ABSTRACT
Obesity-related inflammation promotes cancer development. Tissue resident macrophages affect tumor progression and the tumor micro-environment favors polarization into alternatively activated macrophages (M2) that facilitate tumor invasiveness. Here, we dissected the role of western diet-induced NASH in inducing macrophage polarization in a carcinogen initiated model of hepatocellular carcinoma (HCC). Adult C57BL/6 male mice received diethyl nitrosamine (DEN) followed by 24 weeks of high fat–high cholesterol–high sugar diet (HF-HC-HSD). We assessed liver MRI and histology, serum ALT, AFP, liver triglycerides, and cytokines. Macrophage polarization was determined by IL-12/TNFα (M1) and CD163/CD206 (M2) expression using flow cytometry. Role of hif-1α-induced IL-10 was dissected in hepatocyte specific hif-1αKO and hif-1αdPA (over-expression) mice. The western diet-induced features of NASH and accelerated HCC development after carcinogen exposure. Liver fibrosis and serum AFP were significantly increased in DEN + HF–HC–HSD mice compared to controls. Western diet resulted in macrophage (F4/80+CD11b+) infiltration to liver and DEN + HF–HC–HSD mice showed preferential increase in M2 macrophages. Isolated hepatocytes from western diet fed mice showed significant upregulation of the hypoxia-inducible transcription factor, hif-1α, and livers from hif-1α over-expressing mice had increased proportion of M2 macrophages. Primary hepatocytes from wild-type mice treated with DEN and palmitic acid in vitro showed activation of hif-1α and induction of IL-10, a M2 polarizing cytokine. IL-10 neutralization in hepatocyte-derived culture supernatant prevented M2 macrophage polarization and silencing hif-1α in macrophages blocked their M2 polarization. Therefore, our data demonstrate that NASH accelerates HCC progression via upregulation of hif-1α mediated IL-10 polarizing M2 macrophages.
Abbreviations
| αSMA | = | alpha smooth muscle actin |
| AFP | = | alpha fetoprotein |
| ALT | = | alanine aminotransferase |
| CCND1 | = | cyclin D1 |
| DEN | = | n,n diethyl nitrosa mine |
| EMT | = | epithelial-mesenchymal transition |
| HCC | = | hepatocellular cancer |
| HIF-1α | = | hypoxia inducible factor-1α |
| IFNγ | = | interferon gamma |
| IL-10 | = | interleukin-10 |
| LMNCs | = | liver mononuclear cells |
| MCP-1 | = | monocyte chemoattractant protein-1 |
| MRI | = | magnetic resonance imaging |
| PA | = | palmitic acid |
| siRNA | = | small interference RNA |
| TNFα | = | tumor necrosis factor α |
| TGFβ | = | transforming growth factor β |
| VEGF | = | vascular endothelial growth factor |
Introduction
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related mortality globally.Citation1 In the United States, the incidence of HCC has tripled over the last two decades.Citation2 Recently large-scale epidemiological studies have pointed out that overweight and obese individuals possess substantial increase in cancer risk.Citation3,4 Although the obesity-associated increase in cancer risk is dependent on the type of cancer, the largest increase is seen for HCC.Citation5 Several mechanisms are proposed to explain how obesity increases HCC risk, including the prevalence of type II diabetes, Non Alcoholic Steato-Hepatitis (NASH), and cellular inflammation.Citation6
NASH is caused by excessive caloric intake and metabolic syndrome. The natural history of NASH indicates that while steatosis has an almost negligible effect on liver-related mortality, steatohepatitis is a risk factor for the development of cirrhosis and HCC.Citation7 Pro-inflammatory cytokines produced by immune cells that are implicated in steatohepatitis also play a critical role in promoting cancer invasion and metastasis.Citation8,9 Inflammation in the liver microenvironment also contributes to carcinogenesis.Citation10 Depending on the tissue environment, macrophages can become polarized into M1 (inflammatory) or M2 (alternatively activated) phenotype. We previously reported predominance of M1, inflammatory, macrophages in the liver in NASH.Citation11
Tumor-associated macrophages (TAMs) promote tumor angiogenesis and metastasis. Citation12 Alternatively activated M2 macrophages bear a functional similarity to TAMs and have been shown to play a role in promoting tumor angiogenesis and tissue remodeling.Citation13 M2 macrophages are induced in vitro by IL-4, IL-10, and IL-13, downregulate MHC class II and IL-12 expression and show increased expression of the anti-inflammatory cytokine IL-10, scavenger receptor A, and arginase.Citation12
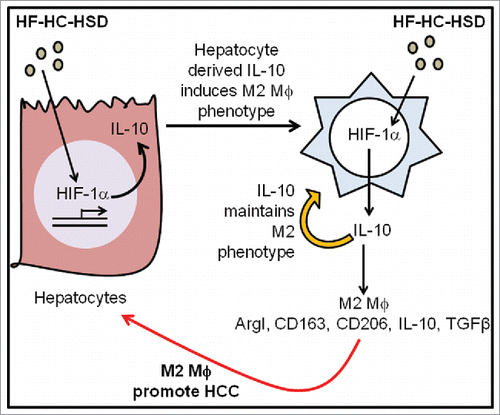
Considering the important role of M2 macrophages in tumor development, we hypothesized that selective induction of M2 polarization in liver macrophage can accelerate HCC progression in NASH livers in mice. In this study, we show that HF–HC–HSD induced steatohepatitis accelerated chemical carcinogen induced HCC. We found that mice injected with DEN and fed with HF–HC–HSD had a significantly higher percent of M2 macrophages in the liver. We discovered that NASH-related increase in hif-1α, a pivotal transcription factor in HCC, regulates IL-10, which is secreted by hepatocytes which aids in maintaining the M2 phenotype of macrophages in the HCC tumor microenvironment.
Results
Long-term western diet induces pathological features of NASH in mice
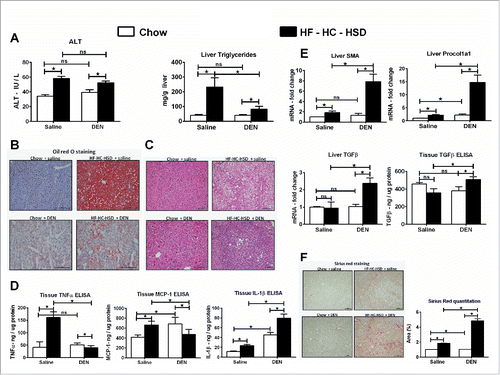
To induce HCC in mice, we administered six repeated injections of DEN, a well-characterized hepatocarcinogen. To evaluate the effect of western diet in accelerating the progression of HCC, we fed these mice with HF–HC–HSD, a diet composition similar to the western diet.Citation14 We found that 24 weeks of HF–HC–HSD feeding resulted in a significant increase in serum ALT as compared to chow fed mice (). Each DEN injection results in a transient spike in ALT levels but those levels returned to baseline in chow + DEN mice, whereas the DEN-induced increase in ALT levels were sustained in mice with HF–HC–HSD (Fig. S2A). Mice on HF–HC–HSD showed progressive weight gain compared to all other experimental groups (Fig. S2B). The HF–HC–HSD mice had significantly higher liver to body weight ratio compared to chow fed mice (Fig. S2C). Interestingly, the DEN + HF–HC–HSD mice never gained weight during the length of the feeding but still had significantly higher liver to body weight ratio at sacrifice (Figs. S2B and C). The mice on HF–HC–HSD alone showed increased accumulation of lipids on liver histology (), by Oil Red O staining (, Fig. S2D), and showed a significant elevation in liver triglyceride levels () consistent with features of NASH. Yet, the DEN + HF–HC–HSD mice had significantly attenuated lipid accumulation and triglyceride levels compared to HF–HC–HSD alone () pointing toward advanced liver disease. Next, we assessed the pro-inflammatory cytokines in liver. TNFα, MCP-1, and IL-1β showed significant induction at the protein levels () and mRNA (Fig. S3) in mice receiving HF–HC–HSD alone compared to mice on chow diet. In addition to inflammation and steatosis, we assessed the markers of fibrosis. Liver αSMA, procol1a1, and TGFβ were significantly induced at RNA level in DEN + HF–HC–HSD mice compared to all other groups (). Similarly, TGFβ tissue protein levels were significantly elevated in DEN + HF–HC–HSD mice compared to all other experimental groups () indicating fibrogenesis. The Sirius Red staining also showed higher positive staining in DEN + HF–HC–HSD mice suggesting advanced fibrosis (). Taken together the HF–HC–HSD induces all pathological features of NASH such as significant induction of ALT, increase in pro-inflammatory cytokines, elevated steatosis, and fibrosis. The same diet in DEN-injected mice resulted in less steatosis and inflammation but more prominent fibrosis indicating advanced liver disease.
Figure 1. HF–HC–HSD induces liver injury, steatosis, and fibrosis. (A) Serum ALT levels, liver triglycerides. (B) Representative Oil Red O staining of hepatic sections from all experimental groups. Bars inside images indicate 100 µm. (C) Representative H & E-stained liver sections from all treatment groups. Bars inside images indicate 100 µm. (D) Tissue protein levels of pro-inflammatory cytokines. (E) Fold changes in mRNA levels of fibrosis markers and tissue protein levels of TGFβ. (F) Representative Sirius Red staining images from all experimental groups. Bars inside images indicate 100 µm. For quantification, at least three different microscope fields were scored for each mouse. Bar graph shows percent Sirius Red positive area quantified using ImageJ. In graphs, values are given as mean ± SEM, Dunnett's multiple comparison were used to compare means of multiple groups; (n ≥ 6 mice per group, *p < 0.05, ns—not significant).
Western diet accelerates DEN-induced HCC in mice
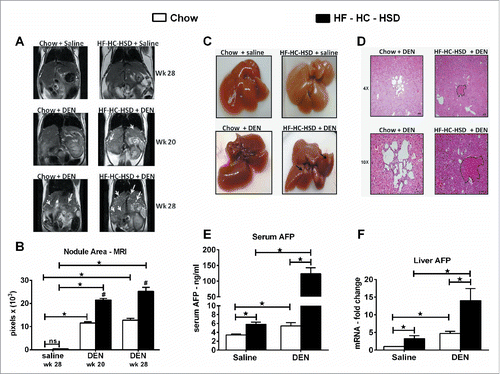
Next, we assessed the effect of western diet on DEN initiated HCC. MRI analysis performed at week 28 of age showed significantly higher number of tumor nodules in livers of DEN + HF–HC–HSD fed mice compared to chow + DEN mice (). Of note, the DEN + HF–HC–HSD showed tumor nodules as early as 20 weeks of age (). Gross macroscopic images taken at sacrifice showed liver tumors in 100 % mice in DEN + HF–HC–HSD group whereas none were found in chow + DEN mice (). Further, the hematoxylin and eosin (H & E) stained sections confirmed the HCC in the DEN + HF–HC–HSD mice (). The chow + DEN mice showed presence of biliary cysts, characteristic of DEN exposure by histopathology though none of these mice had any macroscopic tumor nodules at sacrifice (). Serum AFP levels () and liver AFP mRNA () were significantly elevated in DEN + HF–HC–HSD mice (∼25-fold higher than DEN alone group). This data suggests that the western diet significantly accelerates DEN-induced HCC development in mice.
Figure 2. HF–HC–HSD accelerates HCC development after DEN insult. (A) T2-weighted MRI of liver in coronal section at week 20 and 28. For the non-DEN groups only week 28 is shown. Arrows denote HCC nodules. (B) Quantification of nodule area using ImageJ. (C) Macroscopic images of liver tissue from all experimental groups. Arrows denote macroscopic HCC nodules. (D) Representative H & E-stained liver sections from DEN groups. Bars inside the images indicate 100 µm. (E) Serum AFP levels. (F) Fold changes in liver AFP mRNA levels. In graphs, values are given as mean ± SEM, Dunnett's multiple comparison were used to compare means of multiple groups; (*p < 0.05, n ≥ six mice per group, ns—not significant).
The combination of DEN and western diet causes EMT, induces cell cycle proteins and stemness markers
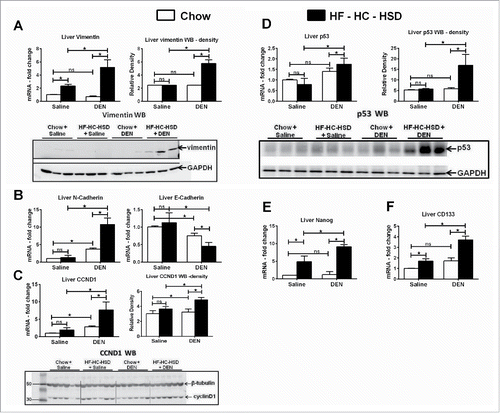
During epithelial-mesenchymal transition (EMT), epithelial cells lose their cell polarity and cell–cell contact, and gain migratory and invasive properties to become mesenchymal stem cells.Citation15 EMT has also been shown to occur in fibrosis and in the initiation of metastasis for cancer progression.Citation16 We recently showed vimentin, that plays an important role in EMT,Citation16 was upregulated in an MCD model of NASH.Citation17 In the present study, DEN + HF–HC–HSD mice showed significantly higher levels of vimentin RNA and protein compared to other groups (). Further, N-cadherin was significantly upregulated in DEN + HF–HC–HSD mice compared to all other groups (). Loss of E-cadherin has been associated with induction of sclerosing cholangitis and promoting HCC.Citation18 E-cadherin was significantly reduced in DEN + HF–HC–HSD mice compared all other groups (). Proliferation- and cell cycle-associated proteins, CyclinD1 () and p53 (), were significantly upregulated at both the RNA and protein levels. Finally, nanog and CD133, stem cell markers were highly induced in DEN + HF–HC–HSD fed mice (). These data suggested that the combination of DEN and western diet causes EMT, induces cell cycle proteins and stemness markers in mouse livers.
Figure 3. HF–HC–HSD upregulates EMT, cell cycle, and stemness markers. (A) Liver mRNA and protein levels of vimentin with relative density of vimentin protein. GAPDH used as loading control. (B) Liver mRNA levels of N-cadherin and E-cadherin. (C) Liver mRNA and protein levels of cyclin D1. (D) Liver mRNA and protein levels of p53. β-tubulin and GAPDH used as loading control. Relative density of signal is shown for each protein blot. (E) Fold changes in mRNA levels of stem cell marker Nanog and (F) CD133 in liver tissue. In graphs, values are given as mean ± SEM, Dunnett's multiple comparison were used to compare means of multiple groups; (n ≥ 6 mice per group, *p < 0.05, ns—not significant).
Western diet induces hypoxia in hepatocytes and promotes HCC
Long-term western diet-induced NASH has been associated with hypoxia.Citation19 Hypoxia is also the driver of several molecular processes in HCC progression and hif-1α, the hypoxia-induced transcription factor has been recognized as a pharmacological target in HCC.Citation20 We found a significant increase in liver hif-1α and VEGF expression after HF–HC–HSD compared to chow-fed mice (). Further, the hif-1α and VEGF mRNA and hif-1α DNA-binding activity were significantly higher in DEN + HF–HC–HSD mice compared to HF–HC–HSD or DEN alone ().
Figure 4. HF–HC–HSD induces hif-1α in hepatocytes and LMNCs from hep-hif-1αdPA mice show increased M2 phenotype. (A) mRNA levels of hif-1α and its target VEGF in liver. (B)Representative EMSA gel picture showing the upregulation in DNA-binding activity of hif-1α in liver. For the cold competition reaction, a 20-fold excess of same, unlabeled, double-stranded oligonucleotide was added to the reaction mixture before adding the labeled oligonucleotide probe. (C) mRNA levels of hif-1α and its target VEGF in isolated hepatocytes. (D) mRNA and tissue protein levels of IL-10 in liver. (E) Isolated hepatocytes from WT, HepHIF1KO, and HepHIF1dPA mice were stimulated with DEN or PA or DEN + PA and IL-10 secreted in culture supernatant was estimated by ELISA. (F) Baseline mRNA levels of M1/M2 marker genes in LMNCs isolated from 6 mo old WT, HepHIF1KO, and HepHIF1dPA mice. In graphs, values are given as mean ± SEM, Dunnett's multiple comparison were used to compare means of multiple groups; (n ≥ 6 mice per group, *p < 0.05 vs. different genotype, $p < 0.05 vs. different stimulations within same group, ns—not significant).
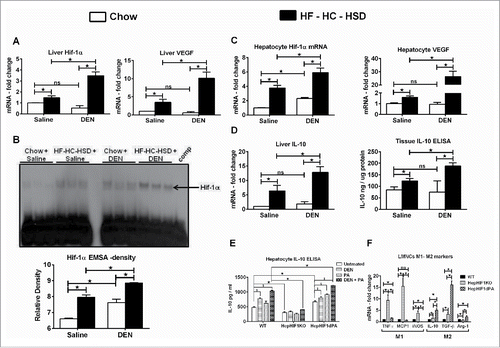
Next, we assessed hif-1α expression in isolated primary hepatocytes. Hif-1α activation was shown to promote fat accumulation in the liver, specifically in hepatocytes.Citation21 Here, we found that hif-1α levels were significantly higher in isolated hepatocytes from HF–HC–HSD alone mice compared to chow fed mice. In addition, hepatocyte hif-1α levels were even higher in DEN + HF–HC–HSD fed mice compared all other groups (). A similar trend was observed in VEGF, the hif-1α target, in isolated hepatocytes with the highest VEGF expression in DEN + HF–HC–HSD mice (). Recently, hif-1α has been shown to bind to the promoter of IL-10 in cardiac myocytes.Citation22 Thus, we measured the expression of IL-10 in the liver and found a robust upregulation of IL-10 both at mRNA and tissue protein levels in DEN + HF–HC–HSD mice () compared to any other groups.
To unravel the potential role of hif-1α in IL-10 induction in hepatocytes, we isolated primary hepatocytes from wild-type, alb-cre hif-1α KO and alb-cre hif-1αdPA (constitutively activated hif-1α in hepatocytes) mice and treated them with DEN or palmitic acid (PA) or DEN + PA in vitro. The hepatocytes isolated from wild-type mice secreted significantly higher amounts of IL-10 after DEN + PA treatment compared to untreated cells (). In contrast, hepatocytes from alb-cre hif-1α KO mice secreted significantly lower amounts of IL-10 as compared to wild-type hepatocytes at baseline (untreated) and failed to induce IL-10 production even after DEN + PA treatment (). Interestingly, hepatocytes from alb-cre hif-1α dPA mice had significantly higher expression of IL-10 as compared to wild-type mice at baseline (untreated), that was further increased upon DEN + PA treatment (). This is the first report of IL-10 production by mouse primary hepatocytes in vitro.
Finally, we also isolated LMNCs from these three different mouse lines and assessed macrophage polarization markers. Interestingly, LMNCs isolated from the alb-cre hif-1αdPA mice that have constitutive HIF-1α activation, showed increased expression of M2 Mφ markers suggesting that increased IL-10 production due to hif-1α over-expression in hepatocytes may induce M2 polarization in liver macrophages ().
Combination of DEN and western diet promotes M2 polarization
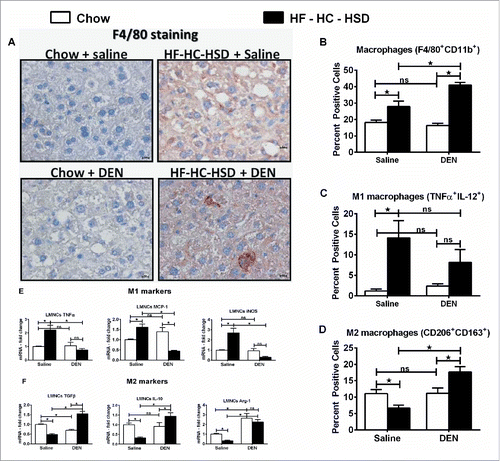
NASH is characterized by macrophage infiltration and activation in the liver.Citation23 Consistent with this, we found increased pro-inflammatory cytokine induction in HF–HC–HSD fed mice while it was intriguing that the levels of TNFα and MCP-1 were lower in DEN + HF–HC–HSD mice (). Hence we analyzed macrophages, the most prominent innate immune cell population in the liver. The DEN + HF–HC–HSD mice showed the highest frequency of F4/80 positive cells in the liver (). Next, we isolated the LMNCs and analyzed them by flow cytometry. The total number of F4/80 CD11b double positive cells was increased in HF–HC–HSD mice compared to chow-fed controls; however, it was the highest in DEN + HF–HC–HSD mouse livers (). Further, there was a significant increase in M1 MØ in HF–HC–HSD mice with a drastic reduction in number of M1 macrophages () in DEN + HF–HC–HSD and a concurrent significant increase in M2 macrophages (). M1 markers such as TNFα, MCP-1, and iNOS were significantly lower in LMNCs isolated from DEN + HF–HC–HSD fed mice () while M2 markers including Arg-1, IL-10, and TGFβ were significantly increased in LMNCs from DEN + HF–HC–HSD fed mice compared to chow + DEN and HF–HC–HSD alone mice (). This data was consistent with the increased expression of TGFß () and the significantly increased liver IL-10 mRNA and protein expression in the liver of DEN + HF–HC–HSD mice compared to HF–HC–HSD alone (), suggesting the presence of a higher number of M2 macrophages and M2-polarizing cytokines in livers of DEN + HF–HC–HSD fed mice.
Figure 5. HF–HC–HSD induces inflammation, macrophage infiltration while the DEN + HF–HC–HSD mice show increased number of M2 macrophages. (A) Representative immunostaining images for F4/80 showing more positive cells in DEN + HF–HC–HSD mice. Bars inside the images indicate 10 µm. (B) Percent F4/80 CD11b double positive LMNCs assessed by flow cytometry. (C) Percent positive M1 macrophages. F4/80/CD11b dual positive cells were further gated on IL-12/TNFα double positive cells, defined as M1 macrophages. (D) Percent positive M2 macrophages. F4/80/CD11b dual positive cells were further gated on CD206/CD163 double positive cells, defined as M2 macrophages. (E) mRNA levels of M1 markers in isolated LMNCs. (F) mRNA levels of M2 markers in isolated LMNCs. In graphs, values are given as mean ± SEM, Dunnett's multiple comparison were used to compare means of multiple groups; (n ≥ 3 mice per group, *p < 0.05, ns—not significant).
Hif-1α activation in hepatocytes promotes M2 polarization in liver macrophages
Macrophage polarization is regulated by the tissue environment and cytokines.Citation24 In the tumor microenvironment, IL-10 has a dual pro-tumorigenic role and can also polarize macrophages toward an M2 phenotype.Citation24 Our data in showed significantly elevated levels of IL-10 in HF–HC–HSD mice. In addition, the LMNCs isolated from alb-cre hif-1αdPA mice showed increased expression of M2 markers at baseline (), raising the possibility that hepatocyte derived, hif-1α dependent factors may be responsible for macrophage polarization in vivo.
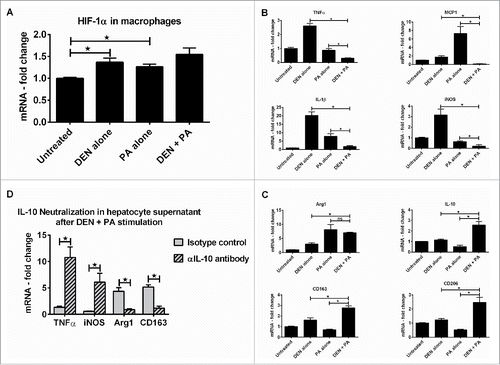
To better understand the role of hepatocyte-derived signals in macrophage polarization in vivo, we isolated primary hepatocytes from wild-type mice and treated them with DEN or PA or DEN + PA in vitro and transferred their culture supernatants onto RAW macrophages. The RAW macrophages cultured in hepatocyte supernatant from DEN or PA or DEN + PA showed induction in hif-1α expression (). Further, these macrophages showed decreased expression of M1 markers () and upregulation of M2 markers (). Our novel data in showed that wild-type hepatocytes secreted higher amounts of IL-10 after DEN + PA treatment and the untreated hepatocytes from alb-cre hif-1αdPA mice produced higher amounts of IL-10 compared to wild-type hepatocytes. Further, the LMNCs isolated from alb-cre hif-1αdPA mice showed increased expression of M2 markers at baseline (). Based on this data, we hypothesized that hif-1α-induced IL-10 secreted by hepatocytes may contribute to this M2 macrophage polarization. Thus, we neutralized the IL-10 secreted in the hepatocyte culture supernatant before transferring it onto RAW macrophages. The RAW macrophages exposed to IL-10 neutralized hepatocyte culture supernatant showed a loss of M2 markers (), suggesting a direct role for hif-1α-induced IL-10 from hepatocytes in macrophage polarization.
Figure 6. Hif-1α in hepatocytes induces M2 polarization in liver macrophages via upregulation of IL-10 in hepatocytes. Primary hepatocytes from WT mice were stimulated with DEN or PA or DEN + PA and culture supernatant was transferred onto RAW macrophages. (A) hif-1α mRNA levels in RAW macrophages. (B) mRNA levels of M1 marker genes in RAW macrophages at the end of incubation. (C) mRNA levels of M2 marker genes in RAW macrophages at the end of incubation. (D) Primary hepatocytes from WT mice were stimulated with DEN or PA or DEN + PA and the IL-10 in culture supernatant was neutralized using an αIL-10 antibody. This culture supernatant was transferred onto RAW macrophages and mRNA levels of M1/M2 marker genes are shown. In graphs, values are given as mean ± SEM, Dunnett's multiple comparison were used to compare means of multiple groups; (*p < 0.05, ns—not significant).
DEN + HF–HC–HSD also induces hif-1α and IL-10 upregulation in macrophages that aids maintenance of the pro-tumorigenic M2 phenotype
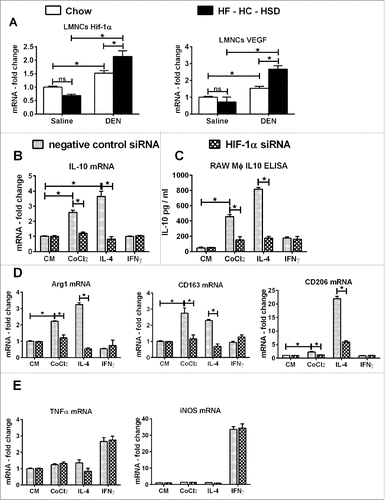
Finally, we dissected the direct effect of HF–HC–HSD on hif-1α and IL-10 in macrophage polarization. While there was no significant difference in hif-1α and VEGF levels between LMNCs isolated from chow and HF–HC–HSD alone mice, LMNCs from DEN + HF–HC–HSD fed mice had significantly higher levels of both hif-1α and VEGF (). Further, in vitro knock-down of hif-1α in macrophages resulted in significantly lower IL-10 expression () and IL-10 protein secretion (). Hif-1α knock-down in RAW macrophages in vitro showed a significant decrease in M2 marker expression (), while the M1 markers were unchanged (). This data suggested a direct role for hif-1α regulated IL-10 in maintaining an M2 phenotype of macrophages.
Figure 7. Hif-1α aids in maintaining M2 phenotype of macrophages. (A) mRNA levels of hif-1α and its target VEGF in primary LMNCs. Hif-1α siRNA or negative control siRNA was transfected in RAW macrophages and were stimulated with CoCl2 (hif-1α inducer) or IL-4 (M2 inducer) or IFNγ (M1 inducer). (B) mRNA levels of IL-10 in RAW macrophages after hif-1α knock-down. (C) IL-10 in culture supernatant was estimated by ELISA after hif-1α knock-down. (D) mRNA levels of M2 marker genes in RAW macrophages after hif-1α knock-down. (E) mRNA levels of M1 marker genes in RAW macrophages after hif-1α knock-down. In graphs, values are given as mean ± SEM, Dunnett's multiple comparison were used to compare means of multiple groups; (n ≥ 3 mice per group, *p < 0.05, ns—not significant).
Taken together, our results suggest that in the DEN + HF–HC–HSD induced HCC microenvironment there is a dual signal for macrophage polarization to M2 phenotype first, via hepatocyte-derived IL-10 and second, due to the autocrine IL-10 induction via HIF-1α activation in MØ ().
Discussion
Overweight and obese individuals possess a substantial increase in HCC risk.Citation4,25 Several mechanisms have been proposed to explain how obesity increases HCC risk, with the primary underlying condition being NASH.Citation6 We have previously shown that extended feeding of the HF–HC–HSD alone results in NASH.Citation11 In this report, we show that HF–HC–HSD feeding for 24 weeks leads to NASH, which accelerates HCC development in DEN-injected mice. The HF–HC–HSD by itself induced steatosis and cellular inflammation, the typical pathological attributes of NASH. We found that features of advanced liver disease indicated by lowered steatosis and increased fibrosis were present in DEN + HF–HC–HSD mice along with early HCC. Our experiments also showed a significant increase in early diagnostic indicators such as serum AFP, factors involved in epithelial-mesenchymal transition (vimentin and cadherins) and markers of stemness (CD133 and nanog). The lower levels of pro-inflammatory mediators in DEN + HF–HC–HSD mice led us to investigate the role of various cell types in our experimental model. We discovered that IL-10 secreted by transformed hepatocytes is responsible for inducing M2 polarization in liver resident macrophages which are similar to TAMs that aid HCC progression (). In addition, we dissected the molecular signaling induced by HF–HC–HSD which is responsible for this polarization and found that hif-1α / IL-10 induction in macrophages promotes the M2 phenotype.
Tissue resident macrophages are the most important drivers of cellular inflammation in NASH.Citation26 Recent literature suggests that macrophages can be further classified into the spectrum of pro-inflammatory M1 or anti-inflammatory M2 macrophages.Citation27 The M1 phenotype is characterized by the expression of high levels of pro-inflammatory cytokines, high production of reactive nitrogen and oxygen intermediates, promotion of Th1 response, and strong microbicidal and tumoricidal activity.Citation24 The M2 macrophages, also known as anti-inflammatory or alternatively activated macrophages are induced by IL-4, IL-13, IL-10, TGF-β, and a wide range of fungal pathogens.Citation28 Our data in showed a significant increase in total macrophage population after HF–HC–HSD feeding, mostly represented by M1 macrophages. However, the DEN + HF–HC–HSD mice showed lowered inflammation () and a significant increase in M2 macrophages. Specifically, we observed a significant induction of M2 markers (CD163 and CD206) by flow cytometry. In addition, the same LMNCs isolated from the DEN + HF–HC–HSD mice showed significantly higher expression of M2 markers such as TGF-β, IL-10, and Arg-1. Based on stimuli and the observed transcriptional changes, the M2 macrophages can be further classified into M2a, M2b, and M2c subtypes.Citation27,29 The M2a activation is a response to IL-4 and IL-13, the M2b to immune complexes and bacterial lipopolysaccharide (LPS), and the M2c to IL-10, glucocorticoids, and TGF-β.Citation30 The M2c macrophage phenotype is characterized by high expression of IL-10 and TGF-β,Citation31 which is similar to the phenotype we found in mice with DEN + HF–HC–HSD. Interestingly, M2c macrophages play a prominent role in suppressing immune responses and promoting tissue remodeling.Citation32 Recently these M2c macrophages were shown to promote tumor invasion and growth in a lung carcinoma model.Citation33 Importantly, these M2c macrophages have been shown to promote epithelial–mesenchymal transition in pancreatic cancer cells, partially through TLR4/IL-10 signaling pathway.Citation34 Consistent with this, we found a profound upregulation in EMT markers (vimentin, N-cadherin) in DEN + HF–HC–HSD mouse livers that showed the M2 macrophage phenotype. Based on their IL-10high IL-12low profile, the M2 macrophages in our experimental model resemble the M2c subtype. Here, for the first time we show the role of M2c macrophages in promoting HCC progression in an animal model of NASH-associated HCC.
Because of their immune-suppressive functions, the M2 macrophages are often referred to as TAMs.Citation35 TAMs represent the major inflammatory component of the stroma of liver cancer and can affect different aspects of the neoplastic transformation of the tissue. CD163 is routinely used a marker of TAMs as it is abundant, sensitive, and accurate.Citation36,37 Our data in showed a significantly higher percentage of CD163/CD206 positive cells in DEN + HF–HC–HSD mice suggesting that these M2 macrophages indeed represent the TAMs in our experimental model. Apart from CD163, several other TAM markers such as IL-10, iNOS, and VEGF have been used for diagnosis of tumor staging in lung,Citation38 breast,Citation39 and colon cancers.Citation40 The upregulation of CD163, VEGF, and IL-10 and the downregulation of iNOS in LMNCs indicates the presence of M2-like TAMs in our experimental model. Several reports indicate that TAMs perform multiple M2-associated functions including promotion of angiogenesis, matrix remodeling, and suppression of adaptive immunity.Citation13,41 These M2 macrophages have been already shown to promote angiogenesis in vivo,Citation42 which may be accelerated by the high VEGF seen in the livers of DEN + HF–HC–HSD mice.
The importance of angiogenesis in cancer progression is well established and the molecules in angiogenesis pathways are being targeted as novel cancer therapy.Citation43,44 Hypoxia-inducible factor 1 (HIF-1) controls oxygen delivery (via angiogenesis) to cancer tissues.Citation45,46 Our previous data using hif-1αdPA mice showed that hif-1α over-expression in hepatocytes led to increased steatosis, while hif-1α deletion in hepatocytes protected mice from alcohol-induced liver injury.Citation21 More importantly, hif-1α has been shown to suppress apoptosis in HCC.Citation47
Apart from angiogenesis, hif-1α has been shown to regulate EMT, cell cycle,Citation48 and macrophage polarization.Citation24 Recent studies have shown that hif-1α can mediate the effects of tumor-derived lactic acidCitation49 and cytokines (Oncostatin M and Eotaxin)Citation50 to promote M2 phenotype in tumor microenvironment. Given this recently reported role of hif-1α in macrophage polarization and our findings in hepatocytes here, we speculated that hif-1α in hepatocytes may be crucial in polarizing macrophages in HCC microenvironment. The LMNCs isolated from 6 mo old HepHIF-1dPA mice showed significant upregulation of M2 markers while the LMNCs from HepHIF1KO mice were predominantly M1. These effects of hif-1α on liver macrophage polarization after deletion and over-expression in hepatocytes are novel. The isolated hepatocytes in our experimental model showed significant induction of hif-1α and its target VEGF after HF-HC-HSD feeding. Further, the culture supernatant collected from DEN + PA treated wild-type primary hepatocytes secreted significantly higher amounts of IL-10 compared to control, which may be responsible for polarization of liver resident macrophages to M2c subtype. We also proved a pivotal role of hif-1α in macrophage polarization in our model by in vitro knock-down of hif-1α in raw macrophages. The hif-1α KD macrophages were unable to polarize to M2 phenotype in response to IL-4 indicating that hif-1α-induced IL-10 was the driving factor in macrophage polarization.
In conclusion, we have shown that in a NASH-associated model of HCC, the HF–HC–HSD induces hif-1α which in turn drives the expression of IL-10 from hepatocytes that triggers M2 polarization of the liver macrophages. The HF–HC–HSD diet by itself may directly induce hif-1α in macrophages which aids in maintaining their M2 phenotype. These observations reveal new insights into the immune-pathomechanisms of the clinically increasingly prevalent NASH-related HCC.
Materials and methods
Tumor induction and animal feeding
To assess the role of HF–HC–HSD in macrophage polarization in HCC, 4 week old C57bl/6 male mice were injected with total six doses of DEN (Sigma, MO, USA). As shown in Fig. S1, a dose of 75 mg/kg DEN was administered weekly for the first 3 weeks followed by a dose of 100 mg/kg DEN for next 3 weeks. At week 6, the mice were randomly divided into chow and HF–HC–HSD groups. Age and gender matched group of mice without DEN were included in the study to understand the role of diet alone. At week 30 all the mice were sacrificed, blood and liver tissues were collected for further assays (Fig. S1). At least three mice from each group were perfused for isolation of hepatocytes and liver mononuclear cells (LMNCs). The study protocol was approved by the Institutional Animal Use and Care Committee of the University of Massachusetts Medical School.
Isolation of hepatocytes and LMNCs
Liver cell isolations were performed as described previously.Citation51 Mice received anesthesia with ketamine and xylazine; the livers were perfused with HBSS for 5 min followed by in vivo digestion with collagenase enzyme (900 mg/L) for 5 min. The hepatocytes were separated by centrifugation for 5 min at 300 rpm, while the non-hepatocyte content was loaded on the top of the 70%–40% Percoll gradient and centrifuged for 60 min at 1800 rpm. Hepatocytes were washed in HBSS + 2% FBS and were plated on collagen coated plates. The inter-cushion fraction from the percoll gradient was washed and immediately stained for flow cytometry or lyzed in Qiazol.
Flow cytometry
Isolated LMNCs were resuspended at a concentration of 106 cells/mL in FACS staining buffer-containing anti-mouse CD16/CD32 mAb (2.4G2) from BD PharMingen (San Jose, CA, USA) to block nonspecific binding to FcgRs and incubated for 10 min at 4°C. Cells were stained with Live/Dead Fixable Blue Dead Cell Stain Kit from Life Technologies (Grand Island, NY, USA) to gate out dead cells. LMNCs were immunostained with CD45-PE-Cy7, CD11b-FITC, and F4/80-PerCP-Cy5.5. CD45+ cells were gated, and the percentage of CD11b+ and F4/80+ dual positive cells was determined from the CD45+ population. To determine the frequency of the M1 macrophage population, the LMNCs were stained with TNFα-PE-Cy7, IL-12-eFluor450 (BD Biosciences, San Jose, CA, USA). To determine the frequency of the M2 macrophage population, the LMNCs were stained with unconjugated CD163 antibody from Santa Cruz Biotechnology (Dallas, TX, USA). We stained the cells with CD11b-FITC, F4/80-PerCP-Cy5.5, and CD206-Alexa 647 and incubated for 30 min in the dark at 4°C (BioLegend, San Diego, CA, USA). For the negative control, the cells were stained with isotype-matched control antibodies (BioLegend). The cells were washed and incubated with PE-conjugated secondary antibody, specific for CD163, for another 25 min in the dark at 4°C. The cells were washed with FACS-staining buffer and fixed with 1% paraformaldehyde. The cells were acquired on a BD LSR II instrument (BD Biosciences, San Jose, CA, USA), and the data were analyzed by FlowJo software (Ashland, OR, USA). The data analysis strategy has been outlined in Fig. S4.
Magnetic resonance imaging
Magnetic resonance imaging (MRI) of liver was performed at week 20 and week 28 to monitor tumor growth in liver. Images were obtained using 3T Philips Achieva whole-body MR scanner (Philips Medical Systems, Best, Netherlands) with a custom-made solenoid T/R coil with a diameter of 30 mm. The animals were anesthetized with 5% isoflurane mixed with carbogen (95% O2/5% CO2) and were maintained with 1% to 2% isofluorane. Coronal T2—weighted spin echo images were acquired with respiratory triggering to reduce the motion artifacts. The respiration rate was monitored with an optical probe (Model 1025T Monitoring and Gating System, SA Instruments Inc., Stony Brook, NY). The output signal from the respiration monitor was used to trigger, in real time, the MR acquisition. As a consequence of the triggered acquisition, the TR value of around 2,000 ms, corresponding to the respiration rate of around 30 bpm, was determined. Other imaging parameters were: echo time (TE) of 70 ms, flip angel of 90 degrees, TSE-factor of 8, number of average = 4, matrix size of 148 × 120, field of view of 30 × 25 mm2, slice thickness of 1 mm with no gap, acquisition time around 4 min for 22 slices. Hyperplastic nodules were distinguished from normal liver tissues on basis of differences in homogeneity and signal intensity. Pixel-based nodule area quantitation was performed using ImageJ software.53
Biochemical assays
Serum alanine aminotransferase (ALT) activity was determined using a kinetic method (D-TEK LLC, PA). Serum AFP was assayed by ELISA (R&D systems, Minneapolis, MN). Intracellular cytokine levels were monitored in liver whole-cell lystate using TNFα, TGFβ, MCP-1, IL-1β ELISA kits (Biolegend, San Diego, CA). Liver triglycerides were assayed in liver whole-cell lysates using L-Type TriGlyceride M kit (Wako Diagnostics, Richmond, VA).
RNA extraction and real-time PCR
Total RNA was extracted using the Direct-zol RNA MiniPrep according to the manufacturer's instructions (Zymo Research, Irvin, CA). RNA was quantified using Nanodrop 2000 (Thermo Scientific, Wilmington, DE). cDNA synthesis was performed by reverse transcription of total RNA using the iScript Reverse Transcription Supermix (BIO-RAD, Hercules, CA). Real-time quantitative PCR was performed using the CFX96 real-time detection system (Bio-Rad Laboratories, Hercules, CA). Primers were synthesized by IDT, Inc. (Coralville, IA). Accumulation of PCR products was detected by monitoring the increase in fluorescence of double-stranded DNA-binding dye SYBR Green during amplification. Relative gene expression was calculated by the comparative cycle threshold (Ct) method. The expression of target genes was normalized to the house-keeping gene, 18S rRNA, in each sample and the fold-change in the target gene expression between experimental groups was expressed as a ratio. Melt-curve analysis was used to confirm the authenticity of the PCR products.
Western blotting
Whole-cell lysates were prepared from mouse livers as described previously.Citation51 Proteins of interest were detected by immunoblotting with specific primary antibodies against: cyclin D1 (SC-753; Santacurz), p53 (ab28; Abcam), vimentin (ab92547; Abcam), β-tubulin-HRP (ab185057; Abcam), GAPDH-HRP (ab9482; Abcam). Respective horseradish peroxidase-labeled secondary antibodies were from Santacruz Biotechnology (Dallas, TX). The specific immunoreactive bands of interest were detected by chemiluminescence (Bio-Rad, Hercules, CA). The immunoreactive bands were quantified by densitometric analysis using the UVP System (Bio-Rad Laboratories, Hercules, CA).
EMSA
The DNA-binding activity of hif-1α was assessed by electrophoretic mobility shift assay as described previously.Citation52 Briefly, liver nuclear protein extract (5–10 μg) was incubated with 50,000 cpm γ32P-labeled hif-1α consensus oligonucleotide (sc-2625, Santacruz Biotechnology) at room temperature for 30 min. All reactions were run on a 6% polyacrylamide gel, and the dried gel was exposed to an X-ray film at −80°C for different times. For the cold competition reaction, a 20-fold excess of same, unlabeled, double-stranded oligonucleotide was added to the reaction mixture before adding the labeled oligonucleotide probe.
Histopathological analysis
Sections of formalin-fixed, paraffin-embedded livers were stained with H & E, or Sirius Red and assessed for histological features of carcinoma and fibrosis. To assess steatosis, Oil Red O staining was performed on cryofrozen liver tissue sections (OCT). Immunohistochemistry staining for F4/80 was performed on formalin-fixed, paraffin-embedded livers using anti-F4/80 antibody (AbD Serotec, Raleigh, NC). The H & E stained sections were independently examined by a veterinary pathologist, Dr Garlick in a blinded manner (see Acknowledgments). Images were acquired on a Nikon microscope and the length of the scale bar is mentioned in figure legends. The quantitation of the Oil Red O, Sirius Red staining was performed using ImageJ software.
In vitro experiments
Mouse macrophage RAW 264.7 cell line was maintained in DMEM + 10% FBS. For in vitro experiments, mouse primary hepatocytes were isolated, plated on collagen-coated plates and were rested for at least 6 h before stimulations. Primary hepatocytes were treated with 10 µg/mL DEN for 3 h, the media was changed and cells were treated with 0.33 mM of palmitic acid (PA) for further 18 h. PA was washed; fresh media was added and was collected after 48 h. This media was used for estimation of IL-10 and was transferred onto RAW macrophages for 48 h. For IL-10 neutralization, the culture supernatant from DEN–PA treated hepatocytes was incubated with 1 µg/mL anti-mouse IL-10 neutralizing antibody or isotype control antibody (R&D systems, Minneapolis, MN) for 30 min and was transferred onto RAW macrophages. To dissect the role of hif-1α in macrophage polarization, hif-1α siRNA (SC44308 Santa Cruz Biotechnology, Dallas, TX) was transiently transfected in RAW macrophages. The cells were stimulated with 100 µM CoCl2 or 20 ng/mL IL-4 or IFNγ 24 h prior to hif-1α siRNA transfection. The culture supernatant and RNA was collected 48 h post siRNA transfection.
Statistical analysis
Statistical significance was determined using two-tailed t-test; ANOVA and Dunnett's multiple comparison post-test were used to compare the means of multiple groups. Data are shown as mean ± SEM and were considered statistically significant at p < 0.05. GraphPad Prism 6.02 (GraphPad Software Inc., La Jolla, CA) was used for analysis.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental data for this article can be accessed on the publisher's website.
Ethics statement
All the animals used in this study received humane treatment and the authors complied with the guidelines for animal use set by the Institutional Animal Use and Care Committee of the University of Massachusetts Medical School. The study protocol was approved by the Institutional Animal Use and Care Committee of the University of Massachusetts Medical School.
Author contributions
AA did the DEN injections and the HF–HC–HSD feeding. AA, AS, BG, and PL performed the animal sacrifice and liver cell isolations. BS stained and acquired the flow cytometry samples. KK and DC helped in primary cell isolations. AA performed all in vitro experiments, analyzed, and interpreted all the data. AA and GS conceptualized the project, designed the experiments, and wrote the manuscript. GS obtained the funding and provided overall study supervision.
KONI_A_1221557_supplementary_data.zip
Download Zip (384.3 KB)Acknowledgments
We thank current members of the Szabo lab for their stimulating intellectual discussions and scientific input. We are also grateful to Timea Csak and Michal Ganz, past members of Szabo lab for their insight in HF–HC–HSD formulation and administration. The help provided by Shaokuan Zheng (Department of Radiology) in acquisition of MRI data is acknowledged. We thank Dr. David Garlick of Cancer Biology Department at UMASS for histopathological examination of liver tissue sections and Candice Dufour for assistance with formatting the manuscript. The authors are grateful to services provided by DERC (histology and immunostaining) and CFAR (primer synthesis) core facilities at UMASS Medical School. This work was supported by NIH/ NIAAA grant AA01157 to G.S.
Funding
CFAR core facility is supported by the University of Massachusetts Center for AIDS Research (P30 AI042845). This work was supported by grant number AA01157 to G.S.
References
- El-Serag HB. Hepatocellular carcinoma. N Engl J Med 2011; 365:1118-27; PMID:21992124; http://dx.doi.org/10.1056/NEJMra1001683
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65:87-108; PMID:25651787; http://dx.doi.org/10.3322/caac.21262
- Basen-Engquist K, Chang M. Obesity and cancer risk: Recent review and evidence. Curr Oncol Rep 2011; 13:71-6; PMID:21080117; http://dx.doi.org/10.1007/s11912-010-0139-7
- De Pergola G, Silvestris F. Obesity as a major risk factor for cancer. J Obes 2013; 2013:291546; PMID:24073332; http://dx.doi.org/10.1155/2013/291546
- Calle EE, Rodriguez C, Walker-Thurmond K, Thun MJ. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N Engl J Med 2003; 348:1625-38; PMID:12711737; http://dx.doi.org/10.1056/NEJMoa021423
- Sun B, Karin M. Obesity, inflammation, and liver cancer. J Hepatol 2012; 56:704-13; PMID:22120206; http://dx.doi.org/10.1016/j.jhep.2011.09.020
- Baffy G, Brunt EM, Caldwell SH. Hepatocellular carcinoma in non-alcoholic fatty liver disease: An emerging menace. J Hepatol 2012; 56:1384-91; PMID:22326465; http://dx.doi.org/10.1016/j.jhep.2011.10.027
- Park EJ, Lee JH, Yu GY, He G, Ali SR, Holzer RG, Osterreicher CH, Takahashi H, Karin M. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell 2010; 140:197-208; PMID:20141834; http://dx.doi.org/10.1016/j.cell.2009.12.052
- Michelotti GA, Machado MV, Diehl AM. NAFLD, NASH and liver cancer. Nat Rev Gastroenterol Hepatol 2013; 10:656-65; PMID:24080776; http://dx.doi.org/10.1038/nrgastro.2013.183
- Landskron G, De la Fuente M, Thuwajit P, Thuwajit C, Hermoso MA. Chronic inflammation and cytokines in the tumor microenvironment. J Immunol Res 2014; 2014:149185; PMID:24901008; http://dx.doi.org/10.1155/2014/149185
- Ganz M, Bukong TN, Csak T, Saha B, Park JK, Ambade A, Kodys K, Szabo G. Progression of non-alcoholic steatosis to steatohepatitis and fibrosis parallels cumulative accumulation of danger signals that promote inflammation and liver tumors in a high fat-cholesterol-sugar diet model in mice. J Transl Med 2015; 13:193,015-0552-7; PMID:26077675; http://dx.doi.org/10.1186/s12967-015-0552-7
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell 2010; 140:883-99; PMID:20303878; http://dx.doi.org/10.1016/j.cell.2010.01.025
- Capece D, Fischietti M, Verzella D, Gaggiano A, Cicciarelli G, Tessitore A, Zazzeroni F, Alesse E. The inflammatory microenvironment in hepatocellular carcinoma: A pivotal role for tumor-associated macrophages. Biomed Res Int 2013; 2013:187204; PMID:23533994; http://dx.doi.org/10.1155/2013/187204
- Kohli R, Kirby M, Xanthakos SA, Softic S, Feldstein AE, Saxena V, Tang PH, Miles L, Miles MV, Balistreri WF et al. High-fructose, medium chain trans fat diet induces liver fibrosis and elevates plasma coenzyme Q9 in a novel murine model of obesity and nonalcoholic steatohepatitis. Hepatology 2010; 52:934-44; PMID:20607689; http://dx.doi.org/10.1002/hep.23797
- Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev 2013; 27:2192-206; PMID:24142872; http://dx.doi.org/10.1101/gad.225334.113
- Heerboth S, Housman G, Leary M, Longacre M, Byler S, Lapinska K, Willbanks A, Sarkar S. EMT and tumor metastasis. Clin Transl Med 2015; 4:6,015-0048-3. eCollection 2015; PMID:25852822; http://dx.doi.org/10.1186/s40169-015-0048-3
- Csak T, Bala S, Lippai D, Satishchandran A, Catalano D, Kodys K, Szabo G. microRNA-122 regulates hypoxia-inducible factor-1 and vimentin in hepatocytes and correlates with fibrosis in diet-induced steatohepatitis. Liver Int 2015; 35:532-41; PMID:25040043; http://dx.doi.org/10.1111/liv.12633
- Nakagawa H, Hikiba Y, Hirata Y, Font-Burgada J, Sakamoto K, Hayakawa Y, Taniguchi K, Umemura A, Kinoshita H, Sakitani K et al. Loss of liver E-cadherin induces sclerosing cholangitis and promotes carcinogenesis. Proc Natl Acad Sci U S A 2014; 111:1090-5; PMID:24395807; http://dx.doi.org/10.1073/pnas.1322731111
- Byrne CD. Hypoxia and non-alcoholic fatty liver disease. Clin Sci (Lond) 2009; 118:397-400; PMID:19900166; http://dx.doi.org/10.1042/CS20090565
- Wu XZ, Xie GR, Chen D. Hypoxia and hepatocellular carcinoma: The therapeutic target for hepatocellular carcinoma. J Gastroenterol Hepatol 2007; 22:1178-82; PMID:17559361
- Nath B, Levin I, Csak T, Petrasek J, Mueller C, Kodys K, Catalano D, Mandrekar P, Szabo G. Hepatocyte-specific hypoxia-inducible factor-1alpha is a determinant of lipid accumulation and liver injury in alcohol-induced steatosis in mice. Hepatology 2011; 53:1526-37; PMID:21520168; http://dx.doi.org/10.1002/hep.24256; 10.1002/hep.24256
- Cai Z, Luo W, Zhan H, Semenza GL. Hypoxia-inducible factor 1 is required for remote ischemic preconditioning of the heart. Proc Natl Acad Sci U S A 2013; 110:17462-7; PMID:24101519; http://dx.doi.org/10.1073/pnas.1317158110
- Marra F, Lotersztajn S. Pathophysiology of NASH: Perspectives for a targeted treatment. Curr Pharm Des 2013; 19:5250-69; PMID:23394092
- Sica A, Mantovani A. Macrophage plasticity and polarization: In vivo veritas. J Clin Invest 2012; 122:787-95; PMID:22378047; http://dx.doi.org/10.1172/JCI59643
- Basen-Engquist K, Chang M. Obesity and cancer risk: Recent review and evidence. Curr Oncol Rep 2011; 13:71-6; PMID:21080117; http://dx.doi.org/10.1007/s11912-010-0139-7
- Bieghs V, Trautwein C. The innate immune response during liver inflammation and metabolic disease. Trends Immunol 2013; 34:446-52; PMID:23668977; http://dx.doi.org/10.1016/j.it.2013.04.005
- Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000Prime Rep 2014; 6:13; PMID:24669294; http://dx.doi.org/10.12703/P6-13
- Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014; 41:14-20; PMID:25035950; http://dx.doi.org/10.1016/j.immuni.2014.06.008
- Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol 2004; 25:677-86; PMID:15530839; http://dx.doi.org/ S1471-4906(04)00295-9 [pii]
- Ferrante CJ, Pinhal-Enfield G, Elson G, Cronstein BN, Hasko G, Outram S, Leibovich SJ. The adenosine-dependent angiogenic switch of macrophages to an M2-like phenotype is independent of interleukin-4 receptor alpha (IL-4Ralpha) signaling. Inflammation 2013; 36:921-31; PMID:23504259; http://dx.doi.org/10.1007/s10753-013-9621-3
- Roszer T. Understanding the mysterious M2 macrophage through activation markers and effector mechanisms. Mediators Inflamm 2015; 2015:816460; PMID:26089604; http://dx.doi.org/10.1155/2015/816460
- Hao NB, Lu MH, Fan YH, Cao YL, Zhang ZR, Yang SM. Macrophages in tumor microenvironments and the progression of tumors. Clin Dev Immunol 2012; 2012:948098; PMID:22778768; http://dx.doi.org/10.1155/2012/948098
- Yuan A, Hsiao YJ, Chen HY, Chen HW, Ho CC, Chen YY, Liu YC, Hong TH, Yu SL, Chen JJ et al. Opposite effects of M1 and M2 macrophage subtypes on lung cancer progression. Sci Rep 2015; 5:14273; PMID:26399191; http://dx.doi.org/10.1038/srep14273
- Liu CY, Xu JY, Shi XY, Huang W, Ruan TY, Xie P, Ding JL. M2-polarized tumor-associated macrophages promoted epithelial-mesenchymal transition in pancreatic cancer cells, partially through TLR4/IL-10 signaling pathway. Lab Invest 2013; 93:844-54; PMID:23752129; http://dx.doi.org/10.1038/labinvest.2013.69
- Quatromoni JG, Eruslanov E. Tumor-associated macrophages: Function, phenotype, and link to prognosis in human lung cancer. Am J Transl Res 2012; 4:376-89; PMID:23145206
- Lau SK, Chu PG, Weiss LM. CD163: A specific marker of macrophages in paraffin-embedded tissue samples. Am J Clin Pathol 2004; 122:794-801; PMID:15491976
- Pettersen JS, Fuentes-Duculan J, Suarez-Farinas M, Pierson KC, Pitts-Kiefer A, Fan L, Belkin DA, Wang CQ, Bhuvanendran S, Johnson-Huang LM et al. Tumor-associated macrophages in the cutaneous SCC microenvironment are heterogeneously activated. J Invest Dermatol 2011; 131:1322-30; PMID:21307877; http://dx.doi.org/10.103/jid.2011.9
- Ohri CM, Shikotra A, Green RH, Waller DA, Bradding P. The tissue microlocalisation and cellular expression of CD163, VEGF, HLA-DR, iNOS, and MRP 8/14 is correlated to clinical outcome in NSCLC. PLoS One 2011; 6:e21874; PMID:21799753; http://dx.doi.org/10.1371/journal.pone.0021874
- Medrek C, Ponten F, Jirstrom K, Leandersson K. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer 2012; 12:306,2407-12-306; PMID:22824040; http://dx.doi.org/10.1186/1471-2407-12-306
- Shabo I, Olsson H, Elkarim R, Sun XF, Svanvik J. Macrophage infiltration in tumor stroma is related to tumor cell expression of CD163 in colorectal cancer. Cancer Microenviron 2014; 7:61-9; PMID:24771466; http://dx.doi.org/10.1007/s12307-014-0145-7
- Sica A, Schioppa T, Mantovani A, Allavena P. Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: Potential targets of anti-cancer therapy. Eur J Cancer 2006; 42:717-27; PMID:16520032; http://dx.doi.org/ S0959-8049(06)00040-2 [pii]
- Jetten N, Verbruggen S, Gijbels MJ, Post MJ, De Winther MP, Donners MM. Anti-inflammatory M2, but not pro-inflammatory M1 macrophages promote angiogenesis in vivo. Angiogenesis 2014; 17:109-18; PMID:24013945; http://dx.doi.org/10.1007/s10456-013-9381-6
- Nishida N, Yano H, Nishida T, Kamura T, Kojiro M. Angiogenesis in cancer. Vasc Health Risk Manag 2006; 2:213-9; PMID:17326328
- Wang Z, Dabrosin C, Yin X, Fuster MM, Arreola A, Rathmell WK, Generali D, Nagaraju GP, El-Rayes B, Ribatti D et al. Broad targeting of angiogenesis for cancer prevention and therapy. Semin Cancer Biol 2015; 35 Suppl:S224-43; PMID:25600295; http://dx.doi.org/10.1016/j.semcancer.2015.01.001
- Semenza GL. HIF-1 and tumor progression: Pathophysiology and therapeutics. Trends Mol Med 2002; 8:S62-7; PMID:11927290; http://dx.doi.org/ S1471491402023171 [pii]
- Liao D, Johnson RS. Hypoxia: A key regulator of angiogenesis in cancer. Cancer Metastasis Rev 2007; 26:281-90; PMID:17603752; http://dx.doi.org/10.1007/s10555-007-9066-y
- Xu Z, Chen X, Peng C, Liu E, Li Y, Li C, Niu J. Hypoxia-inducible factor-1alpha suppressed hepatocellular carcinoma cell apoptosis through influencing on Omi/HtrA2 expression and its releasing from the mitochondrion. Oncol Res 2012; 20:213-20; PMID:23581228
- Luo D, Wang Z, Wu J, Jiang C, Wu J. The role of hypoxia inducible factor-1 in hepatocellular carcinoma. Biomed Res Int 2014; 2014:409272; PMID:25101278; http://dx.doi.org/10.1155/2014/409272
- Colegio OR, Chu NQ, Szabo AL, Chu T, Rhebergen AM, Jairam V, Cyrus N, Brokowski CE, Eisenbarth SC, Phillips GM et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 2014; 513:559-63; PMID:25043024; http://dx.doi.org/10.1038/nature13490
- Tripathi C, Tewari BN, Kanchan RK, Baghel KS, Nautiyal N, Shrivastava R, Kaur H, Bhatt ML, Bhadauria S. Macrophages are recruited to hypoxic tumor areas and acquire a pro-angiogenic M2-polarized phenotype via hypoxic cancer cell derived cytokines oncostatin M and eotaxin. Oncotarget 2014; 5:5350-68; PMID:25051364
- Ambade A, Catalano D, Lim A, Kopoyan A, Shaffer SA, Mandrekar P. Inhibition of heat shock protein 90 alleviates steatosis and macrophage activation in murine alcoholic liver injury. J Hepatol 2014; 61:903-11; PMID:24859453; http://dx.doi.org/10.1016/j.jhep.2014.05.024
- Ambade A, Satishchandran A, Szabo G. Alcoholic hepatitis accelerates early hepatobiliary cancer by increasing stemness and miR-122-mediated HIF-1alpha activation. Sci Rep 2016; 6:21340; PMID:26888602; http://dx.doi.org/10.1038/srep21340