 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Immune checkpoint inhibitors blocking the interaction between programmed death-1 (PD-1) and PD-1 ligand-1 (PD-L1) are revolutionizing the cancer immunotherapies with durable clinical responses. Although high expression of PD-L1 in tumor tissues has been implicated to correlate with the better response to the anti-PD-1 therapies, this association has been controversial. In this study, to characterize immune microenvironment in tumors, we examined mRNA levels of immune-related genes and characterized T cell repertoire in the tumors of 13 melanoma patients before and after nivolumab treatment. We found that, in addition to the PD-L1 (p = 0.03), expression levels of PD-1 ligand-2 (PD-L2), granzyme A (GZMA) and human leukocyte antigen-A (HLA-A) in the pre-treatment tumors were significantly higher (p = 0.04, p = 0.01 and p = 0.006, respectively) in responders (n = 5) than in non-responders (n = 8). With nivolumab treatment, tumors in responders exhibited a substantial increase of CD8, GZMA and perforin 1 (PRF1) expression levels as well as increased ratio of TBX21/GATA3, suggesting dominancy of helper T cell type 1 (Th1) response to type 2 (Th2) response. T cell receptor β (TCR-β) repertoire analysis revealed oligoclonal expansion of tumor-infiltrating T lymphocytes (TILs) in the tumor tissues of the responders. Our findings suggest that melanoma harboring high PD-1 ligands (PD-L1 and PD-L2), GZMA and HLA-A expression may respond preferentially to nivolumab treatment, which can enhance Th1-skewed cellular immunity with oligoclonal expansion of TILs.
Abbreviations
| CDR3 | = | complementarity determining region 3 |
| CTLs | = | cytotoxic T lymphocytes |
| HLA | = | human leucocyte antigen |
| PD-1 | = | programmed death-1 |
| PD-L1 | = | PD-1 ligand-1 |
| PD-L2 | = | PD-1 ligand-2 |
| TCR | = | T cell receptor |
| TILs | = | tumor-infiltrating lymphocytes |
Introduction
Immune checkpoint blockades, such as inhibitors of programmed death 1 (PD-1) or PD-1 ligand-1 (PD-L1), enhance antitumor immunity with the prominent clinical responses in a wide range of cancers, including melanoma, renal cell carcinoma, bladder cancer, hematologic malignancies, and non-small cell lung cancer.Citation1-4 For example, an anti-PD-1 antibody, nivolumab, has been recently approved in the United States and Japan for the treatment of unresectable or metastatic melanoma patients.Citation5 Therapeutic anti-PD-1 antibodies inhibit the interaction of PD-1 and its ligands, such as PD-L1 and PD-1 ligand-2 (PD-L2), then effectively restore the antitumor activity of tumor-infiltrating lymphocytes (TILs).Citation2,4,6,7 Identification of a predictive biomarker(s) is critically important for the precision medicine, which can select patients likely to have clinical benefit, reduce an unnecessary medical cost, and avoid autoimmune adverse events. High expression levels of PD-L1 in tumor cells and infiltration of a high number of immune cells have been suggested to correlate with clinical responses to anti-PD-1 therapy,Citation1,8,9 but several studies have shown clinical benefit of PD-1/PD-L1 antibodies even in patients lacking the PD-L1 expression in cancer tissues.Citation2,4,10
TILs play an essential role in orchestrating the host immune response against cancer cells, and therefore characteristics of TILs may represent the anti- (or pro-) tumor immune response. A higher number of TILs in melanoma tissues were correlated with prolonged patient survival and reduced risk of metastasis,Citation11 and persistent existence of clonal cytotoxic T lymphocytes (CTLs) has been documented in melanoma tumors that responded well to adoptive T cell infusion therapy.Citation12,13 Therefore, comprehensive characterization of TILs and immune microenvironment in tumor tissues is imperative for better understanding of cancer immunotherapies. Recently, we have established T cell receptor (TCR) repertoire analysis which allowed us to characterize therapy-induced temporal changes and clonal expansion of TILs using mRNAs isolated from cancer tissues with next-generation sequencer.Citation14-18
In this study, we analyzed mRNA expression levels of immune-related genes in metastatic melanoma tumors of patients who were treated with anti-PD-1 antibody (nivolumab) and found high expression levels of PD-1 ligands (PD-L1 and PD-L2), GZMA, and HLA-A to be correlated with better clinical response. Along with increase of CTLs and Th1-dominant cellular immunity markers, we also detected oligoclonal expansion of TILs in post-treatment tissues in the responders.
Results
Response-based classification of melanoma patients treated with nivolumab
We obtained pre- and post-treatment tumor tissues from 13 metastatic melanoma patients who were treated with nivolumab. According to the RECIST criteria,Citation19 2 cases were judged as partial response (PR), 6 cases as stable disease (SD), and 5 cases as progressive disease (PD) (). The autoimmune-like adverse events were observed in 3 cases; grade 1 hypothyroiditis in 1 PR case, grade 2 psoriasiform dermatitis in 1 PR case, and grade 4 myasthenia gravis with systemic myocarditis and myositis in 1 SD case (). To examine effect of immunological parameters on clinical outcome, we defined “responders” (n = 5) as patients who achieved PR or those who revealed SD with progression-free survival (PFS) of greater than a median period (233 d) of this cohort, and “non-responders” (n = 8) as patients who revealed PD or those who revealed SD with PFS of less than the median ().
Table 1. Patient demographic data.
Expression of PD-1 ligands and response to nivolumab treatment
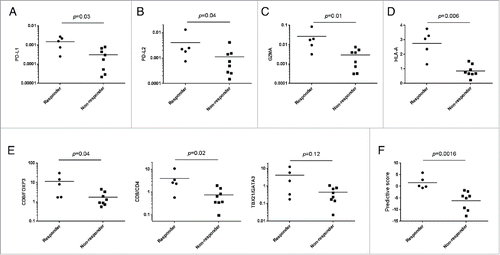
High expression levels of PD-L1 in melanoma have been suggested to be associated with better response to anti-PD-1 therapies,Citation1,8 but some results did not support this association. Another PD-1 ligand, PD-L2 (B7-DC) is also known to suppress cytotoxic activity of T cells in tumor and thereby high PD-L2 expression could be associated with the treatment response.Citation20 Hence, we first examined PD-L1 and PD-L2 mRNA levels in the pre-treatment tumor tissues and found that expression levels of both PD-L1 and PD-L2 were significantly higher in pre-treatment tumors of responders (n = 5) than those of non-responders (n = 8) (p = 0.03 for PD-L1 and p = 0.04 for PD-L2) (); a strong correlation between the expression levels of these two molecules was also observed (Fig. S1A). Because responders had higher expression levels of CD8 and PD-1 in tumors than non-responders (Fig. S1B), we assumed that the responders had a higher number of pre-existing intratumoral CD8+ T cells where the PD-1-PD-L1/2-dependent inhibitory mechanism was likely to be dominant.
Figure 1. Comparison of expression levels of multiple immune-related genes in pre-treatment tumors between responders and non-responders. (A–D) Expression levels of PD-L1 (A), PD-L2 (B), GZMA (C), and HLA-A (D) genes in surgically resected pre-treatment tumors in responders and non-responders are shown. The y-axis indicates expression level of each gene relative to that of GAPDH. (E) The expression ratios of CD8/FOXP3, CD8/CD4 and TBX21/GATA3 are presented. (F) Predictive scores for individual patients were calculated based on expression levels of PD-L1, GZMA, and HLA-A which were significantly higher in the tumors of responders compared with those of non-responders. Horizontal lines represent the means. The Mann–Whitney U test was used to examine statistical significance.
Other immune biomarkers to predict response of nivolumab treatment
We then hypothesized that the pre-existing CD8+ TILs in responder tissues might have higher cytolytic activity, which could be restored by nivolumab treatment. As expected, mRNA level of granzyme A (GZMA), a cytolytic granule in CTLs, was significantly higher in pre-treatment tumors of responders than those of non-responders (p = 0.01) (). The expression level of GZMA was significantly correlated with those of PD-L1 (p = 0.001) and PD-L2 (p = 0.002) (Fig. S1C). In addition, it is well known that HLA class I molecules must be expressed in tumor cells to be recognized by CTLs. Hence, we examined mRNA level of HLA-A, one of the three major HLA class I molecules, and found that it was significantly higher in the pre-treatment tumors in responders than those in non-responders (p = 0.006) (). We further examined expression levels of additional immune-related genes including CD4, FOXP3, interleukin-10 (IL-10), TBX21, and GATA3 as well as ratios of CD8/FOXP3, CD8/CD4, and TBX21/GATA3 in the pre-treatment tumors. Although there were no statistically significant differences in expression levels of CD4, FOXP3, IL-10, TBX21 and GATA3 between the two groups (Fig. S2), the CD8/FOXP3 and CD8/CD4 ratios were significantly higher in tumors in responders than those in non-responders (p = 0.04 and 0.02, respectively), and the TBX21/GATA3 ratio showed higher tendency in responders, compared with non-responders (p = 0.12) (). Based on statistical significance of each gene, we selected three possible predictive markers, PD-L1, GZMA, and HLA-A, and applied to construction of a predictive scoring system (see Materials and Methods) for nivolumab treatment. The scoring system using three possible biomarkers clearly distinguished the responders from non-responders (p = 0.0016) ().
Nivolumab-driven immunological changes in the tumor microenvironment
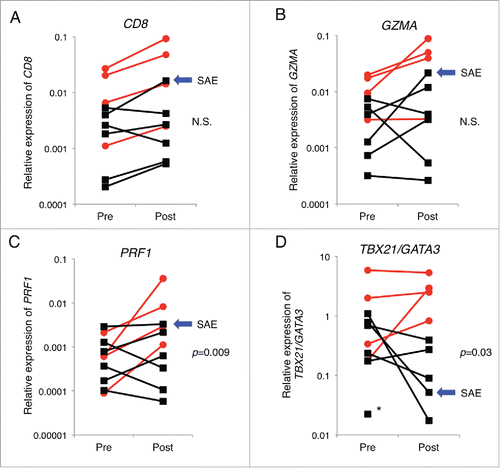
It is likely that blocking the PD-1 immune checkpoint can reinvigorate the pre-existing CTLs and enhance antitumor immune responses. We therefore examined immunological changes between pre- and post-treatment melanoma tissues and found that nivolumab treatment increased the number of CD8 T cells as indicated by the increase of CD8 mRNA in tumor tissues in most of the cases (). Particularly, in the cancer tissue of patient M11, who was a non-responder but manifested severe autoimmune adverse eventsCitation21 displayed the most significant degree (4.1-folds) of CD8 increase (, blue arrow). Furthermore, we examined mRNA levels of GZMA and perforin 1 (PRF1), cytolytic activity markers of T cells, in tumor tissues. Although changes of GZMA expression between the responder and non-responder groups were not statistically significant (), we observed significant changes of PRF1 expression levels between the two groups (p = 0.009) (). These data implying the enhancement of cytotoxic T cell activity might also be supported by the Th1-dominant cellular immunity in tumor tissues. We thereby investigated fold change of expression ratio of TBX21/GATA3 between before and after the nivolumab treatment, which was likely to represent intratumoral Th1/Th2 balance. The expression ratio of TBX21/GATA3 was significantly increased in responders than non-responders (p = 0.03) (), suggesting that nivolumab treatment predominantly facilitated Th1-mediated adaptive cellular immune response in melanoma tissues.
Figure 2. Nivolumab treatment-induced changes in the intratumoral expression of immune-related genes. Expression levels of CD8 (A), GZMA (B), PRF1 (C), and expression ratio of TBX21/GATA3 (D) in pre-treatment (pre) and post-treatment (post) tumors are presented. The y-axis indicates expression level of each gene relative to that of GAPDH. Red circles and black squares indicate tumor samples from responders and non-responders, respectively. Blue arrows indicate a patient who manifested severe adverse events (SAE). Asterisk indicates a patient whose expression of TBX21 in the tumor of post-treatment was undetectable. The Mann–Whitney U test was used to examine statistical significance.
Oligoclonal T cell expansion in tumor tissues after nivolumab treatment
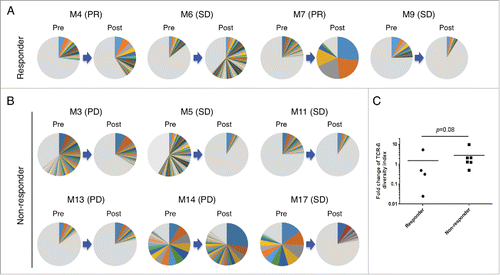
To further characterize the infiltrated T cells in tumors, we performed TCR repertoire analysis using our next-generation sequencing method.Citation14,18 Among 13 melanoma patients, 1 responder (M10) and 2 non-responders (M8 and M15) were omitted for this analysis because their post-treatment tumors were not available. After quantification of individual TCR-β clonotypes based on unique V-D-J combinations with complementarity-determining region 3 (CDR3) sequences, we compared TCR-β repertoire in paired tumors of pre- and post-nivolumab treatment (n = 10). Through cDNA sequencing of TCR-β, we obtained total sequence reads of 1,040,729 ± 708,603 (average ± standard deviation) and unique CDR3 clonotypes of 32,880 ± 30,631 in individual tissues (Table S1). Interestingly, we found that TCR-β clonotypes with the read frequency of >0.5% were drastically increased by nivolumab treatment (). For example, the proportion of TCR-β clonotypes with the read frequency of > 0.5% tend to be increased in the post-treatment tumor tissues of responders, compared with those of non-responders; the proportions in 3 responders (M4, M6 and M7) were 41.4%, 61.9% and 87.9%, respectively, while average and standard deviation of those in non-responders were 29.1% ± 28.1%. Although one remaining responder, M9 did not show oligoclonal expansion of TILs by the treatment, a tumor in this patient revealed the highest expression levels of CD8, GZMA, and PRF1 in both pre- and post-treatment tumors among all patients; this might imply a possibility that polyclonal T cells with high cytolytic activity were present in the tumors of this patient. To compare overall changes in TCR-β repertoire between responders and non-responders, we calculated the diversity index (DI) of TCR-β repertoire to represent clonality of TILs and found a tendency of the decrease of TCR-β DI in tumors of responders, compared with those of non-responders ().
Figure 3. Nivolumab treatment-induced changes of TCR-β repertoires in tumors. Pie charts illustrate distribution of unique CDR3 sequences of TCR-β, detected in paired tumor samples of pre- and post-treatment, from responders (n = 4) (A) or non-responders (n = 6) (B). As each pie chart was separately colored according to CDR3s frequency ranks, the same color between pie charts does not represent an identical CDR3 sequence. Gray color zone indicates combined portion of all clonotypes with the frequencies of less than 0.5%. (C) Fold changes in the diversity index (DI) of TCR-β in tumors are shown according to the patient's response to nivolumab treatment. Horizontal lines represent the means, and the Mann–Whitney U test was used to examine statistical significance.
Discussion
The PD-1/PD-L1 pathway functions as an immune evasion mechanism in tumor, and anti-PD-1 therapies have shown effective and durable antitumor responses in multiple types of solid tumor, including melanoma.Citation1-4 Overexpression of PD-L1 was suggested as a predictive biomarker for the response to the anti-PD-1 therapies, but this association was controversial probably due to the variability in the used antibodies and the definition of positivity among the studies. Hence, at this point, PD-L1 staining is considered to be insufficient to select appropriate patients for the therapies.Citation22 Considering application of this type of treatment to various cancer types as well as its very expensive cost, it is crucially important to develop a better predictive system to the anti-PD-1 therapies.
In this paper, we demonstrated significant positive correlations between expression levels of PD-L1 and PD-L2, and also between those of PD-1 ligands (PD-L1 and PD-L2) and GZMA in melanoma tissues. PD-L2 is one of the ligands for the PD-1 receptor and indicated to inhibit the effector function of T cells, but its expression in tumors was reported to be less frequent (in only 8 of 38 specimens examined),Citation8 although PD-L1 and PD-L2 expression levels were well correlated in our samples. Another immune escape mechanism of cancer cell is loss or decreased expression of HLA class I molecules, which was frequently observed in a range of human malignancies including melanoma.Citation23-25 The pre-treatment tumors from responders exhibited significantly higher expression levels of PD-L1, GZMA, and HLA-A than those from non-responders. When we made a scoring system using these three markers, we were able to separate two groups, responders and non-responders relatively clearly. These results indicated that pre-existence of cytolytic T cells and high expression of HLA class I molecules in tumors might be good predictive markers for better clinical response to nivolumab treatment. Since, this kind of approach often causes over-fitting, further investigation to examine the expression levels of PD-L1, GZMA and HLA-A in pre-treatment tumors of an independent cohort of melanoma patients treated with nivolumab will be definitely needed. In addition, pre-treatment tumors from responders showed a tendency of increased expression levels of CD8 and PD-1, which was consistent with previous results showing that the density of CD8 cells and PD-1+ cells in melanoma of pre-treatment biopsy samples was significantly higher in responders than non-responders.Citation9 Therefore, statistical significance may be attained by further analysis of more patients and consideration of additional immunologic factors.
Secondly, we examined nivolumab-induced changes in the immune microenvironment by assessing expression levels of immune-related molecules and clonality of tumor-infiltrated T cells. Our results demonstrated that all of four paired tumor tissues from responders showed substantial increase of cytotoxic T cells reflected by the increase of CD8, GZMA, and PRF1, compared with those from non-responders; particularly significant increase of PRF1 indicates activation of CD8 T cells. Although both granzyme and perforin are essential for cytolytic activity of CTLs, it was reported that expression of perforin was suppressed in the PD-1+ T cells.Citation26,27 Therefore, our results implied that the high level of perforin expression could be restored by blocking PD-1 pathway with nivolumab treatment, which subsequently conferred higher cytolytic activity of CTLs in the tumors of responders. Another important finding was significant elevation of Th1 cellular immunity, as indicated by increased TBX21/GATA3 expression ratio, in the nivolumab-treated tumors of responders. These results implied that nivolumab treatment could increase Th1 cell-mediated adaptive cellular immunity and cytolytic activity of CTLs, leading to effective generation/expansion of antitumor CTLs in tumors.
Since our results from gene expression analysis indicated that nivolumab treatment restored the CTL activity in tumor, we attempted to characterize T cell repertoire in tumors through TCR deep sequencing analysis. Consequently, we found that nivolumab treatment induced oligoclonal enrichment of certain TCR-β clonotypes in the tumor tissues of responders, which was consistent with a previous report of anti-PD-1 (pembrolizumab)-treated melanoma showing that tumors of better responders displayed significant clonal expansion of TCR-β by the treatment.Citation9
Immune checkpoint inhibitors have revolutionized the cancer immunotherapies, but the tumor–host immune interaction is not yet completely elucidated in the molecular level. Integration of whole exome/genome sequencing, characterization of the immune microenvironment through transcriptome analysis and TCR sequencing will confer better understating in the mechanisms of action for cancer immunotherapies.Citation28 Indeed, results from whole-exome sequencing of non-small cell lung cancers treated with pembrolizumab revealed that higher number of non-synonymous mutations and predicted neoantigens (generated from non-synonymous mutation) were associated with better clinical outcome.Citation29 This finding implicated that presence of abundant and immunogenic tumor antigens might be another indicator for effective anti-PD-1 therapy. Future studies aiming identification of such potent tumor antigens and corresponding TCR sequences will contribute to development of genetically engineered T cells for adoptive T cell therapy.
In summary, we have indicated that not only expression levels of PD-1 ligands but also those of GZMA and HLA-A in melanoma tissues might predict nivolumab treatment response and that nivolumab treatment enhanced Th1-skewed cellular immunity in tumor and induced oligoclonal expansion of TILs with cytolytic activity. The results presented here could provide new insights in the development of novel cancer immunotherapies.
Materials and methods
Patient samples
A total of 13 metastatic melanoma patients (M3, M4, M6, M7, M8, M9, M10, M11, M13, M14, M15, and M17), who had received dacarbazine-based chemotherapy and/or other immunotherapy (interferon-β), were enrolled in Kumamoto University Hospital, Shinshu University Hospital, Tsukuba University Hospital, Nagoya University Hospital, Nagoya City University Hospital, and Kyoto Prefectural University of Medicine Hospital. Detailed clinical information of melanoma patients is summarized in . Metastatic skin melanoma tissues of pre- and post-nivolumab treatment, in which pre- and post-treatment tumors were surgically resected from different skin lesions, were collected according to a study protocol approved by Institutional Review Board of each institution. Informed consents were collected from all the patients. All samples were transferred to the University of Chicago, and subsequently TCR-β repertoire was analyzed under the University of Chicago IRB protocol 13-0797. For TCR repertoire analysis using paired pre- and post-treatment tumors, 1 responder (M10) and 2 non-responders (M8 and M15) among all 13 patients were excluded for this analysis because their post-treatment tumors were not available.
Gene expression analysis
To examine expression levels of immune-related genes, total RNA was extracted using RNeasy mini kit (Qiagen) from frozen tissues of tumors. We synthesized cDNA with 5′ rapid amplification of cDNA end (5′-RACE) adapter using SMART cDNA library kit (Clontech). Gene expression analysis was performed on the tumor cDNA samples using TaqMan gene expression assays (Thermo Fisher Scientific) according to the manufacturer's instructions. A housekeeping gene, GAPDH (assay Hs02758991_g1) was used for data normalization. For the predictive score (PS), we used expression levels of three genes (PD-L1, GZMA, and HLA-A) that were significantly higher in the tumors of responders and calculated the PS in each tumor sample using the following formulation: PS = R (PD-L1) + R (GZMA) + R (HLA-A), where the R is defined as Log2 (expression level in each case/average of expression levels in all cases).
TCR-β sequencing
The libraries for TCR-β sequencing were prepared using previously described methods.Citation14,18 Briefly, cDNA with 5′-RACE adapter was synthesized as described above. The TCR-β chains were amplified using a reverse primer specific to the constant region and a forward primer for the SMART adapter. Illumina sequence adapters with barcode sequence were then added using the Nextera XT Index kit (Illumina). Sequencing was performed by 300-bp paired-end reads on the Illumina MiSeq platform, using MiSeq Reagent v3 600-cycle kit (Illumina).
TCR-β repertoire analysis
TCR-β repertoire analysis was performed using Tcrip software.Citation14 Briefly, sequencing reads were mapped to the human TCR-β reference sequences using Bowtie2 aligner, and then decomposed to the V-D-J components of the CDR3s.Citation14 The inverse Simpson's DI value was calculated to quantify the clonality of the TCR-β repertoires according to DI = ,Citation30 where N is the total number of sequences, ni is the number of sequences belonging to the ith clonotype, and K is the total number of clonotypes. To present TCR-β repertoire of each sample, we used the Excel program (Microsoft) to generate bar graphs and pie charts.
Statistical analysis
Gene expression level and TCR-β DI were compared between patient groups using the Mann–Whitney U test (two-tailed). Statistical analysis was carried out using Prism 6 software (GraphPad). p < 0.05 was considered statistically significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
Y.N. designed and supervised the project; H.In. designed the study, conducted experiments, analyzed data, and interpreted data; J.P. and K.K. analyzed and interpreted data; M.Z. conducted experiments; H.In. wrote the manuscript, and Y.N., J.P., and K.K. edited it; A.M., J.M., Y.K., R.O., R.T., Y.F., H.K., A.M., J.A., N.K., K.Y., M.A., H.Ih., and S.F. provided the samples and clinical information.
KONI_A_1204507_supplementary_data.docx
Download MS Word (382.9 KB)Acknowledgments
We thank Drs Rui Yamaguchi, Seiya Imoto, and Satoru Miyano in The University of Tokyo for developing the algorithm of TCR repertoire analysis and helpful support in data management. The super-computing resource (http://sc.hgc.jp/shirokane.html) was provided by Human Genome Center, the Institute of Medical Science, The University of Tokyo.
Funding
This work was also supported by JSPS KAKENHI grant (15K09772).
References
- Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. New Engl J Med 2012; 366:2443-54; PMID:22658127; http://dx.doi.org/10.1056/NEJMoa1200690
- Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. New Engl J Med 2012; 366:2455-65; PMID:22658128; http://dx.doi.org/10.1056/NEJMoa1200694
- Page DB, Postow MA, Callahan MK, Allison JP, Wolchok JD. Immune modulation in cancer with antibodies. Annu Rev Med 2014; 65:185-202; PMID:24188664; http://dx.doi.org/10.1146/annurev-med-092012-112807
- Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, Hassel JC, Rutkowski P, McNeil C, Kalinka-Warzocha E et al. Nivolumab in previously untreated melanoma without BRAF mutation. New Engl J Med 2015; 372:320-30; PMID:25399552; http://dx.doi.org/10.1056/NEJMoa1412082
- Luke JJ, Ott PA. PD-1 pathway inhibitors: the next generation of immunotherapy for advanced melanoma. Oncotarget 2015; 6:3479-92; PMID:25682878; http://dx.doi.org/10.18632/oncotarget.2980
- Gros A, Robbins PF, Yao X, Li YF, Turcotte S, Tran E, Wunderlich JR, Mixon A, Farid S, Dudley ME et al. PD-1 identifies the patient-specific CD8(+) tumor-reactive repertoire infiltrating human tumors. J Clin Invest 2014; 124:2246-59; PMID:24667641; http://dx.doi.org/10.1172/JCI73639
- Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol 2008; 26:677-704; PMID:18173375; http://dx.doi.org/10.1146/annurev.immunol.26.021607.090331
- Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, Chen L, Pardoll DM, Topalian SL, Anders RA. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res 2014; 20:5064-74; PMID:24714771; http://dx.doi.org/10.1158/1078-0432.CCR-13-3271
- Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014; 515:568-71; PMID:25428505; http://dx.doi.org/10.1038/nature13954
- Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014; 515:563-7; PMID:25428504; http://dx.doi.org/10.1038/nature14011
- Clemente CG, Mihm MC, Jr., Bufalino R, Zurrida S, Collini P, Cascinelli N. Prognostic value of tumor infiltrating lymphocytes in the vertical growth phase of primary cutaneous melanoma. Cancer 1996; 77:1303-10; PMID:8608507; http://dx.doi.org/10.1002/(SICI)1097-0142(19960401)77:7%3c1303::AID-CNCR12%3e3.0.CO;2-5
- Ferradini L, Mackensen A, Genevee C, Bosq J, Duvillard P, Avril MF, Hercend T. Analysis of T cell receptor variability in tumor-infiltrating lymphocytes from a human regressive melanoma. Evidence for in situ T cell clonal expansion. J Clin Invest 1993; 91:1183-90; PMID:8450047; http://dx.doi.org/10.1172/JCI116278
- Hinrichs CS, Rosenberg SA. Exploiting the curative potential of adoptive T-cell therapy for cancer. Immunol Rev 2014; 257:56-71; PMID:24329789; http://dx.doi.org/10.1111/imr.12132
- Fang H, Yamaguchi R, Liu X, Daigo Y, Yew PY, Tanikawa C, Matsuda K, Imoto S, Miyano S, Nakamura Y. Quantitative T cell repertoire analysis by deep cDNA sequencing of T cell receptor alpha and beta chains using next-generation sequencing (NGS). Oncoimmunology 2014; 3:e968467; PMID:25964866; http://dx.doi.org/10.4161/21624011.2014.968467
- Liu X, Venkataraman G, Lin J, Kiyotani K, Smith S, Montoya M, Nakamura Y, Kline J. Highly clonal regulatory T-cell population in follicular lymphoma - inverse correlation with the diversity of CD8 T cells. Oncoimmunology 2015; 4:e1002728; PMID:26155390; http://dx.doi.org/10.1080/2162402X.2014.1002728
- Jang M, Yew PY, Hasegawa K, Ikeda Y, Fujiwara K, Fleming GF, Nakamura Y, Park JH. Characterization of T cell repertoire of blood, tumor, and ascites in ovarian cancer patients using next generation sequencing. Oncoimmunology 2015; 4:e1030561; PMID:26451311; http://dx.doi.org/10.1080/2162402X.2015.1030561
- Yew PY, Alachkar H, Yamaguchi R, Kiyotani K, Fang H, Yap KL, Liu HT, Wickrema A, Artz A, van Besien K et al. Quantitative characterization of T-cell repertoire in allogeneic hematopoietic stem cell transplant recipients. Bone Marrow Transplant 2015; 50:1227-34; PMID:26052909; http://dx.doi.org/10.1038/bmt.2015.133
- Choudhury N, Kiyotani K, Yap KL, Campanile A, Antic T, Yew PY, Steinberg G, Park JH, Nakamura Y, O'Donnell P. Low T-cell receptor diversity, high somatic mutation burden, and high neoantigen load as predictors of clinical outcome in muscle-invasive bladder cancer. Eur Urol Focus 2015; in press; http://dx.doi.org/10.1016/j.euf.2015.09.007
- Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009; 45:228-47; PMID:19097774; http://dx.doi.org/10.1016/j.ejca.2008.10.026
- Sharpe AH, Wherry EJ, Ahmed R, Freeman GJ. The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nat Immunol 2007; 8:239-45; PMID:17304234; http://dx.doi.org/10.1038/ni1443
- Kimura T, Fukushima S, Miyashita A, Aoi J, Jinnin M, Kosaka T, Ando Y, Matsukawa M, Inoue H, Kiyotani K et al. Myasthenic crisis and polymyositis induced by one dose of nivolumab. Cancer Sci 2016; 107:1055-8; PMID:27420474; http://dx.doi.org/10.1111/cas.12961
- Meng X, Huang Z, Teng F, Xing L, Yu J. Predictive biomarkers in PD-1/PD-L1 checkpoint blockade immunotherapy. Cancer Treat Rev 2015; 41:868-76; PMID:26589760; http://dx.doi.org/10.1016/j.ctrv.2015.11.001
- Aptsiauri N, Cabrera T, Mendez R, Garcia-Lora A, Ruiz-Cabello F, Garrido F. Role of altered expression of HLA class I molecules in cancer progression. Adv Exp Med Biol 2007; 601:123-31; PMID:17712999; http://dx.doi.org/10.1007/978-0-387-72005-0_13
- Chang CC, Campoli M, Ferrone S. Classical and nonclassical HLA class I antigen and NK Cell-activating ligand changes in malignant cells: current challenges and future directions. Adv Cancer Res 2005; 93:189-234; PMID:15797448; http://dx.doi.org/10.1016/S0065-230X(05)93006-6
- Maeurer MJ, Gollin SM, Storkus WJ, Swaney W, Karbach J, Martin D, Castelli C, Salter R, Knuth A, Lotze MT. Tumor escape from immune recognition: loss of HLA-A2 melanoma cell surface expression is associated with a complex rearrangement of the short arm of chromosome 6. Clin Cancer Res 1996; 2:641-52; PMID:9816214
- Wu X, Zhang H, Xing Q, Cui J, Li J, Li Y, Tan Y, Wang S. PD-1(+) CD8(+) T cells are exhausted in tumours and functional in draining lymph nodes of colorectal cancer patients. Br J Cancer 2014; 111:1391-9; PMID:25093496; http://dx.doi.org/10.1038/bjc.2014.416
- Vigano S, Banga R, Bellanger F, Pellaton C, Farina A, Comte D, Harari A, Perreau M. CD160-associated CD8 T-cell functional impairment is independent of PD-1 expression. PLoS Pathog 2014; 10:e1004380; PMID:25255144; http://dx.doi.org/10.1371/journal.ppat.1004380
- Nakamura Y. Challenges and future directions of immunopharmacogenomics. In: Nakamura Y, ed. Immunopharmacogenomics. Tokyo: Springer, 2015:159-62.
- Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015; 348:124-8; PMID:25765070; http://dx.doi.org/10.1126/science.aaa1348
- Venturi V, Kedzierska K, Turner SJ, Doherty PC, Davenport MP. Methods for comparing the diversity of samples of the T cell receptor repertoire. J Immunol Methods 2007; 321:182-95; PMID:17337271; http://dx.doi.org/10.1016/j.jim.2007.01.019
