ABSTRACT
Thymic stromal lymphopoietin (TSLP) is an epithelial cell-derived cytokine that primes dendritic cells for Th2 induction. It has been implicated in different types of allergic diseases. Recent work suggested that TSLP could play an important role in the tumor microenvironment and influence tumor progression, in particular in breast cancer. In this study we systematically assessed the production of TSLP at the mRNA and protein levels in several human breast cancer cell lines, large-scale public transcriptomics data sets, and primary human breast tumors. We found that TSLP production was marginal, and concerned less than 10% of the tumors, with very low mRNA and protein levels. In most cases TSLP was undetectable and found to be expressed at lower levels in breast cancer as compared to normal breast tissue. Last, we could not detect any functional TSLP receptor (TSLPR) expression neither on hematopoietic cells nor on stromal cells within the primary tumor microenvironment. We conclude that TSLP-TSLPR pathway activity is not significantly detected within human breast cancer. Taken together, these observations do not support TSLP targeting in breast cancer.
Introduction
Within the tumor microenvironment, a diversity of immune-modulating factors can shape antitumor immunity, either by inducing and strengthening it, or by shifting a protective cytotoxic response toward an inappropriate regulatory response.Citation1 In particular, cytokines mediate complex cross talks between tumor cells and immune cells. Immune cell-derived cytokines may affect tumor cell differentiation, invasion and metastasis, hence participating in the oncogenic process. For example inflammatory cell-derived TNF can promote tumor epithelial cell survival through the induction of genes encoding NF-κF—dependent antiapoptotic molecules.Citation2 Conversely tumor cell-derived cytokines are critical to shape the state and effector functions of tumor-infiltrating immune cells. IL6 produced by renal cell carcinoma inhibits dendritic cell function and differentiation.Citation3 Tumor-derived TNF may contribute to different functions related to the type of tumor and the timing.Citation4 We have shown that tumor-derived GM-CSF promotes plasmacytoid pre-dendritic cell (pDC) survival and activation, with subsequent priming of a regulatory Th2 response.Citation5
Thymic stromal lymphopoietin (TSLP) is an epithelial cell-derived cytokine that promotes Th2 polarization through dendritic cell activation.Citation6,7 TSLP is central to the physiopathology of allergic inflammation, such as atopic dermatitis and asthma.Citation6,8,9 Recently, it was shown to contribute to autoimmune inflammation, in particular in psoriasis.Citation10 Because of its role in linking epithelial cells to immune cells, TSLP has been explored in the past few years as potentially contributing to the tumor-immune cell crosstalk.Citation11-14 TSLP investigation in cancer was also motivated by the presence of Th2 type responses in some tumors, raising the hypothesis that Th2 promoting factors may be produced by tumor cells. Based on this rationale, TSLP was suggested to play a role in human breast cancer as a key cytokine produced by tumor epithelial cells and promoting T helper cells to produce IL-13, a prototypical Th2 cytokine.Citation12,13 TSLP was also implicated in pancreatic cancer where it was shown to be secreted by tumor-infiltrating fibroblasts.Citation11 In these two clinical settings and in a mouse model of breast and pancreatic cancer, TSLP was associated with tumor progression and metastasis.Citation13,15 In contrast, a recent study reported systemic TSLP impairs breast cancer and pancreatic development in mice through direct stimulation of Th2 cells.Citation16 In these studies, association between TSLP and prognosis in clinical cohorts of breast cancer patients was not assessed. In addition, results obtained in various skin cancer models suggested that TSLP may be of good prognosis and favor tumor regression.Citation17,18 Hence, the role of TSLP in cancer and the underlying mechanisms of action remain controversial.
In an effort to map TSLP expression in various tumor types, we had initiated several years ago a screening by immunohistochemistry in human primary tumors. Breast cancer was used as a main model for adenocarcinomas and head and neck cancer as a model for epidermoid tumor. While we observed high TSLP expression in head and neck tumors (see Guillot-Delost et al., companion paper), we did not detect TSLP staining in breast adenocarcinomas. Because of the conflicting data published in breast cancer, we sought to analyze in depth, in a systematic manner, the TSLP-TSLP receptor axis in human breast cancer at different levels and using a variety of methods. Although low TSLP was found in few tumor samples, most results indicated a lack of TSLP expression, both at the mRNA and protein levels, ex vivo and in situ. We also observed a lack of TSLP-receptor expression in the breast cancer microenvironment. We therefore, conclude that there is no evidence for TSLP pathway activity in human breast cancer.
Results
Breast cancer cell lines do not express TSLP mRNA
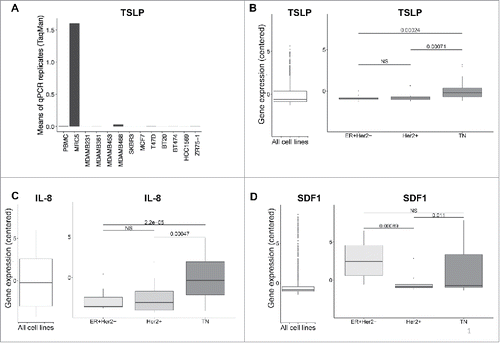
First, we screened TSLP mRNA expression in breast cancer cell lines using quantitative PCR (). The lung fibroblast sarcoma cell line MRC5, which was initially used to clone human TSLP,Citation19 was used as a positive control for TSLP mRNA, with PBMC being our negative control (). In comparison, we analyzed 11 breast cancer cell lines of different molecular subtypes. All breast cancer cell lines were negative for TSLP expression (). In order to further increase the diversity of cell lines in an unbiased manner, we mined the Cancer Cell Line Encyclopedia, CCLE public database, which includes RNAseq expression data of about 1036 human cell lines from different anatomical sites.Citation20 We analyzed TSLP expression, and also assessed IL-8 and SDF1 expression as controls (). TSLP was absent or expressed at very low level in breast cancer cell lines (). Higher levels were observed in triple negative (TN) cell lines, as compared to luminal A (ER+Her2−) and Her2+ cell lines, although expression remained close to detection limit for most cell lines (). In addition, these gene expression levels were marginal as compared to cell lines expressing significant levels (>2 ) of TSLP (). IL-8 expression was detected at low levels in the different cell lines, with again a slightly higher expression in TN breast cancer cell lines (). In comparison, SDF1 was most significantly expressed in luminal A and TN breast cancer cell lines (). In summary, although our own assessments of TSLP expression by quantitative PCR was negative on all 11 breast cancer cell lines tested, data mining of transcriptomic profiles in 58 breast cancer cell lines raised the possibility of a very low TSLP expression in TN breast cancer subtype.
Figure 1. Breast cancer cell lines do not express TSLP mRNA. (A) TSLP mRNA expression quantified by quantitative PCR (TaqMan) in 11 breast cancer cell lines. PBMC and MRC5 correspond to negative and positive control respectively. N = 4. (B, C, D) Boxplots in the left panels represent mRNA expression in 1036 cancer cell lines from CCLE of TSLP, IL-8 and SDF1 respectively. (B, C, D) Boxplots in the right panels show the gene expression of TSLP, IL-8 and SDF1 respectively for breast cancer cell lines, which were grouped according to their corresponding molecular subtype. p values were calculated with a t test comparing different cancer cell subtypes.
Transcriptomic analysis reveals that TSLP mRNA level is higher in normal breast than in primary breast cancer tissue
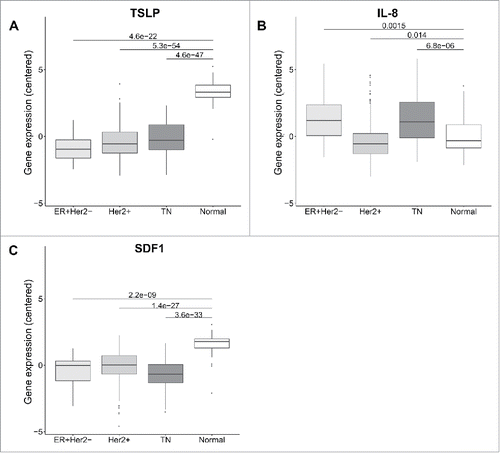
In order to get closer to primary tumors, we went on data mining The Cancer Genome Atlas, TCGA database, comprising transcriptional profiles of about 591 primary breast tumors.Citation21 Importantly, this large-scale data set also included normal breast tissue, which is key to interpret any results related to a neoplastic tissue. Surprisingly, we found significant TSLP expression only in normal breast tissue, which was statistically higher than in any of the three breast cancer molecular subtypes (). In this primary tumor expression dataset, higher TSLP in TN tumors was not observed, since all three tumor types had expression levels close to zero (). By comparison IL-8 expression was significantly higher in luminal A and TN tumors, as compared to Her2+ tumors and normal breast tissue (). SDF1 was higher in normal breast tissue, parallel to TSLP (). These results do not support significant TSLP expression in primary breast cancer, and excluded TSLP expression as a specific feature of the breast tumor inflammation since it was observed at higher levels in normal breast tissue.
Figure 2. TSLP mRNA level is higher in normal breast than breast cancer tissue. (A, B, C) mRNA expression in 58 breast cancer patients from TCGA of TSLP, IL-8 and SDF1 respectively. Boxplots represent data of tumors classified in three different subtypes, namely Luminal (ER+Her2−), Her2+ and Triple negative (TN) and normal breast tissue as indicated. p values were calculated with a t test comparing different clinical groups.
qPCR analysis reveals that TSLP mRNA expression is higher in juxta-tumor tissue than primary breast cancer tissue
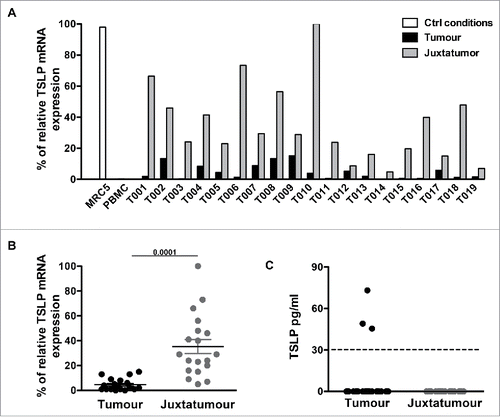
In order to get a more reliable and controlled assessment of TSLP expression in primary breast cancer, we prospectively collected and analyzed primary breast tumors obtained from the Institut Curie Pathology Department. We performed quantitative PCR analysis for TSLP mRNA on 19 independent tumors coupled to their juxta-tumor non-involved counterpart (). As for the cell lines, PBMC were used as negative control and MRC5 as a positive control for TSLP (). TSLP expression was negative or very low (<20 %) in all tumor samples, and was systematically lower in the tumor as compared to the juxta-tumor non-involved counterpart (). This result was in accordance with the higher levels observed in normal breast tissue from the TCGA database (). Considering all tumor samples, higher TSLP levels were observed in the non-involved tumor counterpart in a statistically significant manner ().
Figure 3. TSLP mRNA expression is higher in juxta-tumor tissue than breast cancer tissue. (A) TSLP transcripts were measured by quantitative PCR (TaqMan) in 19 breast cancer tissues (black bars) and 19 corresponding juxta-tumor tissues (gray bars). White bars represent TSLP levels detected in MRC5 and PBMC used as positive and negative control respectively. (B) Quantification of TSLP mRNA transcripts shown as percentage of housekeeping gene expression. Four housekeeping genes were used for these experiments: Actin Beta (ACTB), Hypoxanthine Phosphoribosyltransferase 1 (HPRT1), Ribosomal Protein L31 (RPL31) and Beta-2-Microglobulin (B2M). Lines represent mean +/− the Standard Error of the Mean (SEM). Wilcoxon matched pairs test was used to calculate p value. N = 19. (C) Quantification of soluble TSLP measured by ELISA in the supernatants generated from primary breast tumor tissues and corresponding juxta-tumor samples as described in the Material and Methods section. ELISA sensitivity detection limit, which is represented by the dashed line, was 31 pg/mL as recommended by the manufacturer instructions. N = 40.
Breast cancer tissue do not express detectable amount of TSLP protein
In order to get a first assessment of TSLP protein expression levels, we cultured for 24 h each of the 19 primary tumor samples, as well as their juxta-tumor counterpart, and analyzed the tissue-conditioned supernatants for TSLP secretion by ELISA (). Three tumor samples out of 19 (15%) released TSLP, although at very low levels, slightly above detection limit, which was set by the ELISA manufacturer at 32.5 pg/mL (). Surprisingly, juxta-tumor samples did not release any detectable TSLP protein while they expressed TSLP at the mRNA level. This could be due to TSLP retention within the cytoplasm in this tissue type (), as was noted in skin TSLP studies.Citation22
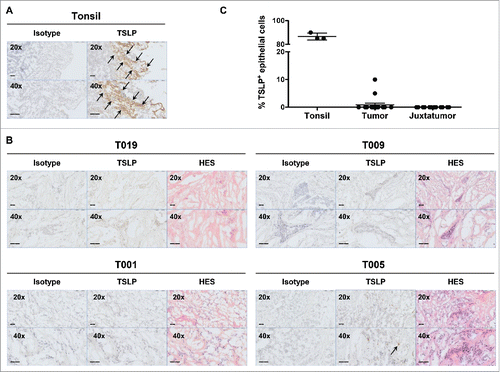
As tissue culture performed in serum-containing medium could artificially alter TSLP expression, we decided to next assess TSLP expression in situ. TSLP expression was assessed by immunohistochemistry on 16 primary breast tumors that were frozen within 15 min following resection. We used a previously validated monoclonal antibody (by us and others).Citation10,23 On human tonsil sections, we could verify that the majority (>80 %) of epithelial cells were TSLP positive, as previously publishedCitation6 (). On the contrary only two out of 16 breast tumors (12.5%) showed a slight positivity for TSLP, with less than 10% of the tumor cells harboring a specific staining pattern (). For each tumor, pathological examination and hematoxylin-eosin-safran (HES) staining in consecutive tissue sections confirmed the presence of epithelial tumor cells (). Thus, using two complementary approaches, we showed that only a minority of tumors was positive for TSLP protein and the TSLP levels we could detect remained marginal.
Figure 4. Breast cancer tissues do not express TSLP. (A) Tonsil sections were used as positive control tissue to validate TSLP staining by immunohistochemistry. TSLP and matched isotype staining was performed in two consecutive tonsil sections. Arrows indicate positive staining. (B) Isotype, TSLP and H&S staining in three consecutive slides of four representative breast cancer specimens. Arrows indicate positive staining. All tissue sections are shown at 20x and 40x magnification. (C) Percentages of TSLP staining in epithelial cells. Each symbol represents a different sample.
TSLP receptor is not expressed in the breast cancer microenvironment
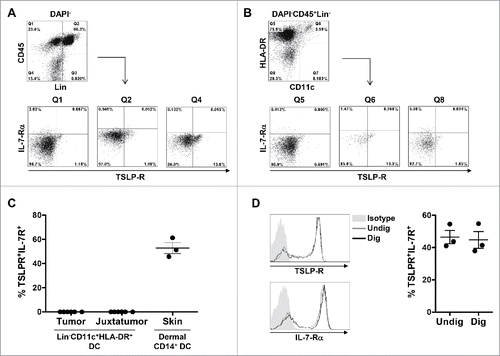
Considering the possibility that low levels of TSLP protein may be secreted in some tumors, downstream function would depend on the expression of the TSLP receptor within the tumor microenvironment. TSLP signals through its receptor only when the TSLP receptor specific chain (TSLP-R) dimerizes with the IL-7 receptor α chain (IL-7-Rα). We thus analyzed by flow cytometry the co-expression of TSLP-R and IL-7-Rα on different cellular compartments. A single cell suspension was obtained from freshly resected primary breast tumors after tissue digestion (). Virtually no IL-7-Rα and TSLP receptor co-expressing cells were detected within the CD45+ compartment, similar to the CD45+Lineage− compartment (). To exclude the possibility that a very low percentage of rare dendritic cells could express the receptor, we also gated on CD45+Lineage−CD11c+HLA-DR+ dendritic cells () and quantified the expression of the TSLP receptor heterodimer in six independent tumors (). TSLP-R and IL-7-Rα double positive dendritic cells ranged from 0 to 0.03%, which we can consider not significant, and close to background (). Similar levels were found on dendritic cells from juxta-tumor non-involved samples (). Comparing digested and non-digested DC showed that the tumor sample digestion protocol did not affect the detection of the two TSLP-R chains (). We conclude that even if few tumors may show low TSLP positivity, the absence of TSLP receptor-expressing cells in the breast tumor microenvironment excludes the possibility of downstream TSLP functionality.
Figure 5. TSLP receptor is not detected in primary breast cancer by flow cytometry. (A–B) Flow cytometry dot plots from one representative human primary breast tumor. (A) Gating strategy used to investigate the expression of TSLP-R and IL-7-Rα in viable CD45+Lin− (Q1), CD45+Lin+ (Q2) and CD45−Lin− (Q4). (B) Gating strategy designed to detect DC in primary breast cancer. DC were defined as DAPI−CD45+Lin−HLA-DRhighCD11chigh. The expression of TSLP-R and IL-7-Rα was assessed in DC (Q6) as well as HLA-DRhighCD11c− (Q5) and HLA-DR−CD11c− (Q8). (C) Percentages of TSLP-R+IL-7-Rα+ DC in tumors, corresponding juxta-tumor tissues and in the dermis of normal skin. Lines represent mean +/− the Standard Error of the Mean (SEM). N = 6 (D) TSLP-R and IL-7-R expression in DC enriched from PBMC without digestion step (gray histogram, Undig) and after mechanical and enzymatic digestion (dark gray histogram, Dig), left panels. One representative background histogram staining is shown in light gray. Quantification of TSLP-R+IL-7-Rα+DC in undigested and digested enriched DC is shown in the left panel. N = 3.
Discussion
In the present study, we systematically assessed the presence of TSLP at the mRNA and protein levels, in several human breast cancer cell lines, large-scale public transcriptomics data sets and human primary breast tumors. We found that TSLP production was marginal, and concerned less than 10% of the tumors, with very low mRNA and protein levels. In most cases, TSLP was undetectable and found to be expressed at lower levels in breast cancer as compared to normal breast tissue. Last, we could not detect any functional TSLPR expression neither on haematopoietic cells nor on stromal cells within the primary tumor microenvironment. We conclude that TSLP-TSLPR pathway activity is not significantly detected within human breast cancer. Those results are in contrast with previous studies reporting TSLP expression in breast cancer.Citation12,13
Expression of immune modulating cytokines in the tumor microenvironment is most of the time interpreted as being associated to the tumoral process, and being part of pro- or antitumor immune mechanisms. This view only stands if the normal tissue counterpart is devoid of expression of that specific cytokine, or harbors a much lower expression, implying that the cytokine expression is a specific feature of the tumor. In our results, we analyzed normal breast TSLP mRNA expression from public databases, and found that TSLP levels were higher than in breast tumors. This is in accordance with recent reports detecting TSLP protein in breast milkCitation24 with a potential role in the intestinal immunity of the neonates.Citation25 The presence of TSLP in normal breast suggests that TSLP may contribute to physiological mechanisms in this context, and is not a feature developed by breast tumors or related to breast cancer inflammation. This is an important aspect to take into consideration in the interpretation of TSLP role in cancer.
TSLP assessment in situ in inflamed tissue has provided a very efficient and unbiased manner to detect TSLP in various diseases.Citation8,9 Using immunohistology, we and others have shown strong TSLP staining in the keratinocytes of atopic dermatitis and psoriasisCitation6,10,26 as well as in the tonsillar epithelial cells.Citation6 Conversely, normal skin has repeatedly been found as negative for any TSLP staining.Citation6,10,27 Such TSLP positive and negative tissue types constitute valuable controls for any study of TSLP expression in disease. In the present study, we have systematically compared TSLP analysis in breast cancer to human tonsils, and the TSLP levels we could detect remained marginal in terms of percentage of TSLP positive epithelial cells, as well as in the intensity of TSLP staining. In a previous study about TSLP role in breast cancer, TSLP expression was analyzed by immunofluorescence and was found positive on most tumor epithelial cells,Citation13 in discrepancy with our own results. However, that study lacked a negative control, and used normal skin as a positive control, when other studies could not detect any TSLP expression in this context.Citation6,10,27 This raises questions on the interpretation of the results. In contrast, in our companion paper, we report that TSLP is highly expressed in head and neck squamous cell carcinoma, at levels similar to atopic dermatitis. This indicates that although TSLP is absent from the breast cancer microenvironment, it can be expressed in other cancer types.
Assessing the protein expression of cytokines in human primary tumors has many intrinsic difficulties, and multiple complementary strategies should be considered. Analysis of tumor-derived supernatants has been used extensively to analyze the soluble tumor microenvironment. It has the potential drawback that tissue culture manipulation may induce the secretion of factors that are not being spontaneously secreted. Culture conditions may also influence the amounts of cytokine production. In this study, we have used basic serum-containing medium without any activator in order to avoid as much as possible artificial induction of cytokine synthesis and secretion. Other studies have used PMA/ionomycin in order to stimulate immune cells, which may generate direct and indirect effects promoting cytokine production within the tumor microenvironment.Citation13 Although such immune activators may be helpful to analyzed T cell-derived cytokines,Citation28 they may also have effects on non-immune cells, either directly or through paracrine activating loops. For example PMA/ionomycin may induce TNF production by T cells,Citation29 which can subsequently activate TSLP production by neighboring epithelial cells.Citation22 Hence, tumor-conditioned media obtained in the presence of any type of activating signal should be interpreted with caution.
Signaling pathway activity requires expression of all its components: ligand, receptor and downstream signaling molecules. In the case of TSLP, a functional pathway would require the presence not only of TSLP but also of TSLP receptor heterodimer (TSLP-R/IL7Rα). However, in previous studies suggesting a TSLP role in human cancer, TSLP receptor heterodimer expression in the tumor microenvironment was not assessedCitation11,13 raising the possibility that this pathway is not functional despite the presence of TSLP itself. Along this line, we show in our companion paper that although head and neck tumors express high levels of TSLP, TSLP-R-expressing cells are absent from the tumor microenvironment. These observations highlight that such a dissociated ligand-receptor expression can result in an inactive TSLP pathway. Here, we report for the first time that TSLP-R is not present in the microenvironment of human breast primary tumors. This suggests that even if low levels of TSLP are present (below the detection threshold, or at low levels in rare cases), TSLP pathway is unlikely to be active as its signaling is disrupted by the absence of TSLP receptor.
Th2 cytokines were reported to contribute to tumorigenesis in several mouse models.Citation30-32 Human breast cancer has also been associated to a Th2 response, which was shown to be promoted by TSLP.Citation13 However, in that study, a large proportion of tumors (61%) were negative for TSLP expression. In our study, TSLP could not be detected in the vast majority of breast tumors and its receptor was also absent. Those observations raise the question of parallel or alternative pathways to induce Th2 response. We have recently shown that GM-CSF produced by tumor epithelial cells activates pDC to promote a regulatory Th2 response in human primary breast tumors.Citation5 Concomitant increase of GM-CSF and pDC was found in 11.8% of breast tumors studied (14 out of 118), and associated to more aggressive breast cancer subtypes. In addition, CCL5 was also reported to promote Th2 polarization in breast cancer. Indeed CCL5 depletion in MMTV-PyMT mouse model leads to a deficit in Th2 cell associated with reduced tumor burden and metastasis.Citation33 Furthermore, CCL5 and IL-4 expression were correlated and associated with aggressiveness of human luminal breast cancer.Citation33 Further studies will be needed to evaluate the relative role of these pathways within the breast cancer microenvironment and their corresponding roles in supporting Th2 cells.
Our work has potential implications for therapy and drug development. Indeed, it does not support TSLP as a relevant target in breast cancer as we do not detect any evidence of TSLP-TSLPR pathway activity in this clinical setting. Although it is almost impossible to completely rule out a biological pathway implication in disease, our negative results should encourage further work to assess TSLP pathway activity in different types of cancer, as well as the relative contribution of Th2-promoting factors in individual tumor samples.
Material and methods
Human samples and patients' characteristics
Tumor and juxta-tumor (adjacent to the tumor and exempt of malignant tumor cells) tissues were collected during standard surgical procedures as surgical residues from untreated breast cancer patients, from the department of Pathology (Institut Curie, Paris). Patients signed an informed consent after approval of the study. This study was approved by the Internal Review Board and Clinical Research Committee of the Institut Curie. Patient characteristics are summarized in .
Table 1. Clinical information of patients included in the study.
Tonsils sections were obtained after surgical resection from children undergoing tonsillar resection (Necker Hospital, Paris) after informed consent of the parents. Tissues were transported in CO2-independant medium (Gibco) and processed within the next 3 h after resection.
Healthy donor human blood buffy coats were obtained from “Etablissement Français du Sang,” Paris, Saint-Antoine Crozatier blood bank through an approved convention with the Institut Curie.
Normal skin samples considered as surgical wastes were obtained from healthy donors undertaking esthetic or reconstructive surgery and processed within 6 h of resection. This discarded human surgical material was obtained anonymously according to the institutional regulations, in compliance with French legislation.
Cell line culture
All cell lines were cultured without stimulation at the density of 0.5 ×106 cells/mL in complete RPMI GlutaMAX (Gibco) containing 10% FBS (HyClone) for 48 h. Cells were then washed with PBS, detached with trypsin (Gibco), pelleted and lysed in RLT buffer (Qiagen) to allow RNA extraction. All cell lines were mycoplasma-free.
Large-scale public database mining
The gene level expression of TSLP, IL8 and SDF1 have been analyzed using the Breast Cancer Cell Lines Encyclopedia. Data were downloaded from the CCLE website (http://www.broadinstitute.org/ccle)Citation20 and normalized with RMA. This data set was composed of 1036 cell lines from 24 tissues. Patient data were retrieve from a sample of 591 breast cancer patients from TCGA (Level 3).Citation34 Breast cancer subtypes were defined using a bimodal mixture of 2 gaussian distributions for ER, PR and HER2 gene expression. TN breast cancer samples were defined by the absence of estrogen and progesterone receptor expression and a lack of HER2 overexpression/amplification. In CCLE dataset, breast cancer cell lines were composed of 31 TN, 12 ER+Her2− and 15 Her2+. Breast tumors from TCGA data set were composed of 26 ER+Her2−, 410 Her2+, 94 TN and 61 Normal.
Primary tumor processing for RNA extraction
Tumor and juxta-tumor tissues were cryopreserved in Tissue-Tek (Sakura Finetek USA, Inc., Torrance, Calif) at −80°C. Tissues were cryosectioned with a Cryostat. Ten sections of 20 μm thickness were collected for every tissue sample. Tissues were lysed in RLT buffer (Qiagen) supplemented with β-mercaptoethanol (Sigma) immediately after cutting. RNA was extracted using RNAeasy mini kit (Qiagen) following manufacturer instructions and processed as described above.
Quantitative PCR
RNA was extracted from cell line and tumor section lysates using RNeasy micro and RNeasy mini kit (Qiagen) respectively following manufacturer instructions including on-column DNase digestion. RNA concentration and absence of protein contamination were determined using the NanoDrop instrument. All RNA samples had 260 nm/280 nm absorbance ratios between 1.9 and 2.1, indicating high purity. RNA quality was assessed using RNA 6000 Nano chips on the Agilent 2100 Bioanalyzer. Only samples with RIN>7 were further processed for reverse transcription. cDNA was synthesized with a mix containing random hexamers (Promega), oligo(dT)15 (Promega) and SuperScript II reverse transcriptase (Invitrogen). TSLP mRNA transcripts, as well as ACTB (Actin β), B2M (Beta-2 Microglobulin), HPRT (hypoxanthine phosphoribosyltransferase 1) and RPL34 (ribosomal protein L34), which were used as housekeeping genes were quantified by real-time quantitative reverse transcription PCR on Light Cycler 480 (Roche) with Applied Biosystems predesigned TaqMan Gene Expression Assays and Absolute qPCR ROXmix (Thermo Fisher Scientific).
Primary breast tumor and juxta-tumor digestion for flow cytometry analysis and generation of tissue-conditioned supernatants
Breast tumor and juxta-tumor tissues were cut in three pieces. One piece was frozen in Tissue-Tek (Sakura Finetek USA, Inc., Torrance, Calif) for further histological analysis. The second piece was carefully minced into smaller pieces in CO2 independent medium (Gibco) containing 5% FBS (HyClone). Those pieces were digested with collagenase (1 mg/mL; Roche) and DNAse (25 μg/mL; Roche) in a total volume of 3 mL CO2 independent medium (Gibco) for 1 h at 37°C under agitation at 180 rpm. Cell suspension was then filtered through a 40 µm nylon cell strainer (Falcon BD) and washed twice in cold PBS containing 5% of human serum (Biowest) and EDTA 2 mM (Gibco). Skin digestion was performed as described in ref.Citation10. Cells were stained with the following mouse anti-human antibodies and corresponding matched isotype controls: CD3-FITC, CD16-FITC, CD45-APC-Cy7, CD11c-PE-Cy5 (BD Biosciences), CD14-FITC, CD20-FITC (Miltenyi Biotec), HLADR-Alexa700, TSLPR-APC (Biolegend), IL-7Rα-PE, (eBiosciences). Dead cells were excluded based on side and forward scatter characteristics and positivity for 4,6-diamidino-2-phenylindole (DAPI) (Invitrogen). Sample acquisition was performed on a LSRII flow cytometer (Becton Dickinson) and data analysis was performed using FlowJo software version 9.4.7. The third piece of tissue was cut in smaller pieces of 40 mg. Each piece of tissue was put in one well of a 48 well plate in 250 μl of complete RPMI GlutaMAX (Gibco) containing 10% FBS (HyClone) without any stimulation. Supernatants were harvested after 24 h of culture and tissues were discarded. Supernatants were spun for 5 min at maximum speed to remove dead cells and debris and stored at −80°C for further ELISA measurements.
Soluble TSLP quantification
Soluble TSLP was quantified using the DuoSet Kit from R&D following manufacturer instructions. Detection limit was set at 31.25 pg/mL as recommended.
Immunohistochemistry
Tissues were embedded in Tissu-Tek (Sakura Finetek USA, Inc., Torrance, Calif) and cryopreserved at −80°C. Acetone-fixed cryosections of 4 μm thickness were stained with monoclonal rat anti-human TSLP 5 µg/mL (kind gift from Pr. Yong-Jun Liu), and corresponding matched isotype control antibody (BD Pharmigen), followed by a biotinylated goat anti-rat secondary antibody (Vector Laboratories). The staining was revealed using a Vectastain ABC peroxidase system (Vector Laboratories) and it was detected using 3-3Œ-diamino-benzidine-tetrahydrochloride (DAB) revelation (Vector Laboratories). The sections were counterstained with haematoxylin and mounted with Perthex mounting media (Histolab). Tonsil sections were treated using the same procedure and they served as positive control for TSLP staining. The staining was performed using the Autostainer 480 (Labvision). Tissue images were taken on the Philips Digital Pathology Ultra-Fast Scanner.
DC enrichment from human blood
Peripheral blood mononuclear cells (PBMC) were isolated using Ficoll-gradient (GE Healthcare). DC were enriched using the EasySep ™ human Pan-DC Pre-Enrichment kit (Stem Cell Technologies) following manufacturer instructions.
Statistical analysis
Unpaired t tests were used to determine statistical significance. Statistical significance was retained for p values lower than 0.05. Symbols used: NS, not significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by funding from Institut National de la Santé et de la Recherche Médicale (BIO2012-02 and BIO2014-08), Fondation pour la Recherche Médicale, Association de la Recherche Contre le Cancer (PJA 20131200436), INCA (2011-1-PL BIO-12-IC-1 and 2012-1-GYN-04-IC-1), ANR-13-BSV1-0024-02, ANR-10-IDEX-0001-02 PSL* and ANR-11-LABX-0043, CIC IGR-Curie 1428.
References
- Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013; 39:1-10; PMID:23890059; http://dx.doi.org/10.1016/j.immuni.2013.07.012
- Luo JL, Maeda S, Hsu LC, Yagita H, Karin M. Inhibition of NF-kappaB in cancer cells converts inflammation- induced tumor growth mediated by TNFalpha to TRAIL-mediated tumor regression. Cancer Cell 2004; 6:297-305; PMID:15380520; http://dx.doi.org/10.1016/j.ccr.2004.08.012
- Song EY, Shurin MR, Tourkova IL, Chatta G, Shurin GV. Human renal cell carcinoma inhibits dendritic cell maturation and functions. Urologe A 2004; 43 Suppl 3:S128-130; PMID:15150693; http://dx.doi.org/10.1007/s00120-004-0599-1
- Balkwill F. Tumour necrosis factor and cancer. Nat Rev Cancer 2009; 9:361-71; PMID:19343034; http://dx.doi.org/10.1038/nrc2628
- Ghirelli C, Reyal F, Jeanmougin M, Zollinger R, Sirven P, Michea P, Caux C, Bendriss-Vermare N, Donnadieu MH, Caly M et al. Breast cancer cell-derived GM-CSF licenses regulatory Th2 induction by plasmacytoid predendritic cells in aggressive disease subtypes. Cancer Res 2015; 75:2775-87; PMID:25977333; http://dx.doi.org/10.1158/0008-5472.CAN-14-2386
- Soumelis V, Reche PA, Kanzler H, Yuan W, Edward G, Homey B, Gilliet M, Ho S, Antonenko S, Lauerma A et al. Human epithelial cells trigger dendritic cell mediated allergic inflammation by producing TSLP. Nat Immunol 2002; 3:673-80; PMID:12055625; http://dx.doi.org/10.1038/nrm910
- Liu YJ, Soumelis V, Watanabe N, Ito T, Wang YH, Malefyt Rde W, Omori M, Zhou B, Ziegler SF. TSLP: an epithelial cell cytokine that regulates T cell differentiation by conditioning dendritic cell maturation. Annu Rev Immunol 2007; 25:193-219; PMID:17129180; http://dx.doi.org/10.1146/annurev.immunol.25.022106.141718
- Shikotra A, Choy DF, Ohri CM, Doran E, Butler C, Hargadon B, Shelley M, Abbas AR, Austin CD, Jackman J et al. Increased expression of immunoreactive thymic stromal lymphopoietin in patients with severe asthma. J Allergy Clin Immunol 2012; 129:104-11 e101-109; PMID:21975173; http://dx.doi.org/10.1016/j.jaci.2011.08.031
- Ying S, O'Connor B, Ratoff J, Meng Q, Mallett K, Cousins D, Robinson D, Zhang G, Zhao J, Lee TH et al. Thymic stromal lymphopoietin expression is increased in asthmatic airways and correlates with expression of Th2-attracting chemokines and disease severity. J Immunol 2005; 174:8183-90; PMID:15944327; http://dx.doi.org/10.4049/jimmunol.174.12.8183
- Volpe E, Pattarini L, Martinez-Cingolani C, Meller S, Donnadieu MH, Bogiatzi SI, Fernandez MI, Touzot M, Bichet JC, Reyal F et al. Thymic stromal lymphopoietin links keratinocytes and dendritic cell-derived IL-23 in patients with psoriasis. J Allergy Clin Immunol 2014; 134:373-81; PMID:24910175; http://dx.doi.org/10.1016/j.jaci.2014.04.022
- De Monte L, Reni M, Tassi E, Clavenna D, Papa I, Recalde H, Braga M, Di Carlo V, Doglioni C, Pia Protti M. Intratumor T helper type 2 cell infiltrate correlates with cancer-associated fibroblast thymic stromal lymphopoietin production and reduced survival in pancreatic cancer. J Exp Med 2011; 208:469-78; PMID:21339327; http://dx.doi.org/10.1084/jem.20101876
- Olkhanud PB, Rochman Y, Bodogai M, Malchinkhuu E, Wejksza K, Xu M, Gress RE, Hesdorffer C, Leonard WJ, Biragyn A. Thymic stromal lymphopoietin is a key mediator of breast cancer progression. J Immunol 2011; 186:5656-62; PMID:21490155; http://dx.doi.org/10.4049/jimmunol.1100463
- Pedroza-Gonzalez A, Xu K, Wu TC, Aspord C, Tindle S, Marches F, Gallegos M, Burton EC, Savino D, Hori T et al. Thymic stromal lymphopoietin fosters human breast tumor growth by promoting type 2 inflammation. J Exp Med 2011; 208:479-90; PMID:21339324; http://dx.doi.org/10.1084/jem.20102131
- Wu TC, Xu K, Banchereau R, Marches F, Yu CI, Martinek J, Anguiano E, Pedroza-Gonzalez A, Snipes GJ, O'Shaughnessy J et al. Reprogramming tumor-infiltrating dendritic cells for CD103+ CD8+ mucosal T-cell differentiation and breast cancer rejection. Cancer Immunol Res 2014; 2:487-500; PMID:24795361; http://dx.doi.org/10.1158/2326-6066.CIR-13-0217
- Lo Kuan E, Ziegler SF. Thymic stromal lymphopoietin and cancer. J Immunol 2014; 193:4283-8; PMID:25326546; http://dx.doi.org/10.4049/jimmunol.1400864
- Demehri S, Cunningham TJ, Manivasagam S, Ngo KH, Moradi Tuchayi S, Reddy R, Meyers MA, DeNardo DG, Yokoyama WM. Thymic stromal lymphopoietin blocks early stages of breast carcinogenesis. J Clin Invest 2016; 126:1458-70; PMID:26927668; http://dx.doi.org/10.1172/JCI83724
- Di Piazza M, Nowell CS, Koch U, Durham AD, Radtke F. Loss of cutaneous TSLP-dependent immune responses skews the balance of inflammation from tumor protective to tumor promoting. Cancer Cell 2012; 22:479-93; PMID:23079658; http://dx.doi.org/10.1016/j.ccr.2012.08.016
- Demehri S, Cunningham TJ, Manivasagam S, Ngo KH, Moradi Tuchayi S, Reddy R, Meyers MA, DeNardo DG, Yokoyama WM. Elevated epidermal thymic stromal lymphopoietin levels establish an antitumor environment in the skin. Cancer Cell 2012; 22:494-505; PMID:23079659; http://dx.doi.org/10.1016/j.ccr.2012.08.017
- Reche PA, Soumelis V, Gorman DM, Clifford T, Liu Mr, Travis M, Zurawski SM, Johnston J, Liu YJ, Spits H et al. Human thymic stromal lymphopoietin preferentially stimulates myeloid cells. J Immunol 2001; 167:336-43; PMID:11418668; http://dx.doi.org/10.4049/jimmunol.167.1.336
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012; 483:603-7; PMID:22460905; http://dx.doi.org/10.1038/nature11003
- Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490:61-70; PMID:23000897; http://dx.doi.org/10.1038/nature11412
- Bogiatzi SI, Fernandez I, Bichet JC, Marloie-Provost MA, Volpe E, Sastre X, Soumelis V. Cutting Edge: Proinflammatory and Th2 cytokines synergize to induce thymic stromal lymphopoietin production by human skin keratinocytes. J Immunol 2007; 178:3373-7; PMID:17339431; http://dx.doi.org/10.4049/jimmunol.178.6.3373
- Fontenot D, He H, Hanabuchi S, Nehete PN, Zhang M, Chang M, Nehete B, Wang YH, Wang YH, Ma ZM et al. TSLP production by epithelial cells exposed to immunodeficiency virus triggers DC-mediated mucosal infection of CD4+ T cells. Proc Natl Acad Sci U S A 2009; 106:16776-81; PMID:19805372; http://dx.doi.org/10.1073/pnas.0907347106
- Macfarlane TV, Seager AL, Moller M, Morgan G, Thornton CA. Thymic stromal lymphopoietin is present in human breast milk. Pediatr Allergy Immunol 2010; 21:e454-456; PMID:20444169; http://dx.doi.org/10.1111/j.1399-3038.2009.00916.x
- Thornton CA, Morgan G. Innate and adaptive immune pathways to tolerance. Nestle Nutr Workshop Ser Pediatr Program 2009; 64:45-57; discussion 57-61, 251-257; PMID:19710514; http://dx.doi.org/10.1159/000235782
- Luo Y, Zhou B, Zhao M, Tang J, Lu Q. Promoter demethylation contributes to TSLP overexpression in skin lesions of patients with atopic dermatitis. Clin Exp Dermatol 2014; 39:48-53; PMID:24341479; http://dx.doi.org/10.1111/ced.12206
- Briot A, Deraison C, Lacroix M, Bonnart C, Robin A, Besson C, Dubus P, Hovnanian A. Kallikrein 5 induces atopic dermatitis-like lesions through PAR2-mediated thymic stromal lymphopoietin expression in Netherton syndrome. J Exp Med 2009; 206:1135-47; PMID:19414552; http://dx.doi.org/10.1084/jem.20082242
- Truneh A, Albert F, Golstein P, Schmitt-Verhulst AM. Early steps of lymphocyte activation bypassed by synergy between calcium ionophores and phorbol ester. Nature 1985; 313:318-20; PMID:3918270; http://dx.doi.org/10.1038/313318a0
- Kim TK, St John LS, Wieder ED, Khalili J, Ma Q, Komanduri KV. Human late memory CD8+ T cells have a distinct cytokine signature characterized by CC chemokine production without IL-2 production. J Immunol 2009; 183:6167-74; PMID:19841187; http://dx.doi.org/10.4049/jimmunol.0902068
- Kobayashi M, Kobayashi H, Pollard RB, Suzuki F. A pathogenic role of Th2 cells and their cytokine products on the pulmonary metastasis of murine B16 melanoma. J Immunol 1998; 160:5869-73; PMID:9637498
- Berzofsky JA, Terabe M. A novel immunoregulatory axis of NKT cell subsets regulating tumor immunity. Cancer Immunol Immunother 2008; 57:1679-83; PMID:18369622; http://dx.doi.org/10.1007/s00262-008-0495-4
- DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, Coussens LM. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell 2009; 16:91-102; PMID:19647220; http://dx.doi.org/10.1016/j.ccr.2009.06.018
- Zhang Q, Qin J, Zhong L, Gong L, Zhang B, Zhang Y, Gao WQ. CCL5-mediated Th2 immune polarization promotes metastasis in luminal breast cancer. Cancer Res 2015; 75:4312-21; PMID:26249173; http://dx.doi.org/10.1158/0008-5472.CAN-14-3590
- Koboldt DC, Fulton RS, McLellan MD, Schmidt H, Kalicki-Veizer J, McMichael JF, Fulton LL, Dooling DJ, Ding L, Mardis ER, et al. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490:61-70; PMID:23000897; http://dx.doi.org/10.1038/nature11412
