ABSTRACT
Myeloid cells including tumor-associated macrophages (TAM) and myeloid-derived suppressor cells (MDSC) are known as important mediators of tumor progression in solid tumors such as pancreatic cancer. Infiltrating myeloid cells have been identified not only in invasive tumors, but also in early pre-invasive pancreatic intraepithelial precursor lesions (PanIN). The functional dynamics of myeloid cells during carcinogenesis is largely unknown. We aimed to systematically elucidate phenotypic and transcriptional changes in infiltrating myeloid cells during carcinogenesis and tumor progression in a genetic mouse model of pancreatic cancer. Using murine pancreatic myeloid cells isolated from the genetic mouse model at different time points during carcinogenesis, we examined both established markers of macrophage polarization using RT-PCR and FACS as well as transcriptional changes focusing on miRNA profiling. Myeloid cells isolated during carcinogenesis showed a simultaneous increase of established markers of M1 and M2 polarization during carcinogenesis, indicating that phenotypic changes of myeloid cells during carcinogenesis do not follow the established M1/M2 classification. MiRNA profiling revealed distinct regulations of several miRNAs already present in myeloid cells infiltrating pre-invasive PanIN lesions. Among them miRNA-21 was significantly increased in myeloid cells surrounding both PanIN lesions and invasive cancers. Functionally, miRNA-21-5p and -3p altered expression of the immune-modulating cytokines CXCL-10 and CCL-3 respectively. Our data indicate that miRNAs are dynamically regulated in infiltrating myeloid cells during carcinogenesis and mediate their functional phenotype by facilitating an immune-suppressive tumor-promoting micro-milieu.
Abbreviations
| BMDM | = | bone marrow-derived macrophages |
| KPC | = | LSL-KrasG12D/+, LSL-Trp53R172H/+ |
| MDSC | = | myeloid-derived suppressor cells |
| PanIN | = | pancreatic intraepithelial precursor lesions |
| PDAC | = | Pancreatic ductal adenocarcinoma |
| PDCD4 | = | Programmed cell death protein 4, Pdx-1-Cre mice |
| TAM | = | tumor-associated macrophages |
Introduction
Pancreatic ductal adenocarcinoma (PDAC), like several other solid tumors, is characterized by an intense inflammatory stromal reaction which has been recognized as an important trigger of tumor progression and therapy resistance.Citation1,2 Infiltrating myeloid cells such as TAM and MDSC represent crucial constituents of the inflammatory stroma reaction.Citation3 Myeloid cells accumulate in primary tumors and metastatic lesions, but can already be found in pre-invasive precursor (PanIN) lesions during early pancreatic carcinogenesis.Citation3 Furthermore, myeloid cells are known to secrete numerous soluble factors which may facilitate metastasis, foster evasion of antitumor immune responses and confer resistance to chemotherapeutic drugs.Citation1,4-6 The molecular mechanisms underlying the attraction of myeloid cells to pre-invasive precursor lesions or to invasive tumor cells and the crosstalk between these cell types during pancreatic carcinogenesis are highly complex and still incompletely understood.Citation7
Increasing evidence indicates that myeloid cells such as TAMs are characterized by a high functional plasticity with diverse phenotypes, which are dependent on the tissue context. Macrophages can be polarized in vitro into opposing functional states that have been named M1 and M2 polarizations in analogy to the TH1 and TH2 phenotypes in lymphocytes, although recent evidence suggests that the M1/M2 classification might be oversimplifying given the complex functional plasticity of TAMs during tumor initiation and progression.Citation7 M1-like TAMs are known to secrete a panel of cytokines which mediate a pro-inflammatory, antitumoral immune response.Citation8 In contrast, M2-like TAMs generally execute tumor-promoting functions by enhancing tissue remodeling, facilitating angiogenesis or regulating immune responses. In most solid tumors including PDAC, TAMs show preferential expression of marker genes associated with an M2-phenotype.Citation9
Recent evidence suggests that miRNAs play an important role in regulating the functional plasticity of TAMs. For example, miR-17 and miR-20a were identified as key regulators of HIF-2α expression in TAMs, thereby promoting the pro-angiogenic effects.Citation10 In contrast, miR-155 was shown to reprogram M2-like macrophages toward classic M1-like activation thereby restraining carcinogenesis.Citation11 In M1-like MRC-1+ macrophages, miR-511-3p has been demonstrated to downregulate tumor-promoting genes, including proteolytic enzymes and other extracellular matrix-remodeling molecules, thereby also exerting antitumoral effects.Citation12 A comprehensive profiling of miRNA expression patterns in infiltrating macrophages during the process of multistep carcinogenesis; however, is lacking so far.
Using genetic mouse models of pancreatic cancer, we aimed to comprehensively analyze the functional dynamics in myeloid cells infiltrating the pancreas which were isolated from the mice at different time points during carcinogenesis. To this extent, we focused on established M1 and M2 markers as well as myeloid cell-derived miRNA patterns and analyzed the functional impact of differentially expressed miRNAs.
Results
Myeloid cells infiltrate PanIN lesions and PDAC of KPC mice
We aimed to analyze myeloid cells from pancreata of two-related genetically engineered mouse models for detailed phenotypic analysis. LSL-KrasG12D/+; Pdx-1-Cre [KC] mice recapitulate early human carcinogenesis which is genetically characterized by an activating mutation of Kras. KC mice show development of pre-invasive pancreatic intraepithelial neoplasias (PanIN) due to pancreas-specific expression of activated Kras. Invasive cancers, however, occur only infrequently and at advanced age.Citation13 When combined with pancreas-specific expression of an inactivating p53 mutation (LSL-KrasG12D/+; LSL-Trp53R172H/+; Pdx-1-Cre [KPC] mice), invasive and metastatic pancreatic adenocarcinomas develop at a high frequency within 6 mo.Citation14
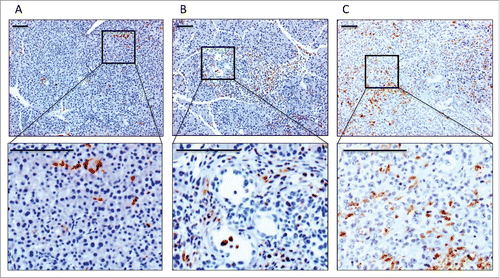
Using this model, we could detect infiltrating positive CD68+ myeloid cells at a low frequency in wild-type pancreas, but with increasing frequencies in areas surrounding pre-invasive PanIN lesions and invasive carcinomas (; Fig. S1). This increase in infiltrating CD68+ myeloid cells during the course of carcinogenesis could also be confirmed in human tissues***rem links (Fig. S2). Based on the presence of infiltrating myeloid cells in early PanIN lesions and invasive tumors, we hypothesized that infiltrating myeloid cells modulate both early carcinogenesis and tumor progression.
Expression of M1 and M2 markers in normal pancreas, PanIN lesions and PDAC
To systematically analyze phenotypic differences between myeloid cell populations at different stages of carcinogenesis we used both KC and KPC mice, with KC mice genetically and histologically recapitulating pre-invasive PanIN stages and KPC mice genetically resembling invasive cancers that histologically evolve sequentially via pre-invasive stages. Wild-type animals were used as controls. We isolated pancreatic CD11b+ myeloid cells from wild-type animals (aged 4–16 weeks), young KC mice without morphological alterations (aged 4–7 weeks), KC mice with PanIN lesions (aged 8–24 weeks), KPC mice with advanced PanIN lesions (aged 12–20 weeks) and KPC mice with invasive PDAC (aged 14–24 weeks). The purity of isolated myeloid cells was confirmed via FACS (Fig. S3). Using these myeloid cell populations, we first examined the dynamics of established markers of M1-polarization and M2-polarization on mRNA and protein levels.
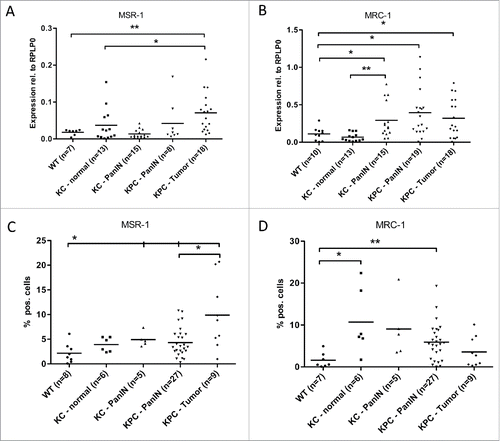
We could show that M2-markers significantly increase during pancreatic carcinogenesis: On mRNA level, MSR-1 (Macrophage scavenger receptor 1/CD204) was significantly upregulated in invasive PDAC without reaching significance in PanIN lesions (). In contrast, expression of MRC-1 (mannose receptor 1/CD206) was significantly elevated in both pre-invasive PanIN lesions and in invasive tumors (). Similar findings could be obtained on protein level by FACS analysis: MSR-1/CD204 expression was increased in both PanIN and PDAC (), MRC-1/CD206 expression was elevated in PanIN, but slightly decreased again on protein level from PanIN stage to PDAC stage ().
Figure 2. Myeloid cells show an increase in M2 marker gene expression during pancreatic carcinogenesis. qRT-PCR analysis of the mRNA levels of the M2 marker genes MSR-1 (A) and MRC-1 (B) in CD11b+ cells isolated from pancreata of WT, KC and KPC mice. (C, D) Protein expression of MSR-1 (C), MRC-1 (D) as detected by FACS. *p < 0.05, **p < 0.01.
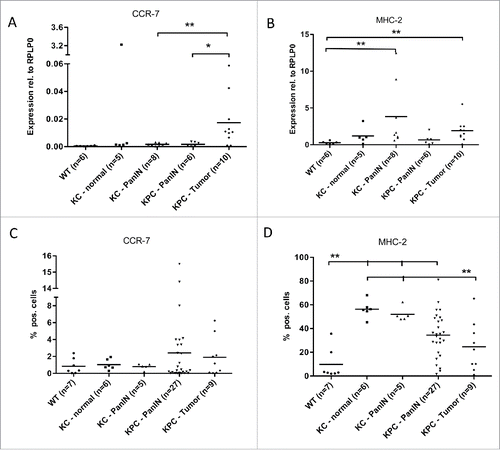
Interestingly, this increase of M2 markers during carcinogenesis was not accompanied by a simultaneous decrease in M1 markers: CCR-7/CD-197 was expressed at low levels and increased on mRNA level in myeloid cells of invasive PDAC on mRNA level (). This upregulation was seen as a trend on protein level, but did not reach significance (). The M1-marker MHC-2 also significantly increased in both PanIN as well as PDAC as compared to normal pancreas (). On protein level, expression was also elevated in PanIN compared to normal pancreas, but slightly decreased again in invasive tumors when compared to PanIN stages ().
Figure 3. Myeloid cells show an increased M1 marker gene expression during pancreatic carcinogenesis. qRT-PCR analysis of the mRNA levels of the M1 marker genes CCR-7 (A) and MHC-2 (B) in CD11b+ cells isolated from pancreata of WT, KC and KPC mice. (C, D) Protein expression of CCR-7 (C) and MHC-2 (D) as detected by FACS. *p < 0.05, **p < 0.01.
Taken together, mRNA and FACS data indicate that M2 markers of macrophage polarization show an increase in expression during pancreatic carcinogenesis. However, selected M1-markers also show a concomitant upregulation in PanIN lesions and PDAC. This suggests that myeloid cells are not unambiguously polarized toward an M2-like phenotype during multi-step carcinogenesis, but rather acquire a mixed M1/M2-like polarization phenotype, as described before in various cancers.Citation15,16
miRNA profiling in myeloid cells from normal pancreas and PDAC
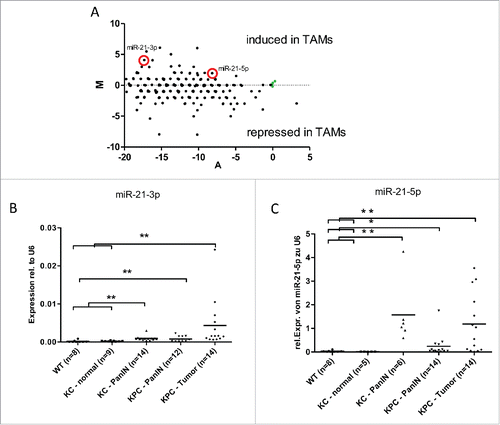
To further elucidate transcriptional changes in infiltrating myeloid cells accompanying the process of carcinogenesis, we focused on miRNA profiling, since miRNAs have been increasingly recognized as modulators of immune cell function.Citation10,11,12 To this extent, we compared expression levels of 770 miRNAs between myeloid cells isolated from normal pancreata in wild-type animals and tumor-associated myeloid cells in mice with invasive PDAC. qPCR-based profiling revealed distinct miRNAs differentially expressed in tumor-associated myeloid cells (, Table S1). Among the upregulated miRNAs, there were several miRNAs which have been implicated in tumor progression, acting directly in tumor cells or detected in the blood of tumor patients, such as miR-107, Citation17 miR-133, Citation18 miR-223, Citation19 miR-155 Citation20 and miR-21.Citation21 In contrast, downregulated miRNAs such as miR-200c Citation22 and miR-99 Citation23 have been implicated in tumor suppression in various cell types. Data on the impact of these miRNAs in tumor-infiltrating myeloid cells, however, are limited.
Figure 4. Expression of miR-21-3p and miR-21-5p increases during pancreatic carcinogenesis. (A) qRT-PCR Profiler for miRNAs in CD11b+ cells isolated from wild-type mice and KPC-mice with invasive PDAC. Green dots represent housekeeping genes. B+C, qRT-PCR analysis of miR-21-3p (B) and miR-21-5p (C) levels in CD11b+ cells isolated from pancreata of WT, KC and KPC mice. *p < 0.05, *p < 0.01.
For further validation and functional analysis we focused on miR-21, since both its 5p and 3p strands were consistently upregulated and recent evidence indicated a role of miR-21 not only in tumor cells but also in inflammatory cells.Citation24
MiR-21 modulates the phenotype of myeloid cells
We could confirm significant increases of both miR-21-3p and miR-21-5p in PanIN-associated as well as in PDAC-associated myeloid cells (). Although the expression levels of the miR-21-3p strand were lower than the 5p strand we continued with functional characterization of both strands since recent studies showed that also the 3p strand of miR-21 can be functionally active.Citation25
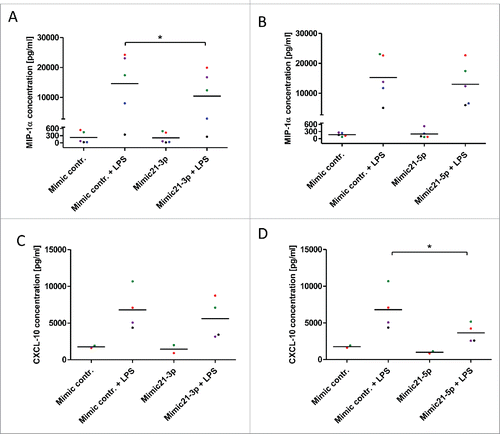
Hypothesizing that upregulation of both strands of miR-21 in myeloid cells influences the functional phenotype of these cells we aimed to analyze miR-21-dependent changes in the secretion of myeloid cell-derived inflammatory cytokines. For this purpose murine bone marrow-derived macrophages (BMDM) were transiently transfected with mimics for miR-21-5p and -3p and subsequently polarized toward an M1- or M2-phenotype by stimulation with LPS or IL-4, respectively. We screened for miR-21-dependent changes in cytokine expression using multiplex ELISAs and ELISAs for individual inflammatory cytokines. Interestingly, cytokine secretion of untreated and IL-4 polarized macrophages was not changed by modulation of miR-21-3p or -5p levels (data not shown). In contrast, upon activation and polarization toward an M1-like phenotype by LPS, significant changes in secreted chemokines were detected (Table S2). Two of the most differentially regulated chemokines were selected for further validation: Chemokine (C-C Motif) Ligand 3 (CCL-3), also known as macrophage inflammatory protein-1α (MIP-1α), and C-X-C motif chemokine 10 (CXCL-10) also known as Interferon gamma-induced protein 10 (IP-10). BMDM transfected with miR-21-3p showed a significantly decreased secretion of CCL-3/MIP-1α, whereas miR-21-5p had no significant effects on this cytokine (). In contrast, miR-21-5p was found to significantly inhibit LPS-induced secretion of CXCL-10/IP-10, whereas miR21-3p did not influence the secretion of this cytokine ().
Figure 5. miR-21 inhibits LPS-induced expression of CCL-3/MIP-1α and CXCL-10/IP-10. (A, B) CCL-3/MIP-1α protein levels determined by ELISA in cell culture supernatants of murine bone marrow-derived macrophages +/− LPS (10 ng/mL) and +/− mimics for miR-21-3p (A) and miR21-5p (B). (C, D) CXCL-10 protein levels determined by ELISA in the cell culture supernatants of murine bone marrow-derived macrophages +/− LPS (10 ng/mL) and +/− mimics for miR-21-3p (C) and miR21-5p (D). *p < 0.05.
MiR-21 mediates an immunosuppressive phenotype in macrophages
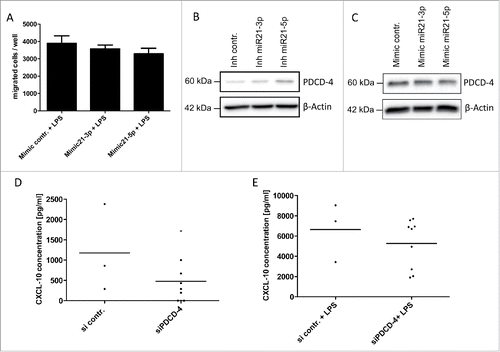
Both miR-21 targets, CCL-3/MIP-1α and CXCL-10/IP-10, have been described as potent attractants for CD8+ cytotoxic T cells and other immune cells.Citation26,27 Based on the high expression of miR-21 observed in tumor-associated myeloid cells, we speculated that miR-21 facilitates an immune-suppressive micro-milieu in the tumor by inhibiting the migration of cytotoxic T cells via suppression of lymphocyte-attractant cytokines such as CCL-3 and CXCL-10. As described in the literature, we could confirm that both CCL-3/MIP-1α (Fig. S4) and CXCL-10/IP-10 Citation27 are able to induce the migration of activated cytotoxic T lymphocytes. Modulating miR-21-3p and miR-21-5p expression in BMDM by miR-mimics resulted in a reproducible decrease in lymphocyte migration in vitro (), which, however, did not reach statistical significance most likely due to limited transfection efficiency in primary BMDM. This indicates that both miR-21 strands mediate the immunosuppressive phenotype in tumor-associated myeloid cells by altering the pattern of secreted cytokines.
Figure 6. miR-21 mediates an immunosuppressive phenotype. (A) CD8+ T-cells isolated from murine spleen and lymph nodes were co-cultured in a transwell assay with conditioned media of murine bone marrow-derived macrophages stimulated with LPS (10 ng/mL) +/− mimics for miR-21-3p or +/− mimics for miR-21-5p. (B, C) Immunoblot analysis of PDCD-4 in murine bone marrow-derived macrophages +/− inhibitor miR-21-5p (B) and +/− mimic miR-21-5p (C). (D, E) CXCL-10 protein levels determined by ELISA in cell culture supernatants of murine bone marrow-derived macrophages +/− LPS (10 ng/mL) and +/− PDCD-4 siRNA. *p < 0.05.
miR-21-5p inhibits CXCL-10 via PDCD-4 repression
To further elucidate the signaling pathways, which mediate the anti-inflammatory effects of miR-21, we examined downstream targets of miR-21 that have been implicated in the regulation of inflammatory signals. Programmed cell death protein-4 (PDCD4) is one of the known targets of miR-21 and has been associated with pro-apoptotic and anti-inflammatory effects.Citation28 We could confirm that miR-21-5p overexpression represses PDCD4, whereas miR-21-5p inhibition induces PDCD4 protein levels (). In contrast, miR-21-3p had no effect on PDCD4 levels (). In line with these findings knock-down of PDCD4 led to reduced levels of secreted CXCL10 under basal conditions () and—to a lesser extent—under LPS-induced conditions (), although statistical significance was not reached. This suggests that CXCL-10 acts as downstream target of PDCD4, mediating its pro-inflammatory effects and identify this chemokine as effector of the miR-21—PDCD4 signaling cascade.
Discussion
In this study, we could show that infiltrating myeloid cells influence carcinogenesis and tumor progression by altering the inflammatory micro-milieu in both early pre-invasive pancreatic precursor lesions (PanIN) and invasive pancreatic cancers of two genetically engineered pancreatic cancer mouse models. Although both, the KC and the KPC mice, develop precursor lesions as well as invasive carcinomas, the incidence of invasive tumors is much lower and occurs later in KC mice compared to KPC mice. This allows a more detailed analysis of pre-invasive stages in KC mice that also genetically resemble human PanIN lesions with Kras activation as predominant alteration. KPC mice with additional inactivation of p53 genetically recapitulate invasive carcinoma. It has been reported that the genetic status in the tumor cells might influence macrophage polarization. Lujambio et al. showed that deficiency of p53 can favor M2-polarization in a non-tumor cell autonomous fashion.Citation29 However, we did not observe significant differences in M1- or M2-marker expression between KC- and KPC-mice with PanIN lesions indicating that morphologically similar precursor lesions exert similar paracrine effects on infiltrating macrophages despite of a different genetic background.
To discriminate pancreatic tissues with PanIN lesions from tissues with invasive cancers in KPC mice used for macrophage isolation, we macroscopically and histologically analyzed adjacent tissues from longitudinal sections throughout the isolated murine pancreas. This technique allowed us to differentiate between pre-invasive and cancerous regions keeping the risk of misclassification of the isolated macrophages in these samples as low as technically feasible. However, we cannot completely rule out small, microscopically invasive tumors in parts of tissue bulks that were histologically classified as PanIN pancreata.
We demonstrate that infiltrating myeloid cells undergo distinct phenotypic changes during carcinogenesis. These changes, however, do not follow an unambiguous polarization from an M1- toward an M2-like phenotype but rather resemble a mixed M1/M2 polarization. Interestingly, some of the investigated markers—such as MHC-2—show higher expression levels in CD11b+ cells isolated from PanIN lesions compared to myeloid cells from established tumors. Since this phenomenon was seen only in some but not all markers and was consistent on mRNA and protein levels, we think that a methodological artifact is unlikely. It might be speculated that immune responses against precursor lesions are overcome by tumor intrinsic mechanisms in established invasive tumors.
Tumor-associated myeloid cells can be subdivided in two main groups: Gr1−CD11b+-macrophages and Gr1+CD11b+-MDSCs (Myeloid Derived Suppressor Cells), both occurring in the stroma of the genetically engineered pancreatic cancer mouse model. CD11b+ macrophages and CD11b+ MDSC share functional properties and are partly overlapping depending on the environmental context. Both macrophages and MDSCs can be polarized in to M1 and M2-phenotypes. Therefore, we focused on CD11b positivity as selection marker for isolation of myeloid cells from primary pancreatic tissues. CD11b+ myeloid cells are encompassing mainly macrophages and MDSCs but to a lesser extent also granulocytes and natural killer (NK) cells.
TAM have been associated with a poor prognosis in numerous solid tumors including pancreatic cancer.Citation9 In particular, high levels of infiltrating macrophages exhibiting M2 markers confer worse overall survival.Citation9 In contrast, in few cancers such as colon cancer dense macrophage infiltration has been associated with improved prognosis.Citation30 Interestingly, the majority of macrophages in colon cancer express M1 markers Citation31, supporting the concept that M1 polarized TAM exert pro-inflammatory, antitumor effects, whereas M2-polarized macrophages enhance carcinogenesis and tumor progression by mediating tumor-promoting immune-suppressive, effects. However, these correlations with clinical outcome are mainly based on immunohistochemical stainings of tissues derived from established tumors. To our knowledge, comprehensive analyses on phenotypic and transcriptional dynamics in infiltrating myeloid cells during carcinogenesis and cancer progression are largely missing. Accumulating evidence indicates that during tumor initiation, pro-inflammatory M1-like mediators secreted by infiltrating myeloid cells might facilitate rather than impede tumor initiation.Citation32,33
Our data indicate that phenotypic changes in infiltrating myeloid cells during pancreatic carcinogenesis do not follow an unequivocal M1- or M2-like polarization. On mRNA and protein levels, representative markers of both M1- and M2-phenotypes were increased in myeloid cells isolated from PanIN lesions and invasive tumors to various extents. These findings are in line with other reports in human pancreatic cancer tissues indicating that infiltrating macrophages display both M1 and M2 features which both impact on tumor progression.Citation32,15 Similarly, in a mouse model of colitis-induced colon cancer, both M1 and M2-polarized peritoneal macrophages co-existed, both of which being able to secrete tumor-promoting cytokines.Citation34
To further dissect the molecular mechanisms underlying the complex phenotypic alterations in myeloid cells during carcinogenesis, we aimed to further characterize changes on a transcriptional level. Given the increasingly recognized role of circulating miRNAs as potential biomarkers,Citation35 we thereby focused on the dynamics of miRNA alterations in infiltrating myeloid cells. We identified differential regulations of distinct myeloid cell-derived miRNAs both in pre-invasive PanIN lesions and in established pancreatic tumors. Several of the upregulated miRNAs such as miR-21, miR-107 and miR-223 have been implicated in cancer progression in various epithelial tumor cells.Citation36 On the other hand, downregulated miRNAs such as miR-99a and miR-200c have been associated with tumor-suppressive functions including inhibition of epithelia-mesenchymal transition (EMT).Citation22,23 MiR-21 and miR-223 have also been identified as upregulated in tumor-infiltrating myeloid cells in a syngeneic tail vein-injection model of breast cancer and melanoma.Citation37 Interestingly, in our profiling experiments, both 3p and 5p strands of miR-21 were found to be upregulated in myeloid cells infiltrating pre-invasive PanIN and invasive PDAC. Therefore, we focused on both strands of this miRNA for further functional characterization in tumor-infiltrating myeloid cells. Both arms of miRNA-21 lead to reduced secretion of the lymphocyte-attractant cytokines CCL-3/MIP-1α (miR-21-3p) and CXCL-10/IP-10 (miR-21-5p), indicating an immune-suppressive, tumor-promoting role of miR-21 in tumor-associated myeloid cells both in early carcinogenesis and tumor progression. Upregulation of miR-21 was also associated with reproducibly reduced migration of activated cytotoxic T lymphocytes in chemotaxis experiments, which however, did not reach statistical significance, most likely due to the limited transfection efficiency in primary murine macrophages used for the conditioned media experiments.
Mechanistically, we could show that miR-21-5p represses the expression of PDCD-4. PDCD-4 acts as tumor suppressor leading to decreased expression of AP-1-target genes like MMP-3, VEGF-C and PLAU, as well decreased pro-inflammatory mediators such as COX-2, IL-1b and IL-6.Citation38 Downregulation of PDCD-4 by miR-21 has been described in macrophages as mediator of an anti-inflammatory phenotype during the resolution of wound inflammation.Citation39 Accordingly, PDCD-4 downregulation led to reduced CXCL-10 levels. In contrast to miR-21-5p the effects of miR-21-3p on CCL-3/MIP-1α were PDCD4-independent.
To our knowledge, this is the first comprehensive study of miRNA dynamics in infiltrating myeloid cells at distinct stages during carcinogenesis in a genetic mouse model of cancer, emphasizing the importance of miRNAs as regulators of immune response in the tumor micro-environment. Our data on miR-21 support its emerging role as a key mediator of the anti-inflammatory response in macrophages.Citation40,41
Upregulation of miR-21-3p and -5p in early PanIN lesions but also invasive tumors underscores the potential diagnostic and therapeutic role of this miRNA not only in tumor cells but also in infiltrating myeloid cells. miR-21 inhibition in infiltrating myeloid cells will promote local inflammation and may therefore represent an interesting target to enhance current therapies for defective immune responses Citation41 in inflammation-triggered tumors such as pancreatic cancer.
Materials and methods
Reagents and cell culture
Primary murine macrophages were generated from bone marrow-derived cells isolated from mice with a mixed 129/SvJae/C57Bl/6 background as described previously.Citation27 In short, bone marrow cells were rinsed from femurs and tibiae and maturated in RPMI 1640 supplemented with 10 % FBS, penicillin-streptomycin and 20 ng/mL recombinant murine M-CSF (Peprotech, Rocky Hill, NJ, USA) for 7 d. All cells were cultured at 37°C in 5% CO2. Recombinant CCL-3/MIP-1α and IL-4 were obtained from Peprotech, Lipopolysaccharides (LPS) were purchased from Sigma-Aldrich (St. Louis, MO, USA).
miRNA and transfection
For transient transfection of primary murine macrophages, TransMessenger™ Transfection Reagent (Qiagen, Hilden, Germany) was used according to manufacturer's instructions. MiR-21 activity was transiently suppressed by mirVana™ miRNA Inhibitor oligonucleotides (Life Technologies) targeting murine miR-21-3p or miR-21-5p respectively. For transient overexpression of miRNA-21-3p and miRNA-21-5p mirVana™ miRNA Mimics (Life Technologies) were used.
Quantitative reverse transcription PCR
For isolation of total RNA and miRNA, the miRNeasy Mini Kit (Qiagen) was used according to the manufacturer's instructions. For detection of mRNA expression quantitative PCR was performed as described previously.Citation42 Sequences of the primers are shown in Table S3. Analysis of miR-21 and U6 expression was performed with Taqman MicroRNA Reverse Transcription Kit, TaqMan® Universal Master Mix II, no UNG and TaqMan® MicroRNA Assays (Life Technologies) according to the manufacturer's instructions.
MiRNA expression profiling comprising 770 miRNAs was performed using the TaqMan® Array Rodent MicroRNA A + B Cards Set v3.0 (Life Technologies) according to the manufacturer's instructions.
A list of the genes analyzed in this profiler is available online (https://www.lifetechnologies.com/order/catalog/product/4444909). Raw data of profiler results are shown in Table S1.
FACS-analysis
Isolated pancreatic macrophages (CD11b+ cells) from wild-type, LSL-KrasG12/D+; Pdx-1-Cre (KC) and LSL-KrasG12/D+; LSL-Tpr53R172H/+; Pdx-1-Cre (KPC) mice were stained for extracellular membrane proteins (CD11b+-, MRC-1+-, MSR-1+-, MHC II+-, CCR-7+). For this purpose, the cells were stained with anti-CD11b-APC (BioLegend), anti-MRC-1-FITC (BioLegend), anti-MSR-1-RPE (Biozol), anti-MHC-2-PE/Cy7 (BioLegend) and anti-CCR-7-PerCP/Cy5.5 (BioLegend) antibodies and analyzed with a BD LSR II flow cytometer (Becton Dickinson, Germany) and FlowJo software (Tree star, Inc.).
Immunoblot analysis and ELISAs
Generation of whole cell lysates and immunoblots were performed as described previously.Citation42,43 Signals were detected by enhanced chemiluminescence (GE Healthcare) in a ChemoCam Imager (Intas, Göttingen, Germany). Rabbit monoclonal anti-PDCD4 was purchased from Cell Signaling, mouse anti-β-actin was obtained from Sigma-Aldrich. CCL-3/MIP-1α secretion by primary murine macrophages was measured in the cell culture supernatant over 24 h using the Mouse MIP-1alpha (CCL-3) Ready-SET-Go! ELISA (eBioscience, San Diego, CA, USA) according to the manufacturer's instructions. Cytokine profiling comprising 23 cytokines was performed using the Bio-Plex Pro™ Mouse Cytokine 23-plex Assay (Bio-Rad Laboratories Ltd, Hercules, CA, USA) on the Bio-Plex 200 System. The levels of analyses were measured in triplicate from the supernatant of primary murine macrophages. Data analysis to measure concentration was performed using Bio-Plex Manager 5.0 Software. A list of the cytokines analyzed in this profiler is available online (http://www.bio-rad.com/en-uk/sku/m60-009rdpd-bio-plex-pro-mouse-cytokine-23-plex-assay). Raw data of profiler results are shown in Table S2.
Immunohistochemistry
CD68 immunostaining was performed using a mouse anti-CD68 (Dako, Hamburg, Germany) as primary antibody as described previously.Citation27
Isolation of macrophages from murine pancreas and spleen
For isolation of pancreatic macrophages from wild-type, LSL-KrasG12/D+; Pdx-1-Cre (KC) and LSL-KrasG12D/+; LSL-Tpr53R172H/+; Pdx-1-Cre (KPC) mice, animals were sacrificed at defined time points between 8–24 weeks of age. Tissues were digested with Collagenase A (Sigma-Aldrich), followed by erythrocyte lysis and filtration through a 30 µm pore-size filter (Miltenyi Biotec, Bergisch Gladbach, Germany). CD11b+ macrophages were isolated by MACS MicroBeads (Miltenyi Biotec) according to manufacturer´s instructions. Purity of isolated cells was determined by FACS analysis.
Murine CD8+ T-cell purification, in vitro stimulation and staining
Murine T cells were isolated from lymph nodes and spleen of C57Bl/6 mice by MACS Cell Separation using the naive CD8a+ T-Cell Isolation Kit (Miltenyi Biotec, Germany) according to manufacturer´s instructions. The purified CD8+ T cells were primed with plate-bound anti-CD3 (5 µg/mL, clone 145-2C11) and soluble anti-CD28 mAb (1 µg/mL, clone 37.51) in the presence of recombinant human (rh) IL-2 (50 U/mL; Novartis) and 5 µg/mL anti-IFNγ (XMG1.2) (CTL conditions) in complete RPMI-1640 medium supplemented with 10% (vol/vol) FBS (Sigma). After 4 d of differentiation, cells were splitted and rested for 3 d in the presence of rhIL-2 (50 U/mL). Subsequently, dead cells were removed by Ficoll gradient centrifugation (Pancoll mouse, density 1.086 g/mL. PANBitoech), live cells were washed twice, counted and used in migration assays. For control stainings, cells were re-stimulated for 4 h with 50 ng/mL PMA and 750 ng/mL ionomycin in the presence of 10 µg/mL brefeldin A (all from Sigma). Thereafter, cells were intracellularly stained with anti-IFNγ (XMG1.2) and anti-TNFα (MP6-XT22) antibodies (eBiosciences) and analyzed with a FACSCalibur and the CellQuest Pro software.
T-Cell migration assay
2,5 × 105 activated T cells were seeded in the upper compartment of a 24-well modified Boyden Chamber 3 µm pore size (Corning) in RPMI supplemented with 2% FCS and 10 mM Hepes. The lower chamber was loaded with conditioned media or recombinant CCL-3/MIP-1α, respectively. After 6 h of incubation at 37°C and 5% CO2, cells migrated to the lower compartment were photographed and counted.
Statistics
Data are presented as mean ± SEM. Two-tailed unpaired Student's t test was used for statistical evaluation of the data. p values < 0.05 were considered as significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KONI_A_1160181_supplementary_materials.zip
Download Zip (1.5 MB)Acknowledgments
This publication reflects only the authors' views. The European Community is not liable for any use that may be made of the information herein.
Funding
This work was supported in part by grants of the European Commisson FP7 grant (Collaborative Project ‘EPC-TM-Net, to PM, AN and TG), Deutsche Forschungsgemeinschaft (DFG) (to PM), the LOEWE initiative of the state of Hessen, the Behring-Roentgen Foundation (to PM), and the Deutsche Krebshilfe (to PM).
References
- Neesse A, Michl P, Frese KK, Feig C, Cook N, Jacobetz MA, Lolkema MP, Buchholz M, Olive KP, Gress TM et al. Stromal biology and therapy in pancreatic cancer. Gut 2011; 60:861-8; PMID:20966025; http://dx.doi.org/10.1136/gut.2010.226092
- Feig C, Gopinathan A, Neesse A, Chan DS, Cook N, Tuveson DA. The pancreas cancer microenvironment. Clin Cancer Res 2012; 18:4266-76; PMID:22896693; http://dx.doi.org/10.1158/1078-0432.CCR-11-3114
- Clark CE, Hingorani SR, Mick R, Combs C, Tuveson DA, Vonderheide RH. Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res 2007; 67:9518-27; PMID:17909062; http://dx.doi.org/10.1158/0008-5472.CAN-07-0175
- Kleeff J, Beckhove P, Esposito I, Herzig S, Huber PE, Lohr JM, Friess H. Pancreatic cancer microenvironment. Int J Cancer Journal international du cancer 2007; 121:699-705; PMID:17534898; http://dx.doi.org/10.1002/ijc.22871
- Beatty GL, Chiorean EG, Fishman MP, Saboury B, Teitelbaum UR, Sun W, Huhn RD, Song W, Li D, Sharp LL et al. CD40 agonists alter tumor stroma and show efficacy against pancreatic carcinoma in mice and humans. Science 2011; 331:1612-6; PMID:21436454; http://dx.doi.org/10.1126/science.1198443
- Bayne LJ, Beatty GL, Jhala N, Clark CE, Rhim AD, Stanger BZ, Vonderheide RH. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell 2012; 21:822-35; PMID:22698406; http://dx.doi.org/10.1016/j.ccr.2012.04.025
- Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity 2014; 41:49-61; PMID:25035953; http://dx.doi.org/10.1016/j.immuni.2014.06.010
- Sica A, Larghi P, Mancino A, Rubino L, Porta C, Totaro MG, Rimoldi M, Biswas SK, Allavena P, Mantovani A. Macrophage polarization in tumour progression. Semin Cancer Biol 2008; 18:349-55; PMID:18467122; http://dx.doi.org/10.1016/j.semcancer.2008.03.004
- Kurahara H, Shinchi H, Mataki Y, Maemura K, Noma H, Kubo F, Sakoda M, Ueno S, Natsugoe S, Takao S. Significance of M2-polarized tumor-associated macrophage in pancreatic cancer. J Surg Res 2011; 167:e211-9; PMID:19765725; http://dx.doi.org/10.1016/j.jss.2009.05.026
- Xu Z, Zhao L, Zhu LY, He M, Zheng L, Wu Y. MicroRNA-17, 20a regulates the proangiogenic function of tumor-associated macrophages via targeting hypoxia-inducible factor 2alpha. PloS One 2013; 8:e77890; PMID:24194900; http://dx.doi.org/10.1371/journal.pone.0077890
- Zonari E, Pucci F, Saini M, Mazzieri R, Politi LS, Gentner B, Naldini L. A role for miR-155 in enabling tumor-infiltrating innate immune cells to mount effective antitumor responses in mice. Blood 2013; 122:243-52; PMID:23487026; http://dx.doi.org/10.1182/blood-2012-08-449306
- Squadrito ML, Pucci F, Magri L, Moi D, Gilfillan GD, Ranghetti A, Casazza A, Mazzone M, Lyle R, Naldini L et al. miR-511-3p modulates genetic programs of tumor-associated macrophages. Cell Rep 2012; 1:141-54; PMID:22832163; http://dx.doi.org/10.1016/j.celrep.2011.12.005
- Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 2003; 4:437-50; PMID:14706336; http://dx.doi.org/10.1016/S1535-6108(03)00309-X
- Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005; 7:469-83; PMID:15894267; http://dx.doi.org/10.1016/j.ccr.2005.04.023
- Karnevi E, Andersson R, Rosendahl AH. Tumour-educated macrophages display a mixed polarisation and enhance pancreatic cancer cell invasion. Immunol Cell Biol 2014; 92:543-52; PMID:24662521; http://dx.doi.org/10.1038/icb.2014.22
- Reinartz S, Schumann T, Finkernagel F, Wortmann A, Jansen JM, Meissner W, Krause M, Schworer AM, Wagner U, Muller-Brusselbach S et al. Mixed-polarization phenotype of ascites-associated macrophages in human ovarian carcinoma: correlation of CD163 expression, cytokine levels and early relapse. Int J Cancer Journal international du cancer 2014; 134:32-42; PMID:23784932; http://dx.doi.org/10.1002/ijc.28335
- Kleivi Sahlberg K, Bottai G, Naume B, Burwinkel B, Calin GA, Borresen-Dale AL, Santarpia L. A serum microRNA signature predicts tumor relapse and survival in triple-negative breast cancer patients. Clin Cancer Res 2015; 21:1207-14; PMID:25547678; http://dx.doi.org/10.1158/1078-0432.CCR-14-2011
- Yu H, Lu Y, Li Z, Wang Q. microRNA-133: expression, function and therapeutic potential in muscle diseases and cancer. Curr Drug Targets 2014; 15:817-28; PMID:24975488; http://dx.doi.org/10.2174/1389450115666140627104151
- Li ZW, Yang YM, Du LT, Dong Z, Wang LL, Zhang X, Zhou XJ, Zheng GX, Qu AL, Wang CX. Overexpression of miR-223 correlates with tumor metastasis and poor prognosis in patients with colorectal cancer. Med Oncol 2014; 31:256; PMID:25270282; http://dx.doi.org/10.1007/s12032-014-0256-5
- Liu Q, Zou R, Zhou R, Gong C, Wang Z, Cai T, Tan C, Fang J. miR-155 Regulates Glioma Cells Invasion and Chemosensitivity by p38 Isforms In Vitro. J Cell Biochem 2015; 116:1213-21; PMID:25535908; http://dx.doi.org/10.1002/jcb.25073
- Yang CH, Pfeffer SR, Sims M, Yue J, Wang Y, Linga VG, Paulus E, Davidoff AM, Pfeffer LM. The oncogenic microRNA-21 inhibits the tumor suppressive activity of FBXO11 to promote tumorigenesis. J Biol Chem 2015; 290:6037-46; PMID:25589783; http://dx.doi.org/10.1074/jbc.M114.632125
- Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol 2009; 11:1487-95; PMID:19935649; http://dx.doi.org/10.1038/ncb1998
- Permuth-Wey J, Chen YA, Fisher K, McCarthy S, Qu X, Lloyd MC, Kasprzak A, Fournier M, Williams VL, Ghia KM et al. A Genome-Wide Investigation of MicroRNA Expression Identifies Biologically-Meaningful MicroRNAs That Distinguish between High-Risk and Low-Risk Intraductal Papillary Mucinous Neoplasms of the Pancreas. PloS One 2015; 10:e0116869; PMID:25607660; http://dx.doi.org/10.1371/journal.pone.0116869
- Li L, Zhang J, Diao W, Wang D, Wei Y, Zhang CY, Zen K. MicroRNA-155 and MicroRNA-21 promote the expansion of functional myeloid-derived suppressor cells. J Immunol 2014; 192:1034-43; PMID:24391219; http://dx.doi.org/10.4049/jimmunol.1301309
- Doberstein K, Bretz NP, Schirmer U, Fiegl H, Blaheta R, Breunig C, Muller-Holzner E, Reimer D, Zeimet AG, Altevogt P. miR-21-3p is a positive regulator of L1CAM in several human carcinomas. Cancer Lett 2014; 354:455-66; PMID:25149066; http://dx.doi.org/10.1016/j.canlet.2014.08.020
- Bernardini G, Sciume G, Bosisio D, Morrone S, Sozzani S, Santoni A. CCL3 and CXCL12 regulate trafficking of mouse bone marrow NK cell subsets. Blood 2008; 111:3626-34; PMID:18227348; http://dx.doi.org/10.1182/blood-2007-08-106203
- Kuhnemuth B, Muhlberg L, Schipper M, Griesmann H, Neesse A, Milosevic N, Wissniowski T, Buchholz M, Gress TM, Michl P. CUX1 modulates polarization of tumor-associated macrophages by antagonizing NF-kappaB signaling. Oncogene 2015; 34:177-87; PMID:24336331; http://dx.doi.org/10.1038/onc.2013.530
- Krug S, Huth J, Goke F, Buchholz M, Gress TM, Goke R, Lankat-Buttgereit B. Knock-down of Pdcd4 stimulates angiogenesis via up-regulation of angiopoietin-2. Biochimica et biophysica acta 2012; 1823:789-99; PMID:22289349; http://dx.doi.org/10.1016/j.bbamcr.2012.01.006
- Lujambio A, Akkari L, Simon J, Grace D, Tschaharganeh DF, Bolden JE, Zhao Z, Thapar V, Joyce JA, Krizhanovsky V, Lowe SW. Non-cell-autonomous tumor suppression by p53. Cell 2013; 153(2):449-60; PMID:23562644; http://dx.doi.org/10.1016/j.cell.2013.03.020
- Forssell J, Oberg A, Henriksson ML, Stenling R, Jung A, Palmqvist R. High macrophage infiltration along the tumor front correlates with improved survival in colon cancer. Clin Cancer Res 2007; 13:1472-9; PMID:17332291; http://dx.doi.org/10.1158/1078-0432.CCR-06-2073
- Edin S, Wikberg ML, Dahlin AM, Rutegard J, Oberg A, Oldenborg PA, Palmqvist R. The distribution of macrophages with a M1 or M2 phenotype in relation to prognosis and the molecular characteristics of colorectal cancer. PloS One 2012; 7:e47045; PMID:23077543; http://dx.doi.org/10.1371/journal.pone.0047045
- Helm O, Held-Feindt J, Grage-Griebenow E, Reiling N, Ungefroren H, Vogel I, Kruger U, Becker T, Ebsen M, Rocken C et al. Tumor-associated macrophages exhibit pro- and anti-inflammatory properties by which they impact on pancreatic tumorigenesis. Int J Cancer Journal international du cancer 2014; 135:843-61; PMID:24458546; http://dx.doi.org/10.1002/ijc.28736
- Sebens S, Bauer I, Geismann C, Grage-Griebenow E, Ehlers S, Kruse ML, Arlt A, Schafer H. Inflammatory macrophages induce Nrf2 transcription factor-dependent proteasome activity in colonic NCM460 cells and thereby confer anti-apoptotic protection. J Biol Chem 2011; 286:40911-21; PMID:21990354; http://dx.doi.org/10.1074/jbc.M111.274902
- Wang W, Li X, Zheng D, Zhang D, Huang S, Zhang X, Ai F, Wang X, Ma J, Xiong W et al. Dynamic changes of peritoneal macrophages and subpopulations during ulcerative colitis to metastasis of colorectal carcinoma in a mouse model. Inflamm Res [et al] 2013; 62:669-80; PMID:23625042; http://dx.doi.org/10.1007/s00011-013-0619-y
- Challagundla KB, Fanini F, Vannini I, Wise P, Murtadha M, Malinconico L, Cimmino A, Fabbri M. microRNAs in the tumor microenvironment: solving the riddle for a better diagnostics. Expert Rev Mol Diagn 2014; 14:565-74; PMID:24844135; http://dx.doi.org/10.1586/14737159.2014.922879
- Song YQ, Ma XH, Ma GL, Lin B, Liu C, Deng QJ, Lv WP. MicroRNA-107 promotes proliferation of gastric cancer cells by targeting cyclin dependent kinase 8. Diagn Pathol 2014; 9:164; PMID:25163571; http://dx.doi.org/10.1186/s13000-014-0164-1
- Mathsyaraja H, Thies K, Taffany DA, Deighan C, Liu T, Yu L, Fernandez SA, Shapiro C, Otero J, Timmers C et al. CSF1-ETS2-induced microRNA in myeloid cells promote metastatic tumor growth. Oncogene 2014; PMID:25241894; http://dx.doi.org/10.1038/onc.2014.294
- Dikshit B, Irshad K, Madan E, Aggarwal N, Sarkar C, Chandra PS, Gupta DK, Chattopadhyay P, Sinha S, Chosdol K. FAT1 acts as an upstream regulator of oncogenic and inflammatory pathways, via PDCD4, in glioma cells. Oncogene 2013; 32:3798-808; PMID:22986533; http://dx.doi.org/10.1038/onc.2012.393
- Das A, Ganesh K, Khanna S, Sen CK, Roy S. Engulfment of apoptotic cells by macrophages: a role of microRNA-21 in the resolution of wound inflammation. J Immunol 2014; 192:1120-9; PMID:24391209; http://dx.doi.org/10.4049/jimmunol.1300613
- Dorner BG, Scheffold A, Rolph MS, Huser MB, Kaufmann SH, Radbruch A, Flesch IE, Kroczek RA. MIP-1alpha, MIP-1beta, RANTES, and ATAC/lymphotactin function together with IFN-gamma as type 1 cytokines. Proc Natl Acad Sci U S A 2002; 99:6181-6; PMID:11972057; http://dx.doi.org/10.1073/pnas.092141999
- Sheedy FJ. Turning 21: Induction of miR-21 as a Key Switch in the Inflammatory Response. Front Immunol 2015; 6:19; PMID:25688245; http://dx.doi.org/10.3389/fimmu.2015.00019
- Ripka S, Neesse A, Riedel J, Bug E, Aigner A, Poulsom R, Fulda S, Neoptolemos J, Greenhalf W, Barth P et al. CUX1: target of Akt signalling and mediator of resistance to apoptosis in pancreatic cancer. Gut 2010; 59:1101-10; PMID:20442202; http://dx.doi.org/10.1136/gut.2009.189720
- Baumgart S, Glesel E, Singh G, Chen NM, Reutlinger K, Zhang J, Billadeau DD, Fernandez-Zapico ME, Gress TM, Singh SK et al. Restricted heterochromatin formation links NFATc2 repressor activity with growth promotion in pancreatic cancer. Gastroenterology 2012; 142:388-98 e1-7; PMID:22079596; http://dx.doi.org/10.1053/j.gastro.2011.11.001