ABSTRACT
PTX3 is a component of the humoral arm of innate immunity and an extrinsic oncosuppressor gene taming tumor-promoting inflammation. Here, we show that two enhancers differently regulate PTX3 expression: enhancer 1, located 230 kb upstream of PTX3 promoter, mediated the action of inflammatory transcription factors; and enhancer 2, encompassing PTX3 second exon, was implicated in pre-initiation complex assembly. Polycomb repressive complex 2 silenced these regulatory elements and the promoter in basal condition. Enhancer 1 was epigenetically inactivated in early colorectal cancer (CRC) stages, while the promoter and enhancer 2 showed increasingly DNA methylation during CRC progression from adenomas to stage II and III CRC. Inhibition of DNA methylation rescued PTX3 expression in CRC. Finally, enhancer 1 acquired the binding of STAT3 in stage I CRC, and inhibition of STAT3 phosphorylation restored PTX3 activity and decreased enhancer 1 methylation. Thus, the expression of PTX3 is under the control of two enhancers, which emerge as important fine regulators of PTX3 expression in inflammation and cancer.
Introduction
The long pentraxin-3 (PTX3) is a component of the humoral arm of innate immunity, playing non-redundant roles in innate resistance against selected microbes and in the regulation of inflammation and tissue remodeling.Citation1-4 PTX3 is produced by different cell types, including various myeloid cells (dendritic cells, monocytes, macrophages, neutrophils), epithelial cells, vascular and lymphatic endothelial cells, and mesenchymal cells (e.g., fibroblasts and adipocytes), upon exposure to inflammatory signals (e.g., TNFα, IL-1β) and TLR agonists (e.g., LPS, OmpA).Citation2,4-6 The kinetics of PTX3 induction is rapid and transient in most in vitro and in vivo inflammatory conditions.Citation2,4,7 Recently, it was found that PTX3 has a key role in cancer-related inflammation (CRI), acting as an extrinsic oncosuppressor gene, dampening CRI through the regulation of the complement cascade.Citation8 PTX3-deficient mice showed increased susceptibility to carcinogenesis, and PTX3 gene was silenced in selected human tumors (e.g., CRC and leiomyosarcoma) by DNA methylation of the promoter and a putative enhancer.Citation8 However, the mechanisms underlying the regulation of PTX3 expression by these regulatory regions have not been addressed yet.
Enhancers are fundamental DNA elements that can determine tissue-specific gene expression. Differentiated cells have a unique repertoire of enhancersCitation9-11 dictated by the activation of different TFs that work in a cell-type-specific fashion and under specific conditions.Citation12,13 They act in cis or in trans, forming a chromatin loop with the promoter of the target genes and promoting the recruitment of the transcriptional machinery to activate genes.Citation14 Recent studies revealed that enhancers are involved in many aspects of the immune response, such as defining tissue-resident macrophage identity,Citation15,16 and mediating the effect of STATs in helper T cell differentiation.Citation17
In an effort to characterize the mechanisms underlying the regulation of PTX3 expression by regulatory regions, we characterized PTX3 enhancers and investigated their mode of action. Here, we show that the expression of PTX3 in inflammation is governed by a promoter and two enhancers, which play different roles in gene transcription: enhancer 1 amplifies the effects of inflammatory transcription factors (TFs), while enhancer 2 regulates PTX3 transcription through the recruitment of a key component of the pre-initiation complex. Silencing of these regulatory elements through DNA methylation occurs in a stage-specific manner in CRC and is responsible for downregulation of PTX3. In addition, STAT3 is responsible of DNA methylation of enhancer 1 in an early stage of cancer progression.
Thus, this study shows the role of enhancers and their epigenetic modifications in modulating PTX3 expression in healthy and pathological conditions.
Results
Identification of two enhancers regulating PTX3 expression in inflammatory conditions
We previously showed that PTX3 expression is silenced in selected human cancers through methylation of the promoter and a putative enhancer encompassing the second exon.Citation8 In order to address the role of enhancers in regulating PTX3 expression, we examined a chromatin immunoprecipitation (ChIP)-sequence (ChIP-seq) data set for H3K4me1 and H3K27Ac, two histone modifications that define active enhancers, in CRC compared to healthy epithelium.Citation18 We then used the Genomic Regions Enrichment of Annotations Tool (GREAT), a web-tool that assigns biological meaning to a set of non-coding genomic regions,Citation19 and we found two active enhancers potentially associated with PTX3 gene in healthy colon epithelium. The first, named enhancer 1, is located 230 kb upstream of the PTX3 gene promoter (chr3:156,893,570-156,894,644). The second, named enhancer 2, is located 350 bp downstream the PTX3 Transcription Start Site (TSS) and encompasses the second PTX3 exon (chr3:157,154,796-157,156,606) (Fig. S1A).
The analysis of these two enhancers revealed that the central region of enhancer 1 is well conserved among species (frog, chicken, opossum, dog, rat, mouse, rhesus macaque, and human), and the enhancer 2 is conserved in mouse, rhesus macaque and dog (Fig. S1B).
To address the functional activity of these enhancers and confirm the association suggested by GREAT, we examined the profile of histone markers of with these regions in PTX3-expressing cells. Human primary macrophages, the fibrosarcoma cell line 8387, and colonic epithelial HCoEpiC cells were treated for 4 h with TNFα, a pro-inflammatory cytokine rapidly inducing PTX3 in several cell types,Citation2,20 and then ChIP assays was performed to determine the levels of the histone markers characterizing active (H3K4me1, H3K27Ac) and inactive (H3K27me3) enhancersCitation11,21 ( and Figs. S2A and B). For enhancer 1, we analyzed the distribution of these histone markers in three different regions: in a central region (chr3: 156893578–156893704), the most conserved among species, and two 500 bp flanking regions ( and Fig. S2A); seven consecutive regions were analyzed in enhancer 2. For the promoter, a CG-enriched region and the TSS were analyzed and the enrichment of markers that characterize active promoters (H3K4me3, H3K9Ac, and H3K27Ac) was used as positive control of the experiment. The enrichment of histone markers in genomic regions nearby those of interest was considered the negative control. As shown in and Figs. S2A and B, we found that the two enhancers switched from an inactive (H3K4me1low, H3K27Aclow, H3K27me3high) to an active state (H3K4me1high, H3K27Achigh, H3K27me3low) after treatment with TNFα in the three cell types analyzed. Also the promoter switched to an active state (H3K4me3high, H3K9Achigh, and H3K27Achigh) upon TNFα treatment. In enhancer 1, the central region (chr3: 156,893,578–156,893,704) had higher levels of histone markers compared with the two flanking regions, indicating that the central region is the effective enhancer, as also suggested by evolutionary conservation of this genomic region (Fig. S1B).
Figure 1. Identification of PTX3 enhancers by histone modification analysis. (A) Analysis of H3K4me1, H3K4me3, H3K9Ac, H3K27Ac, and H3K27me3 histone modifications at enhancer 1, promoter and enhancer 2 by ChIP in human macrophages in basal and inflammatory conditions (TNFα 20 ng/mL, 4h). Results are expressed as fold change relative to IgG (N = 3 experiments). *p ≤ 0.05; Student's t-test. (B) Analysis by RT-qPCR of PTX3 mRNA expression in HCoEpic and 8387 cells and in human macrophages in basal condition and after TNFα treatment (20 ng/mL, 4h). PTX3 mRNA expression is expressed as mean ± SEM (N = 2 experiments). *p ≤ 0.05, **p ≤ 0.01; Student's t-test. (C) Correlation analysis between the enrichment of H3K27Ac on enhancer 1 and the expression of PTX3 mRNA in HCoEpic and 8387 cells and in macrophages. N = 4 experiments for macrophages and 8387 cells and N = 2 for normal human colonic cells.
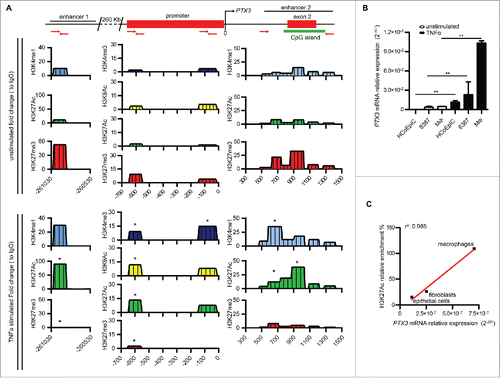
Furthermore, we found a strong and positive correlation between PTX3 RNA expression in the three cell types analyzed after stimulation with TNFα and H3K27ac enrichment in the enhancer 1 (r = 0.9992; p = 0.0176) ( and ), but not in enhancer 2 and promoter (not shown), suggesting a major role of enhancer 1 in mediating the effects of inflammatory TFs on gene transcription.
Taken together, these results suggest that the two enhancers identified are involved in the induction of PTX3 expression in response to sterile inflammation.
Characterization of the factors interacting with PTX3 enhancers
In order to define the mode of action of the two PTX3 enhancers, we next evaluated the presence of TF-binding motifs within these regulatory elements. In silico analysis revealed binding motifs for several TFs on enhancer 1, and among them, many involved in the activation of inflammatory and immune responses, including the NF-κB subunit RelA, c-Jun, c-Fos, PU.1, and SP.1, known to regulate the activity of PTX3 promoter.Citation22,23 To validate the functional role of these predicted binding motifs, we analyzed by ChIP assay the presence of these TFs in PTX3 enhancers and promoter in human macrophages and 8387 cells () in basal conditions and after TNFα stimulation. As shown in , TNFα treatment induced the binding of all the inflammatory TFs tested to the PTX3 promoter. The binding of TFs to enhancer 1 differed in the two cell types analyzed: SP.1 and PU.1 were enriched on enhancer 1 only in macrophages, while c-Jun was found only in 8387 cells. Moreover, an enrichment of NF-κB on enhancer 2 was observed in macrophages but not in 8387 cells (), suggesting that this enhancer, although not having binding motifs for this TF, could be under control of NF-κB in this cell type through indirect binding.
Figure 2. Analysis of inflammatory TFs and transcription preinitiation complexes regulating PTX3 enhancer activity. (A) ChIP assay for inflammatory TFs [NF-κB (p65), c-Fos, c-Jun, Sp1, and Pu.1] in macrophages (top panel) and 8387 cells (bottom panel) in basal and inflammatory (TNFα 20 ng/mL, 4 h) conditions. (B) ChIP assay for TAF1 and RNA Pol II in macrophages in basal and inflammatory conditions. Regions analyzed are reported in the upper part of the panels. Results are expressed as fold change relative to IgG and as mean ± SEM (N = 2 experiments). *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001; Student's t-test.
![Figure 2. Analysis of inflammatory TFs and transcription preinitiation complexes regulating PTX3 enhancer activity. (A) ChIP assay for inflammatory TFs [NF-κB (p65), c-Fos, c-Jun, Sp1, and Pu.1] in macrophages (top panel) and 8387 cells (bottom panel) in basal and inflammatory (TNFα 20 ng/mL, 4 h) conditions. (B) ChIP assay for TAF1 and RNA Pol II in macrophages in basal and inflammatory conditions. Regions analyzed are reported in the upper part of the panels. Results are expressed as fold change relative to IgG and as mean ± SEM (N = 2 experiments). *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001; Student's t-test.](/cms/asset/9a7036df-1a2f-4581-a9c9-59457a9bc1b7/koni_a_1333215_f0002_oc.gif)
Enhancers are transcriptional activators that can regulate the recruitment of RNA Polymerase (Pol) II also through the direct interaction with the components of the preinitiation complex (known also as PIC complex).Citation24,25 Thus, we investigated the possible direct interaction of PTX3 enhancers with PIC by analyzing the binding of the transcription initiation factor TAF1 (also named TAF II p250), the main Tata Binding Protein (TBP)-associated factor, by ChIP assay. We found that TNFα induced the binding of TAF1 only on enhancer 2, suggesting a role of this regulatory element in the assembly of PIC (). We also observed enrichment of RNA Pol II, a marker of active enhancers and promoters,Citation26 on both enhancers and promoter of PTX3 after treatment with TNFα (), in agreement with the activation of these regulatory elements in the presence of an inflammatory stimulus.
In basal conditions, PTX3 regulatory elements were enriched in H3K27me3 (), a repressive histone mark. Polycomb repressive complex 2 (PRC2) is a main player in the deposition of this histone mark,Citation27 and PRC2 inactivation was shown to be required for an inflammatory response to microbial products.Citation28 We thus analyzed the role of this complex in silencing the regulatory regions of PTX3 in basal condition by addressing the binding of PRC2 to PTX3 regulatory elements in macrophages and 8387 cells. ChIP assay performed using antibodies targeting SUZ12 and EZH2, the two major Polycomb group (PcG) subunits, revealed an enrichment of PCR2 subunits on PTX3 enhancers and promoter in basal condition, which was significantly reduced in TNFα-treated macrophages () and 8387 cells (Fig. S3).
Figure 3. PRC2 complex regulates RNA Polymerase II binding on PTX3 regulatory regions. (A) ChIP assay for EZH2 and Suz12 in macrophages in basal and inflammatory (TNFα 20 ng/mL, 4 h) conditions. (B) ChIP assay for H3K27me3, RNA Pol II and TAF1 in 8387 cells in basal condition and after stimulation with GSK343 1 μM for 48 h. Regions analyzed are reported in the upper part of the panels. Results are expressed as fold change relative to IgG and as mean (N = 2 experiments). *p ≤ 0.05, **p ≤ 0.01; Student's t-test.
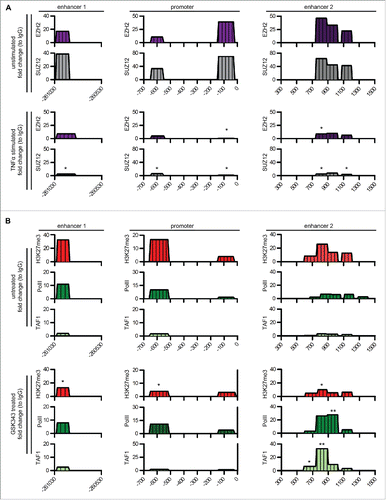
To confirm the functional role of PRC2 in the regulation of PTX3 expression in basal conditions, we treated 8387 cells with GSK343, an inhibitor of EZH2 activity.Citation29,30 GSK343 treatment significantly increased PTX3 mRNA levels (from 2.3 × 10−3 ± 5.8 × 10−4 2ˆ.ΔCt to 7 × 10−3 ± 2.5 × 10−3 2ˆ.ΔCt, p = 0.01). Furthermore, GSK343 treatment significantly reduced the levels of H3K27me3 on both enhancers and the promoter and increased the enrichment of RNA Pol II and TAF1 on enhancer 2 ().
To further assess the importance of PTX3 enhancer 2 in the regulation of PTX3, we addressed whether PTX3 single-nucleotide polymorphisms (SNPs) (rs2305619, rs3816527, rs1840680) located in and close to this regulatory region, and previously reported to influence PTX3 expression,Citation31,32 affected histone mark levels on enhancer 2. In monocyte-derived macrophages, these SNPs in enhancer 2 affected the expression of PTX3 in basal condition (Fig. S4A) and the enrichment of TAF1 and H3K27AC in the central region of enhancer 2 (Figs. S4B and C).
Taken together, these findings provide evidence that enhancer 1 mediates the effect of inflammatory TFs on PTX3 expression and enhancer 2 is implicated in the assembly of PIC complex, and that in the absence of inflammatory stimuli, PRC2 silences these regulatory elements and the PTX3 promoter.
PTX3 enhancers are inactivated by DNA methylation in colorectal cancer
We recently reported that PTX3 is silenced in many tumors through DNA methylation of the promoter and one regulatory region that we have identified as enhancer 2.Citation8 Here, we investigated whether DNA methylation impaired also the activity of enhancer 1 in tumors, and defined whether this epigenetic modification was associated with a specific stage of carcinogenesis. To this aim, we analyzed the methylation level of enhancer 1 in six different mesenchymal and epithelial human tumors () and in CRC at different stages of progression (low- and high-grade adenomas, CRC stage I, II, and III) (). Methylated-CpG island recovery assay (MIRA) revealed that enhancer 1, unmethylated (5%) in the healthy condition, was hypermethylated (46%) in low-grade adenoma, high-grade adenoma and stage I CRC (). In contrast, enhancer 1 showed a hypomethylated (10.3% and 7.5%) pattern in patients with CRC at stage II and III (). This pattern of DNA methylation was different from that observed in enhancer 2 that showed hypomethylation (1.46% and 7.36%,) in low- and high-grade adenoma, and hypermethylation (79.5%, 129.2%, and 113.3%) in CRC stages I, II, and III ().
Figure 4. PTX3 epigenetic modifications in human cancers. (A, B) Analysis by Methylated CpG Island Recovery Assay (MIRA) of the percentage of methylation enrichment of PTX3 regulatory regions in human mesenchymal and epithelial tumors (A) and CRC (B) and in their healthy counterparts. Results are expressed as percentage of enrichment relative to input DNA normalized on a positive control and represented as mean ± SEM. (N = 6 samples for human healthy colon, N = 3 samples for low-grade colon adenoma, N = 5 samples for high-grade adenoma, N = 2 for human healthy counterpart, N = 12–14 for CRC tumor stages). *p <0.05, **p <0.01, Student's t-test. (C) Analysis of H3K4me1, H3K27Ac, and H3K27me3 histone modifications by ChIP in human CRC stages I, II, and III. Results are expressed as fold change relative to IgG and as mean ± SEM. (N = 3 experiments). *p ≤ 0.05, **p ≤ 0.01; Student's t-test. (A–C) Regions analyzed are reported in the upper part of the panels.
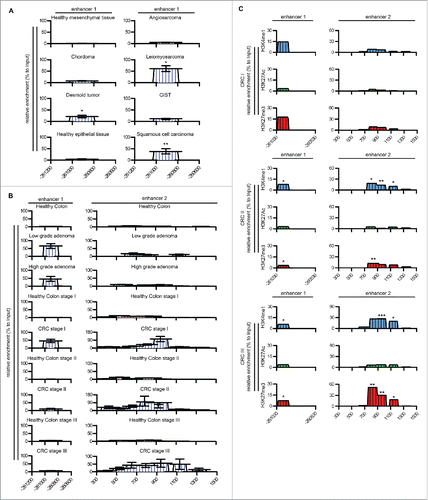
The analysis of the distribution of H3K4me1, H3K27me3, and H3K27ac in CRC samples also showed that the activity of the two enhancers was modified during tumor progression: enhancer 1 was highly enriched in the repressive histone mark H3K27me3 in patients with CRC at stage I, whereas enhancer 2 acquired this epigenetic mark in CRC stage II and III. In all tumor stages, both enhancers had low levels of H3K27Ac (). To address the relevance of these epigenetic modifications in PTX3 silencing, we next treated CRC cells (RKO and HCT116 cell lines) with the inhibitor of DNA methylation 5-Aza-2′-deoxycytidine (5′AZA-dC) and with the inhibitor of histone deacetylases trichostatin A (TSA). As shown in , the treatment with 5′AZA-dC rescued PTX3 mRNA expression in both CRC cell lines to levels similar to those detected in healthy colon cells in basal conditions and strongly upregulated it in TNFα-treated cells. In contrast, TSA did not affect PTX3 mRNA levels in CRC cells in basal or inflammatory conditions, suggesting a dominant role of DNA methylation on histone deacetylation in silencing PTX3 enhancers. 5′AZA-dC was sufficient to induce the activation of enhancer 2 in basal conditions, as shown by enrichment of H3K27Ac and H3K4m1 in ChIP assay, whereas we did not observe changes in the activity of enhancer 1 and promoter after treatment with 5′AZA-dC alone ( and Fig. S5). However, enhancer 1 became active after co-treatment with 5′AZA-dC and TNFα in RKO cells, showing an enrichment of H3K4me1 and H3K27Ac and NF-κB (13.13%, 29.2%, and 32%, respectively), and enhancer 2 was further enriched of H3K27Ac ( and Fig. S5). Furthermore, we observed an enrichment of H3K4me3, H3K9Ac, and H3K27Ac, and of inflammatory TFs (NF-κB, c-Fos, and c-Jun) on the promoter ( and Fig. S6A).
Figure 5. Effect of methylation inhibition on PTX3 mRNA expression and enhancer activity. (A) PTX3 mRNA expression by RKO (left panel) and HCT116 (right panel) cells upon treatment with TSA (150 nM for 48 h) or 5-AZA-dC (15 μM for 72 h) and TNFα (20 ng/mL). Results are expressed as mean ± SEM (N = 2 experiments). *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001; one-way ANOVA. (B) ChIP assay for H3K4me1, H3K4me3, H3K9Ac, H3K27Ac, H3K27me3 in RKO cells upon treatment with TNFα and 5′AZA-dC. Regions analyzed are reported in the upper part of the panels. Results are expressed as fold change relative to IgG and as mean (N = 2 experiments). *,ˆ,$p ≤ 0.05; **,ˆˆ,$$p ≤ 0.01; ***p <0.001. Student's t-test. ˆ: unstimulated vs. 5′AZA-dC; $: 5′AZA-dc vs. 5′AZA-dC + TNFα; *TNFα vs. 5′AZA-dC + TNFα.
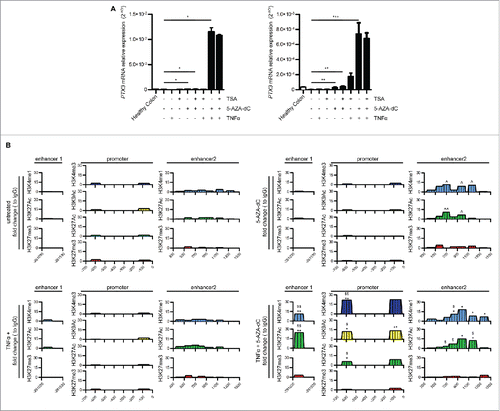
These findings indicate that the repression of PTX3 in CRC is associated with hypermethylation at different regulatory regions depending on the stage of tumor progression, and that the silencing of enhancer 1 could be a critical effector in this process in the early stages of carcinogenesis.
Role of STAT3 in PTX3 gene silencing in CRC
As shown in , enhancer 1 is the first PTX3 regulatory element which undergoes epigenetic modifications and inactivation during CRC progression by DNA hypermethylation. To address the molecular mechanism that causes hypermethylation of enhancer 1 at early stages of intestinal tumorigenesis, we searched for putative-binding motifs for potentially involved proteins present in enhancer 1 sequence. Among the proteins identified using JASPAR matrix models, a putative STAT3-binding site was predicted in enhancer 1 (gtcctgggtaa; chr3:156,893,620-156,893,630) (threshold > 80%). We considered STAT3 a good candidate, because it promotes tumor proliferation, survival, invasion, and immunosuppression, mediates tumor-promoting inflammation, and has been reported to recruit the DNA methyltransferase DNMT1.Citation33-35 To test the role of STAT3 in silencing enhancer 1, we first evaluated its binding to enhancer 1 in RKO and HCT116 CRC cells ( and Fig. S7) and in CRC samples at different stages of progression (stage I, II, and III) (). As negative controls we used samples of healthy colon epithelial cells of CRC patients at each tumor stage and 8387 fibroblastic cells. ChIP assay showed that STAT3 bound to enhancer 1 in RKO (33% enrichment) and HCT116 CRC cells (65%), but not in 8387 cells (1.3% enrichment) ( and Fig. S7). In addition, paraffin-embedded pathology tissue ChIP (PAT-ChIP) assay showed that STAT3 bound to enhancer 1 in samples from stage I CRC (23% enrichment), at lower percentage in stage II samples (9.5%), but not in stage III samples (1.7%). STAT3 enrichment on enhancer 1 correlated (r = 0.9982, p = 0.02) with the relative enrichment of methylated DNA of enhancer 1 in tumor stages I–III. In agreement with in silico analysis of putative TF-binding sites, we did not observe STAT3 binding to the enhancer 2 ( and Fig. S7). Next, to address the causal effect of STAT3 binding to enhancer 1, we treated RKO and HCT116 CRC cells with Stattic, a selective inhibitor of STAT3,Citation36,37 and analyzed PTX3 expression and the methylation status of the regulatory elements. As shown in , treatment with Stattic in combination with TNFα for 4 h rescued PTX3 expression in RKO cells, but was not as efficient as 5′AZA-dC. The treatment did not further increase PTX3 expression in 8387 cells (). Furthermore, treatment with Stattic alone or in combination with TNFα of RKO cells decreased DNA methylation levels of PTX3 enhancer 1 (p = 0.02) but not of promoter and enhancer 2 ().
Figure 6. Role of STAT3 in PTX3 epigenetic modification. (A) ChIP assay for STAT3 in RKO and 8387 cells in basal condition and in CRC stage I, II, and III and in their normal counterparts. Regions analyzed are reported in the upper part of the panels. Results are expressed as fold change relative to IgG and as mean ± SEM (N = 2 experiments for the cell lines and CRC stage II, N = 2 healthy colon samples for each tumor stage and N = 4 for CRC stage I and II). *p ≤ 0.05, **p ≤ 0.01, Student's t-test. (B) PTX3 mRNA expression in RKO and 8387 cells treated with 5-AZA-dC 15 μM for 72 h, Stattic 1 μM for 48 h, and TNFα 20 ng/mL for 4 h, as specified. Results are expressed as mean ± SEM and as fold change of basal conditions (N = 2 experiments). *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001; one-way ANOVA (left panel) or Student's t-test (right panel). (C) Analysis by MIRA of the percentage of methylation enrichment of PTX3 regulatory regions in RKO cells after treatment with stattic and TNFα as specified. Results are expressed as percentage of enrichment relative to input DNA normalized on a positive control and represented as mean ± SEM. (N = 2 experiments) *p < 0.05. Student's t-test.
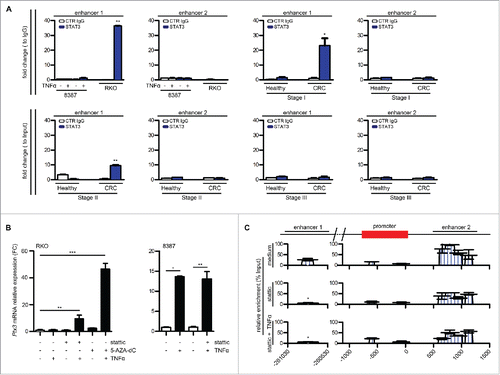
These findings indicated that STAT3 is involved in silencing PTX3 at early stages of CRC through the interaction with enhancer 1, causing its hypermethylation and thus, its inactivation.
Discussion
The long pentraxin PTX3 is a component of the humoral innate immunity that plays non-redundant roles in the recognition and clearance of microbes and in regulating inflammatory responses. PTX3 expression is induced by inflammatory or microbial stimuli in several cell types.Citation2,38 Methylation of the promoter and a putative regulatory region of the long pentraxin PTX3 gene was recently associated with silencing of PTX3 expression in specific human cancers.Citation8 Since PTX3 has emerged as an oncosuppressor gene acting as a regulator of CRI, these results suggest that epigenetic regulation of PTX3 expression is relevant in human pathology. In this study, we further characterized PTX3 regulatory regions and defined their mode of action in different PTX3 expressing cell types, and finally extended the analysis of PTX3 epigenetic modifications associated with colorectal cancer.
By analyzing ChIP-seq data sets comparing histone modifications in CRC and healthy epithelium through the GREAT web-tool,Citation19 in addition to the previously described regulatory region encompassing the second PTX3 exon, we identified a new enhancer located 230 kb upstream of the PTX3 gene promoter. These two putative enhancers are phylogenetically conserved in fugu, frog, chicken, opossum, dog, mouse, rat, and Rhesus macaque, which suggests their relevance in the control of PTX3 expression. Their functional activity was experimentally confirmed by the analysis of the profile of histone markers associated with these regions. The analysis was focused on human primary macrophages, since they are a major source of PTX3 in inflammatory conditions, on a fibroblastic cell line, which constitutively produces PTX3 and overexpresses it upon inflammatory stimulation, and in a cell line of normal colon epithelium, which was previously used to compare PTX3 epigenetic modification occurring upon neoplastic transformation.Citation2,8,20 The results showed that the two enhancers and the promoter switched from an inactive (H3K4me1low, H3K27Aclow, H3K27me3high) to an active state (H3K4me1high, H3K27Achigh, H3K27me3low) in the three different PTX3 producing cell types after the treatment with TNFα, suggesting their involvement in PTX3 gene expression in inflammatory conditions.
Inflammatory cytokines such as IL-1β and TNFα, and microbial moieties interacting with TLRs activate the transcription of PTX3 gene, promoting the binding of TFs, such as NF-κB, SP.1, and PU.1 on PTX3 promoter.Citation22,23 In order to define the mode of action of the two PTX3 enhancers, we thus focused on TFs involved in inflammatory and immune responses and predicted to bind with these regulatory elements. NF-κB subunit RelA, c-Jun, c-Fos, PU.1, and SP.1 that were predicted to interact with the promoter and enhancer 1, but not with enhancer 2, were indeed all associated with the PTX3 promoter in macrophages and fibroblasts upon inflammatory stimulation, whereas the binding to enhancer 1 differed in the two cell types analyzed, with an enrichment of SP.1 and PU.1 in macrophages, and of c-Jun in fibroblasts. An enrichment of NF-κB on enhancer 2 in macrophages suggested that this enhancer may be under the control of NF-κB through indirect binding in specific cell types.Citation24 Thus, the binding of these TFs on enhancer 1 and the positive correlation observed between PTX3 RNA expression and H3K27ac enrichment in the enhancer 1, but not in enhancer 2, in the different cell types after stimulation with TNFα, suggest a major role for enhancer 1, together with the promoter, in regulating gene transcription in inflammatory conditions. The enhancer 2, although activated after stimulation with TNFα, was not found to bind TFs implicated in inflammation, suggesting that its activity is not related to these regulatory proteins.
Recent reports showed that enhancers can be involved in the recruitment of the transcription preinitiation complex (known also as PIC) on promoters, and proposed that only upon formation of PIC on enhancers, it moves along the promoter leading to gene transcription initiation.Citation39 Our results show that enhancer 2 bound TAF1, a component of PIC in macrophages and fibroblasts, in basal condition and after TNFα treatment. Thus, a possible role for enhancer 2 could be the recruitment of PIC during the activation of PTX3 expression. However, we cannot rule out the possibility that enhancer 2 is also under control of TFs. Indeed, we observed the binding of NF-κB on enhancer 2 after stimulation with TNFα in macrophages, but not in fibroblasts or epithelial cells, which suggests that this regulatory element could mediate the action of lineage-specific TFs.Citation39
PRC2 promotes gene transcription repression, catalyzing the trimethylation of lysine 27 of histone H3 (H3K27me3).Citation27,40 The involvement of PRC2 in the modulation of inflammatory responses has recently been proposed. For instance, a H3K27me3 demethylase expressed in macrophages in response to bacterial products and inflammatory cytokines was shown to bind PcG target genes and regulate their H3K27me3 levels and transcriptional activity, thus reprogramming the epigenome in response to inflammation.Citation28 We found in both enhancers enrichment in basal conditions of H3K27me3, and of SUZ2 and EZH2, the two main subunits of PRC2. In addition, we observed that pharmacological inhibition of EZH2 reduced the levels of H3K27me3 on both enhancers and promoter, increased the enrichment of Pol II and TAF1 on enhancer 2 and was associated with increased PTX3 expression. These results suggest a direct role for PRC2 in repressing PTX3 expression in basal conditions, and support previous findings on the role of PRC2 in regulating gene expression changes associated with inflammatory responses.Citation41
We have recently observed that PTX3 promoter and enhancer 2 are hypermethylated in selected human cancers, leading to silencing of PTX3 expression.Citation8 The analysis of PTX3 epigenetic modification associated to human cancer has been extended here and showed that enhancer 1 was unmethylated in healthy conditions, hypermethylated in low- and high-grade adenoma, and stage I CRC, and hypomethylated in stage II and III CRC, whereas progressively increased promoter and enhancer 2 DNA methylation was observed in adenomas and CRC stages I and II. The inhibition of DNA methylation by 5′AZA-dC treatment of CRC cell lines was sufficient to rescue PTX3 mRNA expression both in basal and inflammatory conditions, thus suggesting a prominent role of DNA methylation in silencing PTX3 expression in CRC. The rescue was associated with the activation of enhancer 2 already in basal conditions and of enhancer 1 and promoter in inflammatory conditions.
STAT3 is a major TF associated with tumor-promoting inflammation and immunosuppression,Citation34 and has been reported to recruit the DNA methyltransferase DNMT1.Citation33,42,43 The results reported here indicate that STAT3 was potentially involved in PTX3 methylation and silencing at early stages of intestinal cancer development. Indeed, the binding of STAT3 to enhancer 1 in CRC cell lines and in stage I, II, and III CRC samples correlated with enhancer 1 DNA methylation, and treatment with stattic, a selective inhibitor of STAT3 phosphorylation,Citation36,37 increased PTX3 production by CRC cell lines in inflammatory conditions and reduced enhancer 1 DNA methylation. These findings indicate that the repression of PTX3 in CRC is associated with hypermethylation of different regulatory regions depending on the stage of tumor progression, and that STAT3-dependent hypermethylation and silencing of enhancer 1 could be a critical effector in this process in the early stages of carcinogenesis.
Three PTX3 SNPs neighboring enhancer 2 are associated with susceptibility to specific diseases, such as pulmonary tuberculosis, Pseudomonas aeruginosa infection in cystic fibrosis patients, aspergillosis, and other fungal infections.Citation31,44,45 In specific conditions, namely aspergillosis, meningococcus sepsis (T. Sprong, unpublished result), and myocardial infarction, a specific haplotype of these three SNPs is also associated with reduced expression and production of PTX3.Citation31,32 Several reports showed that SNPs of enhancers could impact on gene expression by influencing the activity of these regulatory elements.Citation46,47 Our results, even if preliminary, indicate that this PTX3 haplotype affects the expression of PTX3 in basal condition by influencing the activity of the enhancer 2 and suggest that this mechanism could impact on the susceptibility to specific pathologic conditions associated with PTX3 genetic variants. Further studies on larger populations will be needed to confirm this potential mechanism of regulation of PTX3 expression associated with PTX3 haplotypes.
PTX3 plays a regulatory role in acute and chronic inflammatory conditions and in tissue repair processes, by interacting with P-selectin,Citation3 modulating complement activation,Citation48 and remodeling provisional fibrin-rich extracellular matrix.Citation4 Future studies are required to assess whether epigenetic modifications of the enhancers characterized here are responsible of PTX3 gene silencing in specific chronic inflammatory conditions or in specific patients.
The results presented here show that the expression of PTX3 is under the control of two enhancers, and that their hypermethylation is responsible for silencing of PTX3 in CRC in stage specific manner. Thus, these genetic elements emerge as important players in the fine regulation of PTX3 expression in physiological and pathological conditions.
Materials and methods
Bioinformatics tools
Putative cis-regulatory regions of PTX3 were identified with GREAT.Citation19 PTX3 enhancer conservation was investigated with Evolutionary Conserved Regions (ECRs) browser.Citation49
We used the matrix-scan program of regulatory sequence analysis tools (RSAT) to detect transcription factor binding sites (TFBS) (http://rsat.sb-roscoff.fr/) that reports individual motifs and cis-regulatory-elements-enriched regions (CRERs). Individual sites were predicted using the position weight matrix (PWM) of five selected TF motifs and filtered using a threshold on a p-value of 2.5e−04. CRERs 30 and 500 bp-long and with a significance of at least two [−log10(p-value)] were analyzed. For matrix-scan, we chose first-order Markov chain background model learned on human gene upstream no-orf sequences. TFBS motifs in the regions of our interest were also identified using Encode database (https://genome.ucsc.edu/ENCODE/).
Human monocytes and cell lines
Human monocytes were obtained from buffy coats of healthy donors as describedCitation32 and differentiated to macrophages by adding 100 ng/mL M-CSF. When specified, monocytes were collected from healthy donors previously genotyped for the PTX3 SNPs, as described.Citation32
The 8387 fibrosarcoma and microsatellite instable CRC cell lines (RKO and HCT116) were from ATCC; HCoEpiC was from ScienceCell Research Laboratories. When specified, cells were treated with the epigenetic modifier 5-AZA-dC (15 μM) for 72 h, TSA (150 nM) for 48 h, GSK343 (1 μM) (all from Sigma-Aldrich) for 48 h, then washed and stimulated with TNFα 20 ng/mL for 4 h.
Gene expression analysis
Total RNA (1 μg) extracted using TRIzol (Ambion) was reverse transcribed using the high-capacity cDNA Reverse Transcription kit (Applied Biosystems). PTX3 gene expression was quantified by RT-qPCR performed in triplicate in the presence of the SYBR green PCR master mix (Applied Biosystems) and specific oligonucleotides (see Table S1), using the 7900HT Fast RT-PCR System (Applied Biosystems). Data were analyzed with the Δ2CT method (SDS 2.2.2 software, Applied Biosystems) and normalized on GAPDH expression.
ChIP, PAT-ChIP and MIRA assays
ChIP and PAT-ChIP assays were performed as described.Citation50,51 Briefly, the chromatin obtained from 12 × 106 cells or 5 × 10-µm-thick FFPE tissue sections was sonicated to obtain 200−400 bp chromatin fragments, then immunoprecipitated at 4°C overnight with the following antibodies: anti-H3, anti-H3K27ac and anti-H3K4me1 (Abcam), anti-H3K4me3 (Active Motif), anti-H3K27me3, anti-NF-κB p65 (RelA), anti-EZH2 and anti-SUZ12 (Millipore-Upstate), anti-Pol II (N-20), anti TAF II p250, and anti STAT3 (C-20) (Santa Cruz biochemistry). Rabbit IgG (Millipore) was used as negative control. The immunoprecipitated DNA was analyzed by RT-PCR, using the primers listed in Table S1. Signals obtained from the ChIP samples were normalized on signals obtained from corresponding input, according to the formula 100 × 2ˆ(input Ct−sample Ct), and then expressed as fold change relative to IgG. MIRA assay was performed as described,Citation8 using primers listed in Table S1.
Tissue samples
Paraffin-embedded tissues from oncologic Caucasian patients who underwent resection surgery at Humanitas Clinical and Research Center were examined for PTX3 gene epigenetic modification. The study included the following cases: 3 angiomyosarcomas, 5 leiomyosarcomas, 5 synovial sarcomas, 5 solitary fibrous tumors, 5 chordomas, 5 gastrointestinal stromal tumors, 3 desmoid tumors, 6 squamous cell carcinoma, 5 high-grade adenomas, 10 stage I, 10 stage II, 10 stage III, and 10 stage IV CRC. The Institutional Review Board approved this study.
Adenoma laser microdissection was performed as described.Citation8
Statistical analysis
Statistical analysis was performed using the Student's t-test and ANOVA; a value of p < 0.05 was considered statistically significant. In DNA methylation studies on cancer samples, outliers were identified and eliminated using GraphPad Prism version 6. Statistics were calculated with GraphPad Prism version 6 (GraphPad Software).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
M.R. designed and performed most experiments, analyzed the data, and drafted the manuscript; C.M.G. and F.P. performed some experiments; P.K. and S.S. performed bioinformatics studies and analyzed data; G.B., M.R., and L.L. provided human samples; A.M. critically revised the manuscript; R.P. and C.G. conceived the study, directed research, designed experiments, analyzed data, and wrote the manuscript.
Supplementary_materials.pdf
Download PDF (6 MB)Acknowledgments
We thank Nadia Corrado for help with PAT-ChIP assays.
Funding
This work was supported by the European Commission (ERC to AM project PHII-669415; FP7-HEALTH-2011-ADITEC-N°280873 to AM; PK and RP were supported by ERC Advanced Grant to Gianluigi Condorelli CardioEpigen-294609); Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR) (project FIRB RBAP11H2R9; project PRIN 2015YYKPNN); the Italian Ministry of Health (RF-2013-02355470 to CG); and Associazione Italiana Ricerca sul Cancro (AIRC and AIRC 5 × 1000).
References
- Garlanda C, Bottazzi B, Bastone A, Mantovani A. Pentraxins at the crossroads between innate immunity, inflammation, matrix deposition, and female fertility. Annu Rev Immunol 2005; 23:337-66; PMID:15771574; https://doi.org/10.1146/annurev.immunol.23.021704.115756
- Bottazzi B, Doni A, Garlanda C, Mantovani A. An integrated view of humoral innate immunity: Pentraxins as a paradigm. Annu Rev Immunol 2010; 28:157-83; PMID:19968561; https://doi.org/10.1146/annurev-immunol-030409-101305
- Deban L, Russo RC, Sironi M, Moalli F, Scanziani M, Zambelli V, Cuccovillo I, Bastone A, Gobbi M, Valentino S et al. Regulation of leukocyte recruitment by the long pentraxin PTX3. Nat Immunol 2010; 11:328-34; PMID:20208538; https://doi.org/10.1038/ni.1854
- Doni A, Musso T, Morone D, Bastone A, Zambelli V, Sironi M, Castagnoli C, Cambieri I, Stravalaci M, Pasqualini F et al. An acidic microenvironment sets the humoral pattern recognition molecule PTX3 in a tissue repair mode. J Exp Med 2015; 212:905-25; PMID:25964372; https://doi.org/10.1084/jem.20141268
- Jaillon S, Moalli F, Ragnarsdottir B, Bonavita E, Puthia M, Riva F, Barbati E, Nebuloni M, Cvetko Krajinovic L, Markotic A et al. The humoral pattern recognition molecule PTX3 is a key component of innate immunity against urinary tract infection. Immunity 2014; 40:621-32; PMID:24745336; https://doi.org/10.1016/j.immuni.2014.02.015
- Jaillon S, Peri G, Delneste Y, Fremaux I, Doni A, Moalli F, Garlanda C, Romani L, Gascan H, Bellocchio S et al. The humoral pattern recognition receptor PTX3 is stored in neutrophil granules and localizes in extracellular traps. J Exp Med 2007; 204:793-804; PMID:17389238; https://doi.org/10.1084/jem.20061301
- Latini R, Maggioni AP, Peri G, Gonzini L, Lucci D, Mocarelli P, Vago L, Pasqualini F, Signorini S, Soldateschi D et al. Prognostic significance of the long pentraxin PTX3 in acute myocardial infarction. Circulation 2004; 110:2349-54; PMID:15477419; https://doi.org/10.1161/01.CIR.0000145167.30987.2E
- Bonavita E, Gentile S, Rubino M, Maina V, Papait R, Kunderfranco P, Greco C, Feruglio F, Molgora M, Laface I et al. PTX3 is an extrinsic oncosuppressor regulating complement-dependent inflammation in cancer. Cell 2015; 160:700-14; PMID:25679762; https://doi.org/10.1016/j.cell.2015.01.004
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 2010; 107:21931-6; PMID:21106759; https://doi.org/10.1073/pnas.1016071107
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 2009; 459:108-12; PMID:19295514; https://doi.org/10.1038/nature07829
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 2011; 470:279-83; PMID:21160473; https://doi.org/10.1038/nature09692
- Stergachis AB, Haugen E, Shafer A, Fu W, Vernot B, Reynolds A, Raubitschek A, Ziegler S, LeProust EM, Akey JM et al. Exonic transcription factor binding directs codon choice and affects protein evolution. Science 2013; 342:1367-72; PMID:24337295; https://doi.org/10.1126/science.1243490
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 2009; 457:854-8; PMID:19212405; https://doi.org/10.1038/nature07730
- Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature 2009; 461:199-205; PMID:19741700; https://doi.org/10.1038/nature08451
- Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 2014; 159:1312-26; PMID:25480296; https://doi.org/10.1016/j.cell.2014.11.018
- Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 2014; 159:1327-40; PMID:25480297; https://doi.org/10.1016/j.cell.2014.11.023
- Vahedi G, Takahashi H, Nakayamada S, Sun HW, Sartorelli V, Kanno Y, O'Shea JJ. STATs shape the active enhancer landscape of T cell populations. Cell 2012; 151:981-93; PMID:23178119; https://doi.org/10.1016/j.cell.2012.09.044
- Akhtar-Zaidi B, Cowper-Sal-lari R, Corradin O, Saiakhova A, Bartels CF, Balasubramanian D, Myeroff L, Lutterbaugh J, Jarrar A, Kalady MF et al. Epigenomic enhancer profiling defines a signature of colon cancer. Science 2012; 336:736-9; PMID:22499810; https://doi.org/10.1126/science.1217277
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 2010; 28:495-501; PMID:20436461; https://doi.org/10.1038/nbt.1630
- Doni A, Michela M, Bottazzi B, Peri G, Valentino S, Polentarutti N, Garlanda C, Mantovani A. Regulation of PTX3, a key component of humoral innate immunity in human dendritic cells: Stimulation by IL-10 and inhibition by IFN-gamma. J Leukoc Biol 2006; 79:797-802; PMID:16461742; https://doi.org/10.1189/jlb.0905493
- Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res 2011; 21:1273-83; PMID:21632746; https://doi.org/10.1101/gr.122382.111
- Basile A, Sica A, d'Aniello E, Breviario F, Garrido G, Castellano M, Mantovani A, Introna M. Characterization of the promoter for the human long pentraxin PTX3. Role of NF-kappaB in tumor necrosis factor-alpha and interleukin-1beta regulation. J Biol Chem 1997; 272:8172-8; PMID:9079634; https://doi.org/10.1074/jbc.272.13.8172
- Altmeyer A, Klampfer L, Goodman AR, Vilcek J. Promoter structure and transcriptional activation of the murine TSG-14 gene encoding a tumor necrosis factor/interleukin-1-inducible pentraxin protein. J Biol Chem 1995; 270:25584-90; PMID:7592730; https://doi.org/10.1074/jbc.270.43.25584
- Spitz F, Furlong EE. Transcription factors: From enhancer binding to developmental control. Nat Rev Genet 2012; 13:613-26; PMID:22868264; https://doi.org/10.1038/nrg3207
- Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J 2007; 26:2041-51; PMID:17380126; https://doi.org/10.1038/sj.emboj.7601654
- Savic D, Roberts BS, Carleton JB, Partridge EC, White MA, Cohen BA, Cooper GM, Gertz J, Myers RM. Promoter-distal RNA polymerase II binding discriminates active from inactive CCAAT/ enhancer-binding protein beta binding sites. Genome Res 2015; 25:1791-800; PMID:26486725; https://doi.org/10.1101/gr.191593.115
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat Rev Mol Cell Biol 2009; 10:697-708; PMID:19738629; https://doi.org/10.1038/nrm2763
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 2007; 130:1083-94; PMID:17825402; https://doi.org/10.1016/j.cell.2007.08.019
- Tripathy MK, McManamy ME, Burch BD, Archin NM, Margolis DM. H3K27 demethylation at the proviral promoter sensitizes latent HIV to the effects of vorinostat in ex vivo cultures of resting CD4+ T Cells. J Virol 2015; 89:8392-405; PMID:26041287; https://doi.org/10.1128/JVI.00572-15
- Ding M, Zhang H, Li Z, Wang C, Chen J, Shi L, Xu D, Gao Y. The polycomb group protein enhancer of zeste 2 is a novel therapeutic target for cervical cancer. Clin Exp Pharmacol Physiol 2015; 42:458-64; PMID:25739318; https://doi.org/10.1111/1440-1681.12382
- Cunha C, Aversa F, Lacerda JF, Busca A, Kurzai O, Grube M, Löffler J, Maertens JA, Bell AS, Inforzato A et al. Genetic PTX3 deficiency and aspergillosis in stem-cell transplantation. N Engl J Med 2014; 370:421-32; PMID:24476432; https://doi.org/10.1056/NEJMoa1211161
- Barbati E, Specchia C, Villella M, Rossi ML, Barlera S, Bottazzi B, Crociati L, d'Arienzo C, Fanelli R, Garlanda C et al. Influence of pentraxin 3 (PTX3) genetic variants on myocardial infarction risk and PTX3 plasma levels. PLoS One 2012; 7:e53030; PMID:23285251; https://doi.org/10.1371/journal.pone.0053030
- Zhang Q, Wang HY, Woetmann A, Raghunath PN, Odum N, Wasik MA. STAT3 induces transcription of the DNA methyltransferase 1 gene (DNMT1) in malignant T lymphocytes. Blood 2006; 108:1058-64; PMID:16861352; https://doi.org/10.1182/blood-2005-08-007377
- Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat Rev Cancer 2014; 14:736-46; PMID:25342631; https://doi.org/10.1038/nrc3818
- Kang HJ, Yi YW, Hou SJ, Kim HJ, Kong Y, Bae I, Brown ML. Disruption of STAT3-DNMT1 interaction by SH-I-14 induces re-expression of tumor suppressor genes and inhibits growth of triple-negative breast tumor. Oncotarget 2015; PMID:26675255; https://doi.org/10.18632/oncotarget.4054
- Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Stattic: A small-molecule inhibitor of STAT3 activation and dimerization. Chem Biol 2006; 13:1235-42; PMID:17114005; https://doi.org/10.1016/j.chembiol.2006.09.018
- Pan Y, Zhou F, Zhang R, Claret FX. Stat3 inhibitor stattic exhibits potent antitumor activity and induces chemo- and radio-sensitivity in nasopharyngeal carcinoma. PLoS One 2013; 8:e54565; PMID:23382914; https://doi.org/10.1371/journal.pone.0054565
- Jaillon S, Bonavita E, Gentile S, Rubino M, Laface I, Garlanda C, Mantovani A. The long pentraxin PTX3 as a key component of humoral innate immunity and a candidate diagnostic for inflammatory diseases. Int Arch Allergy Immunol 2014; 165:165-78; PMID:25531094; https://doi.org/10.1159/000368778
- Koch F, Fenouil R, Gut M, Cauchy P, Albert TK, Zacarias-Cabeza J, Spicuglia S, de la Chapelle AL, Heidemann M, Hintermair C et al. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat Struct Mol Biol 2011; 18:956-63; PMID:21765417; https://doi.org/10.1038/nsmb.2085
- Sawarkar R, Paro R. Interpretation of developmental signaling at chromatin: The polycomb perspective. Dev Cell 2010; 19:651-61; PMID:21074716; https://doi.org/10.1016/j.devcel.2010.10.012
- Liu Y, Zhang Q, Ding Y, Li X, Zhao D, Zhao K, Guo Z, Cao X. Histone lysine methyltransferase Ezh1 promotes TLR-triggered inflammatory cytokine production by suppressing Tollip. J Immunol 2015; 194:2838-46; PMID:25687760; https://doi.org/10.4049/jimmunol.1402087
- Thomas NS. The STAT3-DNMT1 connection. JAKSTAT 2012; 1:257-60; PMID:24058781; https://doi.org/10.4161/jkst.22436
- Zhang Q, Wang HY, Marzec M, Raghunath PN, Nagasawa T, Wasik MA. STAT3- and DNA methyltransferase 1-mediated epigenetic silencing of SHP-1 tyrosine phosphatase tumor suppressor gene in malignant T lymphocytes. Proc Natl Acad Sci U S A 2005; 102:6948-53; PMID:15870198; https://doi.org/10.1073/pnas.0501959102
- Olesen R, Wejse C, Velez DR, Bisseye C, Sodemann M, Aaby P, Rabna P, Worwui A, Chapman H, Diatta M et al. DC-SIGN (CD209), pentraxin 3 and vitamin D receptor gene variants associate with pulmonary tuberculosis risk in West Africans. Genes Immun 2007; 8:456-67; PMID:17611589; https://doi.org/10.1038/sj.gene.6364410
- Chiarini M, Sabelli C, Melotti P, Garlanda C, Savoldi G, Mazza C, Padoan R, Plebani A, Mantovani A, Notarangelo LD et al. PTX3 genetic variations affect the risk of Pseudomonas aeruginosa airway colonization in cystic fibrosis patients. Genes Immun 2010; 11:665-70; PMID:20927127; https://doi.org/10.1038/gene.2010.41
- Matsumura K, Saito T, Takahashi Y, Ozeki T, Kiyotani K, Fujieda M, Yamazaki H, Kunitoh H, Kamataki T. Identification of a novel polymorphic enhancer of the human CYP3A4 gene. Mol Pharmacol 2004; 65:326-34; PMID:14742674; https://doi.org/10.1124/mol.65.2.326
- Liu H, Zhai J, Luo K, Liu L. Chromatin structure is distinct between coding and non-coding single nucleotide polymorphisms. BMC Mol Biol 2014; 15:22; PMID:25282079; https://doi.org/10.1186/1471-2199-15-22
- Inforzato A, Doni A, Barajon I, Leone R, Garlanda C, Bottazzi B, Mantovani A. PTX3 as a paradigm for the interaction of pentraxins with the complement system. Semin Immunol 2013; 25:79-85; PMID:23747040; https://doi.org/10.1016/j.smim.2013.05.002
- Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: A tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res 2004; 32:W280-6; PMID:15215395; https://doi.org/10.1093/nar/gkh355
- Papait R, Cattaneo P, Kunderfranco P, Greco C, Carullo P, Guffanti A, Viganò V, Stirparo GG, Latronico MV, Hasenfuss G et al. Genome-wide analysis of histone marks identifying an epigenetic signature of promoters and enhancers underlying cardiac hypertrophy. Proc Natl Acad Sci U S A 2013; 110:20164-9; PMID:24284169; https://doi.org/10.1073/pnas.1315155110
- Fanelli M, Amatori S, Barozzi I, Minucci S. Chromatin immunoprecipitation and high-throughput sequencing from paraffin-embedded pathology tissue. Nat Protoc 2011; 6:1905-19; PMID:22082985; https://doi.org/10.1038/nprot.2011.406
