadin ME, Vajdic CM, Monnereau A, Maynadié M, Chiu BC, Marcos-Gragera R et al. InterLymph hierarchical classification of lymphoid neoplasms for epidemiologic research based on the WHO classification (2008): update and future directions. Blood 2010; 116:e90-8; PMID:20699439; http://dx.doi.org/10.1182/blood-2010-06-289561
- Shaffer AL, 3rd, Young RM, Staudt LM. Pathogenesis of human B cell lymphomas. Annu Rev Immunol 2012; 30:565-610; PMID:22224767; http://dx.doi.org/10.1146/annurev-immunol-020711-075027
- Motta G, Cea M, Moran E, Carbone F, Augusti V, Patrone F, Nencioni A. Monoclonal antibodies for non-Hodgkin's lymphoma: state of the art and perspectives. Clin Dev Immunol 2010; 2010:428253; PMID:21437222; http://dx.doi.org/10.1155/2010/428253
- Henry C, Deschamps M, Rohrlich PS, Pallandre JR, Remy-Martin JP, Callanan M, Traverse-Glehen A, GrandClément C, Garnache-Ottou F, Gressin R et al. Identification of an alternative CD20 transcript variant in B-cell malignancies coding for a novel protein associated to rituximab resistance. Blood 2010; 115:2420-9; PMID:20089966; http://dx.doi.org/10.1182/blood-2009-06-229112
- McLaughlin P, Grillo-Lopez AJ, Link BK, Levy R, Czuczman MS, Williams ME, Heyman MR, Bence-Bruckler I, White CA, Cabanillas F et al. Rituximab chimeric anti-CD20 monoclonal antibody therapy for relapsed indolent lymphoma: half of patients respond to a four-dose treatment program. J Clin Oncol 1998; 16:2825-33; PMID:9704735
- Czuczman MS, Olejniczak S, Gowda A, Kotowski A, Binder A, Kaur H, Knight J, Starostik P, Deans J, Hernandez-Ilizaliturri FJ. Acquirement of rituximab resistance in lymphoma cell lines is associated with both global CD20 gene and protein down-regulation regulated at the pretranscriptional and posttranscriptional levels. Clin Cancer Res 2008; 14:1561-70; PMID:18316581; http://dx.doi.org/10.1158/1078-0432.CCR-07-1254
- Kuppers R, Klein U, Hansmann ML, Rajewsky K. Cellular origin of human B-cell lymphomas. N Engl J Med 1999; 341:1520-9; PMID:10559454; http://dx.doi.org/10.1056/NEJM199911113412007
- Stevenson F, Sahota S, Zhu D, Ottensmeier C, Chapman C, Oscier D, Hamblin T. Insight into the origin and clonal history of B-cell tumors as revealed by analysis of immunoglobulin variable region genes. Immunol Rev 1998; 162:247-59; PMID:9602369; http://dx.doi.org/10.1111/j.1600-065X.1998.tb01446.x
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000; 403:503-11; PMID:10676951; http://dx.doi.org/10.1038/35000501
- Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med 2002; 346:1937-47; PMID:12075054; http://dx.doi.org/10.1056/NEJMoa012914
- Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol 2006; 6:836-48; PMID:17063185; http://dx.doi.org/10.1038/nri1961
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012; 12:252-64; PMID:22437870; http://dx.doi.org/10.1038/nrc3239
- Ame-Thomas P, Tarte K. The yin and the yang of follicular lymphoma cell niches: role of microenvironment heterogeneity and plasticity. Semin Cancer Biol 2014; 24:23-32; PMID:23978491; http://dx.doi.org/10.1016/j.semcancer.2013.08.001
- Kridel R, Sehn LH, Gascoyne RD. Pathogenesis of follicular lymphoma. J Clin Invest 2012; 122:3424-31; PMID:23023713; http://dx.doi.org/10.1172/JCI63186
- Scott DW, Gascoyne RD. The tumour microenvironment in B cell lymphomas. Nat Rev Cancer 2014; 14:517-34; PMID:25008267; http://dx.doi.org/10.1038/nrc3774
- Thieblemont C, Delfau-Larue MH, Coiffier B. Lenalidomide in diffuse large B-cell lymphoma. Adv Hematol 2012; 2012:861060; PMID:23251161; http://dx.doi.org/10.1155/2012/861060
- List AF, Spier CM, Miller TP, Grogan TM. Deficient tumor-infiltrating T-lymphocyte response in malignant lymphoma: relationship to HLA expression and host immunocompetence. Leukemia 1993; 7:398-403; PMID:7680400
- Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, Wells VA, Grunn A, Messina M, Elliot O et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet 2011; 43:830-7; PMID:21804550; http://dx.doi.org/10.1038/ng.892
- Ramsay AG, Clear AJ, Kelly G, Fatah R, Matthews J, Macdougall F, Lister TA, Lee AM, Calaminici M, Gribben JG. Follicular lymphoma cells induce T-cell immunologic synapse dysfunction that can be repaired with lenalidomide: implications for the tumor microenvironment and immunotherapy. Blood 2009; 114:4713-20; PMID:19786615; http://dx.doi.org/10.1182/blood-2009-04-217687
- Yang ZZ, Grote DM, Ziesmer SC, Niki T, Hirashima M, Novak AJ, Witzig TE, Ansell SM. IL-12 upregulates TIM-3 expression and induces T cell exhaustion in patients with follicular B cell non-Hodgkin lymphoma. J Clin Invest 2012; 122:1271-82; PMID:22426209; http://dx.doi.org/10.1172/JCI59806
- Yang ZZ, Novak AJ, Stenson MJ, Witzig TE, Ansell SM. Intratumoral CD4+CD25+ regulatory T-cell-mediated suppression of infiltrating CD4+ T cells in B-cell non-Hodgkin lymphoma. Blood 2006; 107:3639-46; PMID:16403912; http://dx.doi.org/10.1182/blood-2005-08-3376
- Lee NR, Song EK, Jang KY, Choi HN, Moon WS, Kwon K, Lee JH, Yim CY, Kwak JY. Prognostic impact of tumor infiltrating FOXP3 positive regulatory T cells in diffuse large B-cell lymphoma at diagnosis. Leuk Lymphoma 2008; 49:247-56; PMID:18231910; http://dx.doi.org/10.1080/10428190701824536
- Ame-Thomas P, Le Priol J, Yssel H, Caron G, Pangault C, Jean R, Martin N, Marafioti T, Gaulard P, Lamy T et al. Characterization of intratumoral follicular helper T cells in follicular lymphoma: role in the survival of malignant B cells. Leukemia 2012; 26:1053-63; PMID:22015774; http://dx.doi.org/10.1038/leu.2011.301
- Gertner-Dardenne J, Fauriat C, Orlanducci F, Thibult ML, Pastor S, Fitzgibbon J, Bouabdallah R, Xerri L, Olive D. The co-receptor BTLA negatively regulates human Vgamma9Vdelta2 T-cell proliferation: a potential way of immune escape for lymphoma cells. Blood 2013; 122:922-31; PMID:23692853; http://dx.doi.org/10.1182/blood-2012-11-464685
- Xerri L, Chetaille B, Serriari N, Attias C, Guillaume Y, Arnoulet C, Olive D. Programmed death 1 is a marker of angioimmunoblastic T-cell lymphoma and B-cell small lymphocytic lymphoma/chronic lymphocytic leukemia. Hum Pathol 2008; 39:1050-8; PMID:18479731; http://dx.doi.org/10.1016/j.humpath.2007.11.012
- Sedy JR, Gavrieli M, Potter KG, Hurchla MA, Lindsley RC, Hildner K, Scheu S, Pfeffer K, Ware CF, Murphy TL et al. B and T lymphocyte attenuator regulates T cell activation through interaction with herpesvirus entry mediator. Nat Immunol 2005; 6:90-8; PMID:15568026; http://dx.doi.org/10.1038/ni1144
- Cartron G, Dacheux L, Salles G, Solal-Celigny P, Bardos P, Colombat P, Watier H. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood 2002; 99:754-8; PMID:11806974; http://dx.doi.org/10.1182/blood.V99.3.754
- Clynes RA, Towers TL, Presta LG, Ravetch JV. Inhibitory Fc receptors modulate in vivo cytotoxicity against tumor targets. Nat Med 2000; 6:443-6; PMID:10742152; http://dx.doi.org/10.1038/74704
- Aldemir H, Prod'homme V, Dumaurier MJ, Retiere C, Poupon G, Cazareth J, Bihl F, Braud VM. Cutting edge: lectin-like transcript 1 is a ligand for the CD161 receptor. J Immunol 2005; 175:7791-5; PMID:16339512; http://dx.doi.org/10.4049/jimmunol.175.12.7791
- Rosen DB, Bettadapura J, Alsharifi M, Mathew PA, Warren HS, Lanier LL. Cutting edge: lectin-like transcript-1 is a ligand for the inhibitory human NKR-P1A receptor. J Immunol 2005; 175:7796-9; PMID:16339513; http://dx.doi.org/10.4049/jimmunol.175.12.7796
- Germain C, Meier A, Jensen T, Knapnougel P, Poupon G, Lazzari A, Neisig A, Håkansson K, Dong T, Wagtmann N et al. Induction of lectin-like transcript 1 (LLT1) protein cell surface expression by pathogens and interferon-gamma contributes to modulate immune responses. J Biol Chem 2011; 286:37964-75; PMID:21930700; http://dx.doi.org/10.1074/jbc.M111.285312
- Germain C, Bihl F, Zahn S, Poupon G, Dumaurier MJ, Rampanarivo HH, Padkjær SB, Spee P, Braud VM. Characterization of Alternatively Spliced Transcript Variants of CLEC2D Gene. J Biol Chem 2010; 285:36207-15; PMID:20843815; http://dx.doi.org/10.1074/jbc.M110.179622
- van Zelm MC, Szczepanski T, van der Burg M, van Dongen JJ. Replication history of B lymphocytes reveals homeostatic proliferation and extensive antigen-induced B cell expansion. J Exp Med 2007; 204:645-55; PMID:17312005; http://dx.doi.org/10.1084/jem.20060964
- Caron G, Le Gallou S, Lamy T, Tarte K, Fest T. CXCR4 expression functionally discriminates centroblasts versus centrocytes within human germinal center B cells. J Immunol 2009; 182:7595-602; PMID:19494283; http://dx.doi.org/10.4049/jimmunol.0804272
- Victora GD, Dominguez-Sola D, Holmes AB, Deroubaix S, Dalla-Favera R, Nussenzweig MC. Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood 2012; 120:2240-8; PMID:22740445; http://dx.doi.org/10.1182/blood-2012-03-415380
- Fergusson JR, Smith KE, Fleming VM, Rajoriya N, Newell EW, Simmons R, Marchi E, Björkander S, Kang YH, Swadling L et al. CD161 defines a transcriptional and functional phenotype across distinct human T cell lineages. Cell Rep 2014; 9:1075-88; PMID:25437561; http://dx.doi.org/10.1016/j.celrep.2014.09.045
- Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 1999; 401:708-12; PMID:10537110; http://dx.doi.org/10.1038/44385
- Takahashi T, Dejbakhsh-Jones S, Strober S. Expression of CD161 (NKR-P1A) defines subsets of human CD4 and CD8 T cells with different functional activities. J Immunol 2006; 176:211-6; PMID:16365412; http://dx.doi.org/10.4049/jimmunol.176.1.211
- Ueno H, Banchereau J, Vinuesa CG. Pathophysiology of T follicular helper cells in humans and mice. Nat Immunol 2015; 16:142-52; PMID:25594465; http://dx.doi.org/10.1038/ni.3054
- Visco C, Li Y, Xu-Monette ZY, Miranda RN, Green TM, Tzankov A, Wen W, Liu WM, Kahl BS, d'Amore ES et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Leukemia 2012; 26:2103-13; PMID:22437443; http://dx.doi.org/10.1038/leu.2012.83
- Coutinho R, Clear AJ, Owen A, Wilson A, Matthews J, Lee A, Alvarez R, Gomes da Silva M, Cabeçadas J, Calaminici M et al. Poor concordance among nine immunohistochemistry classifiers of cell-of-origin for diffuse large B-cell lymphoma: implications for therapeutic strategies. Clin Cancer Res 2013; 19:6686-95; PMID:24122791; http://dx.doi.org/10.1158/1078-0432.CCR-13-1482
- Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A 2003; 100:9991-6; PMID:12900505; http://dx.doi.org/10.1073/pnas.1732008100
- Liu H, Bebu I, Li X. Microarray probes and probe sets. Front Biosci (Elite Ed) 2010; 2:325-38; PMID:20036881; http://dx.doi.org/10.2741/e93
- Liu H, Zeeberg BR, Qu G, Koru AG, Ferrucci A, Kahn A, Ryan MC, Nuhanovic A, Munson PJ, Reinhold WC et al. AffyProbeMiner: a web resource for computing or retrieving accurately redefined Affymetrix probe sets. Bioinformatics 2007; 23:2385-90; PMID:17660211; http://dx.doi.org/10.1093/bioinformatics/btm360
- Monti S, Savage KJ, Kutok JL, Feuerhake F, Kurtin P, Mihm M, Wu B, Pasqualucci L, Neuberg D, Aguiar RC et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood 2005; 105:1851-61; PMID:15550490; http://dx.doi.org/10.1182/blood-2004-07-2947
- Westin JR, Chu F, Zhang M, Fayad LE, Kwak LW, Fowler N, Romaguera J, Hagemeister F, Fanale M, Samaniego F et al. Safety and activity of PD1 blockade by pidilizumab in combination with rituximab in patients with relapsed follicular lymphoma: a single group, open-label, phase 2 trial. Lancet Oncol 2014; 15:69-77; PMID:24332512; http://dx.doi.org/10.1016/S1470-2045(13)70551-5
- Exley M, Garcia J, Balk SP, Porcelli S. Requirements for CD1d recognition by human invariant Valpha24+ CD4-CD8- T cells. J Exp Med 1997; 186:109-20; PMID:9207002; http://dx.doi.org/10.1084/jem.186.1.109
- Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL−6. Science 1997; 276:589-92; PMID:9110977; http://dx.doi.org/10.1126/science.276.5312.589
- Fukuda T, Yoshida T, Okada S, Hatano M, Miki T, Ishibashi K, Okabe S, Koseki H, Hirosawa S, Taniguchi M et al. Disruption of the Bcl6 gene results in an impaired germinal center formation. J Exp Med 1997; 186:439-48; PMID:9236196; http://dx.doi.org/10.1084/jem.186.3.439
- Ye BH, Cattoretti G, Shen Q, Zhang J, Hawe N, de Waard R, Leung C, Nouri-Shirazi M, Orazi A, Chaganti RS et al. The BCL−6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet 1997; 16:161-70; PMID:9171827; http://dx.doi.org/10.1038/ng0697-161
- Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol 2012; 30:429-57; PMID:22224772; http://dx.doi.org/10.1146/annurev-immunol-020711-075032
- Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol 2008; 8:22-33; PMID:18097447; http://dx.doi.org/10.1038/nri2217
- DeFranco AL, Rookhuizen DC, Hou B. Contribution of Toll-like receptor signaling to germinal center antibody responses. Immunol Rev 2012; 247:64-72; PMID:22500832; http://dx.doi.org/10.1111/j.1600-065X.2012.01115.x
- Hou B, Saudan P, Ott G, Wheeler ML, Ji M, Kuzmich L, Lee LM, Coffman RL, Bachmann MF, DeFranco AL. Selective utilization of Toll-like receptor and MyD88 signaling in B cells for enhancement of the antiviral germinal center response. Immunity 2011; 34:375-84; PMID:21353603; http://dx.doi.org/10.1016/j.immuni.2011.01.011
- Travert M, Ame-Thomas P, Pangault C, Morizot A, Micheau O, Semana G, Lamy T, Fest T, Tarte K, Guillaudeux T. CD40 ligand protects from TRAIL-induced apoptosis in follicular lymphomas through NF-kappaB activation and up-regulation of c-FLIP and Bcl−xL. J Immunol 2008; 181:1001-11; PMID:18606651; http://dx.doi.org/10.4049/jimmunol.181.2.1001
- Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Müller-Hermelink HK, Campo E, Braziel RM, Jaffe ES et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 2004; 103:275-82; PMID:14504078; http://dx.doi.org/10.1182/blood-2003-05-1545
- Jedema I, van der Werff NM, Barge RM, Willemze R, Falkenburg JH. New CFSE-based assay to determine susceptibility to lysis by cytotoxic T cells of leukemic precursor cells within a heterogeneous target cell population. Blood 2004; 103:2677-82; PMID:14630824; http://dx.doi.org/10.1182/blood-2003-06-2070
