ABSTRACT
Chronic lymphocytic leukemia (CLL) is associated with a profound dysregulation of the immune system. Loss of T cell function is frequently caused in cancer by sustained signaling of inhibitory receptors. Here, we analyzed the role of the novel inhibitory receptor Ig-like transcript 2 (ILT2) in the pathogenesis of CLL. We observed that ILT2 expression was markedly reduced on leukemic cells, whereas it was increased on CD8 and CD4 T cells from CLL patients, particularly in those patients harboring chromosome 11q deletion, which includes the ATM gene. A deep dysregulation of ILT2 ligands expression in leukemia cells was also observed. ILT2 impaired the activation and proliferation of CD4 and CD8 T cells in CLL patients, but it had no effect in leukemic cells. ILT2 downregulated the production of IL-2 by CD4 T cells of CLL patients and induced the expression of cytokines that promote the survival of leukemic cells, such as IFN-γ, by T cells. Importantly, ILT2 blockade restored the activation, proliferation and cytokine production of T cells. In conclusion, we describe a novel immune inhibitory pathway that is upregulated in CLL and delineate a new potential target to be explored in this disease.
Introduction
Chronic lymphocytic leukemia (CLL) is a heterogeneous disease with variable genetic signature, clinical presentation and evolution, which is characterized by an accumulation of functionally incompetent mature malignant B cells.Citation1,2 CLL is associated with a profound dysregulation of the immune system. T cells are increased at diagnosis and may mediate anti-tumor responses that affect disease progression and survival.Citation3-5 However, the evolution of the disease is associated with a progressive immunosuppression that is the major cause of morbidity and mortality in these patients.Citation6,7 In CLL, B cell defects and hypogammaglobinemia are associated with an impairment of the activity of other immune cells, including T cells, NK cells and dendritic cells.Citation6,8,9 CD4 and CD8 T cells show functional defects and, particularly, there is a marked impairment of T cell helper activity.Citation8,9 Such a tumor-immunosuppressive environment may contribute to disease progression and may be further exacerbated by common cytotoxic therapies. Besides, it may also block the effectiveness of current immunotherapy approaches.
In the context of cancer, the loss of T cell function is frequently caused by sustained signaling of multiple inhibitory receptors that dampen the patient's immune responses and inhibit tumor elimination.Citation10-11 Sustained signaling via these checkpoint proteins, such as programmed cell death protein 1 (PD1), programmed cell death ligand 1 (PDL1) or cytotoxic T lymphocyte antigen 4 (CTLA-4), results in functional exhaustion of T cells, which lose the capability of proliferating, secreting cytokines and lysing tumor cells.Citation11 Targeting these inhibitory checkpoints has achieved noteworthy benefit in multiple cancers by enabling patients to restore an effective antitumor response.Citation11 These promising clinical data have created a recent excitement in the field of tumor immunology and immunotherapy. Thus, there is an intense search for new inhibitory molecules involved in cancer immunosuppression.
In this direction, Ig-like transcript 2 (ILT2) (also named CD85j, LILRB1, or LIR-1) is an inhibitory receptor expressed by B cells, T cells, NK cells, dendritic cells, and other immune cells.Citation12,13 ILT2 interacts with classical and non-classical MHC class I molecules, including HLA-E, HLA-F and HLA-G.Citation14-16 Specifically, ILT2 binds to HLA-G with a 3- to 4-fold higher affinity than to classical MHC class I molecules.Citation17 ILT2 contains 4 immunoreceptor tyrosine-based inhibition motifs (ITIMs) in its cytoplasmic domain that are involved in negative signaling through the recruitment of Src homology 2 (SH2) domain-containing proteins, such as SHP-1.Citation18 The interaction of ILT2 with its ligands impairs the function and effector activity of T and B cells.Citation19-23 Thus, engagement of ILT2 on T cells has been shown to inhibit T cell antigen receptor (TCR) signaling and downstream events such as actin reorganization.Citation23
HLA-G expression correlated with advanced disease and poor clinical outcome in several types of cancer, including hepatocellular carcinomaCitation24 and CLL,Citation25-28 and it has been described as a tumor-escape mechanism favoring cancer progression.Citation29 Nevertheless, despite the fact that an intense research in the function of inhibitory immune molecules in cancer is being made, the role of ILT2 in the pathogenesis of cancer has achieved less attention than other inhibitory proteins.
In this study, we report a novel mechanism of immune suppression in CLL characterized by a sustained ILT2-dependent inhibitory signaling that down-regulates the activation, proliferation and modulates cytokine production of T cells, suggesting that ILT2 could be explored as a new target for immunotherapy in CLL.
Results
The surface expression of ILT2 is reduced on leukemic cells and increased on T cells from CLL patients
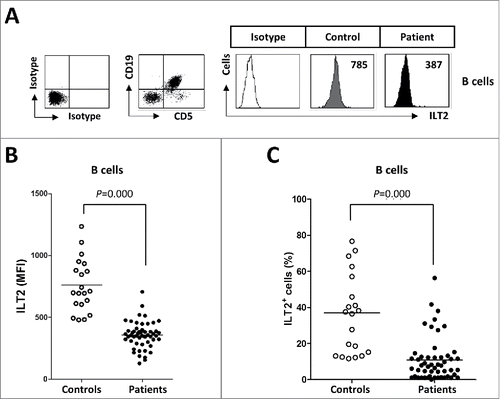
ILT2 is expressed on B cells and it is a critical inhibitor of their proliferation, differentiation and functional activity.Citation19,22 In this study, the expression of ILT2 on B cells was analyzed in 52 CLL patients and 20 age-matched healthy donors () by flow cytometry. Compared with B cells from controls, ILT2 expression (mean of mean fluorescence intensity (MFI): 357 ± 107 vs. 760 ± 214, P<0.001) and the percentage of ILT2+ cells (11 ± 10% vs. 36 ± 22%, P<0.001) was significantly reduced on leukemic cells ().
Table 1. Clinical characteristics of CLL patients.
Figure 1. ILT2 expression is reduced on the surface of leukemic cells. (A) PBMCs from 52 CLL patients and 20 healthy donors were stained with CD19-, CD5- and ILT2-conjugated antibodies and analyzed by flow cytometry. The histogram shows the ILT2 expression in B cells from a healthy donor and leukemic cells (CD19+CD5+) from a patient. (B) The comparison between the MFI ± SEM of ILT2 surface expression on B cells from controls and patients is shown. (C) The comparison between percentage ± SEM of ILT2+ B cells from controls and patients is shown. Horizontal bars represent the mean.
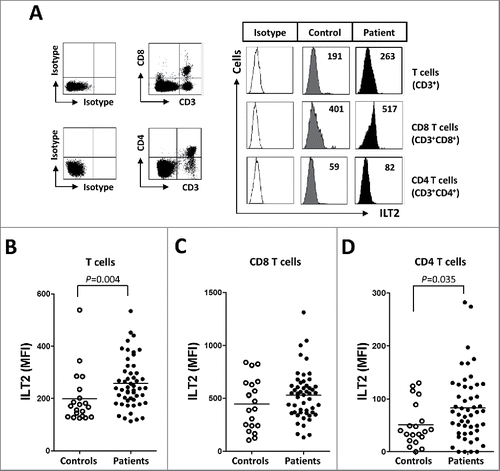
ILT2 is an inhibitory receptor also expressed by T cells.Citation12,13,23,30 In our study, lower expression of ILT2 was detected in T cells compared with B cells; and in contrast with B cells, the expression of ILT2 was increased in T cells of CLL patients, and specifically in CD4 T cells (mean of the MFI: 82 ± 63 vs. 51 ± 40, P<0.05) ().
Figure 2. ILT2 is overexpressed on T cells from CLL patients. (A) PBMCs were obtained from 52 CLL patients and 20 healthy donors and the expression of ILT2 on T cells, and CD8 and CD4 T cell subsets was determined by staining the cells with CD3-, CD4-, CD8-, and ILT2-conjugated antibodies. Dot plots show the cytometric prolife of a CLL patient. Histograms in the right show flow cytometry profiles of a healthy donor and a representative patient. The comparison of the MFI of ILT2 surface expression on T cells (B), CD8 T cells (C) and CD4 T cells (D) between controls and patients is shown.
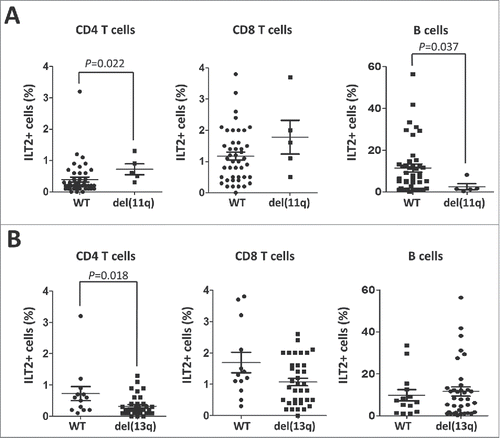
Of note, significant clinical association with ILT2 expression was found (). Patients harboring del(11q), which has been associated with a poor clinical outcome in CLL,Citation31-33 showed higher levels of ILT2+ CD4 T cells (P<0.05) and lower levels of ILT2+ B cells (P<0.05) (). ILT2+ CD8 T cells were not significantly increased in del(11q) patients. Contrasting these data, ILT2+ CD4 T cells (P<0.05) were significantly reduced in CLL patients with del(13q), which is associated with more favorable clinical outcomeCitation34 ().
Figure 3. ILT2 expression correlates with cytogenetic abnormalities that are markers of the progression of the disease. (A) Comparison between ILT2+ CD8 T cells, ILT2+ CD4 T cells, and ILT2+ B cells from CLL patients stratified by the presence of chromosome 11q deletion. Horizontal bars represent the mean ± SEM. (B) The comparison between ILT2+ CD4 T cells, ILT2+ CD8 T cells and ILT2+ B cells from CLL patients with or without chromosome 13q deletion is shown.
Surface expression of ILT2 ligands on leukemic cells
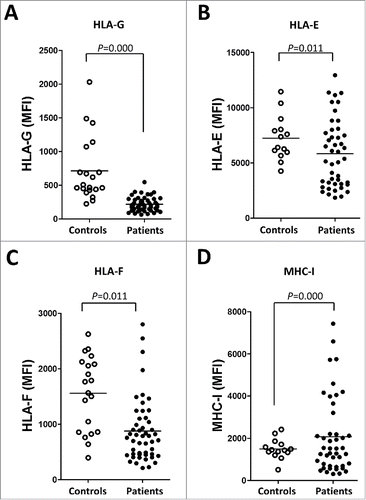
The expression of ILT2 ligands was also profoundly dysregulated in leukemic cells. Leukemic cells expressing HLA-G (215 ± 14 vs. 712 ± 106, P<0.001), HLA-E (7248 ± 537 vs. 5827 ± 455 P<0.05) and HLA-F (1556 ± 149 vs. 874 ± 81, P<0.001) were decreased in patients compared with B cells from controls (). The expression of HLA-G in B cells from healthy controls was further confirmed by reverse transcription PCR (Fig. S1). Classical MHC class I molecules were broadly expressed on B cells, but their expression was increased in CLL patients (mean of MFI: 2335 ± 1590.6 vs. 1433.4 ± 437.5; P<0.05) (). Therefore, a deep dysregulation of the expression of ILT2 receptor and its ligands is observed in CLL patients.
ILT2 impairs T cell activation
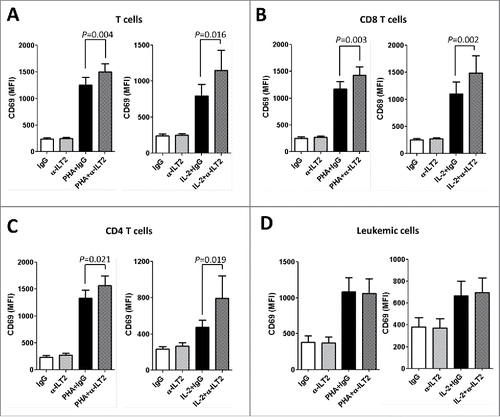
Next, the role of ILT2 in the activation of the different lymphocyte subsets was studied by using the HP-F1 antibody, that blocks the interaction between ILT2 and its ligands and promotes the functional activity of T cells.Citation13,20 Pheripheral Blood Monoclear Cells (PBMCs) obtained from 14 CLL patients and 7 healthy donors stimulated with PHA or IL-2 were treated with anti-ILT2 blocking antibody and the expression of the early immune activation marker CD6935 in different lymphocyte subsets was analyzed by flow cytometry. In CLL, ILT2 blockade significantly increased the activation of stimulated CD4 and CD8 T cells (). Remarkably, no effect on CD69 expression was observed in leukemic cells upon ILT2 blockade (). Similarly, ILT2 blockade induced in lesser extent the expression of CD69 in T cells of healthy donors (Fig. S2). Moreover, contrasting leukemic cells, ILT2 blockade also activated healthy B cells. Importantly, PHA or IL-2 stimulation did not alter the expression of ILT2 in T and B cells obtained from CLL patients and donors (not shown). The expression of HLA-E in B cells and leukemic cells was the only one among ILT2 ligands to be induced in response to IL-2 and PHA (Fig. S3). These observations suggest that ILT2 has a co-inhibitory effect on T cell function, but this effect was not observed in leukemic cells.
Figure 5. ILT2 blockade promotes T-cell activation. Flow cytometry analyses were conducted to evaluate the expression of CD69 in PBMCs from 14 CLL patients stimulated with IL-2 (50 U/mL) or PHA (5 µg/mL) and incubated in the presence of anti-ILT2 blocking antibody or irrelevant IgG1 (10 μg/mL) for 48 hours. The figure shows the comparison between the surface expression of CD69 detected on T cells (A), CD8 T cells (B), CD4 T cells (C) and leukemic cells (D) from CLL patients. Bars represent the mean MFI of CD69 ± SEM for each condition.
The effect of MHC class I molecules and HLA-G on the activation of T cells was further analyzed by using specific blocking antibodies. Only MHC class I blockade had a significant effect on the activation of CD8 T cells (Fig. S4). No significant effect on the activation of CD4 T cells and leukemic cells was observed.
ILT2 inhibits T cell proliferation in CLL
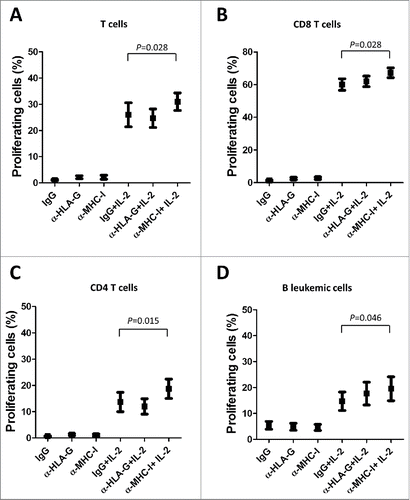
T cells from CLL patients exhibit features of exhaustion, showing functional defects in proliferation.Citation36 We next analyzed whether the dysregulation of ILT2 expression may affect B or T cell proliferation using the CFSE assay. Thus, stimulated PBMCs obtained from 12 CLL patients and 7 healthy donors were treated with the anti-ILT2 blocking antibody, and its effect on the proliferation of T and B cells was determined by flow cytometry analysis (). As shown in , ILT2 blockade significantly induced the proliferation of T cells (P<0.05), CD8 T cells (P<0.05) and CD4 T cells (P<0.05) in patients and controls. ILT2 blockade also stimulated the proliferation of healthy B cells (P<0.05); but no effect in leukemic cell proliferation was observed (). The effect of MHC class I molecules and HLA-G on the proliferation of immune cells was further analyzed using specific blocking antibodies. As shown in , only MHC class I blockade had a significant effect on the proliferation of CD4 T cells, CD8 T cells, and leukemic cells, but no effect with HLA-G blockade was observed.
Figure 6. ILT2 inhibits T cell proliferation in CLL and healthy donors. (A) IL-2-stimulated (50 U/mL) or unstimulated PBMCs obtained from 12 CLL patients and 7 healthy donors were stained with CFSE and cultured in the presence of anti-ILT2 blocking antibody (10 μg/ml) or irrelevant IgG1 for 7 days, and the proliferation of different cell subsets was analyzed by flow cytometry. Histograms show the flow cytometry profiles corresponding to CFSE expression in T cells, CD8 T cells, CD4 T cells and leukemic cells of a representative CLL patient. (B-E) Comparison of the percentage of proliferating T cells (B), CD8 T cells (C), CD4 T cells (D) and B cells (E) from patients and donors between the different experimental conditions analyzed are shown. Bars represent the mean ± SEM from samples analyzed.
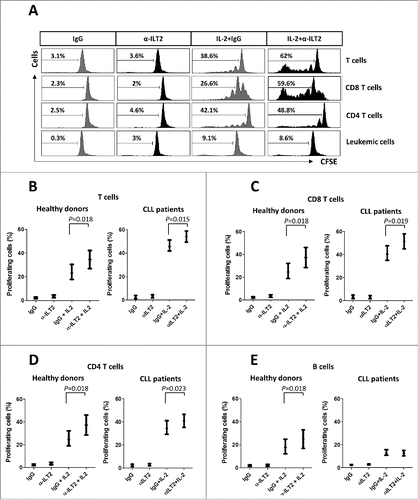
Figure 7. MHC class I molecules inhibits T and B cell proliferation in CLL. IL-2-stimulated or unstimulated PBMCs obtained from 6 CLL patients were stained with CFSE and cultured in the presence of anti-HLA-G blocking antibody, anti-MHC-I blocking antibody (both 10 μg/ml) or irrelevant IgG for 7 days, and the proliferation of different cell subsets was analyzed by flow cytometry. The comparison of the percentage of proliferating T cells (A), CD8 T cells (B), CD4 T cells (C) and B cells (D) between the different experimental conditions analyzed are shown. Bars represent the mean ± SEM from samples analyzed.
ILT2 impairs IL-2 production and enhances IFN-γ production in CLL
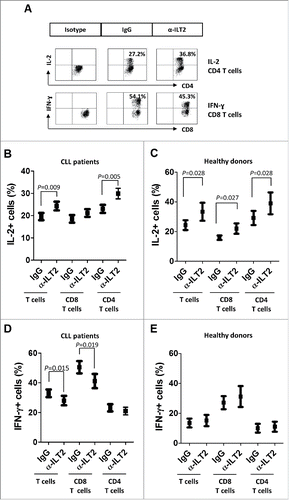
The effect of ILT2 blockade in the functional activity of T cells was next analyzed. No effect was observed in the cytotoxic activity mediated by CD8 T cells as determined by the CD107a degranulation assay (not shown). However, a significant effect on the production of key cytokines by T cells was detected (). The production of IL-2 has been reported to be diminished in CLL,Citation37 whereas IFN-γ and TNF-α are increased in CLL.Citation38 Further, IFN-γ has been shown to promote the survival of leukemic cells.Citation39 In our study, the blockade of ILT2 had no effect in the production of TNF-α by T cells obtained from 15 CLL patients and 6 healthy donors (not shown). However, it significantly enhanced the production of IL-2 by CD4 T cells (P<0.01) in both patients () and donors (). In CLL, ILT2 blockade significantly inhibited the expression of IFN-γ in T cells (P<0.05), and particularly in CD8 T cells (P<0.05) (); but no effect on the production of IFN-γ by healthy donors was observed (). In leukemic cells and healthy B cells ILT2 blockade did not significantly modulate the expression of these cytokines (not shown).
Figure 8. ILT2 blockade increases IL-2 levels and reduces IFN-γ production by T cells in CLL patients. (A) PBMCs from 15 CLL patients and 6 donors were cultured in the presence of anti-ILT2 blocking antibody or irrelevant IgG1 for 48 hours, and the intracytoplasmic expression of IL-2 and IFN-γ was analyzed in T and B cells by flow cytometry. Dot plots show the level of IL-2 expression in CD4 T cells and IFN-γ expression in CD8 T cells from a CLL patient. The mean of percentage ± SEM of intracytoplasmic levels of IL-2 in patients (B) and healthy donors (C) is shown. The intracytoplasmic expression of IFN-γ in patients (D) and healthy donors (E) is shown.
Overall, our results show that ILT2 blockade induced the proliferation, activation and cytokine production of T cells of CLL patients, without affecting leukemic cell functional activity, suggesting that ILT2 may be a potential therapeutic target to be explored in CLL.
Discussion
CLL is associated with multiple alterations of the immune system; being the immunosuppression the most typical feature.Citation6,7 The severity of the immunodeficiency increases with the time from diagnosis, suggesting that leukemic cells may directly cause this immunosuppression. In fact, direct T cell inhibition by leukemic cells has been broadly documented.Citation6,39 One of the relevant mechanisms that account for the progressive loss of T cell function in cancer is the sustained signaling of multiple inhibitory receptors, such as PD1 and CTLA-4, which down-regulate the antitumor immune response of patients.Citation10,11 Targeting these immunosuppressive mechanisms may restore the antitumor activity of T cells, providing a great benefit for cancer patients.Citation10,11 As a result, there is an intense search for new inhibitory molecules involved in cancer immunosuppression.
ILT2 is an inhibitory receptor involved in negative signaling in B cells, T cells, NK cells and other immune cells.Citation12,13,20,40 However, its role in cancer has been poorly defined. The present study shows that the expression of this receptor and its ligands are deeply dysregulated in leukemic cells and T cells of CLL patients. ILT2 was overexpressed in T cells from CLL patients; whereas its expression was considerably decreased in leukemic cells. Of note, despite the fact that only a minor population of ILT2+ T cells is detected in CLL patients, the blockade of ILT2 has a strong effect on the promotion of T cell function. Thus, a detailed characterization of the phenotype and the regulatory characteristics of this population of ILT2+ T cells warrants further investigation. Additionally, the analysis of other ILT2+ immune cells, such as NK cells and dendritic cells, is needed to completely elucidate the role of ILT2 in CLL.
We also observed that the expression of non-classical MHC class I molecules, including HLA-G, was significantly reduced and the expression of the classical ones was increased in CLL patients. Little information regarding the role of ILT2, HLA-E and HLA-F in CLL is currently available. Nevertheless, our study contrasts with previous studies suggesting that HLA-G may be related with progression-free survival in CLL. Different studies reported that the amount of leukemic cells expressing HLA-G varies from 1 to 34%Citation25,41,42; and a relation between HLA-G expression and markers of poor clinical outcome, such as CD38 expression and advanced Binet stage, was described.Citation25,42 However, our findings are in agreement with other studies which reported low or no expression of HLA-G in CLL and no correlation with the expression of well-established prognostic factors such as ZAP-70 and CD38.Citation26,42-44
ILT2 is an inhibitory receptor that impairs the proliferation, differentiation and functional activity of B and T cells.Citation13,19,20,23 In our study, we found that the dysregulation of ILT2 expression was enhanced in patients with bad prognostic markers. Of special interest was the distribution of ILT2 expression in patients carrying a deletion of chromosome 11q, which often exhibit bulky adenopathy and experience an aggressive clinical course, with shorter survival and poor response to the treatment.Citation31,32 The minimal region of deletion on 11q22.3–23.1 often involves the Radixin (RDX) and Ataxia telangiectasia mutated (ATM) genes. Given that ATM is crucial for DNA damage repair, it is likely that the deletion of this gene would lead to enhanced clonal aggressiveness and worse evolution, due to the acquisition of novel genomic variants.Citation45 In our study, a significant increase of ILT2 expression in CD4 T cells and a lower expression in B cells were observed in patients carrying the del(11q), which may suggest a greater immunosuppression in these patients and could also account for their poor clinical outcome. Nevertheless, due to the low frequency of the chromosome 11q22–23 deletion, which is detected in 5–20% of CLL patients,Citation46,47 further specific studies in larger populations are needed to draw definite conclusions.
T cells from CLL patients exhibit characteristics of exhaustion, showing defects in proliferation and cytotoxicity.Citation36 Our findings suggest that ILT2 may be involved in the impairment of T cell activation and proliferation observed in CLL patients and this effect is higher in T cells of CLL patients than in healthy donors. Importantly, ILT2 blockade showed no effect on the activation or proliferation of leukemic cells, although it has a significant effect in healthy B cells. Further, ILT2 blockade has been reported to promote the proliferation of tumor B cell lines.Citation22 This could be explained by the fact that peripheral B cells cannot be considered as the normal counterpart of leukemic cells of CLL,Citation48 and compared with B cells and other B cell-derived cancers, CLL cells show very low proliferative activity.
ILT2 interacts with both classical and non-classical MHC-I molecules.Citation14-16 However, the blockade of HLA-G did not show significant effects in the activation and proliferation of T and B lymphocytes in CLL. Conversely, our experiments show that classical MHC class I molecules are upregulated in the leukemia cells of CLL patients, and blocking experiments in CLL showed increased activation and proliferation of T cells, suggesting that classical MHC class I molecules may play a more prominent role in this effect than HLA-G.
We observed that ILT2 blockade did not enhance the cytotoxic activity of CD8 T cells of CLL patients. This may be related with the multiple defects of T cells observed in CLL patients and the low immunogenicity of leukemic cells, which are well known for their inability to process and present antigens and to form a proper synapse with T cells.Citation6 Additionally, CD8 T cells from CLL patients had increased expression of exhaustion markers such as CD244, CD160, and PD1 and showed functional defects in proliferation and cytotoxicity due to impaired granzyme packaging into vesicles and nonpolarized degranulation.Citation49 These data suggest that to increase the efficacy of ILT2 blocking in CLL should be combined with therapies that increase the activity of cytotoxic T cells.Citation50
ILT2 signaling was involved in the modulation of the production of key cytokines by T cells. The production of the T cell-stimulating cytokine IL-2 by CD4 T cells is diminished in CLLCitation37 and our findings suggests that ILT2 may be involved in this impairment. Conversely, IFN-γ production has been reported to be increased in CLL and it may be involved in the promotion of the survival of the leukemic clone.Citation38 In agreement, our study shows that T cells of CLL patients express higher levels of IFN-γ than healthy donors, and we also found that ILT2 is involved in the upregulation of IFN-γ observed in CLL patients. This effect seems to be a specific feature of CLL patients, since no effect on IFN-γ production in healthy donors was observed. Moreover, ILT2 has been reported to interfere with CD8 T cell antiviral function by suppressing IFN-γ production.Citation30 Thus, specific signaling pathways that regulate ILT2 in CLL warrant further investigation.
Overall, our findings describe a novel immune inhibitory pathway that is upregulated in CLL and support that ILT2 may represent a new potential target for immunotherapy to be investigated in this disease.
Patients, materials and methods
Patients and samples
52 consecutive non-treated CLL patients from the Hospital Universitario Central de Asturias and Hospital Universitario de Cabueñes (Spain) fulfilling the diagnostic criteria for CLL (), and 20 age-matched healthy donors (mean age 59.5 years) were studied. All methods and procedures were performed following the Declaration of Helsinki. This study was approved by the Ethics Committee of our Institution (Hospital Universitario Central de Asturias), and informed consent was obtained from all patients and controls. Both clinical and immunological characteristics of patients were analyzed when patients were enrolled in this study. The patient mean age was 68 years; 53% were male and 47% female. The median time from diagnosis to study was 62.3 months (range, 0 to 275 months). Patients did not receive cytoreductive treatment within 18 months before the investigation. Clinical and laboratory evaluation at visit included history and physical examination, standard clinical laboratory evaluation, evaluation for ZAP-70 by flow cytometry (20% cut-off), characterization of CD38 expression; and standard metaphase karyotype. Karyotype was categorized as complex, single abnormality or diploid. FISH analysis for 17p deletion, 11q deletion, trisomy 12, and chromosome 13q deletion was also performed. A total of 200 cells in interphase were analyzed for each probe. Positive patient cases were those with 5% or more cells with the abnormality. IGHV mutation status was characterized by direct sequencing method, and patients were categorized as unmutated (IGHV 98% germline homology) or mutated (98% homology). PBMCs were isolated by Ficoll density gradient centrifugation. The mean of the percentage of leukemic cells in PBMCs obtained from CLL patients was 51.4% (range 11.9–92%).
Antibodies and material
Recombinant human interleukin 2 (IL-2) (Peprotech), phytohemagglutinin (PHA, Sigma-Aldrich), etoposide (Sigma-Aldrich), monensin (Sigma-Aldrich), phorbol 12-myristate 13-acetate (PMA, Sigma-Aldrich), ionomycin (Sigma-Aldrich), brefeldin A (BD Biosciences), HP-F1 anti-ILT2/CD85j blocking antibody (IgG1), (provided by Miguel López-Botet, Universitat Pompeu Fabra, Spain) 87G anti-HLA-G blocking antibody (Invitrogen), pan-HLA-I blocking antibody (IgG2a) (clone W6/32), an and an irrelevant IgG1 antibody (provided by JR de los Toyos-González, Universidad de Oviedo, Spain) were used in this study.
Flow cytometry
To determine immune cell subsets, cells were stained with anti-CD3-FITC, anti-CD4-PerCP, anti-CD8-APC and anti-CD19-APC (all from Immunostep), anti-CD3-PECy7 (eBioscience), and isotype-matched control conjugates. T cells were defined as CD3+; CD4 T cells as CD3+CD4+; CD8 T cells as CD3+CD8+; and leukemic cells as CD19+CD5+. Anti-ILT2-PE (eBioscience), anti-HLA-F rabbit antibody (Abgent), anti-HLA-G-PE (Biolegend, clone 87G), anti-HLA-E-PE (Biolegend) and a pan-anti-HLA-I antibody (clone W6/32) were used to measure the expression of ILT2 and its ligands on different immune populations. Cells were analyzed on a BD Biosciences FACSCanto II cytometer (Beckton Dickinson).
Reverse transcription PCR
mRNA was obtained from purified B cells of healthy controls using the High Pure RNA Isolation Kit (Roche). Reverse transcription PCR was performed using reverse transcribed cDNA from these samples and the level of GAPDH gene and HLA-G gene was determined using the following primers: GAPDH sense: 5′-CGG AGT CAA CGG ATT TGG TC-3′, GAPDH antisense: 5′-AAT CAT ATT GGA ACA TGT AAA CCA TGT AGT-3′. HLA-G sense: 5′-CTG CTT GTC CTT GCA GCT GTA G-3′; HLA-G antisense: 5′-CCT TTT CAA TCT GAG CTC TTC TTT CT-3′.
Immune cell activation
Freshly isolated PBMCs obtained from CLL patients and healthy donors were cultured in the presence of anti-ILT2 blocking antibody (HP-F1) (10 μg/mL), anti-HLA-G blocking antibody (87G)(10 μg/mL), pan-anti-MHC class I blocking antibody (clone W6/32) (10 μg/mL), or irrelevant mouse IgG1, and stimulated with IL-2 (50 U/mL) or PHA (5 µg/mL) for 48 hours. The expression of CD69 was evaluated by flow cytometry. The effect of IL-2 and PHA on the expression of ILT2 and its ligands in B and T cells was also analyzed by flow cytometry.
Proliferation assays
PBMCs were labeled with 1 µM 5,6-carboxyfluorescein diacetate succinimidyl ester (CFSE) (Sigma-Aldrich) and cultured in the presence of the anti-ILT2 blocking antibody, anti-HLA-G blocking antibody, pan-HLA-I blocking antibody and IL-2 for 7 d. CFSE staining was analyzed by flow cytometry.
Analysis of T cell cytotoxicity
To analyze T cell cytotoxicity, a CD107a degranulation assay was performed. PBMCs were stimulated for 5 hours with plate-coated anti-CD3 antibody (Clone HIT3a, BD Biosciences, 10 µg/mL) and the anti-ILT2 blocking antibody or control IgG; and in the presence of monensin and anti-CD107a-PE. Finally, cells were analyzed by flow cytometry.
Intracellular cytokine staining
To study the intracellular production of interferon-γ (IFN-γ), tumor necrosis factor-α (TNF-α) and IL-2, PBMCs were incubated with anti-ILT2 blocking antibody or irrelevant mouse IgG1 antibody for 48 hours. Cells were then washed and incubated for 4 hours in the presence of 50 ng/mL PMA and 500 ng/mL ionomycin, with 1 µM brefeldin A or 1 µM monensin. Intracellular cytokine expression was then analyzed by flow cytometry.
Statistics
The relationship between continuous and categorical prognostic variables was evaluated by Mann–Whitney U test, and Wilcoxon Matched-Pairs Signed Ranks test was used for intra-group comparisons.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Authorship
Contribution: MVA collaborated in the design of the study, designed and performed the experiments, analyzed the data, and wrote and edited the manuscript. SLH performed experiments and analyzed data. APGR, ARP and EGG diagnosed the patients, made the clinical evaluation and laboratory analyses of the patients, collected and analyzed the clinical data, and collaborated with the manuscript edition. LHZ and ALS participated in the experimental design and development of the study, and collaborated with the manuscript edition. SG designed the study, analyzed the data, and wrote and edited the manuscript.
KONI_A_1353856_Supplementary_materials.pdf
Download PDF (491 KB)Acknowledgments
The authors would like to thank Miguel López-Botet for providing the HP-F1 monoclonal antibody and Juan Ramón de los Toyos-González for providing the mouse IgG1 antibody. This work was supported by the Spanish grants of Instituto de Salud Carlos III (PI12/01280 and PI16/01485) and FEDER European Union. MVA and SLH hold a Severo Ochoa Grant (BP-12076 and BP14–150). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Zenz T, Mertens D, Kuppers R, Dohner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer 2010; 10:37-50; PMID:19956173; https://doi.org/10.1038/nrc2764
- Puente XS, Pinyol M, Quesada V, Conde L, Ordóñez GR, Villamor N, Escaramis G, Jares P, Beà S, González-Díaz M, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011; 475(7354):101-5; PMID:21642962; https://doi.org/10.1038/nature10113
- Gonzalez-Rodriguez AP, Contesti J, Huergo-Zapico L, López-Soto A, Fernández-Guizán A, Acebes-Huerta A, Gonzalez-Huerta AJ, Gonzalez E, Fernandez-Alvarez C, Gonzalez S. Prognostic significance of CD8 and CD4 T cells in chronic lymphocytic leukemia. Leuk Lymphoma 2010; 51:1829-36; PMID:20846097; https://doi.org/10.3109/10428194.2010.503820
- Palmer S, Hanson CA, Zent CS, Porrata LF, LaPlant B, Geyer SM, Markovic SN, Call TG, Bowen DA, Diane F, et al. Prognostic importance of T and NK-cells in a consecutive series of newly diagnosed patients with chronic lymphocytic leukaemia. Br J Haematol 2008; 141:607-14; PMID:18384436; https://doi.org/10.1111/j.1365-2141.2008.07070.x
- Huergo-Zapico L, Acebes-Huerta A, Gonzalez-Rodriguez AP, Contesti J, Gonzalez-García E, Payer AR, Villa-Álvarez M, Fernández-Guizán A, López-Soto A, Gonzalez S. Expansion of NK cells and reduction of NKG2D expression in chronic lymphocytic leukemia. Correlation with progressive disease. PLoS One 2014; 9:e108326; PMID:25286418; https://doi.org/10.1371/journal.pone.0108326
- Riches JC, Gribben JG. Understanding the immunodeficiency in chronic lymphocytic leukemia: Potential clinical implications. Hematol Oncol Clin North Am 2013; 27:207-35; PMID:23561470; https://doi.org/10.1016/j.hoc.2013.01.003
- Moreira J, Rabe KG, Cerhan JR, Kay NE, Wilson JW, Call TG, Leis JF, Jelinek DF, Schwager SM, Bowen DA, et al. Infectious complications among individuals with clinical monoclonal B-cell lymphocytosis (MBL): A cohort study of newly diagnosed cases compared to controls. Leukemia 2013; 27:136-41; PMID:22781591; https://doi.org/10.1038/leu.2012.187
- Ramsay AG, Johnson AJ, Lee AM, Gorgün G, Le Dieu R, Blum W, Byrd JC, Gribben JG. Chronic lymphocytic leukemia T cells show impaired immunological synapse formation that can be reversed with an immunomodulating drug. J Clin Invest 2008; 118:2427-37; PMID:18551193; https://doi.org/10.1172/JCI35017
- Hamblin AD, Hamblin TJ. The immunodeficiency of chronic lymphocytic leukaemia. Br Med Bull 2008; 87:49-62; PMID:18755702; https://doi.org/10.1093/bmb/ldn034
- Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell 2015; 27:450-61; PMID:25858804; https://doi.org/10.1016/j.ccell.2015.03.001
- Weber J. Immune checkpoint proteins: A new therapeutic paradigm for cancer–preclinical background: CTLA-4 and PD-1 blockade. Semin Oncol 2010; 37:430-9; PMID:21074057; https://doi.org/10.1053/j.seminoncol.2010.09.005
- Cosman D, Fanger N, Borges L, Kubin M, Chin W, Peterson L, Hsu ML. A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity 1997; 7:273-82; PMID:9285411; https://doi.org/10.1016/S1074-7613(00)80529-4
- Colonna M, Navarro F, Bellón T, Llano M, García P, Samaridis J, Angman L, Cella M, López-Botet M. A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells. J Exp Med 1997; 186:1809-18; PMID:9382880; https://doi.org/10.1084/jem.186.11.1809
- Lamar DL, Weyand CM, Goronzy JJ. Promoter choice and translational repression determine cell type-specific cell surface density of the inhibitory receptor CD85j expressed on different hematopoietic lineages. Blood 2010; 115:3278-86; PMID:20194892; https://doi.org/10.1182/blood-2009-09-243493
- Liang S, Zhang W, Horuzsko A. Human ILT2 receptor associates with murine MHC class I molecules in vivo and impairs T cell function. Eur J Immunol 2006; 36:2457-71; PMID:16897816; https://doi.org/10.1002/eji.200636031
- Navarro F, Llano M, Bellón T, Colonna M, Geraghty DE, López-Botet M. The ILT2(LIR1) and CD94/NKG2A NK cell receptors respectively recognize HLA-G1 and HLA-E molecules co-expressed on target cells. Eur J Immunol 1999; 29:277-83; PMID:9933109; https://doi.org/10.1002/(SICI)1521-4141(199901)29:01<277::AID-IMMU277>3.0.CO;2-4
- Shiroishi M, Tsumoto K, Amano K, Shirakihara Y, Colonna M, Braud VM, Allan DS, Makadzange A, Rowland-Jones S, Willcox B, et al. Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc Natl Acad Sci U S A 2003; 100:8856-61; PMID:12853576; https://doi.org/10.1073/pnas.1431057100
- Bellon T, Kitzig F, Sayos J, Lopez-Botet M. Mutational analysis of immunoreceptor tyrosine-based inhibition motifs of the Ig-like transcript 2 (CD85j) leukocyte receptor. J Immunol 2002; 168:3351-9; PMID:11907092; https://doi.org/10.4049/jimmunol.168.7.3351
- Naji A, Menier C, Morandi F, Agaugué S, Maki G, Ferretti E, Bruel S, Pistoia V, Carosella ED, Rouas-Freiss N. Binding of HLA-G to ITIM-bearing Ig-like transcript 2 receptor suppresses B cell responses. J Immunol 2014; 192:1536-46; PMID:24453251; https://doi.org/10.4049/jimmunol.1300438
- Saverino D, Fabbi M, Ghiotto F, Merlo A, Bruno S, Zarcone D, Tenca C, Tiso M, Santoro G, Anastasi G, et al. The CD85/LIR-1/ILT2 inhibitory receptor is expressed by all human T lymphocytes and down-regulates their functions. J Immunol 2000; 165:3742-55; PMID:11034379; https://doi.org/10.4049/jimmunol.165.7.3742
- Cosman D, Fanger N, Borges L. Human cytomegalovirus, MHC class I and inhibitory signalling receptors: more questions than answers. Immunol Rev 1999; 168:177-85; PMID:10399074; https://doi.org/10.1111/j.1600-065X.1999.tb01292.x
- Naji A, Menier C, Maki G, Carosella ED, Rouas-Freiss N. Neoplastic B-cell growth is impaired by HLA-G/ILT2 interaction. Leukemia 2012; 26:1889-92; PMID:22441169; https://doi.org/10.1038/leu.2012.62
- Dietrich J, Cella M, Colonna M. Ig-like transcript 2 (ILT2)/leukocyte Ig-like receptor 1 (LIR1) inhibits TCR signaling and actin cytoskeleton reorganization. J Immunol 2001; 166:2514-21; PMID:11160312; https://doi.org/10.4049/jimmunol.166.4.2514
- Lin A, Chen HX, Zhu CC, Zhang X, Xu HH, Zhang JG, Wang Q, Zhou WJ, Yan WH. Aberrant human leukocyte antigen-G expression and its clinical relevance in hepatocellular carcinoma. J Cell Mol Med 2010; 14:2162-71; PMID:19799650; https://doi.org/10.1111/j.1582-4934.2009.00917.x
- Attia MA, Nosair NA, Gawally A, Elnagar G, Elshafey EM. HLA-G expression as a prognostic indicator in B-cell chronic lymphocytic leukemia. Acta Haematol 2014; 132:53-8; PMID:24557341; https://doi.org/10.1159/000353757
- Nuckel H, Rebmann V, Durig J, Duhrsen U, Grosse-Wilde H. HLA-G expression is associated with an unfavorable outcome and immunodeficiency in chronic lymphocytic leukemia. Blood 2005; 105:1694-8; PMID:15466928; https://doi.org/10.1182/blood-2004-08-3335
- Maki G, Hayes GM, Naji A, Tyler T, Carosella ED, Rouas-Freiss N, Gregory SA. NK resistance of tumor cells from multiple myeloma and chronic lymphocytic leukemia patients: Implication of HLA-G. Leukemia 2008; 22:998-1006; PMID:18288133; https://doi.org/10.1038/leu.2008.15
- Godal R, Bachanova V, Gleason M, McCullar V, Yun GH, Cooley S, Verneris MR, McGlave PB, Miller JS. Natural killer cell killing of acute myelogenous leukemia and acute lymphoblastic leukemia blasts by killer cell immunoglobulin-like receptor-negative natural killer cells after NKG2A and LIR-1 blockade. Biol Blood Marrow Transplant 2010; 16:612-21; PMID:20139023; https://doi.org/10.1016/j.bbmt.2010.01.019
- Rouas-Freiss N, Moreau P, LeMaoult J, Carosella ED. The dual role of HLA-G in cancer. J Immunol Res 2014; 2014:359748; PMID:24800261; https://doi.org/10.1155/2014/359748
- Ince MN, Harnisch B, Xu Z, Lee SK, Lange C, Moretta L, Lederman M, Lieberman J. Increased expression of the natural killer cell inhibitory receptor CD85j/ILT2 on antigen-specific effector CD8 T cells and its impact on CD8 T-cell function. Immunology 2004; 112:531-42; PMID:15270723; https://doi.org/10.1046/j.1365-2567.2004.01907.x
- Greipp PT, Smoley SA, Viswanatha DS, Frederick LS, Rabe KG, Sharma RG, Slager SL, Van Dyke DL, Shanafelt TD, Tschumper RC, et al. Patients with chronic lymphocytic leukaemia and clonal deletion of both 17p13.1 and 11q22.3 have a very poor prognosis. Br J Haematol 2013; 163:326-33; PMID:24032430; https://doi.org/10.1111/bjh.12534
- Guièze R, Robbe P, Clifford R, de Guibert S, Pereira B, Timbs A, Dilhuydy MS, Cabes M, Ysebaert L, Burns A, et al. Presence of multiple recurrent mutations confers poor trial outcome of relapsed/refractory CLL. Blood 2015; 126:2110-7; PMID:26316624; https://doi.org/10.1182/blood-2015-05-647578
- Knittel G, Liedgens P, Reinhardt HC. Targeting ATM-deficient CLL through interference with DNA repair pathways. Front Genet 2015; 6:207; PMID:26113859; https://doi.org/10.3389/fgene.2015.00207
- Chiorazzi N. Implications of new prognostic markers in chronic lymphocytic leukemia. Hematology Am Soc Hematol Educ Program 2012; 2012:76-87; PMID:23233564; https://doi.org/10.1182/asheducation-2012.1.76
- Sancho D, Gomez M, Sanchez-Madrid F. CD69 is an immunoregulatory molecule induced following activation. Trends Immunol 2005; 26:136-40; PMID:15745855; https://doi.org/10.1016/j.it.2004.12.006
- Riches JC, Davies JK, McClanahan F, Fatah R, Iqbal S, Agrawal S, Ramsay AG, Gribben JG. T cells from CLL patients exhibit features of T-cell exhaustion but retain capacity for cytokine production. Blood 2013; 121:1612-21; PMID:23247726; https://doi.org/10.1182/blood-2012-09-457531
- Acebes-Huerta A, Huergo-Zapico L, Gonzalez-Rodriguez AP, Fernandez-Guizan A, Payer AR, López-Soto A, Gonzalez S. Lenalidomide induces immunomodulation in chronic lymphocytic leukemia and enhances antitumor immune responses mediated by NK and CD4 T cells. Biomed Res Int 2014; 2014:265840; PMID:25313353; https://doi.org/10.1155/2014/265840
- Buschle M, Campana D, Carding SR, Richard C, Hoffbrand AV, Brenner MK. Interferon gamma inhibits apoptotic cell death in B cell chronic lymphocytic leukemia. J Exp Med 1993; 177:213-8; PMID:7678114; https://doi.org/10.1084/jem.177.1.213
- Forconi F, Moss P. Perturbation of the normal immune system in patients with CLL. Blood 2015; 126:573-81; PMID:26084672; https://doi.org/10.1182/blood-2015-03-567388
- Favier B, Lemaoult J, Lesport E, Carosella ED. ILT2/HLA-G interaction impairs NK-cell functions through the inhibition of the late but not the early events of the NK-cell activating synapse. FASEB J 2010; 24:689-99; PMID:19841038; https://doi.org/10.1096/fj.09-135194
- Rizzo R, Audrito V, Vacca P, Rossi D, Brusa D, Stignani M, Bortolotti D, D'Arena G, Coscia M, Laurenti L, et al. HLA-G is a component of the chronic lymphocytic leukemia escape repertoire to generate immune suppression: Impact of the HLA-G 14 base pair (rs66554220) polymorphism. Haematologica 2014; 99:888-96; PMID:24362551; https://doi.org/10.3324/haematol.2013.095281
- Ozet G, Falay M, Dagdas S, Ceran F. Determination of HLA-G Expression and evaluation of its role as a prognostic factor in chronic lymphocytic leukemia. J Clin Lab Anal 2016; 30(5):399-403; PMID:26303056
- Erikci AA, Karagoz B, Ozyurt M, Ozturk A, Kilic S, Bilgi O. HLA-G expression in B chronic lymphocytic leukemia: A new prognostic marker? Hematology 2009; 14:101-5; PMID:19298722; https://doi.org/10.1179/102453309X385197
- Perez-Chacon G, Rosado S, Rebolleda N, Losada-Fernandez I, Vargas JA, Morado M, Jorda J, Perez-Aciego P. Prognostic irrelevance of HLA-G in B-cell chronic lymphocytic leukemia. Int J Lab Hematol 2009; 31:327-37; PMID:18241213; https://doi.org/10.1111/j.1751-553X.2008.01030.x
- Stankovic T, Skowronska A. The role of ATM mutations and 11q deletions in disease progression in chronic lymphocytic leukemia. Leuk Lymphoma 2014; 55:1227-39; PMID:23906020; https://doi.org/10.3109/10428194.2013.829919
- Döhner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L, Döhner K, Bentz M, Lichter P. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med 2000; 343:1910-6; PMID:11136261; https://doi.org/10.1056/NEJM200012283432602
- Marasca R, Maffei R, Martinelli S, Fiorcari S, Bulgarelli J, Debbia G, Rossi D, Rossi FM, Rigolin GM, Martinelli S, et al. Clinical heterogeneity of de novo 11q deletion chronic lymphocytic leukaemia: Prognostic relevance of extent of 11q deleted nuclei inside leukemic clone. Hematol Oncol 2013; 31:88-95; PMID:23027683; https://doi.org/10.1002/hon.2028
- Seifert M, Sellmann L, Bloehdorn J, Wein F, Stilgenbauer S, Dürig J, Küppers R. Cellular origin and pathophysiology of chronic lymphocytic leukemia. J. Exp. Med 2012; 209(12):2183-98; PMID:23091163; https://doi.org/10.1084/jem.20120833
- Riches JC, Davies JK, McClanahan F, Fatah R, Iqbal S, Agrawal S, Ramsay AG, Gribben JG. T cells from CLL patients exhibit features of T-cell exhaustion but retain capacity for cytokine production. Blood 2013; 121(9):1612-21; PMID:23247726; https://doi.org/10.1182/blood-2012-09-457531
- Riches JC, Gribben JG. Immunomodulation and immune reconstitution in chronic lymphocytic leukemia. Semin Hematol 2014; 51(3):228-34; PMID:25048786; https://doi.org/10.1053/j.seminhematol.2014.05.006