ABSTRACT
A prospective analysis of natural killer (NK) cell phenotype and function was performed on fresh peripheral blood samples from untreated patients with B-cell chronic lymphocytic leukemia (CLL) or small lymphocytic lymphoma (SLL). Compared to healthy controls, CD56dim NK cells in CLL patients displayed reduced expression of the NKG2D activating receptor and increased CD27 expression, which indicates declines in mature cells. In addition, NK cells from CLL patients showed reduced degranulation responses toward transformed B cells alone or with rituximab and were more sensitive to activation-induced cell death. We further noted a striking reduction in the frequency and viability of NK cells expressing the inhibitory killer cell Ig-like receptors (KIR)2DL1 and/or KIR3DL1, which progressed over time in most patients. Comparisons between a CLL patient and healthy monozygotic twin were consistent with our results in the larger cohorts. Functional and biomarker alterations were less pronounced on NK cells from SLL patients, which have lower tumor burden in peripheral blood than CLL, but significant reduction in degranulation under ADCC conditions and lower frequency and viability of KIR-expressing NK cells were still evident in SLL. We conclude that mature KIR-expressing NK cells respond to the high circulating B cell tumor burden in CLL, but undergo activation-induced apoptosis. Consequently, CLL patients may benefit from therapies that augment NK cell survival and function.
Introduction
Chronic lymphocytic leukemia (CLL) is the most prevalent adult leukemia and is characterized by clonal expansion of mature B lymphocytes.Citation1 The disease has a variable clinical course, with some patients living essentially normal lives with indolent disease for many years, while others succumb with rapidly developing complications. About 15,000 CLL cases are diagnosed yearly in the United States, resulting in >100,000 patients currently living with the disease.Citation2 Progression of CLL is characterized by increasing peripheral blood lymphocytosis of monoclonal B cells, as well as lymphadenopathy, hepatosplenomegaly, anemia, and thrombocytopenia. CLL is considered incurable by standard chemotherapy, so treatment is often withheld until disease is progressive and symptomatic.Citation3 Treatments vary from single alkylating agents to combinations of chemotherapy and immunotherapy, particularly anti-CD20 monoclonal antibodies and targeted therapies affecting B cell antigen receptor signaling pathways, as well as promising chimeric antigen receptor-expressing T cells.Citation1 Unmet clinical needs are identifying patients who will require early therapy and tailoring therapy to disease status, including exploration of relatively non-toxic, immune-modulating therapies that could maintain the indolent phase of disease.
Small lymphocytic lymphoma (SLL) is less common, but similar to CLL in morphology, surface marker phenotype, and gene expression signature.Citation4 SLL patients present with monoclonal B cells that predominantly infiltrate tissues, especially lymph nodes, bone marrow, and spleen, instead of circulating in peripheral blood. This distinct localization of SLL and CLL appears to result from their differential expression of chemokine receptors and/or adhesion molecules.Citation4-7
Deficits in the functions of circulating T cells have been widely recognized in CLL patients. T cells in these patients display increased expression of markers of activation and exhaustion (PD-1, CD244, and CD160), as well as greater incidence of immunosuppressive regulatory T (Treg) cells.Citation8-10 Dysregulated cytotoxicity by CD8+ T cells in CLL has been associated with reduced polarization of granzyme B,Citation8 and defects in cytoskeleton remodeling and immune synapse formation, which is driven by expression of numerous inhibitory ligands (CD200, CD270, CD276, and PD-L1) on CLL tumor cells.Citation11 T cells from CLL patients also have significantly reduced expression of genes involved in cytoskeletal regulation, cytotoxicity responses, and TCR signaling.Citation12,Citation13
Although defects in cytolytic responses by NK cells in CLL patients were first reported in the early 1980s,Citation14-17 mechanistic understanding of these functional deficits is surprisingly limited. Decreased expression of several NK cell activating receptors, such as NKp30 and NKp46, has been reported in CLL patients in some studies,Citation18-20 but these findings have been inconsistent and may in fact reflect higher incidence of “memory-like” or “adaptive” NK cells associated with Cytomegalovirus (CMV)-infected individuals, which have similar expression profiles.Citation21-23 It has also been reported that NK cells from CLL patients express higher levels of the CD85j/ILT2 inhibitory receptor, while CLL tumors abnormally express its ligand, HLA-G, which was found to suppress NK cell cytotoxicity and was associated with poor prognosis.Citation16,Citation24,Citation25 In addition, high expression levels of glucocorticoid-induced TNFR-related protein ligand (GITR) on CLL tumor cells and ROS production by monocytes can both reportedly reduce rituximab-mediated ADCC responses by NK cells.Citation26,Citation27 Competent ADCC by NK cells can be important, since it has been shown to play a significant role in the therapeutic efficacy of certain antibodies, including rituximab in treating CLL and other B cell malignancies.Citation28,Citation29 Accordingly, lenalidomide is being tested in combination with rituximab to improve NK cell cytolytic and ADCC responses in CLL, and recent work suggests that the B-cell activating factor (BAFF) inhibitor, belimumab, may also be effective to improve rituximab efficacy.Citation11,Citation30-32
In view of the need for better understanding of the basis of NK cell defects in CLL patients, we performed a comprehensive prospective analysis of their status in previously untreated cohorts of adult patients with CLL or SLL, as compared with healthy controls of similar median age. Since NK cells are found more prevalently in peripheral blood than lymph nodes, we hypothesized that NK cells in blood would be more significantly influenced by persistent encounter with the more significant circulating tumor mass in CLL patients, as compared with SLL patients. To avoid the influences of therapeutics, we chose to study untreated indolent patients to assess the impacts of tumor burden on the circulating pool of NK cells before therapeutic intervention. Our results demonstrate that NK cells from CLL patients have defects in degranulation, reduced NKG2D expression, and enhanced susceptibility to activation-induced cell death (AICD). A reduction in fully mature NK cells was evident, and the fraction of NK cells expressing inhibitory killer cell Ig-like receptors (KIR) was significantly diminished and less viable. Alterations in peripheral NK cells were more pronounced in patients with CLL, as compared with SLL, suggesting that they are indeed more significantly impacted by continuous exposure to the higher burden of circulating tumor cells.
Results
Increased frequency and decreased maturity of NK cells in CLL patients
A prospective analysis of NK cell frequencies, phenotypes, and function was performed on fresh peripheral blood mononuclear cells (PBMC) from 25 untreated CLL patients, 10 untreated SLL patients, and 17 healthy controls of similar median age. We observed that high B cell counts correlated with a significant increase in numbers of NK cells in the blood of CLL, but not SLL patients, as compared with healthy donors ( and ). While our primary focus was NK cells, T cells were also analyzed in parallel, and increased frequencies of CD4+ and CD8+ T cells were also observed only in the CLL patients (Fig. S1A–D). The increased frequencies of T and NK cells in CLL patients confirm previous reports,Citation13,Citation33,Citation34 and these increases have been reported to correlate with slower disease progression.Citation35
Figure 1. Increased NK cell counts and changes in surface receptors in CLL patients. (A) NK cells/µL were quantified in whole blood of different donor groups. (B) Correlation between NK cell counts and B cell counts for SLL (open circles) and CLL (filled circles) patients. Vertical and horizontal lines mark the global medians, diagonal line marks a least squares fit analysis, and statistics were calculated with a Spearman test. (C–H) NK cells were gated as viable (propidium iodide-negative), CD45+CD3−CD56(dim or bright) and assessed by mean fluorescence intensity (MFI) of staining for expression of (C, D) CD27 or (E, F) NKG2D. For panels (A) and (C–F), filled icons designate monzygotic twins. Horizontal lines designate median values, and statistics were calculated with an unpaired Wilcoxon rank-sum test.
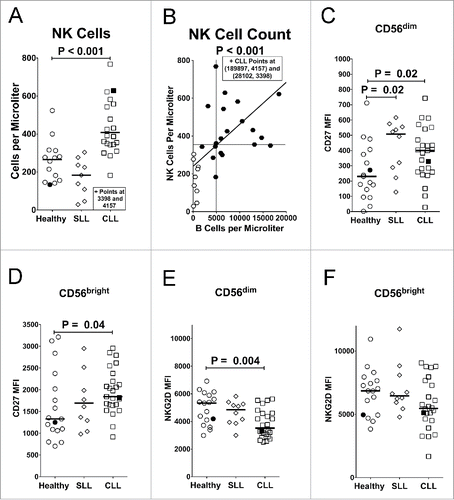
NK cells can be subdivided based upon their level of CD56 expression. CD56dim cells constitute about 90–95% of the NK cells in peripheral blood and primarily mediate cellular cytotoxicity responses, while CD56bright NK cells are less mature and primarily specialized toward cytokine production.Citation36 The ratio of CD56bright to CD56dim NK cells did not differ between the SLL, CLL, or healthy cohorts (data not shown). On the other hand, expression of CD27, which is downregulated on mature effector T and NK cells,Citation37,Citation38 was significantly increased on CD56dim NK cells from SLL patients and on both CD56dim and CD56bright NK cell subsets from CLL patients ( and ), suggesting expansion of immature NK cells or reduction in mature NK cells. In contrast, CD27 expression on T cells did not differ between SLL, CLL, and healthy donors (Fig. S1E). One CLL patient had a healthy monozygotic twin, and the pair provided parallel samples. The statistically significant changes between healthy donors and CLL patient cohorts were also evident in the twin samples (filled symbols in all figures).
Evidence of immune suppression in CLL patients
Several signs of immune suppression were noted in CLL patients, as compared with healthy controls. While virtually all NK cells expressed the NKG2D activating receptor, expression levels were significantly reduced on the cytolytic CD56dim subset from CLL patients, but not SLL patients (). On the other hand, NKG2D expression in CLL patients was not significantly reduced on the cytokine-producing CD56bright NK cell subset () or T cells expressing NKG2D (Fig. S1F and G).
CD4+ T cells and CD8+ T cells from CLL patients were also found to express significantly higher levels of the immunosuppressive receptor PD-1 than healthy individuals or SLL patients (Fig. S1H and I). We also noted that Treg cells (CD3+CD4+CD25highFOXP3+) were significantly elevated in CLL patients (Fig. S1J), as shown in previous studies.Citation10,Citation39 Treg cells were also increased in SLL patients, but the increase was just short of statistical significance (Fig. S1J).
Defective NK cell degranulation associated with increased activation-induced cell death
Surface expression of LAMP-1 (Lysosomal Associated Membrane Protein 1; CD107a) was used to measure NK cell degranulation upon exposure to the EBV-transformed B cell line 721.221, either alone or with rituximab [antibody-dependent cellular cytotoxicity (ADCC) conditions]. LAMP-1 surface expression has been shown to directly correlate with NK cell-mediated cytotoxicity.Citation40,Citation41 A small but significant reduction in basal LAMP-1 expression was observed on unstimulated CD56dim NK cells from CLL patients (), suggesting a hyporesponsive state.Citation15,Citation17 Despite substantial numbers of CD20+ tumor cells, however, the addition of rituximab alone to PBMC of CLL patients did not increase LAMP-1 expression any more than treatment of PBMC from healthy donors (A). Degranulation by NK cells from CLL patients, but not SLL patients, was significantly lower when exposed to 721.221 target cells alone or with rituximab (). Although we did not directly measure target cytotoxicity, previous studies have shown reduced cytotoxicity by purified NK cells from CLL patients using chromium release or Europium release assays.Citation15,Citation17,Citation20 A moderate reduction in degranulation by NK cells from SLL patients exposed to target cells alone was not statistically significant, consistent with a recent report by Parry et al. showing reduced cytotoxicity of K562 and primary B-CLL target cells by purified NK cells from CLL, but not SLL patients.Citation20 However, we did observe statistically significant suppression of degranulation by NK cells from SLL patients under ADCC conditions (). Importantly, our results demonstrate degranulation defects in NK cells from both CLL and SLL patients under ADCC conditions using an antibody commonly used to treat these diseases.
Figure 2. Suppressed degranulation and increased sensitivity to activation-induced cell death (AICD) by NK cells from CLL patients. PBMC were incubated alone, with rituximab (RXB), with 721.221 target cells (721), or with 721.221 targets and RXB for 2 h at 37 °C and stained for (A) Percent LAMP-1 (CD107a) expression or (B) viability (propidium iodide negative). NK cells were gated as CD56dimCD3− after the viability gate in panel (A), and before the viability gate in panel (B) from the same experiments. Filled icons represent monozygotic twins. Horizontal lines mark median values, and statistics were calculated with an unpaired Wilcoxon rank-sum test.
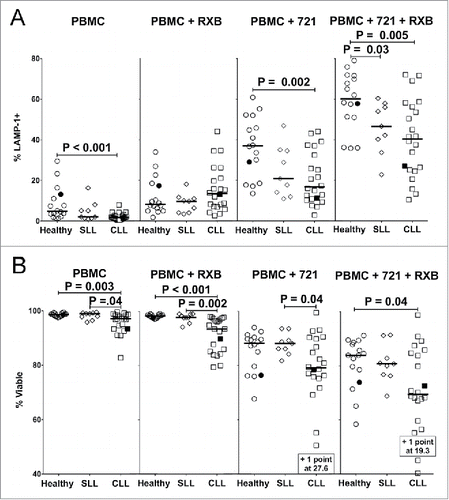
Basal viability of NK cells was also significantly reduced in CLL, but not SLL patients, as assessed by propidium iodide uptake (). While adding rituximab did not alter the viability of NK cells from healthy donors or SLL patients, rituximab significantly reduced the viability of NK cells from CLL patients (), possibly contributing to their limited degranulation capacity (). Exposure to 721.221 target cells substantially reduced the viability of NK cells from both healthy donors and patients (), consistent with AICD that commonly occurs during healthy NK cell cytolytic responses.Citation42-45 NK cell viability was further reduced upon exposure to the combination of 721.221 target cells and rituximab, and this reduction was greater in CLL patients, as compared with healthy controls (). Viability of NK cells from SLL patients was similar to healthy donors in these experiments, suggesting greater sensitivity to AICD by peripheral NK cells from CLL patients may be causally related to their persistent exposure to circulating tumor cells.
Selective loss of NK cells expressing inhibitory KIR
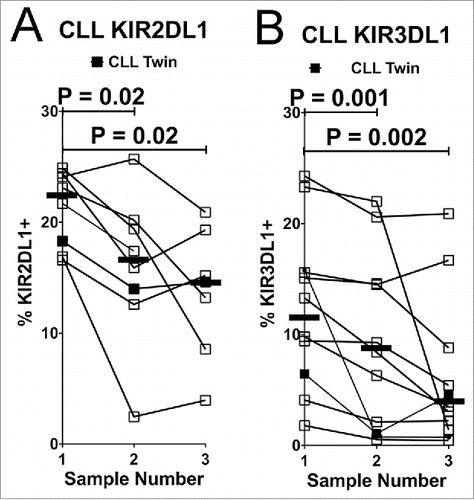
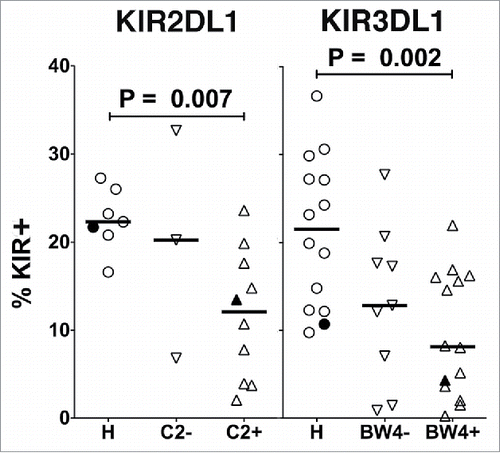
The expression of KIR2DL1 and KIR3DL1 was monitored on the surface of NK cells, since these KIR can be directly measured by specific mAbs. KIR are differentially expressed on distinct CD56dim NK cells in an individual,Citation46 and the percentage of CD56dim NK cells expressing KIR2DL1 or KIR3DL1 was found to be significantly reduced in both SLL and CLL patients compared with healthy controls ( and ). This prompted us to compare the viability of CD56dim NK cells expressing or lacking these inhibitory KIR. Interestingly, viability was significantly lower in KIR2DL1+ or KIR3DL1+ NK cells from the patients, especially with CLL, as compared with healthy controls ( and ). In fact, viability was most significantly reduced in NK cells co-expressing both KIR (). We further noted a progressive reduction in the percentage of NK cells expressing KIR2DL1 or KIR3DL1 in serial blood samples from most CLL patients, which was statistically significant when compared with initial samples ( and ). In contrast, progressive reductions in KIR expression were not evident in healthy donors (Fig. S2).
Figure 3. Reduced expression of inhibitory KIR on NK cells in CLL patients is associated with reduced viability of KIR+ cells. Viable CD45+CD3−CD56dim NK cells from PBMC of the different donor groups were analyzed by flow cytometry for fraction of cells expressing KIR2DL1 (A) or KIR3DL1 (B). Viability was determined in CD56dim NK cells expressing (+, filled icons) or lacking (-, open icons) KIR2DL1 (C) or KIR3DL1 (D). (E–H) Percentage viability of subsets of CD45+CD3−CD56dim NK cells with the indicated KIR2DL1 and KIR3DL1 expression profiles was analyzed by flow cytometry. The viability of NK cells in each quadrant was determined by propidium iodide staining. Data from healthy donors are displayed as circles, SLL patients as diamonds, and CLL patients as squares. Only donors confirmed by genotyping to express the indicated KIR were included in these panels. Horizontal lines indicate median values and statistics were calculated with an unpaired Wilcoxon rank-sum test. Filled icons are from monozygotic twins in panels A, B, and E-H.
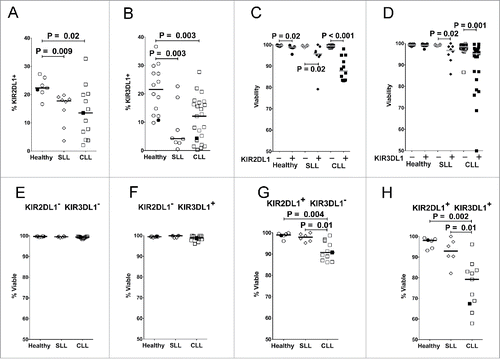
Figure 4. NK cells expressing inhibitory KIR decline over time in CLL patients. Viable CD45+CD3−CD56dim NK cells were analyzed for expression of inhibitory KIR on consecutive blood samples from CLL patients. Lines connect the fraction of NK cells expressing KIR2DL1 (A) or KIR3DL1 (B) from individual donors. KIR2DL1 and KIR3DL1 expression data are only shown from donors confirmed by genotyping. Serial sampling of SLL patients is not shown due to insufficient data points. Horizontal lines designate median values, and statistics were calculated with a paired Wilcoxon rank-sum test, with the initial and subsequent sample from each donor constituting a pair.
Effects of ligands for inhibitory KIR
We next examined whether the presence of ligand influences the negative impact of inhibitory KIR expression on CD56dim NK cells in the CLL patients. KIR2DL1 and KIR3DL1 recognize subsets of human MHC class I molecules known as HLA-C2, and HLA-Bw4, respectively, and genes for KIR and their ligands are differentially inherited by individuals.Citation46 Therefore, we genotyped the patients and categorized those expressing KIR2DL1 or KIR3DL1 based upon whether they lack or express at least one allele of their HLA ligands. Compared to healthy controls, a statistically significant reduction was noted in the percentage of NK cells expressing either KIR2DL1 or KIR3DL1 in CLL patients expressing their cognate ligands ().
Figure 5. Effect of inhibitory KIR ligands on expression of KIR. Healthy donors (H) or CLL patients confirmed by genotyping to express either KIR2DL1 (left) or KIR3DL1 (right) were compared for percent of NK cells (gated as viable, CD45+CD3−CD56dim) expressing these KIR (HLA-C2 for KIR2DL1 or HLA-Bw4 for KIR3DL1). Horizontal lines mark the median values, and statistical significance was determined by an unpaired Wilcoxon rank-sum test. Filled icons are from monozygotic twins.
We also compared degranulation responses of CD56dim NK cells in the CLL patients based upon their expression of KIR and their HLA ligands. As compared with healthy controls, the statistically significant deficits in degranulation against 721.221 target cells alone or with rituximab were restricted to CLL patients expressing KIR2DL1 or KIR3DL1 in combination with their ligands, but responses were not significantly different between patients lacking or bearing their ligands (Fig. S3). Furthermore, NK cells from CLL patients expressing either KIR showed reduced viability under resting conditions in a manner that was independent of ligand expression (Fig. S4). Taken together, our data demonstrate dysfunction in NK cells expressing inhibitory KIR in CLL patients, but do not entirely support a role for KIR ligands in driving the dysfunction. A significantly larger cohort and analysis of all KIR/ligand combinations may be necessary to confirm that ligands are critical elements in the process.
Further mechanistic insights from the monozygotic twins
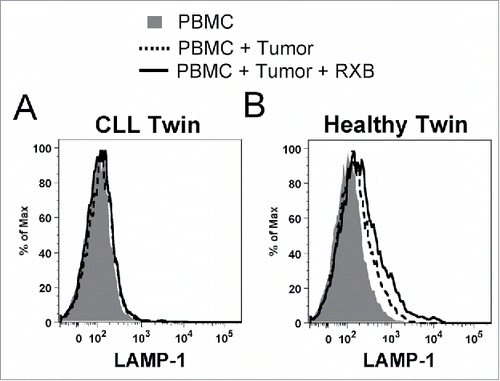
In the previous figures, the NK cell phenotype of the twins paralleled the full cohorts in most assays. Although only one healthy and CLL pair of twins was analyzed, the results reinforce our conclusions. In contrast, NK cells in blood samples from a pair of healthy monozygotic twins neither reveal differences in expression of KIR, NKG2D or CD27, nor viability of KIR-expressing NK cells (Fig. S5). The genetically identical donors provided a unique opportunity to test whether NK cells from the healthy twin could degranulate when exposed to PBMC from the CLL twin, which contained high tumor burden (60% B cells). In this experiment, NK cells from the CLL twin failed to degranulate when reconstituted with autologous tumor cells, even in the presence of rituximab (). However, healthy twin NK cells exhibited substantial degranulation when exposed to sibling tumor cells (), which increased further in the presence of rituximab. This demonstrates that healthy NK cells have innate capacity to respond to CLL tumor cells and implies that NK cell hyporesponsiveness in the CLL twin is due to an acquired functional deficit and/or altered repertoire. Furthermore, in the degranulation assays, CD56dim NK cells from the CLL twin showed significantly elevated annexin V staining after exposure to 721.221 targets, indicating that the reduced viability in the total patient cohort results from greater sensitivity to activation-induced apoptosis (Fig. S6). In contrast, apoptosis did not differ on NK cells from the twins when PBMC were cultured alone or with rituxumab (Fig. S6).
Figure 6. NK cells from the healthy monozygotic twin can degranulate when exposed to sibling CLL tumor. Freshly isolated effector NK cells from each twin were challenged with fresh tumor-containing PBMC from the twin with CLL as target cells in degranulation assays. NK cells were partially purified from the CLL patient to achieve similar effector cell concentrations (7.5%), and target PBMC were prestained with CellTracker Blue to gate out of flow cytometry analysis. LAMP-1 (CD107a) expression was subsequently measured on CD56dim NK cells from the (A) CLL and (B) healthy twins. Histogram plots are shown for PBMC alone (shaded), with CLL tumor cells (dashed line), and with CLL tumor cells and rituximab (solid line).
Discussion
Taken together, our data demonstrate that CLL patients undergo an expansion of NK cells in peripheral blood in parallel with increased B cell tumor burden, which is accompanied by a selective decrease in viability and loss of the NK cells that are CD56dimCD27dim/−NKG2DbrightKIR2DL1/KIR3DL1+. This biomarker phenotype is characteristic of the most mature NK cells, which typically elicit cytolytic responses toward tumors,Citation36 and could explain at least part of the reduced degranulation response by patient NK cells. While we did not observe increased numbers of NK cells in SLL patients, these patients did also exhibit increased expression of CD27 and reduced incidence of KIR2DL1 or KIR3DL1 expression on CD56dim NK cells, which correlated with reduced viability of the KIR-expressing cells. Despite a similar overall reduction of the most mature NK cells in both SLL and CLL patients, most of the other phenotypic characteristics of NK cells in SLL patients were comparable to those of healthy donors.
Increased CD27 expression on CD56dim NK cells in SLL and both CD56dim and CD56bright NK cell subsets in CLL indicates a decline in the more mature cells, since CD27 is more densely expressed on the less mature NK cells.Citation37,Citation38 Many CLL tumors reportedly express the CD27 ligand, CD70,Citation47 and the CD27/CD70 interaction has been reported to stimulate IFNγ production by NK cells.Citation48 We show that NK cells from a healthy monozygotic twin can degranulate in response to sibling CLL tumor cells, but NK cells from CLL patients exhibit suppressed degranulation and increased susceptibility to AICD. It should be noted that we did not directly measure reduced cytotoxicity of tumor target cells by NK cells in our study, which can be considered a limitation. Nonetheless, our degranulation assays did reveal increased susceptibility to AICD. We conclude that the most mature circulating CD56dim NK cells generate a relentless response to prevalent circulating tumor cells in CLL patients, but they become overwhelmed and apoptose by AICD. In contrast, the tumor cells in SLL patients are predominantly confined to lymph nodes, which are primarily populated with CD56bright NK cells.Citation36 This may explain the modest impacts on phenotype and function of peripheral blood NK cells in SLL patients. Increased IFNγ production by NK cells can result from prolonged target cell conjugation associated with a failed cytolytic event,Citation49 which is consistent with our observation of reduced degranulation. Hence, the decreased degranulation capacity may possibly be associated with a shift of circulating CD56dim NK cells in CLL patients from cytolytic effectors to cytokine production, which may actually benefit tumor survival, since IFNγ can reportedly protect CLL cells from apoptosis.Citation50,Citation51
Reduced expression of NKp30 and NKp44 activating receptors has been reported on NK cells in CLL patients with high tumor burden, while expression of NKp46 was unchanged.Citation18,Citation19 A recent report found reduced expression of CD16, DNAM-1, NKp30, and NKp46 activating receptors on NK cells from CLL patients.Citation20 CMV infection status of the patients was not reported in the latter study, however, and the observed reduction of NKp30, NKp46, and coincidental reductions of mRNAs for FcϵRI-γ and Syk are indicative of expression profiles found in adaptive/memory-like NK cells from CMV seropositive individuals.Citation22,Citation23 A soluble form of the NKp30 ligand BAG6/BAT3 was also reported in the serum of at least some CLL patients, which correlated with suppressed NK cell function.Citation19,Citation52 Our study reproduced the degranulation defect in CLL and SLL patients, but did not confirm any reduced expression of NKp44, perforin, or granzyme B by NK cells (data not shown; NKp30 and NKp46 were not assayed). Suppressed NK cell-mediated ADCC responses could be particularly problematic in CLL and SLL patients, since several antibody therapies (e.g., rituximab, alemtuzumab) rely at least partially on NK cell-mediated ADCC responses through FcγRIIIA (CD16).Citation29 To exploit NK cell-mediated ADCC, obinutuzumab was developed as an anti-CD20 antibody glycoengineered with higher Fc affinity for CD16, and this antibody was found to be superior to rituximab in recent clinical trials in CLL patients.Citation53
We also found selective reduction of NKG2D expression levels on CD56dim NK cells from CLL patients, but not CD56bright NK cells or T cells in CLL patients or CD56dim NK cells from SLL patients. Our result is consistent with a recent report describing reduced expression of NKG2D on total NK cells in CLL, but not SLL patients.Citation20,Citation34 Previous studies have reported that CLL tumors express NKG2D ligands, and increased soluble forms of NKG2D ligands have been observed in the serum of CLL patients, which correlated with poor treatment-free survival.Citation19,Citation52,Citation54,Citation55 Soluble NKG2D ligands can down modulate surface expression of NKG2D,Citation55 but soluble ligand would be expected to affect NKG2D equivalently on both CD56dim and CD56bright NK, as well as T cells. Alternatively, an interesting report by Nakamura et al. demonstrated that mouse NK cells can acquire NKG2D ligands from tumors by trogocytosis, thereby making themselves susceptible to fratricidal attack by other NK cells.Citation56 Trogocytosis of NKG2D ligands required direct contact with tumor cells and resulted in clathrin-dependent down modulation of NKG2D, which could explain the selective loss on the cytolytic CD56dim NK cells in CLL through their subsequent death by fratricide.
It is particularly intriguing that inhibitory KIR-expressing CD56dim NK cells were progressively lost in our CLL patients. In addition to their classic role in tolerizing NK cells to normal MHC-I-expressing cells in the body, self-recognition of MHC-I by KIR and other inhibitory receptors promotes the education of developing NK cells to achieve a fully functional state.Citation57 NK cells expressing KIR2DL1 and/or KIR3DL1 were found at strikingly low frequency and reduced viability in the peripheral blood of CLL patients. Unfortunately, the surface expression levels of other inhibitory KIR could not be specifically analyzed in our study, due to the lack of specificity of available anti-KIR antibodies. Our data demonstrate, however, that the mature inhibitory KIR+ NK cells are most sensitive to AICD, but their MHC-I ligands are not obviously driving NK cell dysfunction (, S3 and S4). Fully assessing a role for education by KIR ligands on NK cell dysfunction may, however, require a larger cohort of CLL patients and systematic analysis of all inhibitory KIR as more specific antibodies become available. Although not assessed in our study, a recent report found no differences in expression of the NKG2A/CD94 inhibitory receptor on NK cells when comparing CLL patients and healthy controls, but demonstrated upregulation of its ligand, HLA-E, on CLL tumor cells as a potential NK cell evasion mechanism.Citation58
Our data suggest that immunotherapies that promote NK cell survival and function could improve immune responses toward CLL, especially treatments that enhance function or improve viability of the mature KIR+ NK cells. NK cell boosting therapies may be especially beneficial to combat minimal residual disease after tumor-depleting therapies, or perhaps to prevent progression of indolent disease. Therapies to potentiate NK cell function in CLL patients could include lenalidomide, bortezomib, and cytokines.Citation59-62
Patients and methods
Study design
These studies were conducted in a laboratory that operates under exploratory research principles using established laboratory protocols. NK and T cell frequencies, phenotypes, and function were analyzed by multiparameter flow cytometry in fresh PBMC from 25 untreated CLL patients, 10 untreated SLL patients, and 17 healthy controls of similar median age (details in Table S1). Each patient was assayed up to three times at 3–6 mo intervals. Since the expression of most biomarkers did not vary appreciably in serial samples, mean values from each patient were used for comparisons to improve precision.
Patient diagnosis
Untreated CLL or SLL patients (Table S1) were diagnosed in the Fox Chase Cancer Center (FCCC) Clinical Pathology Flow Cytometry Laboratory based on six-color flow cytometry, which identified kappadim or lambdadim CD5+CD19+CD20dimCD22dimCD79adimCD10− neoplastic cells. CLL was diagnosed with >5 × 109/L mature small monoclonal B cells, while SLL was defined by monoclonal B cells with the same immunophenotype found on biopsy of abnormal lymph node, bone marrow, or other tissue, with <5 × 109/L circulating tumor cells, after review by an experienced hematopathologist (T.A-S.), using established guidelines.Citation3 Cases of monoclonal B lymphocytosis were excluded.
Blood sample preparation and genotyping
Fresh blood samples (20 mL) were drawn by venipuncture from patients and healthy donors into heparinized tubes, kept at room temperature, and processed within 6 h. Written informed consent followed HIPAA-compliant procedures approved by the FCCC Institutional Review Board. Whole blood (20 mL) was mixed in equal proportions with complete RPMI 1640 medium [supplemented with 10% fetal bovine serum (pre-tested by the FCCC Cell Culture Facility to support optimal growth rates of seven cell lines), 100 μg/mL penicillin/streptomycin, 2 mM L-glutamine, 10 mM HEPES, 1 mM sodium pyruvate, and 50 μM 2-mercaptoethanol], and PBMC were isolated on Lymphoprep (Axis-shield POC AS, Oslo, Norway) as described.Citation63 The buffy coat was removed, resuspended with staining buffer (RPMI 1640 without biotin or phenol red, and supplemented with 2.0 g/L NaHCO3 and 2.4 g/L HEPES, pH 7.0), and viable cells were counted in the presence of trypan blue dye by microscopic analysis using a hemocytometer. Viability of total lymphocytes by trypan blue exclusion was always >95% for healthy and patient samples. The FCCC Genomics Facility performed KIR genotyping with SSP kits (Invitrogen) and HLA-C/-B genotyping with SSP UniTray kits (Life Technologies) using DNA prepared from total PBMC. Only donors with positive genotype for either KIR2DL1 or KIR3DL1 were incorporated into data sets describing their expression. Since the “KIR2DL1-specific” antibody (clone #143221) also stains KIR2DS5,Citation64 KIR2DL1 and KIR2DL1-ligand data sets only include KIR2DL1+KIR2DL5− individuals (, , , and S2–S4).
Monozygotic twins
One CLL patient has a healthy monozygotic twin [confirmed by GFI Lab (Somerset, PA)], who also participated as a healthy donor. Two concurrent samples from these siblings were processed together, including additional assays. First, annexin V was added to the degranulation assays to assess apoptosis. Second, to determine if the healthy twin's NK cells degranulate toward tumor from the leukemic sibling, PBMC from the CLL twin were stained with CellTracker Blue (Life Technologies) and mixed at a 1:1 leukocyte ratio with effector cells from each twin in degranulation assays. Here, PBMC of the CLL twin were pre-enriched for NK cells using a MACS negative-selection kit (Miltenyi, San Diego, CA) to achieve a similar percentage of NK cells in effector cell populations from both twins (7.5 ± 1.5%). Data points from the twin donors are indicated in the figures by filled symbols.
Antibodies and cell staining
One million cell aliquots were stained with antibodies (Table S2) in approximately 200 µL staining buffer (RPMI-1640 without biotin or phenol red and supplemented with 2.0 g/L NaHCO3 and 2.4 g/L HEPES, pH 7.0) on ice for 20 min and rinsed twice in ice cold staining buffer supplemented with 100 ng/mL propidium iodide (Invitrogen) in the second rinsing step, as described previously.Citation63 Cell staining protocols were optimized in previous experiments and performed according to a consistent SOP. Absolute lymphocyte counts in whole blood were determined using a BD Multitest IMK kit (#340503) and Treg cells were quantified with the eBioscience FoxP3 fix/perm kit according to manufacturer's instructions. Some stains were added to the panel after the study began and therefore have fewer data points collected. Representative staining and gating are shown in Fig. S7.
Degranulation assay
The degranulation assay was modified from Bryceson et al.Citation41 Freshly isolated PBMC (106 cells/sample) were incubated in complete RPMI + 10% autologous plasma for 2 h at 37 °C in a 7% CO2 incubator, either alone (negative control), with 1 μg/mL rituximab, with 106 721.221 EBV-transformed human B cell line targets, or with rituximab and 721.221 cells. A control tube containing only 721.221 target cells was also included, and the final volume in all tubes was 200μL culture medium. Staining reagents (Table S2) were added during the last 30 min, then cells were centrifuged, rinsed twice (100 ng/mL propidium iodide in second rinse), and analyzed by flow cytometry. NK cell degranulation was quantified as percentage of cells staining for surface expression of LAMP-1 (Lysosomal-Associated Membrane Protein 1; CD107a), with the LAMP-1 expression based on a gate in which <0.1% of the unstimulated CD56bright NK cells (negative control) showed positive LAMP-1 staining.
Flow cytometry instrumentation and data analysis
Stained cells were analyzed on a BD FACSAria II flow cytometer containing 4 diode lasers with excitation wavelengths at 633 nm, 488 nm, 405 nm, and 355 nm (355 nm laser was not used for these experiments), and absolute lymphocyte counts were analyzed on a BD FACSCalibur flow cytometer, as described previously.Citation63 The cytometers are located in the FCCC Clinical Pathology Flow Cytometry Laboratory, where they are CLIA- and CAP-certified, and calibrated daily with fluorescent BD CompBeads for clinical diagnostic use. Between 100,000 and 400,000 events were acquired from each sample at 500–2,500 events/sec using 70 psi pressure. Compensation and PMT voltage settings were consistent for acquisition of all samples, as optimized at the beginning of the project based upon analysis of unstained, single stained, and multi-stained PBMC samples. Data were collected with BD FACS Diva software version 6 and were analyzed with FlowJo (v9.2 or later; Tree Star, Inc.), Microsoft Excel (v12), GraphPad Prism (v5.0d; GraphPad Software, Inc.), and R (The R Foundation; www.r-project.org). Positive staining was determined either by selecting the expressing subset of a clearly bimodal population, or by comparison to cellular subsets in the same tube that do not express the marker in question. All raw data were processed manually to ensure data consistency and raw data can be provided per request. Mean values used for statistical analysis are provided in Table S4. Biomarker parameters were quantified as percentage of expressing cells or mean fluorescence intensity (MFI) of staining (Table S3), and these were compared between the three groups (healthy controls, SLL patients, and CLL patients) to identify statistically significant differences.
Statistical methods
Unpaired Wilcoxon rank-sum tests were used for statistical comparisons between donor groups. A paired rank-sum test was used to determine the significance of change over time, with the initial and subsequent samples from each donor constituting a pair. The Spearman test was used to determine statistically significant correlations between parameters. p values < 0.05 were considered to be significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KONI_A_1330235_supplementary_data.zip
Download Zip (6.7 MB)Acknowledgments
We thank JoEllen Weaver, Mary Gilroy, Barbara Dettore, Loretha Moore, and Ryan Winters from the FCCC Biosample Repository for coordinating patient consenting and donations, Drs. Richard Hardy, Poliana Patah, and Valentin Robu for advice, and support from the FCCC Cell Culture, Genomics, Flow Cytometry, and Biostatistics & Bioinformatics Facilities. This manuscript is dedicated to the memory of Dr. Elena Gitelson, who inspired our successful search for a patient with a healthy identical twin.
Funding
This work was supported by the FCCC Keystone Program in Blood Cell Development and Cancer, the Main Line and Bucks County Board of Associates, the National Institutes of Health under Grants CA083859 (K.S.C.) and CA06927 (FCCC), and a Health Research Formula Fund (CURE) grant from the PA Department of Health (K.S.C.).
References
- Rai KR, Jain P. Chronic lymphocytic leukemia (CLL) – then and now. Am J Hematol 2016; 91:330–40; PMID:26690614; https://doi.org/10.1002/ajh.24282
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: Cancer J Clin 2015; 65:5–29; PMID:25559415; https://doi.org/10.3322/caac.21254
- Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H, Hillmen P, Keating MJ, Montserrat E, Rai KR et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International workshop on chronic lymphocytic leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood 2008; 111:5446–56; PMID:18216293; https://doi.org/10.1182/blood-2007-06-093906
- Santos FP, O'Brien S. Small lymphocytic lymphoma and chronic lymphocytic leukemia: are they the same disease? Cancer J 2012; 18:396–403; PMID:23006943; https://doi.org/10.1097/PPO.0b013e31826cda2d/bib>
- Ghobrial IM, Bone ND, Stenson MJ, Novak A, Hedin KE, Kay NE, Ansell SM. Expression of the chemokine receptors CXCR4 and CCR7 and disease progression in B-cell chronic lymphocytic leukemia/ small lymphocytic lymphoma. Mayo Clin Proc 2004; 79:318–25; PMID:15008605; https://doi.org/10.4065/79.3.318
- Trentin L, Cabrelle A, Facco M, Carollo D, Miorin M, Tosoni A, Pizzo P, Binotto G, Nicolardi L, Zambello R et al. Homeostatic chemokines drive migration of malignant B cells in patients with non-Hodgkin lymphomas. Blood 2004; 104:502–8; PMID:15001469; https://doi.org/10.1182/blood-2003-09-3103
- Wong S, Fulcher D. Chemokine receptor expression in B-cell lymphoproliferative disorders. Leuk Lymphoma 2004; 45:2491–6; PMID:15621766; https://doi.org/10.1080/10428190410001723449/bib>
- Riches JC, Davies JK, McClanahan F, Fatah R, Iqbal S, Agrawal S, Ramsay AG, Gribben JG. T cells from CLL patients exhibit features of T-cell exhaustion but retain capacity for cytokine production. Blood 2013; 121:1612–21; PMID:23247726; https://doi.org/10.1182/blood-2012-09-457531
- Nunes C, Wong R, Mason M, Fegan C, Man S, Pepper C. Expansion of a CD8(+)PD-1(+) replicative senescence phenotype in early stage CLL patients is associated with inverted CD4:CD8 ratios and disease progression. Clin Cancer Res 2012; 18:678–87; PMID:22190592; https://doi.org/10.1158/1078-0432.CCR-11-2630
- Beyer M, Kochanek M, Darabi K, Popov A, Jensen M, Endl E, Knolle PA, Thomas RK, von Bergwelt-Baildon M, Debey S et al. Reduced frequencies and suppressive function of CD4+CD25hi regulatory T cells in patients with chronic lymphocytic leukemia after therapy with fludarabine. Blood 2005; 106:2018–25; PMID:15914560; https://doi.org/10.1182/blood-2005-02-0642
- Ramsay AG, Clear AJ, Fatah R, Gribben JG. Multiple inhibitory ligands induce impaired T-cell immunologic synapse function in chronic lymphocytic leukemia that can be blocked with lenalidomide: establishing a reversible immune evasion mechanism in human cancer. Blood 2012; 120:1412–21; PMID:22547582; https://doi.org/10.1182/blood-2012-02-411678
- Gorgun G, Holderried TA, Zahrieh D, Neuberg D, Gribben JG. Chronic lymphocytic leukemia cells induce changes in gene expression of CD4 and CD8 T cells. J Clin Invest 2005; 115:1797–805; PMID:15965501; https://doi.org/10.1172/JCI24176
- Christopoulos P, Pfeifer D, Bartholome K, Follo M, Timmer J, Fisch P, Veelken H. Definition and characterization of the systemic T-cell dysregulation in untreated indolent B-cell lymphoma and very early CLL. Blood 2011; 117:3836–46; PMID:21270444; https://doi.org/10.1182/blood-2010-07-299321
- Katrinakis G, Kyriakou D, Papadaki H, Kalokyri I, Markidou F, Eliopoulos GD. Defective natural killer cell activity in B-cell chronic lymphocytic leukaemia is associated with impaired release of natural killer cytotoxic factor(s) but not of tumour necrosis factor-alpha. Acta Haematologica 1996; 96:16–23; PMID:8677756; https://doi.org/10.1159/000203709
- Kay NE, Zarling JM. Impaired natural killer activity in patients with chronic lymphocytic leukemia is associated with a deficiency of azurophilic cytoplasmic granules in putative NK cells. Blood 1984; 63:305–9; PMID:6607080.
- Maki G, Hayes GM, Naji A, Tyler T, Carosella ED, Rouas-Freiss N, Gregory SA. NK resistance of tumor cells from multiple myeloma and chronic lymphocytic leukemia patients: implication of HLA-G. Leukemia 2008; 22:998–1006; PMID:18288133; https://doi.org/10.1038/leu.2008.15
- Ziegler HW, Kay NE, Zarling JM. Deficiency of natural killer cell activity in patients with chronic lymphocytic leukemia. Int J Cancer 1981; 27:321–7; PMID:6169660; https://doi.org/10.1002/ijc.2910270310
- Costello RT, Knoblauch B, Sanchez C, Mercier D, Le Treut T, Sebahoun G. Expression of natural killer cell activating receptors in patients with chronic lymphocytic leukaemia. Immunology 2012; 135:151–7; PMID:22044312; https://doi.org/10.1111/j.1365-2567.2011.03521.x
- Veuillen C, Aurran-Schleinitz T, Castellano R, Rey J, Mallet F, Orlanducci F, Pouyet L, Just-Landi S, Coso D, Ivanov V et al. Primary B-CLL resistance to NK cell cytotoxicity can be overcome in vitro and in vivo by priming NK cells and monoclonal antibody therapy. J Clin Immunol 2012; 32:632–46; PMID:22318393; https://doi.org/10.1007/s10875-011-9624-5
- Parry HM, Stevens T, Oldreive C, Zadran B, McSkeane T, Rudzki Z, Paneesha S, Chadwick C, Stankovic T, Pratt G et al. NK cell function is markedly impaired in patients with chronic lymphocytic leukaemia but is preserved in patients with small lymphocytic lymphoma. Oncotarget 2016; 7:68513–26; PMID:27655680; https://doi.org/10.18632/oncotarget.12097
- Le Garff-Tavernier M, Decocq J, de Romeuf C, Parizot C, Dutertre CA, Chapiro E, Davi F, Debré P, Prost JF, Teillaud JL et al. Analysis of CD16+CD56dim NK cells from CLL patients: evidence supporting a therapeutic strategy with optimized anti-CD20 monoclonal antibodies. Leukemia 2011; 25:101–9; PMID:20975664; https://doi.org/10.1038/leu.2010.240
- Lee J, Zhang T, Hwang I, Kim A, Nitschke L, Kim M, Scott JM, Kamimura Y, Lanier LL, Kim S. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity 2015; 42:431–42; PMID:25786175; https://doi.org/10.1016/j.immuni.2015.02.013
- Schlums H, Cichocki F, Tesi B, Theorell J, Beziat V, Holmes TD, Han H, Chiang SC, Foley B, Mattsson K et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity 2015; 42:443–56; PMID:25786176; https://doi.org/10.1016/j.immuni.2015.02.008
- Wlasiuk P, Tomczak W, Zajac M, Dmoszynska A, Giannopoulos K. Total expression of HLA-G and TLR-9 in chronic lymphocytic leukemia patients. Hum Immunol 2013; 74:1592–7; PMID:23994589; https://doi.org/10.1016/j.humimm.2013.08.277
- Nuckel H, Rebmann V, Durig J, Duhrsen U, Grosse-Wilde H. HLA-G expression is associated with an unfavorable outcome and immunodeficiency in chronic lymphocytic leukemia. Blood 2005; 105:1694–8; PMID:15466928; https://doi.org/10.1182/blood-2004-08-3335
- Buechele C, Baessler T, Wirths S, Schmohl JU, Schmiedel BJ, Salih HR. Glucocorticoid-induced TNFR-related protein (GITR) ligand modulates cytokine release and NK cell reactivity in chronic lymphocytic leukemia (CLL). Leukemia 2012; 26:991–1000; PMID:22064350; https://doi.org/10.1038/leu.2011.313
- Werlenius O, Aurelius J, Hallner A, Akhiani AA, Simpanen M, Martner A, Andersson PO, Hellstrand K, Thorén FB. Reactive oxygen species induced by therapeutic CD20 antibodies inhibit natural killer cell-mediated antibody-dependent cellular cytotoxicity against primary CLL cells. Oncotarget 2016; 7:32046–53; PMID:27097113; https://doi.org/10.18632/oncotarget.8769
- Dall'Ozzo S, Tartas S, Paintaud G, Cartron G, Colombat P, Bardos P, Watier H, Thibault G. Rituximab-dependent cytotoxicity by natural killer cells: influence of FCGR3A polymorphism on the concentration-effect relationship. Cancer Res 2004; 64:4664–9; PMID:15231679; https://doi.org/10.1158/0008-5472.CAN-03-2862
- Hatjiharissi E, Xu L, Santos DD, Hunter ZR, Ciccarelli BT, Verselis S, Modica M, Cao Y, Manning RJ, Leleu X et al. Increased natural killer cell expression of CD16, augmented binding and ADCC activity to rituximab among individuals expressing the FcγRIIIa-158 V/V and V/F polymorphism. Blood 2007; 110:2561–4; PMID:17475906; https://doi.org/10.1182/blood-2007-01-070656
- Wild J, Schmiedel BJ, Maurer A, Raab S, Prokop L, Stevanovic S, Dörfel D, Schneider P, Salih HR. Neutralization of (NK-cell-derived) B-cell activating factor by belimumab restores sensitivity of chronic lymphoid leukemia cells to direct and rituximab-induced NK lysis. Leukemia 2015; 29:1676–83; PMID:25710310; https://doi.org/10.1038/leu.2015.50
- Wu L, Adams M, Carter T, Chen R, Muller G, Stirling D, Schafer P, Bartlett JB. lenalidomide enhances natural killer cell and monocyte-mediated antibody-dependent cellular cytotoxicity of rituximab-treated CD20+ tumor cells. Clin Cancer Res 2008; 14:4650–7; PMID:18628480; https://doi.org/10.1158/1078-0432.CCR-07-4405
- Chavez JC, Piris-Villaespesa M, Dalia S, Powers J, Turba E, Nodzon L, Komrokji R, Sokol L, Locke FL, Lancet J et al. Results of a phase II study of lenalidomide and rituximab for refractory/relapsed chronic lymphocytic leukemia. Leuk Res 2016; 47:78–83; PMID:27285853; https://doi.org/10.1016/j.leukres.2016.05.012
- Gonzalez-Rodriguez AP, Contesti J, Huergo-Zapico L, Lopez-Soto A, Fernandez-Guizan A, Acebes-Huerta A, Gonzalez-Huerta AJ, Gonzalez E, Fernandez-Alvarez C, Gonzalez S. Prognostic significance of CD8 and CD4 T cells in chronic lymphocytic leukemia. Leuk Lymphoma 2010; 51:1829–36; PMID:20846097; https://doi.org/10.3109/10428194.2010.503820/bib>
- Huergo-Zapico L, Acebes-Huerta A, Gonzalez-Rodriguez AP, Contesti J, Gonzalez-Garcia E, Payer AR, Villa-Alvarez M, Fernández-Guizán A, López-Soto A, Gonzalez S. Expansion of NK cells and reduction of NKG2D expression in chronic lymphocytic leukemia. Correlation with progressive disease. PLoS One 2014; 9:e108326; PMID:25286418; https://doi.org/10.1371/journal.pone.0108326
- Palmer S, Hanson CA, Zent CS, Porrata LF, Laplant B, Geyer SM, Markovic SN, Call TG, Bowen DA, Jelinek DF et al. Prognostic importance of T and NK-cells in a consecutive series of newly diagnosed patients with chronic lymphocytic leukaemia. Br J Haematol 2008; 141:607–14; PMID:18384436; https://doi.org/10.1111/j.1365-2141.2008.07070.x
- Caligiuri MA. Human natural killer cells. Blood 2008; 112:461–9; PMID:18650461; https://doi.org/10.1182/blood-2007-09-077438
- Silva A, Andrews DM, Brooks AG, Smyth MJ, Hayakawa Y. Application of CD27 as a marker for distinguishing human NK cell subsets. Int Immunol 2008; 20:625–30; PMID:18326863; https://doi.org/10.1093/intimm/dxn022
- Vossen MT, Matmati M, Hertoghs KM, Baars PA, Gent MR, Leclercq G, Hamann J, Kuijpers TW, van Lier RA. CD27 defines phenotypically and functionally different human NK cell subsets. J Immunol 2008; 180:3739–45; PMID:18322179; https://doi.org/10.4049/jimmunol.180.6.3739
- Giannopoulos K, Schmitt M, Kowal M, Wlasiuk P, Bojarska-Junak A, Chen J, Rolinski J, Dmoszynska A. Characterization of regulatory T cells in patients with B-cell chronic lymphocytic leukemia. Oncology Rep 2008; 20:677–82; PMID:18695923; https://doi.org/10.3892/or_00000059
- Alter G, Malenfant JM, Altfeld M. CD107a as a functional marker for the identification of natural killer cell activity. J Immunol Methods 2004; 294:15–22; PMID:15604012; https://doi.org/10.1016/j.jim.2004.08.008
- Bryceson YT, Fauriat C, Nunes JM, Wood SM, Bjorkstrom NK, Long EO, Ljunggren HG. Functional analysis of human NK cells by flow cytometry. Methods Mol Biol 2010; 612:335–52; PMID:20033652; https://doi.org/10.1007/978-1-60761-362-6_23
- Eischen CM, Schilling JD, Lynch DH, Krammer PH, Leibson PJ. Fc receptor-induced expression of Fas ligand on activated NK cells facilitates cell-mediated cytotoxicity and subsequent autocrine NK cell apoptosis. J Immunol 1996; 156:2693–9; PMID:8609385.
- Jewett A, Cavalcanti M, Giorgi J, Bonavida B. Concomitant killing in vitro of both gp120-coated CD4+ peripheral T lymphocytes and natural killer cells in the antibody-dependent cellular cytotoxicity (ADCC) system. J Immunol 1997; 158:5492–500; PMID:9164972.
- Warren H. Target-induced natural killer cell loss as a measure of NK cell responses. Curr Protocols Immunol 2013; Chapter 14.29.1-21; PMID:23564684; https://doi.org/10.1002/0471142735.im1429s101.
- Poggi A, Massaro AM, Negrini S, Contini P, Zocchi MR. Tumor-induced apoptosis of human IL-2-activated NK cells: role of natural cytotoxicity receptors. J Immunol 2005; 174:2653–60; PMID:15728472; https://doi.org/10.4049/jimmunol.174.5.2653
- Purdy AK, Campbell KS. Natural killer cells and cancer: regulation by the killer cell Ig-like receptors (KIR). Cancer Biol Ther 2009; 8:2211–20; PMID:19923897; https://doi.org/10.4161/cbt.8.23.10455
- Ranheim EA, Cantwell MJ, Kipps TJ. Expression of CD27 and its ligand, CD70, on chronic lymphocytic leukemia B cells. Blood 1995; 85:3556–65; PMID:7540066.
- Jang YS, Kang W, Chang DY, Sung PS, Park BC, Yoo SH, Park YW, Shin EC. CD27 engagement by a soluble CD70 protein enhances non-cytolytic antiviral activity of CD56bright natural killer cells by IFN-gamma secretion. Clin Immunol 2013; 149:379–87; PMID:24211844; https://doi.org/10.1016/j.clim.2013.09.007
- Jenkins MR, Rudd-Schmidt JA, Lopez JA, Ramsbottom KM, Mannering SI, Andrews DM, Voskoboinik I, Trapani JA. Failed CTL/NK cell killing and cytokine hypersecretion are directly linked through prolonged synapse time. J Exp Med 2015; 212:307–17; PMID:25732304; https://doi.org/10.1084/jem.20140964
- Rojas R, Roman J, Torres A, Ramirez R, Carracedo J, Lopez R, Garcia JM, Martin C, Pintado O. Inhibition of apoptotic cell death in B-CLL by interferon gamma correlates with clinical stage. Leukemia 1996; 10:1782–8; PMID:8892682.
- Zaki M, Douglas R, Patten N, Bachinsky M, Lamb R, Nowell P, Moore J. Disruption of the IFN-gamma cytokine network in chronic lymphocytic leukemia contributes to resistance of leukemic B cells to apoptosis. Leuk Res 2000; 24:611–21; PMID:10867137; https://doi.org/10.1016/S0145-2126(00)00022-9
- Reiners KS, Topolar D, Henke A, Simhadri VR, Kessler J, Sauer M, Bessler M, Hansen HP, Tawadros S, Herling M et al. Soluble ligands for NK cell receptors promote evasion of chronic lymphocytic leukemia cells from NK cell anti-tumor activity. Blood 2013; 121:3658–65; PMID:23509156; https://doi.org/10.1182/blood-2013-01-476606
- Goede V, Fischer K, Busch R, Engelke A, Eichhorst B, Wendtner CM, Chagorova T, de la Serna J, Dilhuydy MS, Illmer T et al. Obinutuzumab plus chlorambucil in patients with CLL and coexisting conditions. N Engl J Med 2014; 370:1101–10; PMID:24401022; https://doi.org/10.1056/NEJMoa1313984
- Nuckel H, Switala M, Sellmann L, Horn PA, Durig J, Duhrsen U, Küppers R, Grosse-Wilde H, Rebmann V. The prognostic significance of soluble NKG2D ligands in B-cell chronic lymphocytic leukemia. Leukemia 2010; 24:1152–9; PMID:20428196; https://doi.org/10.1038/leu.2010.74
- Hilpert J, Grosse-Hovest L, Grunebach F, Buechele C, Nuebling T, Raum T, Steinle A, Salih HR. Comprehensive analysis of NKG2D ligand expression and release in leukemia: implications for NKG2D-mediated NK cell responses. J Immunol 2012; 189:1360–71; PMID:22730533; https://doi.org/10.4049/jimmunol.1200796
- Nakamura K, Nakayama M, Kawano M, Amagai R, Ishii T, Harigae H, Ogasawara K. Fratricide of natural killer cells dressed with tumor-derived NKG2D ligand. Proc Natl Acad Sci USA 2013; 110:9421–6; PMID:23690625; https://doi.org/10.1073/pnas.1300140110
- Shifrin N, Raulet DH, Ardolino M. NK cell self-tolerance, responsiveness and missing self-recognition. Semin Immunol 2014; 26:138–44; PMID:24629893; https://doi.org/10.1016/j.smim.2014.02.007
- McWilliams EM, Mele JM, Cheney C, Timmerman EA, Fiazuddin F, Strattan EJ, Mo X, Byrd JC, Muthusamy N, Awan FT. Therapeutic CD94/NKG2A blockade improves natural killer cell dysfunction in chronic lymphocytic leukemia. Oncoimmunology 2016; 5:e1226720; PMID:27853650; https://doi.org/10.1080/2162402X.2016.1226720
- Mentlik James A, Cohen AD, Campbell KS. Combination immune therapies to enhance anti-tumor responses by NK cells. Front Immunol 2013; 4:481; PMID:24391651; https://doi.org/10.3389/fimmu.2013.00481
- Ardolino M, Azimi CS, Iannello A, Trevino TN, Horan L, Zhang L, Deng W, Ring AM, Fischer S, Garcia KC et al. Cytokine therapy reverses NK cell anergy in MHC-deficient tumors. J Clin Invest 2014; 124:4781–94; PMID:25329698; https://doi.org/10.1172/JCI74337
- Eskelund CW, Nederby L, Thysen AH, Skovbo A, Roug AS, Hokland ME. Interleukin-21 and rituximab enhance NK cell functionality in patients with B-cell chronic lymphocytic leukaemia. Leuk Res 2011; 35:914–20; PMID:21354618; https://doi.org/10.1016/j.leukres.2011.02.006
- Laprevotte E, Voisin G, Ysebaert L, Klein C, Daugrois C, Laurent G, Fournie JJ, Quillet-Mary A. Recombinant human IL-15 trans-presentation by B leukemic cells from chronic lymphocytic leukemia induces autologous NK cell proliferation leading to improved anti-CD20 immunotherapy. J Immunol 2013; 191:3634–40; PMID:23997218; https://doi.org/10.4049/jimmunol.1300187
- MacFarlane AW IV, Jillab M, Plimack ER, Hudes GR, Uzzo RG, Litwin S, Dulaimi E, Al-Saleem T, Campbell KS. PD-1 expression on peripheral blood cells increases with stage in renal cell carcinoma patients and is rapidly reduced after surgical tumor resection. Cancer Immunol Res 2014; 2:320–31; PMID:24764579; https://doi.org/10.1158/2326-6066.CIR-13-0133
- Czaja K, Borer AS, Schmied L, Terszowski G, Stern M, Gonzalez A. A comprehensive analysis of the binding of anti-KIR antibodies to activating KIRs. Genes Immun 2014; 15:33–7; PMID:24173145; https://doi.org/10.1038/gene.2013.58
