ABSTRACT
Epithelial-mesenchymal transition (EMT) is a molecular and cellular program in which epithelial cells lose their well-differentiated phenotype and adopt mesenchymal characteristics. This process, which occurs naturally during embryogenesis, has also been shown to be associated with cancer progression and with tumor recurrence following conventional therapies. Brachyury is a transcription factor that mediates EMT during development, and is aberrantly expressed in various human cancers where it promotes tumor cell EMT, metastatic dissemination, and resistance to conventional therapies. We have recently shown that very high expression of brachyury can protect tumor cells against immune cell-mediated cytotoxicity. In seeking to elucidate mechanisms of immunotherapy resistance, we have discovered a novel positive association between brachyury and mucin-1 (MUC1). MUC1 is overexpressed in the majority of carcinomas, and it has been shown to mediate oncogenic signaling and confer resistance to genotoxic agents. We found that MUC1 is concomitantly upregulated in tumor cell lines that highly express brachyury due to an enhancement of MUC1 mRNA stability. Analysis of patient lung tumor tissues also identified a positive association between these two proteins in the majority of samples. Inhibition of MUC1 by siRNA-based gene silencing markedly enhanced the susceptibility of brachyury-expressing cancer cells to killing by tumor necrosis-related apoptosis-inducing ligand (TRAIL) and to perforin/granzyme-dependent lysis by immune cytotoxic cells. These studies confirm a protective role for MUC1 in brachyury-expressing cancer cells, and suggest that inhibition of MUC1 can restore the susceptibility of mesenchymal-like cancer cells to immune attack.
KEYWORDS:
Abbreviations
| AIF | = | apoptosis-inducing factor |
| CMA | = | concanamycin A |
| CTLs | = | cytotoxic T lymphocytes |
| EMT | = | epithelial-mesenchymal transition |
| ΔΨm | = | mitochondrial transmembrane potential |
| MVA | = | modified vaccinia Ankara |
| MUC1 | = | mucin-1 |
| MUC1-N | = | MUC1 N-terminal extracellular subunit |
| MUC1-C | = | MUC1 C-terminal cytoplasmic subunit |
| NK | = | natural killer |
| TRICOM | = | triad of costimulatory molecules |
| TRAIL | = | tumor necrosis factor (TNF)-related apoptosis-inducing ligand. |
Introduction
Metastatic disease is responsible for over 90% of deaths from solid tumors and is the preeminent challenge in clinical oncology.Citation1 Although much remains to be understood about tumor metastasis, in recent years it has become recognized that specific alterations in cellular phenotype represent a major mechanism that contributes to metastatic spread.Citation2 Termed EMT, this process involves the progressive loss of epithelial characteristics in favor of acquiring mesenchymal features that facilitate tumor cell motility and invasiveness.Citation3 Furthermore, EMT has been associated with the resistance to multiple therapeutic modalities, including radiation, chemotherapy, and certain targeted inhibitors,Citation4,5 and may ultimately lead to relapse with treatment-resistant disease.Citation6
The T-box transcription factor brachyury (gene name T) is an important regulator of EMT during vertebrate developmentCitation7 that is absent from the majority of normal adult tissues, but becomes overexpressed in a wide variety of carcinomas,Citation8-14 as well as in chordomasCitation15-18 and hemangioblastomas.Citation19,20 Similar to its role in development, brachyury was found to regulate tumor cell plasticity, induce features of EMT resulting in enhanced metastatic properties,Citation8,9 and confer resistance to chemotherapy and radiation.Citation21,22 Furthermore, brachyury expression is associated with poor prognosis in breast,Citation11 lung,Citation14 prostate,Citation13 and colon cancer.Citation23
Given its pathophysiological significance and cancer-specific expression profile, brachyury is now being exploited as a novel target for two different cancer vaccine platforms designed to elicit T cell-based adaptive immune responses against patient tumors.Citation24,25 However, whereas low levels of brachyury expression enable the recognition and killing of tumor cells by brachyury-specific cytotoxic T lymphocytes (CTLs), very high expression of brachyury was recently found to desensitize tumor cells to killing by both antigen-specific CTLs and innate immune effector cells.Citation26 This resistance was found to be the result of an intrinsic defect in caspase-dependent apoptotic processes only, as these cells remained sensitive to caspase-independent killing by the perforin/granzyme pathway. In light of our recent study, we sought to identify mediators of resistance that could be targeted to sensitize brachyury-high tumor cells to immune attack.
Mucin 1 (MUC1) is a transmembrane glycoprotein that is expressed by normal epithelial cells, but it becomes overexpressed and aberrantly glycosylated in the majority of carcinomas.Citation27 Structurally, MUC1 is a heterodimer composed of an N-terminal extracellular subunit (MUC1-N) and a C-terminal transmembrane cytoplasmic subunit (MUC1-C). Extensive studies have demonstrated that MUC1-C can promote cancer progression by enhancing pro-oncogenic signaling,Citation27-29 inducing EMT,Citation30-33 increasing stem cell characteristics,Citation32,34 and augmenting resistance to genotoxic anticancer agents.Citation35,36 In this study, we report a novel positive association between brachyury and MUC1 in both cell lines and tumor tissues. We further demonstrate that silencing MUC1 sensitized brachyury-expressing tumor cells to killing by TRAIL and to lysis by natural killer (NK) cells by improving mitochondrial apoptosis. These studies confirm that MUC1 is a mediator of brachyury-induced therapy resistance that can be targeted to restore the susceptibility of mesenchymal-like cancer cells to immune attack.
Results
Brachyury enhances MUC1 expression in carcinoma cells
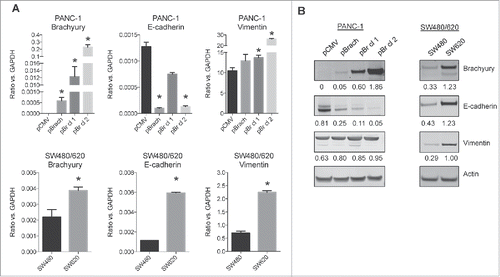
Both brachyuryCitation8,9 and MUC1Citation30-33 have been reported to play a role in EMT, and both are currently being used as targets for cancer vaccine approaches.Citation24,25,37 In order to investigate a potential association between brachyury and MUC1 and their relation to EMT, we first characterized the expression of epithelial and mesenchymal markers in various cancer cell line models. In one model, brachyury was overexpressed in the pancreatic PANC-1 cells generating an isogenic cell pair (pCMV vs. pBrach) and two single cell-derived clonal populations of brachyury-expressing cells, designated as pBr cl 1 and cl 2, which exhibited a range of brachyury mRNA and protein expression (). The second model consisted of two colon cancer cell lines, SW480 and SW620, which were derived from the primary and metastatic sites, respectively, of the same patient.Citation38 These cells exhibited different levels of naturally expressed brachyury mRNA and protein (), and have been previously described as epithelial vs. mesenchymal (SW480 vs. SW620, respectively).Citation39 As shown in , control PANC-1 cells co-expressed epithelial E-cadherin and mesenchymal vimentin, which is associated with an intermediate phenotype.Citation40,41 Brachyury overexpression was able to drive PANC-1 cells toward the mesenchymal phenotype, as manifested by the reduction of E-cadherin and increase of vimentin, a phenomenon correlated to brachyury expression levels. In the colon cancer model, the primary tumor-derived SW480 cells exhibited low levels of brachyury expression, whereas the metastasis-derived SW620 cells appeared to exhibit an intermediate phenotype with significantly higher levels of brachyury, vimentin, and E-cadherin. These observations indicate that high levels of brachyury in tumor cells associate with a gain of mesenchymal traits consistent with EMT.
Figure 1. EMT marker profiling of PANC-1 and SW480/620 cell lines. (A) qRT-PCR analysis of EMT marker transcript expression from the PANC-1 (top) and SW480/620 (bottom) cell lines. Graphs depict the mean±SD from one representative experiment performed in triplicate. *p < 0.05 compared to the PANC-1 pCMV (top) or SW480 (bottom) cell line. (B) Immunoblot images of EMT marker protein expression from the PANC-1 (left) and SW480/620 (right) cell lines. Numbers represent the ratio of the respective protein compared to actin.
To determine whether there was an association between brachyury and MUC1, we assessed MUC1 expression by quantitative PCR (), flow cytometry (), and immunoblot (), and observed significant upregulation in both the PANC-1 brachyury-overexpressing cells and the SW620 cells. In agreement, immunofluorescence of the PANC-1 cells also revealed enhanced MUC1 expression in the brachyury overexpressing cells (). Similar results at the mRNA and protein levels were obtained using other stable brachyury-overexpressing (ONYCAP23 and SW480) and brachyury-silenced (H460) cell line pairs (Fig. S1). In order to investigate whether brachyury and MUC1 could directly interact in tumor cells, immunoprecipitation was conducted with both anti-brachyury and anti-MUC1-C-specific monoclonal antibodies. With both systems, we were unable to detect an interaction between brachyury and MUC1 in the PANC-1 pBr cl 2 cells.
Figure 2. Association of brachyury and MUC1 expression in cancer cell lines. (A) qRT-PCR analysis of MUC1 transcript expression from the PANC-1 (left) and SW480/620 (right) cell lines. *p < 0.05 compared to the PANC-1 pCMV (left) or SW480 (right) cell line. (B) Flow cytometry analysis of MUC1 cell surface expression from the PANC-1 (left) and SW480/620 (right) cell lines. (C) Immunoblot analysis of MUC1-C expression from the PANC-1 (left) and SW480/620 (right) cell lines. Numbers represent the ratio of the MUC1 glycosylated protein (20–25 kDa) compared to actin. (D) Immunofluorescence staining of brachyury and MUC1-C in the PANC-1 cells. DAPI staining (inset) was used to visualize nuclei. Images were acquired at 200X. (E) qRT-PCR analysis of MUC1 transcript expression from the PANC-1 cell lines treated with actinomycin D (5 μg/mL) for the indicated time points. MUC1 levels were normalized to GAPDH expression. Graphs depict the mean±SD from one representative experiment performed in triplicate.
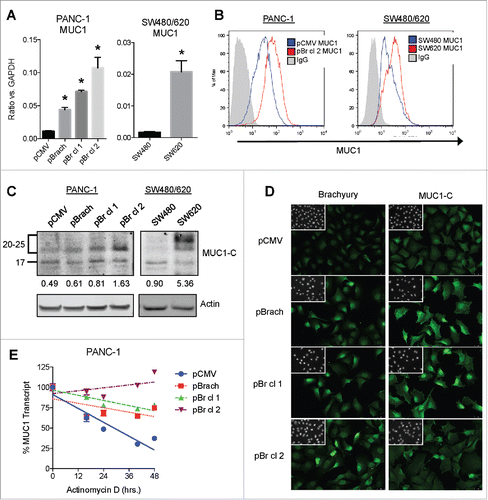
Since our data demonstrated a positive association between brachyury and MUC1 expression, we next turned our attention to elucidating the mechanism of regulation. The analysis of the MUC1 gene promoter failed to identify any canonical brachyury binding site(s) and no significant changes were observed regarding its activity among the various cell lines (not shown). These results led us to analyze the posttranscriptional regulation of MUC1 mRNA, which revealed a marked enhancement in transcript stability in the brachyury-overexpressing PANC-1 cells (). This effect was specific for the MUC1 transcript, as measurement of the unrelated transcript TJP1 (encoding the protein ZO-1) revealed no differences in either its level of expression or stability (Fig. S2A, B). Since transcript stability can be regulated by micro-RNAs (miRNAs), three different miRNAs previously shown to bind and regulate the MUC1 transcript were evaluated (miR-125b,Citation42 miR-145,Citation43 and miR-1226Citation44), however, none of them showed a consistent downregulation in the PANC-1 cell lines (Fig. S2C). Taken together, these data indicate that brachyury overexpression enhances MUC1 transcript stability, though the involved mechanism remains unknown.
Brachyury and MUC1 are positively associated in patient tumor samples
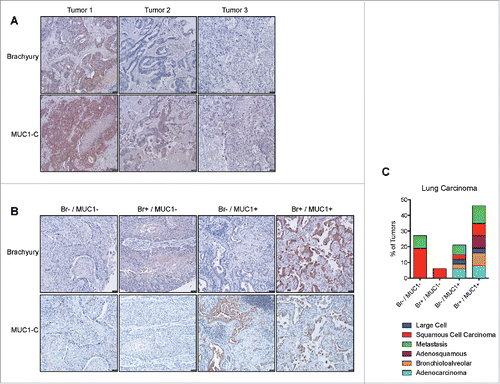
The upregulation of MUC1 by brachyury in vitro suggested that a positive association might also exist in vivo. In order to evaluate this possibility, we assessed the expression of both proteins in colon cancer liver metastasis samples from three separate patients. As shown in , a positive association was observed in which the highest MUC1 expression (detected with a MUC1-C-specific antibody) occurred in the tumor that had the highest brachyury expression (, tumor 1). In order to extend these observations to other tumor types, we assessed a larger panel of 36 lung tumors of diverse histological subtypes. As shown in , four patterns of expression were observed; double negative, brachyury positive, MUC1 positive, and double positive. Similar to the colon cancer samples, the majority (76%) of the lung tumor samples were either double negative (28%) or double positive (47%) for brachyury and MUC1 expression, suggesting that these proteins are positively associated in vivo ().
Figure 3. Association of brachyury and MUC1 expression in patient tumor samples. (A) Immunohistochemistry images of colon carcinoma liver metastasis tumor samples stained for brachyury or MUC1-C, respectively. (B) Representative immunohistochemistry images of lung carcinoma tumor samples stained for brachyury or MUC1-C, respectively, for each of the 4 expression categories as described in the text. Images were acquired at 100X. (C) Quantification of the lung carcinoma tumor samples in each of the four expression categories by histological subtype.
Inhibition of MUC1 overcomes TRAIL-resistance in brachyury-overexpressing cells
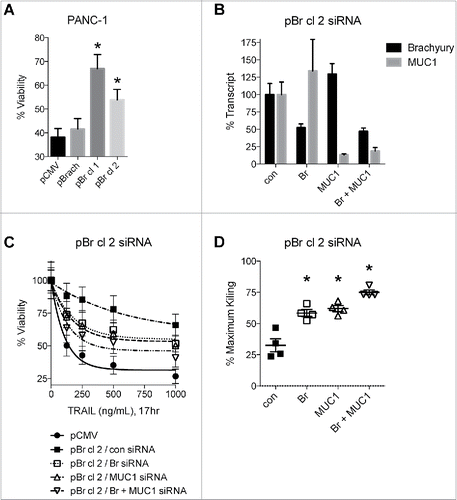
Our previous investigations demonstrated that brachyury protects tumor cells from the cytotoxic effects of multiple agents, including radiation, chemotherapy, and apoptosis-inducing ligands such as TRAIL.Citation11,21,22,26 In agreement, we observed enhanced resistance to TRAIL in the PANC-1 single-cell clones that overexpress high levels of brachyury (pBr cl 1 and 2, ). To determine whether MUC1 could play a functional role in this enhanced resistance to TRAIL, expression of brachyury or MUC1 were reduced either alone or in combination using siRNA-based knockdown of pBr cl 2 (), and then assessed their sensitivity to TRAIL (). Comparison of each cell line at all dose levels revealed that the control siRNA-treated pBr cl 2 cells were only 32.5 ± 5.2% as sensitive to TRAIL as the parental pCMV cells, and that knockdown of brachyury nearly doubled the sensitivity of these cells to 58.4 ± 2.8% as expected (). However, knockdown of MUC1 also enhanced sensitivity to a comparable 62.0 ± 2.4%, and combination-knockdown enhanced sensitivity to a significantly greater degree than either single-knockdown (75.0 ± 1.9%, p < 0.05, ). These results indicate that inhibition of MUC1, either alone or in combination with brachyury inhibition, can reconstitute the killing of TRAIL-resistant brachyury-overexpressing cells.
Figure 4. MUC1 knockdown reconstitutes TRAIL-mediated killing of brachyury-overexpressing cells. (A) Viability of the PANC-1 cell lines treated with TRAIL (1000 ng/mL, 24 h). *p < 0.05 compared to the pCMV cell line. (B) qRT-PCR analysis of transcript levels in pBr cl 2 cells treated with the indicated siRNAs for 72 h. (C) TRAIL-mediated killing of siRNA-treated pCMV and pBr cl 2 cells. (D) Analysis of TRAIL cytotoxicity. Percent maximum killing is the cytotoxicity for the siRNA-treated cell line at each dose relative to the cytotoxicity for the control pCMV cells at each matched dose. Graphs for viability assays depict the mean±SD from one representative experiment performed in triplicate, and graphs for % maximum killing depict the mean±SE. *p < 0.05 compared to the control siRNA-treated cell line.
Inhibition of MUC1 improves TRAIL-sensitivity by reducing mitochondrial integrity in brachyury-overexpressing cells
In order to understand the mechanism involved in the reconstitution of TRAIL killing by MUC1 knockdown in brachyury-expressing cells, we first investigated the cleavage of lamin B1, a nuclear lamin protein that we previously found to be inefficiently cleaved in brachyury-expressing cells.Citation26 In agreement with our previous work, improved cleavage of the full-length 66 kDa lamin B1 in response to TRAIL was observed in the brachyury silenced cells versus the control pBr cl 2 cells (). However, no improvement of lamin B1 cleavage was observed in the MUC1 knockdown cells (), implying that MUC1 does not cooperate with brachyury in protecting against nuclear degradation in response to apoptotic stimuli.
Figure 5. MUC1 knockdown reconstitutes TRAIL-mediated killing by reducing mitochondrial integrity. (A) Immunoblot of lamin B1 cleavage from siRNA-treated PANC-1 pBr cl 2 cells treated with TRAIL (500 ng/mL) for the indicated time points. (B) Immunoblot of caspase-3 cleavage from siRNA-treated PANC-1 pBr cl 2 cells treated with TRAIL (500 ng/mL) for the indicated time points. (C) Immunoblot of caspase-8 cleavage from siRNA-treated PANC-1 pBr cl 2 cells treated with TRAIL (500 ng/mL) for the indicated time points. All numbers represent the ratio of the respective full-length protein compared to actin and then normalized to the control 0 h treatment. (D) Flow cytometry analysis of JC-1 fluorescence from the siRNA-treated PANC-1 pBr cl 2 cells treated with TRAIL (500 ng/mL, 2 h). The top gate represents the ΔΨmhi population and the bottom gate represents the ΔΨmlow population.
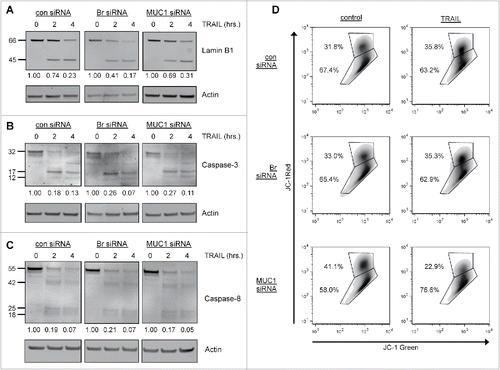
Since MUC1 has been shown to attenuate the cleavage of both the effector caspase-335 and the initiator caspase-845 in response to apoptotic stimuli, we next assessed the cleavage of these caspases in response to treatment with TRAIL. As shown in , treatment of the siRNA-treated cells with TRAIL resulted in the cleavage of the 35 kDa procaspase-3 into the active 17 kDa and 12 kDa fragments, but no substantial differences in the production of these fragments were observed in any of the siRNA-treated cells. TRAIL treatment also resulted in the cleavage of the 55 kDa procaspase-8 into several smaller fragments, including the active 18 kDa fragment. However, as with caspase-3, no substantial differences in the production of these fragments were observed in any of the siRNA-treated cells (). These data imply that the cleavage of these caspases is not affected by MUC1 in brachyury-overexpressing cells.
TRAIL and other apoptotic agents cause mitochondrial permeabilization that induces a loss of mitochondrial transmembrane potential (ΔΨm) and the release of pro-apoptotic mitochondrial-resident proteins.Citation46 Since MUC1 has been shown to prevent mitochondrial permeabilization in response to apoptotic stimuli,Citation35,36 we analyzed the mitochondrial integrity of the siRNA-treated cells by flow cytometry using JC-1. This fluorescent dye selectively enters mitochondria and reversibly changes color from red to green as ΔΨm decreases.Citation47 We found that exposure to TRAIL (500 ng/mL, 2 h) did not induce a population shift from the top gate (ΔΨmhi) to the bottom gate (ΔΨmlow) in the control- or Br-siRNA-treated cells (), indicating that these cells were resistant to mitochondrial depolarization. In contrast, MUC1-siRNA treated cells did exhibit a pronounced population shift, with the ΔΨmhi population decreasing by 44% and the ΔΨmlow population increasing by 32% (). Similar effects were observed at various time points and with even lower doses of TRAIL (Table S1). Altogether, our results suggest that inhibition of MUC1 reconstitutes TRAIL killing by enhancing mitochondrial-mediated apoptosis.
Inhibition of MUC1 overcomes resistance to killing by NK cells
We have previously shown that brachyury-expressing cells are more resistant to lysis by immune effector cells, including NK cells.Citation26 In agreement, brachyury-overexpressing PANC-1 cells were found to be more resistant to killing by NK cells (). Next, we sought to overcome this resistance through siRNA-mediated knockdown of brachyury and MUC1, either alone or in combination, in pBr cl 1 and cl 2 cells (). We observed that pBr cl 1 cells were only 55.0 ± 8.5% as susceptible to NK killing relative to control pCMV cells, and brachyury knockdown enhanced killing to 67.8 ± 7.7%. MUC1 knockdown fully reconstituted NK killing to 109.6 ± 6.8%, and combination knockdown did not lead to any further enhancement beyond MUC1 single knockdown (103 ± 4.5%, , top). For the pBr cl 2 cells, we observed that these cells were only 37.9 ± 2.9% as susceptible to NK killing relative to control pCMV cells, and brachyury and MUC1 knockdown partially reconstituted NK killing to 42.4 ± 6.6% and 71.5 ± 6.2%, respectively (, bottom). However, combination knockdown elicited full reconstitution of killing in the pBr cl 2 cells (100.1 ± 8.9%, , bottom).
Figure 6. MUC1 knockdown reconstitutes NK-mediated lysis of brachyury-overexpressing cells. (A) NK-mediated lysis of the PANC-1 cell lines. E:T ratio = 25:1. *p < 0.05 compared to the pCMV cell line. (B) qRT-PCR of brachyury and MUC1 transcript levels in pBr cl 1 and 2 cells treated with the indicated siRNAs for 72 h. (C) NK-mediated lysis of pCMV cells compared to siRNA-treated pBr cl 1 (top) or pBr cl 2 (bottom) cells. (D) Analysis of NK killing for the siRNA-treated pBr cl 1 (top) and pBr cl 2 (bottom) cells. Percent maximum killing is the killing for the siRNA-treated cell line at each E:T ratio relative to the killing for the control pCMV cells at each matched E:T ratio. *p < 0.05 compared to the control siRNA-treated cell line. (E) NK-mediated lysis of siRNA-treated pBr cl 2 cells using NKs pretreated with or without CMA. Graphs for cytotoxicity assays depict the mean±SD from one representative experiment performed in triplicate, and graphs for % maximum killing depict the mean±SE.
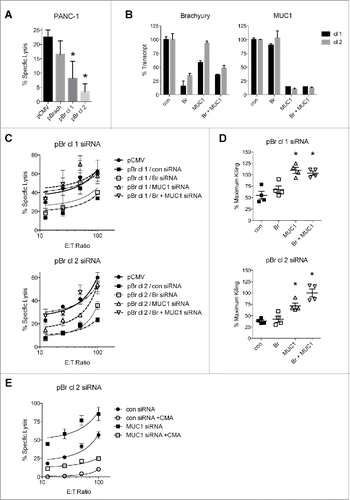
Our previous investigation reported that brachyury protects against caspase-dependent mechanisms of immune killing, but does not protect against lysis by the caspase-independent perforin/granzyme pathway.Citation26 This may explain our result in , as brachyury knockdown would not be expected to enhance killing if the NK cells killed primarily by the perforin/granzyme pathway. In order to determine whether MUC1 knockdown can enhance this mechanism of killing, an NK killing assay that used concanamycin A (CMA) to block the perforin-based cytotoxic activity of the NK cells was performed.Citation48 Once again, knockdown of MUC1 in the pBr cl 2 cells enhanced NK-mediated cytotoxicity compared to the knockdown control cells (average 200 ± 41.7% across all E:T ratios), and this enhancement was almost completely abolished by inhibiting the perforin/granzyme pathway with CMA (). These results indicate that MUC1 knockdown is also capable of enhancing the cytotoxicity of perforin-based death mechanisms.
Discussion
Cancer treatment is being revolutionized by the successful development and implementation of immunotherapy-based approaches that have led to substantial clinical responses. However, as with conventional and targeted therapies, intrinsic or acquired resistance to immunotherapy could emerge as a barrier to the expansion of these approaches to a wider array of patients. Given its prominent role in cancer progression, EMT is now being targeted as a means of simultaneously mitigating metastatic dissemination and treatment insensitivity. We have previously demonstrated a role for brachyury, an EMT-inducing transcription factor, in promoting the resistance of tumor cells to killing by innate and adaptive immune cells. Now, we have expanded upon those findings by implicating MUC1 as one mediator of brachyury-associated immunoresistance. Our results indicate that inhibiting MUC1 may be an attractive strategy to overcome intrinsic tumor immunoresistance driven by brachyury-mediated EMT.
MUC1 was originally identified as an epithelial protein that lubricates and protects epithelial cell surfaces.Citation27 However, it becomes aberrantly overexpressed in carcinoma cells where it induces transformation and transmits growth and survival signals.Citation27 MUC1 was also originally shown to be repressed during EMT,Citation49 but subsequent reports have revealed that overexpression of the MUC1-C subunit in tumor cells plays a role in actually inducing EMT.Citation30-33 In support of these prior observations, we found that MUC1-C was elevated in a variety of tumor cell lines undergoing brachyury-induced EMT. Our results are reminiscent of a similar report in which another EMT transcription factor, Snail, was also found to upregulate MUC1 in renal carcinoma cells.Citation50
We were surprised to find that brachyury did not regulate the transcription of the MUC1 gene, but rather associated with increased MUC1 mRNA stability. We hypothesized that brachyury may downregulate a MUC1-targeting miRNA that would lead to more stable MUC1 transcripts; however, we failed to identify a decrease in any of three reported MUC1-targeting miRNAs. It is possible that other miRNAs target the MUC1 transcript, so further investigation will be needed to elucidate the complete mechanism of MUC1 transcript regulation by brachyury. While we have clearly observed that overexpression of brachyury results in increased MUC1 expression in the context of MUC1 mRNA stabilization, transient siRNA-based targeting of brachyury (as shown in ) did not result in decreased MUC1 levels. While these results could be due to the transient character of the knockdown experiments, it is possible that brachyury upregulation is sufficient for the initiation of MUC1 upregulation but not required for its sustained expression. In this scenario, we hypothesize that once MUC1 is upregulated in tumor cells undergoing EMT via high levels of brachyury, additional signaling events may come into place to sustain MUC1 expression regardless of brachyury expression. These potential regulatory mechanisms will be evaluated in future studies.
Our data reveals that MUC1 plays a functional role in enhancing the resistance of brachyury-overexpressing cells to immune death mechanisms. MUC1 was previously shown to attenuate TRAIL-mediated cell death by preventing the cleavage and activation of caspase-8,Citation45 yet we did not observe any changes in caspase-8 cleavage in our study. However, another key survival role for MUC1 has been described at the level of mitochondrial apoptosis. Under normal circumstances, an apoptotic stimulus activates a cascade of events that ultimately leads to the formation of pores in the outer mitochondrial membrane,Citation46 which causes a loss of transmembrane potential, disruption of oxidative metabolism, and the release of mitochondrial-resident apoptogenic proteins that further spread and affect the apoptotic signal.Citation46 MUC1 was shown to attenuate these responses by inhibiting the dimerization of the pore-forming protein BAX, thereby maintaining mitochondrial integrity.Citation36 In agreement, we found that only MUC1 knockdown enhanced the loss of mitochondrial transmembrane potential in response to TRAIL. These results indicate that whereas brachyury blocks nuclear apoptosis by preventing the CDK1-dependent degradation of nuclear lamins,Citation26 MUC1 blocks mitochondrial apoptosis by preventing mitochondrial permeabilization. A fundamental difference in these cell death pathways appears to be the requirement for caspase activation, as inhibition of MUC1, but not brachyury, was able to enhance caspase-independent cell death mediated by perforin/granzymes. The enhancement of perforin/granzyme killing by MUC1 knockdown may be due to enhanced mitochondrial release of proteins such as endonuclease G and apoptosis-inducing factor (AIF) which induce chromatinolysis in a caspase-independent manner.Citation51,52 Nevertheless, these death pathways appear to complement one another, as combination brachyury and MUC1 knockdown led to a greater reconstitution of killing by both TRAIL and NK cells for the highest brachyury-overexpressing clone.
Our findings demonstrate a positive association between brachyury and MUC1 in patient tumors from various carcinoma subtypes. Although a larger set of samples needs to be evaluated, the existence of this association in various tumor types suggests that the high MUC1 expression associated with brachyury-high tumors might be clinically relevant. MUC1 has been evaluated as a target for vaccine therapy of a range of human tumors, including the use of polypeptides or vector-based vaccines encoding for full-length MUC1.Citation53-56 Recent studies have shown that alterations in the amino acid sequence of putative T cell epitopes of MUC-1, particularly those in the C-terminal region, result in the generation of enhancer agonist epitope(s) that, in turn, lead to enhanced T cell activation, greater cytokine production and improved lysis of MUC1-positive tumor cells.Citation57 The use of these agonist epitopes in various vaccine modalities could thus greatly enhance the efficacy of MUC1-based immune interventions. Regarding brachyury, the feasibility of targeting this EMT transcription factor via vaccination has recently been demonstrated in Phase I clinical trials.Citation24 Currently, two vaccine platforms targeting brachyury are undergoing clinical development; the first is a heat-killed recombinant yeast that expresses full-length human brachyury protein (GI-6301),Citation24,58 and the second is a modified vaccinia Ankara (MVA) poxviral vector encoding human brachyury and a triad of costimulatory molecules (B7.1, ICAM-1, LFA-3, designated TRICOM).Citation25 The results from the present study demonstrating co-expression of brachyury and MUC1 in human tumors provide rationale for future combinations of cancer vaccines targeting both tumor antigens, which is predicted to be therapeutically advantageous over the use of a vaccine targeting a single antigen.
It is known, however, that EMT in general,Citation26,59-61 and brachyury-induced EMT in particular,Citation26 may promote immunoresistance. The results presented here further support this notion, and provide a justification for a therapeutic strategy combining brachyury-targeting vaccines with inhibition of MUC1 in order to render the tumor cells more amenable to immune destruction. A promising MUC1 inhibitor is termed GO-203, which is a cell-penetrating peptide decoy that binds to the juxtamembrane region of MUC1-C thereby preventing dimerization and associated oncogenic signaling.Citation62 GO-203 has been shown not only to induce tumor regression in numerous preclinical studies,Citation29,62,63 but also to block MUC1-C interaction with BAXCitation36 thereby permitting mitochondrial apoptosis. Furthermore, a phase I clinical trial with GO-203 was completed in 2013, and a next generation polymeric nanoparticle-encapsulated GO-203 formulation has been shown in preclinical models to be more effective than the naked peptide at equivalent doses and less frequent dosing.Citation64
In summary, we report for the first time a novel positive association in vitro and in vivo between the EMT-inducing transcription factor brachyury and the oncogenic protein MUC1, thus providing rationale for future combinations of brachyury- and MUC1-targeting anticancer vaccine approaches. We have also demonstrated that this relationship has significant pathophysiological consequences that serve to protect tumor cells from immune-mediated cytotoxicity, indicating that MUC1 inhibition could be used to enhance the effectiveness of antitumor immune responses generated by cancer immunotherapy.
Materials and methods
Cell culture
The following human carcinoma cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA): pancreatic PANC-1; colon SW480 and SW620; and lung H460 and cultured in RPMI supplemented with 10% fetal bovine serum. Human prostate ONYCAP23 cells were provided by Onyvax Ltd.Citation22 Cell lines were used within 6 months of receiving or authenticated by STR profiling. Cells were stably transfected with an empty pcDNA3.1 vector (Thermo Fisher Scientific, Waltham, MA) or a vector encoding the full-length human brachyury protein (pBrach) using nucleofection (Lonza, Basel, Switzerland), and single-cell clones of the PANC-1 pBrach cell line were established as previously described.Citation26 H460 cells were stably transfected with a control or an shRNA vector specific for brachyury as previously described.Citation9
Quantitative real-time PCR
Total RNA was prepared using the PureLink RNA Mini Kit (Thermo Fisher Scientific) and reverse transcribed with the XLAScript cDNA MasterMix (WordWide Life Sciences, Hamilton, NJ). The resulting cDNA (10ng) was amplified in triplicate with the following TaqMan human gene expression assays (Thermo Fisher Scientific) T (brachyury) (Hs00610080_m1), CDH1 (Hs01013959_m1), VIM (Hs00958116_m1), MUC1 (Hs00904314_g1), TJP1 (Hs01551861_m1), GAPDH (4326317E). Expression of each target gene relative to GAPDH was calculated as 2−(Ct(GAPDH) – Ct(target gene).
Immunoblot analysis
Protein lysates from cell lines were prepared with RIPA lysis buffer (Santa Cruz Biotechnology, Dallas, TX) supplemented with 1 mM phenylmethanesulfonyl fluoride (Sigma-Aldrich, St. Louis, MO). The following primary antibodies were used: brachyury (mAb 54–1);Citation65 pan-actin (clone Ab-5, Thermo Fisher Scientific); E-cadherin, vimentin (BD Biosciences, East Rutherford, NJ); MUC1-C (clone CT2, Thermo Fisher Scientific); lamin B1 (Abcam, Cambridge, UK); caspase-3 (Cell Signaling Technology, Danvers, MA), and caspase-8 (AM46, EMD Millipore, Billerica, MA). Membranes were imaged using the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE), and blot quantification was performed using ImageJ (NIH, Bethesda, MD).
Immunofluorescence
Cells cultured on glass coverslips were fixed with 3% paraformaldehyde, permeabilized with 0.05% Triton X-100, and blocked with PBS containing 10% goat serum and 1% BSA. Coverslips were incubated overnight with the following primary antibodies: brachyury 54–1,Citation65 and MUC1-C (clone CD2, KeraFast, Boston, MA). Coverslips were then incubated with an AlexaFluor-488-conjugated secondary antibody (Thermo Fisher Scientific), stained with DAPI (Thermo Fisher Scientific), mounted using Fluorogel (Electron Microscopy Sciences, Hatfield, PA), and imaged utilizing a fluorescence microscope (Leica Microsystems, Wetzlar, Germany).
Flow cytometry
Cells were incubated with FITC-conjugated MUC1 antibody (CD227, BD Biosciences) or isotype control IgG antibody (BD Biosciences) for 40 min at 4°C, washed, and fluorescence was measured using a FACSCalibur cell analyzer (BD Biosciences). Data analysis was performed using FlowJo single cell analysis software (Tree Star, Ashland, OR).
To measure mitochondrial membrane potential, the JC-1 Mitochondrial Membrane Potential Assay Kit (Cayman Chemical, Ann Arbor, MI) was used according to the manufacturer's instructions. Briefly, cells were incubated with JC-1 diluted 1:100 in cell culture media for 15 min at 37°C, washed, and fluorescence was measured by flow cytometry as described above.
Immunohistochemistry
Lung tumor array LC952 was purchased from US Biomax (Rockville, MD) and metastatic colon tumor samples were purchased from Origene. For brachyury antigen retrieval, tissue sections were immersed in Antigen Unmasking Solution (Vector Laboratories, Burlingame, CA) at 90°C for 20 min and stained with brachyury 54–1 antibody as previously described.Citation65 For MUC1 antigen retrieval, tissue sections were immersed in buffer containing 10 mM Tris base, 1 mM EDTA, and 0.05% Tween20 at 100°C for 40 min. Tissue sections were blocked in horse serum (Thermo Fisher Scientific) for 30 min at room temperature and then incubated with primary antibody (MUC1-C clone CD2, KeraFast) diluted in 10% horse serum for 90 min at room temperature. Detection was performed with the ImmPRESS HRP-labeled universal anti-mouse/anti-rabbit peroxidase polymer detection kit (Vector Laboratories) followed by DAB peroxidase substrate (Vector Laboratories) and counterstained with hematoxylin. Staining was assessed for each sample, and samples were scored as negative if focal staining occurred only in a minority (<10%) of the tumor cells.
RNA interference
Cells were transfected with ON-TARGETplus SMARTpool siRNA specific for brachyury, MUC1, and a non-targeting control using DharmaFECT two transfection reagent (GE Healthcare, Little Chalfont, UK) according to the manufacturer's instructions. Cells were incubated for at least 48 h in antibiotic-free medium prior to use.
Viability assay
Cells were seeded in triplicate at 5.0 × 103 cells/well in white-walled 96-well plates. Following attachment, cells were treated with various doses of recombinant active multimeric Superkiller TRAIL (Enzo Life Sciences, Farmingdale, NY) and incubated for 16–24 h. Cell survival was assessed using the CellTiter-Glo luminescent viability assay (Promega) according to the manufacturer's instructions.
NK killing assay
Peripheral blood from healthy donors was obtained from the NIH Blood Bank (Bethesda, MD) under the appropriate Institutional Review Board approval and informed consent. NK cells were isolated using human CD56+ MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany). Target cells were labeled with 111In oxide and seeded in triplicate at 3.0 × 103 cells/well in 96-well round-bottom culture plates with effector NK cells at various effector-to-target (E:T) ratios. Following 16 h incubation, supernatants were harvested and the release of 111In was measured utilizing a gamma counter (PerkinElmer, Waltham, MA). Spontaneous release was determined by incubating the target cells with medium alone, and complete lysis was determined by incubating the target cells with 1% triton X-100 in water. Specific lysis was calculated as follows: % specific lysis = [(observed release – spontaneous release) / (complete release – spontaneous release)] × 100. To inhibit the function of perforin/granzyme, NK cells were preincubated with 200 nM concanamycin A (CMA, Sigma Aldrich) for 2 h at 37°C.
Statistical analysis
Data were analyzed using GraphPad Prism (version 6; GraphPad Software, La Jolla, CA), and p < 0.05 was considered significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KONI_A_1117738_supplementary_information.pdf
Download PDF (536 KB)Acknowledgment
The authors gratefully acknowledge their financial support from the NIH intramural research program.
Funding
This research was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health.
References
- Gupta GP, Massague J. Cancer metastasis: building a framework. Cell 2006; 127:679-95; PMID:17110329; http://dx.doi.org/10.1016/j.cell.2006.11.001
- Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med 2013; 19:1438-49; PMID:24202396; http://dx.doi.org/10.1038/nm.3336
- Christiansen JJ, Rajasekaran AK. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res 2006; 66:8319-26; PMID:16951136; http://dx.doi.org/10.1158/0008-5472.CAN-06-0410
- Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 2009; 138:645-59; PMID:19682730; http://dx.doi.org/10.1016/j.cell.2009.06.034
- Thomson S, Petti F, Sujka-Kwok I, Epstein D, Haley JD. Kinase switching in mesenchymal-like non-small cell lung cancer lines contributes to EGFR inhibitor resistance through pathway redundancy. Clin Exp Metastasis 2008; 25:843-54; PMID:18696232; http://dx.doi.org/10.1007/s10585-008-9200-4
- Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, Rimm DL, Wong H, Rodriguez A, Herschkowitz JI et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A 2009; 106:13820-5; PMID:19666588; http://dx.doi.org/10.1073/pnas.0905718106
- Showell C, Binder O, Conlon FL. T-box genes in early embryogenesis. Dev Dyn 2004; 229:201-18; PMID:14699590; http://dx.doi.org/10.1002/dvdy.10480
- Fernando RI, Castillo MD, Litzinger M, Hamilton DH, Palena C. IL-8 signaling plays a critical role in the epithelial-mesenchymal transition of human carcinoma cells. Cancer Res 2011; 71:5296-306; PMID:21653678; http://dx.doi.org/10.1158/0008-5472.CAN-11-0156
- Fernando RI, Litzinger M, Trono P, Hamilton DH, Schlom J, Palena C. The T-box transcription factor Brachyury promotes epithelial-mesenchymal transition in human tumor cells. J Clin Invest 2010; 120:533-44; PMID:20071775; http://dx.doi.org/10.1172/JCI38379
- Palena C, Polev DE, Tsang KY, Fernando RI, Litzinger M, Krukovskaya LL, Baranova AV, Kozlov AP, Schlom J. The human T-box mesodermal transcription factor Brachyury is a candidate target for T-cell-mediated cancer immunotherapy. Clin Cancer Res 2007; 13:2471-8; PMID:17438107; http://dx.doi.org/10.1158/1078-0432.CCR-06-2353
- Palena C, Roselli M, Litzinger MT, Ferroni P, Costarelli L, Spila A, Cavaliere F, Huang B, Fernando RI, Hamilton DH et al. Overexpression of the EMT driver brachyury in breast carcinomas: association with poor prognosis. J Natl Cancer Inst 2014; 106(5); pii:dju054; PMID:24815864; http://dx.doi.org/10.1093/jnci/dju054
- Roselli M, Fernando RI, Guadagni F, Spila A, Alessandroni J, Palmirotta R, Costarelli L, Litzinger M, Hamilton D, Huang B et al. Brachyury, a driver of the epithelial-mesenchymal transition, is overexpressed in human lung tumors: an opportunity for novel interventions against lung cancer. Clin Cancer Res 2012; 18:3868-79; PMID:22611028; http://dx.doi.org/10.1158/1078-0432.CCR-11-3211
- Pinto F, Pertega-Gomes N, Pereira MS, Vizcaino JR, Monteiro P, Henrique RM, Baltazar F, Andrade RP, Reis RM. T-box transcription factor brachyury is associated with prostate cancer progression and aggressiveness. Clin Cancer Res 2014; 20:4949-61; PMID:25009296; http://dx.doi.org/10.1158/1078-0432.CCR-14-0421
- Haro A, Yano T, Kohno M, Yoshida T, Koga T, Okamoto T, Takenoyama M, Maehara Y. Expression of Brachyury gene is a significant prognostic factor for primary lung carcinoma. Ann Surg Oncol 2013; 20(Suppl 3):S509-16; PMID:23456319; http://dx.doi.org/10.1245/s10434-013-2914-9
- Tirabosco R, Mangham DC, Rosenberg AE, Vujovic S, Bousdras K, Pizzolitto S, De Maglio G, den Bakker MA, Di Francesco L, Kalil RK et al. Brachyury expression in extra-axial skeletal and soft tissue chordomas: a marker that distinguishes chordoma from mixed tumor/myoepithelioma/parachordoma in soft tissue. Am J Surg Pathol 2008; 32:572-80; PMID:18301055; http://dx.doi.org/10.1097/PAS.0b013e31815b693a
- Vujovic S, Henderson S, Presneau N, Odell E, Jacques TS, Tirabosco R, Boshoff C, Flanagan AM. Brachyury, a crucial regulator of notochordal development, is a novel biomarker for chordomas. J Pathol 2006; 209:157-65; PMID:16538613; http://dx.doi.org/10.1002/path.1969
- Kelley MJ, Shi J, Ballew B, Hyland PL, Li WQ, Rotunno M, Alcorta DA, Liebsch NJ, Mitchell J, Bass S et al. Characterization of T gene sequence variants and germline duplications in familial and sporadic chordoma. Hum Genet 2014; 133:1289-97; PMID:24990759; http://dx.doi.org/10.1007/s00439-014-1463-z
- Yang XR, Ng D, Alcorta DA, Liebsch NJ, Sheridan E, Li S, Goldstein AM, Parry DM, Kelley MJ. T (brachyury) gene duplication confers major susceptibility to familial chordoma. Nat Genet 2009; 41:1176-8; PMID:19801981; http://dx.doi.org/10.1038/ng.454
- Barresi V, Ieni A, Branca G, Tuccari G. Brachyury: a diagnostic marker for the differential diagnosis of chordoma and hemangioblastoma vs. neoplastic histological mimickers. Dis Markers 2014; 2014:514753; PMID:24591762; http://dx.doi.org/10.1155/2014/514753
- Barresi V, Vitarelli E, Branca G, Antonelli M, Giangaspero F, Barresi G. Expression of brachyury in hemangioblastoma: potential use in differential diagnosis. Am J Surg Pathol 2012; 36:1052-7; PMID:22446946; http://dx.doi.org/10.1097/PAS.0b013e31824f4ce3
- Huang B, Cohen JR, Fernando RI, Hamilton DH, Litzinger MT, Hodge JW, Palena C. The embryonic transcription factor Brachyury blocks cell cycle progression and mediates tumor resistance to conventional antitumor therapies. Cell Death Dis 2013; 4:e682; PMID:23788039; http://dx.doi.org/10.1038/cddis.2013.208
- Larocca C, Cohen JR, Fernando RI, Huang B, Hamilton DH, Palena C. An autocrine loop between TGF-beta1 and the transcription factor brachyury controls the transition of human carcinoma cells into a mesenchymal phenotype. Mol Cancer Ther 2013; 12:1805-15; PMID:23783250; http://dx.doi.org/10.1158/1535-7163.MCT-12-1007
- Kilic N, Feldhaus S, Kilic E, Tennstedt P, Wicklein D, Wasielewski R, Viebahn C, Kreipe H, Schumacher U. Brachyury expression predicts poor prognosis at early stages of colorectal cancer. Eur J Cancer 2011; 47:1080-5; PMID:21220197; http://dx.doi.org/10.1016/j.ejca.2010.11.015
- Heery CR, Singh BH, Rauckhorst M, Marte JL, Donahue RN, Grenga I, Rodell TC, Dahut W, Arlen PM, Madan R et al. Phase I trial of a yeast-based therapeutic cancer vaccine (GI-6301) targeting the transcription factor brachyury. Cancer Immunol Res 2015; 3(11):1248-56; PMID:26130065; http://dx.doi.org/10.1158/2326-6066.CIR-15-0119
- Safety and Tolerability of a Modified Vaccinia Ankara (MVA)-Based Vaccine Modified to Express Brachyury and T-cell Costimulatory Molecules (MVA-Brachyury-TRICOM). http://www.clinicaltrials.gov/, 2015
- Hamilton DH, Huang B, Fernando RI, Tsang KY, Palena C. WEE1 inhibition alleviates resistance to immune attack of tumor cells undergoing epithelial-mesenchymal transition. Cancer Res 2014; 74:2510-9; PMID:24626094; http://dx.doi.org/10.1158/0008-5472.CAN-13-1894
- Kufe DW. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer 2009; 9:874-85; PMID:19935676; http://dx.doi.org/10.1038/nrc2761
- Kufe DW. MUC1-C oncoprotein as a target in breast cancer: activation of signaling pathways and therapeutic approaches. Oncogene 2013; 32:1073-81; PMID:22580612; http://dx.doi.org/10.1038/onc.2012.158
- Raina D, Kosugi M, Ahmad R, Panchamoorthy G, Rajabi H, Alam M, Shimamura T, Shapiro GI, Supko J, Kharbanda S et al. Dependence on the MUC1-C oncoprotein in non-small cell lung cancer cells. Mol Cancer Ther 2011; 10:806-16; PMID:21421804; http://dx.doi.org/10.1158/1535-7163.MCT-10-1050
- Rajabi H, Alam M, Takahashi H, Kharbanda A, Guha M, Ahmad R, Kufe D. MUC1-C oncoprotein activates the ZEB1/miR-200c regulatory loop and epithelial-mesenchymal transition. Oncogene 2014; 33:1680-9; PMID:23584475; http://dx.doi.org/10.1038/onc.2013.114
- Kharbanda A, Rajabi H, Jin C, Alam M, Wong KK, Kufe D. MUC1-C confers EMT and KRAS independence in mutant KRAS lung cancer cells. Oncotarget 2014; 5:8893-905; PMID:25245423; http://dx.doi.org/10.18632/oncotarget.2360
- Takahashi H, Jin C, Rajabi H, Pitroda S, Alam M, Ahmad R, Raina D, Hasegawa M, Suzuki Y, Tagde A et al. MUC1-C activates the TAK1 inflammatory pathway in colon cancer. Oncogene 2015; 34(40):5187-97; PMID:25659581; http://dx.doi.org/10.1038/onc.2014.442
- Roy LD, Sahraei M, Subramani DB, Besmer D, Nath S, Tinder TL, Bajaj E, Shanmugam K, Lee YY, Hwang SI et al. MUC1 enhances invasiveness of pancreatic cancer cells by inducing epithelial to mesenchymal transition. Oncogene 2011; 30:1449-59; PMID:21102519; http://dx.doi.org/10.1038/onc.2010.526
- Alam M, Rajabi H, Ahmad R, Jin C, Kufe D. Targeting the MUC1-C oncoprotein inhibits self-renewal capacity of breast cancer cells. Oncotarget 2014; 5:2622-34; PMID:24770886; http://dx.doi.org/10.18632/oncotarget.1848
- Ren J, Agata N, Chen D, Li Y, Yu WH, Huang L, Raina D, Chen W, Kharbanda S, Kufe D. Human MUC1 carcinoma-associated protein confers resistance to genotoxic anticancer agents. Cancer Cell 2004; 5:163-75; PMID:14998492; http://dx.doi.org/10.1016/S1535-6108(04)00020-0
- Ahmad R, Alam M, Rajabi H, Kufe D. The MUC1-C oncoprotein binds to the BH3 domain of the pro-apoptotic BAX protein and blocks BAX function. J Biol Chem 2012; 287:20866-75; PMID:22544745; http://dx.doi.org/10.1074/jbc.M112.357293
- Kimura T, Finn OJ. MUC1 immunotherapy is here to stay. Expert Opin Biol Ther 2013; 13:35-49; PMID:22998452; http://dx.doi.org/10.1517/14712598.2012.725719
- Leibovitz A, Stinson JC, McCombs WB, 3rd, McCoy CE, Mazur KC, Mabry ND. Classification of human colorectal adenocarcinoma cell lines. Cancer Res 1976; 36:4562-9; PMID:1000501
- Buck E, Eyzaguirre A, Barr S, Thompson S, Sennello R, Young D, Iwata KK, Gibson NW, Cagnoni P, Haley JD. Loss of homotypic cell adhesion by epithelial-mesenchymal transition or mutation limits sensitivity to epidermal growth factor receptor inhibition. Mol Cancer Ther 2007; 6:532-41; PMID:17308052; http://dx.doi.org/10.1158/1535-7163.MCT-06-0462
- Huang RY, Wong MK, Tan TZ, Kuay KT, Ng AH, Chung VY, Chu YS, Matsumura N, Lai HC, Lee YF et al. An EMT spectrum defines an anoikis-resistant and spheroidogenic intermediate mesenchymal state that is sensitive to e-cadherin restoration by a src-kinase inhibitor, saracatinib (AZD0530). Cell Death Dis 2013; 4:e915; PMID:24201814; http://dx.doi.org/10.1038/cddis.2013.442
- Schliekelman MJ, Taguchi A, Zhu J, Dai X, Rodriguez J, Celiktas M, Zhang Q, Chin A, Wong CH, Wang H et al. Molecular portraits of epithelial, mesenchymal, and hybrid States in lung adenocarcinoma and their relevance to survival. Cancer Res 2015; 75:1789-800; PMID:25744723; http://dx.doi.org/10.1158/0008-5472.CAN-14-2535
- Rajabi H, Jin C, Ahmad R, McClary C, Joshi MD, Kufe D. Mucin 1 oncoprotein expression is suppressed by the miR-125b oncomir. Genes Cancer 2010; 1:62-8; PMID:20729973; http://dx.doi.org/10.1177/1947601909357933
- Sachdeva M, Mo YY. MicroRNA-145 suppresses cell invasion and metastasis by directly targeting mucin 1. Cancer Res 2010; 70:378-87; PMID:19996288; http://dx.doi.org/10.1158/0008-5472.CAN-09-2021
- Jin C, Rajabi H, Kufe D. miR-1226 targets expression of the mucin 1 oncoprotein and induces cell death. Int J Oncol 2010; 37:61-9; PMID:20514397; http://dx.doi.org/10.3892/ijo_00000653
- Agata N, Ahmad R, Kawano T, Raina D, Kharbanda S, Kufe D. MUC1 oncoprotein blocks death receptor-mediated apoptosis by inhibiting recruitment of caspase-8. Cancer Res 2008; 68:6136-44; PMID:18676836; http://dx.doi.org/10.1158/0008-5472.CAN-08-0464
- Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol 2007; 35:495-516; PMID:17562483; http://dx.doi.org/10.1080/01926230701320337
- Cossarizza A, Baccarani-Contri M, Kalashnikova G, Franceschi C. A new method for the cytofluorimetric analysis of mitochondrial membrane potential using the J-aggregate forming lipophilic cation 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolcarbocyanine iodide (JC-1). Biochem Biophys Res Commun 1993; 197:40-5; PMID:8250945; http://dx.doi.org/10.1006/bbrc.1993.2438
- Kataoka T, Shinohara N, Takayama H, Takaku K, Kondo S, Yonehara S, Nagai K. Concanamycin A, a powerful tool for characterization and estimation of contribution of perforin- and Fas-based lytic pathways in cell-mediated cytotoxicity. J Immunol 1996; 156:3678-86; PMID:8621902
- Guaita S, Puig I, Franci C, Garrido M, Dominguez D, Batlle E, Sancho E, Dedhar S, De Herreros AG, Baulida J. Snail induction of epithelial to mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression. J Biol Chem 2002; 277:39209-16; PMID:12161443; http://dx.doi.org/10.1074/jbc.M206400200
- Gnemmi V, Bouillez A, Gaudelot K, Hemon B, Ringot B, Pottier N, Glowacki F, Villers A, Vindrieux D, Cauffiez C et al. MUC1 drives epithelial-mesenchymal transition in renal carcinoma through Wnt/β-catenin pathway and interaction with SNAIL promoter. Cancer Lett 2014; 346:225-36; PMID:24384091; http://dx.doi.org/10.1016/j.canlet.2013.12.029
- Li LY, Luo X, Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature 2001; 412:95-9; PMID:11452314; http://dx.doi.org/10.1038/35083620
- Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, Brothers GM, Mangion J, Jacotot E, Costantini P, Loeffler M et al. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 1999; 397:441-6; PMID:9989411; http://dx.doi.org/10.1038/17135
- Beatty PL, Narayanan S, Gariepy J, Ranganathan S, Finn OJ. Vaccine against MUC1 antigen expressed in inflammatory bowel disease and cancer lessens colonic inflammation and prevents progression to colitis-associated colon cancer. Cancer Prevention Res (Philadelphia, Pa) 2010; 3:438-46; PMID:20332301; http://dx.doi.org/10.1158/1940-6207.CAPR-09-0194
- Butts C, Maksymiuk A, Goss G, Soulieres D, Marshall E, Cormier Y, Ellis PM, Price A, Sawhney R, Beier F et al. Updated survival analysis in patients with stage IIIB or IV non-small-cell lung cancer receiving BLP25 liposome vaccine (L-BLP25): phase IIB randomized, multicenter, open-label trial. J Cancer Res Clin Oncol 2011; 137:1337-42; PMID:21744082; http://dx.doi.org/10.1007/s00432-011-1003-3
- Mohebtash M, Tsang KY, Madan RA, Huen NY, Poole DJ, Jochems C, Jones J, Ferrara T, Heery CR, Arlen PM et al. A pilot study of MUC-1/CEA/TRICOM poxviral-based vaccine in patients with metastatic breast and ovarian cancer. Clin Cancer Res 2011; 17:7164-73; PMID:22068656; http://dx.doi.org/10.1158/1078-0432.CCR-11-0649
- Morse MA, Niedzwiecki D, Marshall JL, Garrett C, Chang DZ, Aklilu M, Crocenzi TS, Cole DJ, Dessureault S, Hobeika AC et al. A randomized phase II study of immunization with dendritic cells modified with poxvectors encoding CEA and MUC1 compared with the same poxvectors plus GM-CSF for resected metastatic colorectal cancer. Annals Surg 2013; 258:879-86; PMID:23657083; http://dx.doi.org/10.1097/SLA.0b013e318292919e
- Jochems C, Tucker JA, Vergati M, Boyerinas B, Gulley JL, Schlom J, Tsang KY. Identification and characterization of agonist epitopes of the MUC1-C oncoprotein. Cancer Immunol Immunotherapy 2014; 63:161-74; PMID:24233342; http://dx.doi.org/10.1007/s00262-013-1494-7
- Hamilton DH, Litzinger MT, Jales A, Huang B, Fernando RI, Hodge JW, Ardiani A, Apelian D, Schlom J, Palena C. Immunological targeting of tumor cells undergoing an epithelial-mesenchymal transition via a recombinant brachyury-yeast vaccine. Oncotarget 2013; 4:1777-90; PMID:24125763; http://dx.doi.org/10.18632/oncotarget.1295
- Kudo-Saito C, Shirako H, Ohike M, Tsukamoto N, Kawakami Y. CCL2 is critical for immunosuppression to promote cancer metastasis. Clin Exp Metastasis 2013; 30:393-405; PMID:23143679; http://dx.doi.org/10.1007/s10585-012-9545-6
- Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell 2009; 15:195-206; PMID:19249678; http://dx.doi.org/10.1016/j.ccr.2009.01.023
- Akalay I, Janji B, Hasmim M, Noman MZ, Andre F, De Cremoux P, Bertheau P, Badoual C, Vielh P, Larsen AK et al. Epithelial-to-mesenchymal transition and autophagy induction in breast carcinoma promote escape from T-cell-mediated lysis. Cancer Res 2013; 73:2418-27; PMID:23436798; http://dx.doi.org/10.1158/0008-5472.CAN-12-2432
- Raina D, Ahmad R, Joshi MD, Yin L, Wu Z, Kawano T, Vasir B, Avigan D, Kharbanda S, Kufe D. Direct targeting of the mucin 1 oncoprotein blocks survival and tumorigenicity of human breast carcinoma cells. Cancer Res 2009; 69:5133-41; PMID:19491255; http://dx.doi.org/10.1158/0008-5472.CAN-09-0854
- Joshi MD, Ahmad R, Yin L, Raina D, Rajabi H, Bubley G, Kharbanda S, Kufe D. MUC1 oncoprotein is a druggable target in human prostate cancer cells. Mol Cancer Ther 2009; 8:3056-65; PMID:19887552; http://dx.doi.org/10.1158/1535-7163.MCT-09-0646
- Hasegawa M, Sinha RK, Kumar M, Alam M, Yin L, Raina D, Kharbanda A, Panchamoorthy G, Gupta D, Singh H et al. Intracellular Targeting of the Oncogenic MUC1-C Protein with a Novel GO-203 Nanoparticle Formulation. Clin Cancer Res 2015; 21:2338-47; PMID:25712682; http://dx.doi.org/10.1158/1078-0432.CCR-14-3000
- Hamilton DH, Fernando RI, Schlom J, Palena C. Aberrant expression of the embryonic transcription factor brachyury in human tumors detected with a novel rabbit monoclonal antibody. Oncotarget 2015; 6:4853-62; PMID:25605015; http://dx.doi.org/10.18632/oncotarget.3086
