ABSTRACT
Vγ9Vδ2 T cells are anti-tumor immune effectors of growing interest in cancer including Pancreatic Ductal Adenocarcinoma (PDAC), an especially aggressive cancer characterized by a hypoxic and nutrient-starved immunosuppressive microenvironment. Since Butyrophilin 3 A (BTN3A) isoforms are critical activating molecules of Vγ9Vδ2 T cells, we set out to study BTN3A expression under both basal and stress conditions in PDAC primary tumors, and in novel patient-derived xenograft and PDAC-derived cell lines. BTN3A2 was shown to be the most abundant isoform in PDAC and was stress-regulated. Vγ9Vδ2 T cells cytolytic functions against PDAC required BTN3A and this activity was strongly enhanced by the agonist anti-BTN3A 20.1 mAb even under conditions of hypoxia. In PDAC primary tumors, we established that BTN3A expression and high plasma levels of soluble BTN3A were strongly associated with a decreased survival. These findings may have important implications in the design of new immunotherapeutic strategies that target BTN3A for treating PDAC.
Introduction
Pancreatic Ductal Adenocarcinoma (PDAC) is an aggressive disease with a 5-year survival rate below 8%,Citation1 for which conventional treatments have had a very limited impact. The efficacy of immunotherapy strategies in melanomaCitation2 and non-small cell lung carcinomaCitation3 has paved the way for their exploration in other solid tumors with poor prognosis. While the association of systemic and intra-tumoral immune status with PDAC prognosticCitation4-6 would suggest it relevant to develop such approaches in PDAC, it is thought that the hostile tumor micro-environment of PDAC may ultimately restrain the efficacy of immunotherapy. Indeed, the poorly vascularized and fibrotic stroma of PDAC gives rise to nutrient deprivation and hypoxia which induces the selection of aggressive tumor cellsCitation7 together with an immunosuppressive tumor microenvironment.Citation8-10 Novel original strategies that take into account these specificities of PDAC are therefore being actively investigated.
The PDAC micro-environment contains γδ T cellsCitation11,12 which constitute up to 75% of PDAC-infiltrating T cells.Citation6 γδ T cells encompass two major cytotoxic subtypes: the Vδ1 subtype that is mainly intra-epithelial and the Vγ9Vδ2 subtype which accounts for 1–10% of human circulating T lymphocytes but also comprises a considerable intra-epithelial compartment.Citation13 Vγ9Vδ2 T cells can migrate towards tumorsCitation14 and are found within various human solid tumors.Citation15,16 The Vγ9Vδ2 T cell receptor (TCR) senses phosphoAntigen, a metabolite of the mevalonate pathway of cholesterol biosynthesis. The blockade of this pathway with pharmacological agents, namely aminobisphosphonates, leads to an accumulation of phoshoAntigen in malignant cells, that triggers Vγ9Vδ2 T cells activation and their MHC-unrestricted cytotoxicity towards tumor cells.Citation17-19 Although intratumor γδ T cells have been shown to display high cytotoxicity against PDAC ex vivo,Citation11 this cytotoxicity has been suggested to be dampened in patients with PDAC.Citation20 Furthermore, recent studies have shown tumor-infiltrating γδ T cells to instead drive the progression of PDAC in miceCitation6,21 by restraining αβ T cells activation. Therefore, novel strategies are necessary to counter these γδ T cells-dependent immune escape mechanisms and restore Vγ9Vδ2 T cells cytolytic functions in PDAC.
TCR agonists including BrHpp (a synthetic phosphoAntigen) and aminobisphosphonates such as Zoledronate, are able to enhance Vγ9Vδ2 T cell anti-tumor functions and to increase the survival of PDAC cell line-xenograft mice.Citation22 In this regard, the Butyrophilin3 A (BTN3A, CD277) subfamily through its TCR-mediated sensing of phosphoAntigens, has been shown to be a critical determinant in the recognition of human tumors by Vγ9Vδ2 T cells.Citation23-25 The BTN3A subfamily belongs to the B7 family of costimulatory molecules and is composed of three isoforms (BTN3A1, BTN3A2 and BTN3A3).Citation26 While expressed by both immune cells and tumor cells,Citation24-27 BTN3A is more highly expressed in epithelial tumor tissues than in normal tissues, and is upregulated by tumor-associated soluble factors.Citation26,28,29 Our team has demonstrated that targeting BTN3A molecules with antagonist anti-BTN3A 103.2 or 108.5 monoclonal antibodies (mAb) can completely abrogate Vγ9Vδ2 T cell mediated lysis of tumors.Citation23,25 Conversely, the anti-BTN3A 20.1 mAb can strongly boost Vγ9Vδ2 T cells cytolytic function. Indeed, BTN3A targeting with this agonist mAb can sensitize tumor cell linesCitation23 and resistant primary leukemic blasts to Vγ9Vδ2 T cells killing including when tested in a xenograft model.Citation25
Together, these data have opened new perspectives in Vγ9Vδ2 T cells based-immunotherapies and shown the potential of BTN3A 20.1 mAb towards enhancing Vγ9Vδ2 T cell anti-tumor functions. In this study, we thus decided to investigate how BTN3A molecules might improve Vγ9Vδ2 T cells-based immunotherapy in PDAC.
Results
BTN3A is overexpressed in human pancreatic tumors
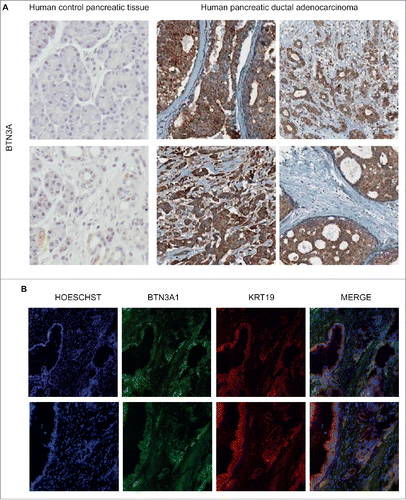
BTN3A expression in primary pancreatic tumors was addressed by comparing to peritumorally obtained control pancreatic tissue. Immunohistochemical analysis of PDAC Tissue Micro Arrays (TMA) confirmed BTN3A expression in all the tested tumor samples (n = 32) (). By contrast, BTN3A expression was either absent or barely detectable in control pancreatic tissue. Immunofluorescence analysis also revealed prominent epithelial staining as determined by a co-localization of BTN3A and Keratin19 staining ().
Figure 1. BTN3A epithelial expression in human pancreatic tumors. (A) Immunohistochemical characterization of BTN3A expression assessed in human control pancreas (peritumoral tissue) and human inflammatory primary pancreatic tumors Tissue Micro Array (TMA). Representative staining with anti-BTN3A mAb. Magnification x10. (B) Immunolocalization of BTN3A and keratin19 (KRT19). Merged images show the co-localization of BTN3A and KRT19 in pancreatic tumor tissue. Magnification x10 (upper panel) and x20 (lower panel).
BTN3A2 is the most abundant isoform expressed by PDAC
BTN3A expression was first studied in cell lines that have been extensively used as PDAC models.Citation30 These included PANC-1, MiaPACA2, and BxPc3 that have been shown to differ in their KRAS, P53, SMAD4 mutational status as well as Patu8902 and Patu8988 t that originate from primary and liver-metastatic PDAC and are respectively highly metastatic and poorly metastatic in mice.Citation31,32 We observed BTN3A surface expression in all these pancreatic tumor cell lines, irrespective of their origin, mutational status or differentiation state (, upper panel).
Figure 2. BTN3A isoform expression in pancreatic cell lines. (A) Representative overlays of BTN3A global surface expression (black line) compared to isotype control (grey line) in pancreatic cell lines (n = 5) as assessed by flow cytometry (upper panel) and in PDX-derived cell lines (n = 4) using an anti-BTN3A mAb (lower panel). (B) Transcriptional expression of BTN3A isoforms. Representative expression of the three BTN3A isoforms in pancreatic- (upper panel) and PDX-derived cell lines (lower panel). qRT PCR analyses were performed on total RNA isolated from pancreatic cell lines. Data were normalized using Peptidylpropyl isomerase A (PPIA) as an endogenous control (ΔCt = CtTarget gene – CtPPIA). Results were expressed as mean 2−ΔCt and shown as percentage of total quantified BTN3A isoforms. (C) BTN3A protein expression. Western Blot analysis of total protein extracts of pancreatic cell lines. Extracts were loaded in 10% SDS PAGE gel and membranes were hybridized with anti-BTN3A 20.1 mAb and anti-Grb2 as a loading control.
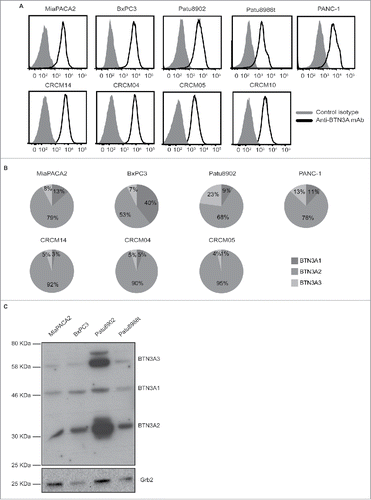
Secondly, we evaluated PDAC patient-derived xenograft cell lines that were derived from fresh tumors with various differentiation states and prognosis and established in our laboratory (n = 18).Citation33 BTN3A was expressed in all of the tested patient-derived cell lines including one derived from a liver-metastasis (CRCM-14) ( lower panel).
As the available anti-BTN3A antibodies (used to confirm BTN3A surface expression) do not discriminate between the 3 BTN3A isoforms, we investigated BTN3A isoform expression at the transcript and protein levels by qRT-PCR and Western Blot. At the transcriptional level, we observed that all three isoforms were expressed with BTN3A2 the most abundant isoform in all of the tested pancreatic ( upper panel) and patient-derived xenograft-derived cell lines ( lower panel). At the protein level, BTN3A2 was also found to be the most abundant isoform (mean density quantification relative to loading control: 8.2 ± 6.2) when compared with BTN3A1 (1.3 ± 0.75) and BTN3A3 (3.7 ± 3.4) ( and ).
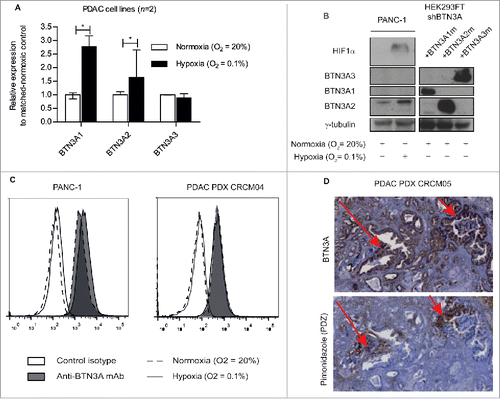
Figure 3. Hypoxia-induced regulation of BTN3A isoforms and BTN3A surface expression. (A) Transcriptional analysis by qRT PCR of the 3 BTN3A isoforms expression in PANC-1 and MiaPACA2 under hypoxia. Data were normalized using Peptidylprolyl isomerase A (PPIA) as an endogenous control; (ΔCt = Ct target gene – Ct PPIA) and fold change (2−ΔΔCt) was established using the expression of the matched-BTN3A isoform in normoxia as a calibrator gene. Results were expressed as median 2−ΔΔCt ± interquartile range and statistical significance was established using Mann Whitney U Test. *p < 0.05. Cumulative data from 2 independent cell lines performed in duplicate. (B) Western-blot analysis of total protein extracts of Pancreatic Ductal Adenocarcinoma (PDAC) cells line PANC-1 in normoxic and hypoxic conditions. Extracts were loaded in 10% SDS PAGE gel and membranes were hybridized with anti-BTN3A mAb, anti-HIF1α and anti-ɣtubulin as a loading control. BTN3A knock-down HEK293FT (sh#284; clone#30) transiently transfected with BTN3A1 m, BTN3A2 m, BTN3A3 m are used as size controls. (C) Flow cytometry analysis of BTN3A surface expression in PANC-1 and in PDAC PDX CRCM 04-derived cell lines, under normoxia (dashed line) and hypoxia (full line). Control isotype (open histogram) and anti-BTN3A mAb (filled histogram) are depicted. Representative data from 3 independent experiments. (D) BTN3A and PDZ staining of hypoxic regions in PDX tumor CRCM05. (Magnification 10X).
Collectively, these data demonstrated BTN3A surface expression in PDAC tumors with BTN3A2 isoform dominant ie. the isoform without the B30.2 cytoplasmic domain
The expression of BTN3A isoforms is modulated under hypoxia in PDAC
We aimed to determine whether BTN3A isoform expression was modified under hypoxic stress in PANC-1 and MiaPACA2 cell lines. BTN3A1 and BTN3A2 transcript levels were higher under hypoxia compared to normoxia (fold increase of 2.78 [2.58–3.18]; P = 0.0294 and 1.64 [1.3–2.65]; P = 0.0265) (). BTN3A isoform expression at the protein level was also assessed by Western Blot (), in parallel with HIF1α expression as a control marker of hypoxia (). BTN3A2 protein expression (36 kDa) was found to be higher under hypoxia in both PANC-1 (0.34 versus 0.22) () and a PDAC patient-derived xenograft (CRCM04) (0.45 versus 0.2) (Supplementary ). In contrast, BTN3A1 and BTN3A3 could not be detected at their respective expected molecular weights of 57 and 65 kDa ().
We next asked whether BTN3A surface expression was influenced by hypoxia. Flow cytometry analysis showed global surface expression of BTN3A in both PANC-1 and a PDAC-patient-derived xenograft (CRCM04) to remain stable under hypoxia when tested in vitro (). To test in vivo, we used a xenograft mouse model transplanted with human PDAC (CRCM05) and looked for BTN3A expression in pimonidazole-stained hypoxic regions. BTN3A was still expressed in the hypoxic region of the PDAC tumor in vivo (). In summary, our data suggest that while hypoxic conditions preferably enhance expression of the BTN3A2 isoform, global surface expression of BTN3A remains stable.
BTN3A expression is also tuned under nutrient starvation
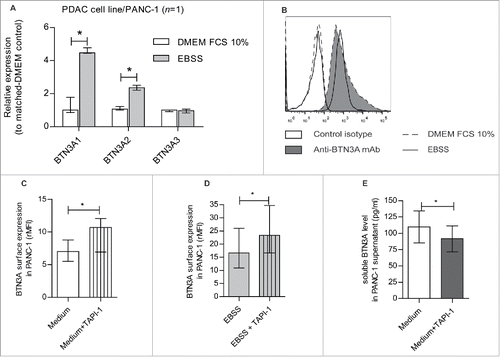
At the transcript level, the expression of the BTN3A1 and BTN3A2 isoforms were increased under nutrient starvation when compared to standard nutrient supply when examined in the PANC-1 cell line (fold increase of 4.47 [4.44–4.77]; P = 0.0294 and 2.35 [2.2–2.51]; P = 0.0294 respectively) (). To determine if this also translated to an increase in global expression at the membrane surface, we also assessed BTN3A expression by flow cytometry in PANC-1 and PDAC patient-derived xenograft-derived (CRCM04) cell lines cultured in basal condition i.e standard media and nutrient supply (DMEM FCS10%) or under conditions of nutrient starvation (EBSS). As illustrated in , surface expression of BTN3A was found to not significantly differ between the two conditions.
Figure 4. Nutrient starvation-induced regulation of BTN3A isoforms, BTN3A surface expression and release under soluble form. (A) Transcriptional analysis by qRT PCR of the 3 BTN3A isoforms expression in PANC-1 cell line under nutrient starvation. Data were normalized using Peptidylprolyl isomerase A (PPIA) as an endogenous control; (ΔCt = Ct target gene –Ct PPIA) and fold change (2−ΔΔCt) was established using the expression of the matched BTN3A isoform in the DMEM FCS 10% condition as a calibrator gene. Results were expressed as median 2−ΔΔCt ± interquartile range and statistical significance was established using Mann Whitney U Test. *p < 0.05. Cumulative data from 2 independent experiments performed in duplicate. (B) BTN3A surface expression under nutrient starvation. Flow cytometry analysis of BTN3A surface expression in PANC-1 cell line and PDAC-PDX CRCM 04, cultured with DMEM FCS 10% (medium, dashed line) or EBSS (nutrient starvation, full line). Control isotype (open histogram) and anti-BTN3A mAb (filled histogram) are depicted. Representative data from 2 independent experiments. (C, D) Effect of MMP inhibitor TAPI-1 on BTN3A surface expression. Flow cytometry analysis of BTN3A expression in PANC-1 cell line treated or not with TAPI-1 (20 µM), under DMEM 10% FCS culture (C) and 24 hours-nutrient starvation (D). Cumulative data from 3 independent experiments. Results are expressed as median of Median Fluorescence Intensity Ratio (rMFI) ± interquartile range and statistical significance was established using paired t-test. *p < 0.05. (E) Effect of MMP inhibitor TAPI-1 on BTN3A release as a soluble form. ELISA analysis of BTN3A in PANC-1 supernatants treated or not with TAPI-1 (20 µM). Concentrations are expressed in pg/ml. Cumulative data of 2 independent experiments performed in duplicate. Statistical significance was established using paired t-test. *p < 0.05.
BTN3A is shed under soluble form via a mechanism partly mediated by MMP
The increased transcript level of BTN3A1 and BTN3A2 observed under nutrient starvation () was not associated with an overall increase in surface expression of BTN3A (). We asked therefore whether MMP cleavage could participate in BTN3A regulation. BTN3A expression was assessed in the PANC-1 cell line cultured under basal or nutrient-starvation conditions in the presence or absence of TAPI-1, a broad MMP inhibitor. Under basal conditions, the addition of TAPI-1 resulted in a significantly higher surface expression of BTN3A (7 [5.5–8.8]) versus 10.7 [6.9–12] for untreated cells; P = 0.0123)) (). Similarly, under nutrient starvation conditions, TAPI-1 also significantly increased BTN3A expression (23.2 [16.6–34.7]) compared to untreated nutrient-starved cells (16.6 [10.9–26]; P = 0.0298) (). These observations indirectly suggest that there is a perpetual BTN3A cleavage by MMP in the PANC-1 cell line irrespective of the culture conditions used.
To determine whether BTN3A is released by PANC-1 cells, we analyzed soluble BTN3A concentrations in recovered supernatants by ELISA. Under basal conditions, the concentrations of BTN3A molecules PANC-1 supernatants were found to be significantly lower level in TAPI-1-treated cells (99.3 ± 27.11 pg/ml versus 81.83 ± 22.1 pg/ml for untreated, P = 0.0355) (). Collectively therefore, our data indicates that BTN3A molecules are released from PANC-1 cells in a process involving MMP.
BTN3A2 participates in Vγ9Vδ2 T cells anti-tumor functions towards PDAC
The role played by BTN3A in the lysis of PDAC by Vγ9Vδ2 T cells was investigated. The lysis of PANC-1 cells by Vγ9Vδ2 T cells was found either to be to be enhanced (P = 0.0156) by BrHpp treatment and reversed by anti-BTN3A 108.5 mAb (P = 0.0313) (). On the contrary, the anti-BTN3A 20.1 mAb significantly increased the lysis of PANC-1 cells (P = 0.0313) (). BrHpp and 20.1 mAb treatment also sensitized BxPC3, Patu8902, Patu8988 t and CRCM04 to Vγ9Vδ2 T cell mediated lysis but had no effect on MiaPACA2 cells (Supplementary ). These findings highlight a key role of BTN3A (when triggered by BrHpp or anti-BTN3A 20.1 mAb) in the enhancement of Vγ9Vδ2 T cell lysis of all of the most poorly-sensitive PDAC cell lines except for MiaPACA2.
Figure 5. Vγ9Vδ2 T cell anti-tumor functions towards PDAC result in BTN3A enhanced PDAC lysis that involve BTN3A2 and are conserved under hypoxia. (A, B) Specific lysis of PANC-1 by effector Vγ9Vδ2 T cells from healthy donors assessed in a standard [51Cr]-release assay in the presence of (A) BrHpp ± anti-BTN3A 108.5 antagonist mAb (n = 6) or (B) agonist anti-BTN3A 20.1 mAb (n = 7). Data are shown for Effector to Target (E:T) ratio 30:1; 10:1 and 1:1. Cumulative data from 3 independent experiments. Results were expressed as median ± interquartile range and statistical significance was established using the non-parametric paired Wilcoxon U-test. *p < 0.05. (C,D,E) Expanded Vγ9Vδ2 T cells from healthy donors (n = 3) were co-cultured with Wild Type BxPc3 or BTN3A2 knock-out BxPc3 cell line and treated with anti-BTN3A agonist (20.1) mAb or BrHpp ± anti-BTN3A antagonist (108.5) mAb or Zoledronate (ZOL). (C) Cytolytic degranulation was assessed by CD107 a/b expression of Vγ9Vδ2 T cells. (D,E) By flow cytometry, TNFα production (D) and IFNγ production (E) were assessed by intracellular staining of Vγ9Vδ2 T cells. Results are expressed as median ± range. Cumulative data from 2 independent experiments. (F,G,H) Expanded Vγ9Vδ2 T cells from healthy donors (n = 7) were co-cultured with PANC-1 cell line and treated with BrHpp ± anti-BTN3A antagonist (108.5) mAb or anti-BTN3A agonist (20.1) mAb in normoxic (20% O2) or hypoxic conditions (0.1% O2). (F) Cytolytic degranulation was assessed by CD107 a/b expression of Vγ9Vδ2 T cells (n = 7). (G,H) By flow cytometry, IFNγ production (G) and TNFα production (H) were assessed by intracellular staining of Vγ9Vδ2 T cells (n = 6). Results are expressed as median ± interquartile range. Cumulative data from 3 independent experiments. Statistical significance was established using the non-parametric paired Wilcoxon U-test. *p < 0.05.
![Figure 5. Vγ9Vδ2 T cell anti-tumor functions towards PDAC result in BTN3A enhanced PDAC lysis that involve BTN3A2 and are conserved under hypoxia. (A, B) Specific lysis of PANC-1 by effector Vγ9Vδ2 T cells from healthy donors assessed in a standard [51Cr]-release assay in the presence of (A) BrHpp ± anti-BTN3A 108.5 antagonist mAb (n = 6) or (B) agonist anti-BTN3A 20.1 mAb (n = 7). Data are shown for Effector to Target (E:T) ratio 30:1; 10:1 and 1:1. Cumulative data from 3 independent experiments. Results were expressed as median ± interquartile range and statistical significance was established using the non-parametric paired Wilcoxon U-test. *p < 0.05. (C,D,E) Expanded Vγ9Vδ2 T cells from healthy donors (n = 3) were co-cultured with Wild Type BxPc3 or BTN3A2 knock-out BxPc3 cell line and treated with anti-BTN3A agonist (20.1) mAb or BrHpp ± anti-BTN3A antagonist (108.5) mAb or Zoledronate (ZOL). (C) Cytolytic degranulation was assessed by CD107 a/b expression of Vγ9Vδ2 T cells. (D,E) By flow cytometry, TNFα production (D) and IFNγ production (E) were assessed by intracellular staining of Vγ9Vδ2 T cells. Results are expressed as median ± range. Cumulative data from 2 independent experiments. (F,G,H) Expanded Vγ9Vδ2 T cells from healthy donors (n = 7) were co-cultured with PANC-1 cell line and treated with BrHpp ± anti-BTN3A antagonist (108.5) mAb or anti-BTN3A agonist (20.1) mAb in normoxic (20% O2) or hypoxic conditions (0.1% O2). (F) Cytolytic degranulation was assessed by CD107 a/b expression of Vγ9Vδ2 T cells (n = 7). (G,H) By flow cytometry, IFNγ production (G) and TNFα production (H) were assessed by intracellular staining of Vγ9Vδ2 T cells (n = 6). Results are expressed as median ± interquartile range. Cumulative data from 3 independent experiments. Statistical significance was established using the non-parametric paired Wilcoxon U-test. *p < 0.05.](/cms/asset/7b19c86c-cd65-4449-82c8-350a5f572292/koni_a_1372080_f0005_b.gif)
The role of BTN3A2 in Vγ9Vδ2 T cell anti-tumor function was also assessed by measuring Vγ9Vδ2 T cell degranulation and TH1 cytokine production towards wild type BxPc3 or CrispR-Cas9 BTN3A2 invalidated-BxPc3 cell lines. Vγ9Vδ2 T cell degranulation (), and the production of TNFα () and IFNγ () were significantly lower towards the BTN3A2 knock out cell line treated with anti-BTN3A 20.1 mAb, BrHpp or aminobisphosphonate Zoledronate (ZOL), than towards wild-type BxPc3 cell line. These data were consistent with the role of BTN3A2 in Vγ9Vδ2 T cell anti-tumor functions towards PDAC.
BTN3A-dependent anti-tumor functions of Vγ9Vδ2 T cells are preserved under hypoxia
We assessed whether hypoxia could affect BTN3A mediated anti-tumor functions of expanded Vγ9Vδ2 T cells from healthy donors. Treatment with the anti-BTN3A agonist 20.1 mAb was associated with a higher Vγ9Vδ2 T cells degranulation capability as determined by CD107 a/b expression (p = 0.0313) (), and a greater production of IFNγ (p = 0.0313) () and TNFα (p = 0.0313) () towards PANC-1 cell line under both normoxic and hypoxic conditions. Treatment with BrHpp also significantly increased degranulation (), and production of IFNγ () and TNFα () by Vγ9Vδ2 T cells towards PANC-1. These anti-tumor functions of BrHpp-treated Vγ9Vδ2 T cells were conserved under hypoxia () and significantly abrogated by anti-BTN3A 108.5 antagonist mAb (P = 0.0313)
Collectively, these data demonstrated the ability of anti-BTN3A 20.1 mAb to boost Vγ9Vδ2 T cell anti-tumor functions even under hypoxic conditions.
BTN3A expression in human primary pancreatic tumors is associated with invasiveness
Immunohistochemistry was used in order to determine whether BTN3A was differentially expressed within PDAC tumors. In PDAC inflammatory tissue micro arrays (n = 34), it was first noted that the percentage of BTN3A staining was significantly higher in the center than in the periphery of the PDAC tumors (5.62 [3.87-7.81] versus 2.84 [1.13–5.49]; P = 0.0036) (, left panel) in PDAC patients. Second, the percentage of BTN3A staining was found to be significantly higher in a group of patients with lymph node involvement (N1) (5.44 [2.87–7.77]; (n = 23)) when compared with patients devoid of lymph node involvement (N0) (2.73 [1.12–4.84]; (n = 9); P = 0.006) (, right panel).
Figure 6. Surface BTN3A expression, BTN3A2 gene expression and soluble BTN3A levels are prognosis marker in patients with Pancreatic Ductal AdenoCarcinoma (PDAC). (A) The median percentage of BTN3A staining was higher in the center than in the periphery of the PDAC Tissue Multi Array (n = 34) (left panel). The median percentage of BTN3A staining was higher in the group of pancreatic tumors with lymph node involvement (N1) compared with the group devoid of lymph node involvement (N0) (n = 32) (right panel). Results were expressed as median percentages ± range. Paired-t test was used to compare differences between the center and the periphery of the tumor and Mann Whitney U test between N0 and N1 groups. 0.001 <**p < 0.01. (B) BTN3A surface expression assessed in PDX-derived cell lines classified in 2 groups: A/short-term survival group (overall survival (OS) ≤ 8 months; n = 6) and B/ long-term survival group (OS > 8 months; n = 12). Surface expression was quantified by Median Fluorescence Intensity minus matched control isotype (MFI). Results are shown as median ± range. (C) Comparative overall survival in patient with respect to BTN3A2 gene expression. The cohort was divided at median of BTN3A2 expression: BTN3A2 Low (n = 85) and BTN3A2 High (n = 85). Data were obtained from TCGA public database. *p <0.05. (D) Correlation between sBTN3A concentrations assessed with “pan-BTN3A” ELISA or “BTN3A1-specific” ELISA in the plasma of PDAC patients' plasmas (n = 45). Data are shown in ng/ml. The correlation was established using the nonparametric Spearman correlation coefficient (rs). ***p < 0.001. (E) Comparative dosage of sBTN3A1 concentration in Healthy Donors (HD) (n = 22), patients with Chronic Calcific Pancreatitis (CCP) (n = 10), Intraductal Papillary Mucinous Neoplasm (IPMN) (n = 8) and PDAC (n = 54). sBTN3A1 levels were assessed by ELISA in the plasma of subjects. Data were expressed in ng/ml, and median concentration of sBTN3A1 ± interquartile range. Significance was established using non-parametric Mann Whitney Test. *p < 0.05. (F, G) Comparative survival in PDAC patients according to (F) sBTN3A (n = 55) and (G) sBTN3A1 dosage (n = 53). Kaplan-Meier curves showing overall survival: (F) in patients with Low sBTN3A (< 8 ng/ml) or High sBTN3A levels (< 8 ng/ml) and (G) in patients with Low sBTN3A1 levels (< 6 ng/ml) or High sBTN3A1 levels (>6 ng/ml). Statistical significance was established with Log-rank (Mantel-Cox) Test. *p <0.05; ***p < 0.0005.
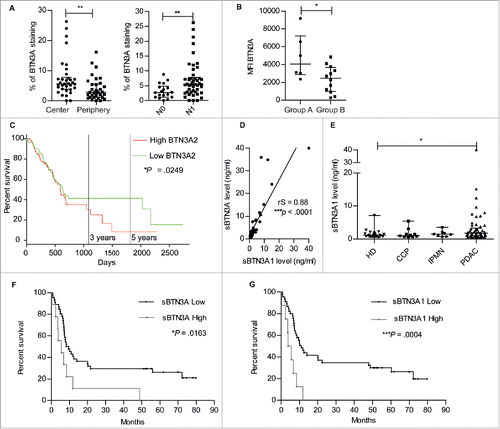
The level of BTN3A surface expression in patient-derived xenograft-derived cell lines was also assessed by flow cytometry. Two different groups of patients were segregated as previously reported (33): 1) a short-term survival group with poorly differentiated tumors with an overall survival less than 8 months and 2) a long-term survival with moderately/well differentiated tumors and an overall survival greater than 8 monthsCitation33 (Supplementary ). We observed that BTN3A expression expressed as Median Fluorescence Intensity, was significantly higher in patients with poorly differentiated tumors and a poor prognosis when compared to patients in the long-term survival group (4057 [2867–7203] versus 2472 [976,5–3660]; P = 0.0441) (). In addition, the available data from TCGA database revealed that patients with higher BTN3A2 gene expression (i.e higher than the median expression of the group; n = 85) had lower overall median survival time than patient with lower BTN3A2 gene expression (n = 85) (596 days versus 634 days; Hazard Ratio = 1.37 [1.04–1.81; P = 0.025) ().
Altogether these data indicate an association of BTN3A expression with tumor progression, and in particular, with poorly differentiated tumors from PDAC patients who exhibited short-term survival.
Soluble BTN3A and BTN3A1 concentration is a prognostic marker in PDAC patients
As BTN3A was found as a released soluble form in pancreatic tumor cell line supernatant, we investigated whether soluble BTN3A was present in plasma samples taken from PDAC patients. We first evaluated whether soluble BTN3A1 concentrations obtained with a “BTN3A1-specific” dosage correlated with concentrations obtained with a “pan-BTN3A” dosage. We found a significant positive correlation between soluble BTN3A and soluble BTN3A1 concentrations (rS = 0.88; P < 0.0001) (). We next compared soluble BTN3A1 concentrations obtained in PDAC patients with concentrations observed in patients with Chronic Calcificating Pancreatitis (CCP) and Intraductal Papillary Mucinous Neoplasm (IPMN). The highest median concentration was observed in PDAC patients (1.78 ng/ml [1.03–3.28]) which was significantly higher than that observed in healthy donors (1.17 ng/ml [0.79–1.7]; P = 0.036) (). With respect to the overall survival of patients with PDAC, soluble BTN3A and BTN3A1 concentrations greater than 8 ng/ml () and 6 ng/ml respectively, were associated with a decreased overall survival (). Altogether these data show the potential of soluble BTN3A and BTN3A1 as biomarkers that reflect the progression and prognosis of PDAC.
Discussion
The identification of novel immunotherapeutic targets while taking into account the specific microenvironment of PDAC is of great concern. We report that the BTN3A subfamily is stress-regulated in PDAC tumors and shed by these cells under basal and nutrient-deprived conditions. We also demonstrate a critical role of BTN3A in mediating Vγ9Vδ2 T cells cytolytic functions against PDAC that is strongly enhanced by the agonist anti-BTN3A 20.1 mAb, even under conditions of hypoxia. Strikingly, we provide evidence that high levels of BTN3A expression in tissues and soluble BTN3A in plasma are prognostic markers in patients with PDAC.
BTN3A expression has rarely been investigated in the primary tumor context.Citation24,25,27,28,29,34 Based on the analysis of human primary tumor samples and novel patient-derived xenograft, we have now established that BTN3A overexpression and a dominant expression of the BTN3A2 isoform is strongly associated with a poor prognosis, in accordance with what was recently described in primary leukemic blastsCitation25 and in gastric cancer.Citation29 In this line, the overexpression of BTN3A2 gene was associated with an increased proliferation and invasion of gastric cancer cell lines.
We show here that BTN3A2 is involved in phosphoAntigen-mediated Vγ9Vδ2 T cells activation towards PDAC. Indeed, the BTN3A subfamily including BTN3A2 has been shown to play a key role in mediating Vγ9Vδ2 T cells sensing of phosphoAntigen-containing tumor cell linesCitation23,24 and primary leukemic blasts.Citation25 Of note, binding of phosphoAntigen by the B30.2 intracellular domain of BTN3A1 has been demonstrated to be critical in this process.Citation23,24,35-37 As a receptor that is devoid of an intracellular domain but can negatively regulate NK cells, it has previously been suggested that BTN3A2 may function as a decoy receptor via the binding of a putative ligand. Hence, we could postulate that the higher level of expression of the BTN3A2 isoform compared to BTN3A1 may facilitate tumor immune escape mechanisms that lead to Vγ9Vδ2 T cells exhaustionCitation38 and/or fail to activate Vγ9Vδ2 T cells in the absence of PAg.
Here, the expression of BTN3A1 and BTN3A2 was higher under stress condition, whereas BTN3A surface expression remained stable. This indicates a tight regulation of membrane BTN3A expression that might occur through the intracellular retention of BTN3A1 and BTN3A2 in the endoplasmic reticulum due to the presence of canonical ER retention/retrieval signals in their intracellular domains (Vantouroux et al., γδ T cell conference 2016) or via BTN3A isoforms shedding. Accordingly, we observed soluble BTN3A isoforms including soluble BTN3A1 in the supernatants of pancreatic cell lines and in the plasma of PDAC patients. In addition, we showed that BTN3A shedding occurred in manner that is in part MMP-dependent, similar to that previously described for the NKG2D ligands MICA/B. We can assume that this increased release might result from enhanced MMP activity and BTN3A expression in PDAC patients. Indeed, both levels of soluble BTN3A and of BTN3A surface expression were associated with a poor prognosis of PDAC patients. Conversely, in patient with inflammatory or non-malignant pancreatic aggression such as chronic calcificating pancreatitis and intraductal papillary mucinous neoplasm, soluble BTN3A1 levels were not different from those found in healthy donors. This suggests that BTN3A1 upregulation might be a specific feature of PDAC associated inflammation.
The present study also confirms BTN3A as a promising therapeutic target: the agonist anti-BTN3A 20.1 mAb was shown to be a potent tool for boosting Vγ9Vδ2 T cell anti-tumor functions against PDAC. BTN3A triggering by the 20.1 mAb occurs mainly through the induction of extra-cellular conformational changes in BTN3A molecules of target cells, resulting in the induction of Vγ9Vδ2 T cell activation.Citation23,39 Interestingly, the anti-BTN3A 20.1 mAb can separately trigger each of the three BTN3A isoforms including BTN3A2.Citation23,39 Hence, the dominant expression of BTN3A2 found in tumors does not preclude the triggering of BTN3A molecules by the agonist 20.1 mAb and the effective activation of Vγ9Vδ2 T cells. Indeed, the anti-BTN3A 20.1 mAb was able to effectively enhance the lysis of various pancreatic cell lines including a novel aggressive patient-derived xenograft cell line. Nevertheless, BTN3A triggering fails to restore the sensitization of MiaPACA2 cell line illustrating the heterogeneous intrinsic sensitivity of pancreatic tumors, partly due to the lack of expression of the adhesion molecule ICAM-1 in this cell line.Citation40
The tumor-induced exhaustion of Vγ9Vδ2 T cells in the PDAC microenvironment has been recently suggested.Citation6 Hence, the 20.1 mAb could be of high therapeutic interest to both restore the cytotoxic functions of intratumoral Vγ9Vδ2 T cells and prevent the exhaustion of adoptively transferred Vγ9Vδ2 T cells. By way of support, we have shown that the agonist anti-BTN3A 20.1 is able to sustain the cytolytic functions of Vγ9Vδ2 T cells towards primary leukemic blasts resulting in their clearance from the bone marrow.Citation25 In addition, this agonist effect on Vγ9Vδ2 T cells anti-tumor function was also maintained under hypoxia conditions, further underscoring its potential as a potent therapeutic tool that can overcome the stress-related characteristics of the PDAC microenvironment.
In conclusion, we have established that BTN3A expression is stress regulated and associated with pancreatic tumor prognosis, and might constitute an immune escape mechanism against Vγ9Vδ2 T cells recognition. However, BTN3A targeting with agonist mAb allows for the sensitization of PDAC to Vγ9Vδ2 T cells lysis even under conditions of hypoxia. Altogether these results underscore a promising pathway to explore in order to improve immunotherapeutic combinatorial strategies to treat PDAC.
Methods
Patients
Thirty-four PDAC samples were formalin-fixed surgical specimens obtained from the Pathology Department of Aix-Marseille University. Demographic data, lymph node status and overall survival corresponding to the length of time from the date of diagnosis of these patients is detailed in Supplementary Table 1.
Plasmas from PDAC patients (n = 67), Chronic Calcific Pancreatitis (n = 12), Intra Papillary Mucinous Neoplasm (n = 8) and Healthy control subjects (n = 23) were collected and provided by Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas (CIBEREHD), Barcelona.
Demographic data, TNM status and overall survival corresponding to the length of time from the date of diagnosis of these patients are provided in Supplementary Table 2.
Healthy pancreatic specimens were taken from peritumoral region during surgery (tissue collection: DC-2013-1857).
Xenograft murine model
Mice were home-bred and maintained under pathogen-free conditions. All animal procedures were in accordance with protocols approved by the local Committee for Animal Experiments.
Patient-derived xenograft murine models were established as previously described.Citation41 Briefly, patient-derived pancreatic tumor pieces (1 mm3) were embedded in Matrigel before being s.c implanted into flank of adult male Swiss nude Mice (Charles River laboratories), carried out under isoflurane anesthesia. Tumors were measured weekly with a caliper until tumor volume reached 1mm.Citation3 At 4 h after intratumoral injection of pimonidazole hydrochloride, pieces of tumor were removed fixed in 4% (wt/vol) formaldehyde or frozen in cold isopentane for further analysis.
Tissue microarrays (TMAs)
The procedure for construction of TMAs was as previously described.Citation42,43 Briefly, cores were punched from the selected paraffin blocks and distributed in new blocks, with 2 cores of 0.6-mm diameter, one from the periphery (P) and one from the center of each tumor (T). TMA serial tissue sections were prepared 24 hours before immunohistochemical processing and stored at 4°C. The dilution of each antibody was determined by pre-screening on the full 4-μm-thick sections before use on TMA sections. The immunoperoxidase procedures were performed using an automated Ventana BenchMark XT autostainer. Measurements of immunoprecipitate densitometry in cores were made for each marker in an individual core after digitization and “cropping” of microscopic images as previously reported.Citation42,43
Immunohistochemistry
Pancreatic sections were fixed in 4% paraformaldehyde and paraffin embedded. Immunohistochemistry was performed using standard procedures. Sections were stained with anti-BTN3A mAb (clone 103.2).
Hypoxic regions in PDAC were revealed using the Hypoxyprobe-1 Plus kit (Hypoxyprobe). Quenching of endogenous peroxidase activity by 3% (vol/vol) H2 O2 was followed by an antigen retrieval step (10 mM sodium citrate, pH 6, 55°C). Finally, sections were incubated with FITC-conjugated anti-pimonidazole monoclonal antibody (1:400, Hypoxyprobe) followed by an incubation with HRP-conjugated anti-FITC antibody (1:100, Hypoxyprobe). Peroxydase activity was revealed using liquid-Diaminobenzidine+ substrate chromogen system (Dako). Counterstaining with Mayer hematoxylin was followed by a bluing step in 0.1% sodium bicarbonate buffer, before final dehydration, clearance, and mounting of the sections.
Confocal microscopy
Ten-micrometer cryosections of pancreatic tissue were dried and fixed with acetone. After nonspecific binding site blockade with 3% bovine serum albumin, 10% fetal calf serum, and 10% goat serum for 30 minutes, tissue sections were labeled 1 hour at room temperature with primary antibodies BTN3A (clone 103.2) and keratin 19 (abnova (clinisciences), Nanterre, France), followed by incubation for 30 minutes at room temperature with secondary antibodies (Alexa Fluor 488 and 568 (Invitrogen, Cergy Pontoise, France)) and SYTOX Blue when nuclei staining was required. Slides were mounted in ProLong Gold (Invitrogen, Cergy Pontoise, France) and observed with a Zeiss LSM 510 confocal microscope. Images were analyzed using Adobe Photoshop 7.0.
Pancreatic cell lines and culture system
MiaPACA2, PANC-1, and BxPC-3 cells were obtained from the American Type Culture Collection. Patu8902 and Patu8988 t were obtained from the Leibniz Institute DSMZ-German Collection of microorganisms and cell cultures. Patient-derived xenograft-derived cell lines were established as previously described.Citation33 All cell lines were periodically tested for Mycoplasma contamination and were Mycoplasma-Free. Pancreatic cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% FCS at 37°C with 5% CO2. To avoid any supplementary stress, all media were preheated at 37°C before rinsing or changing media. Nutrient starvation was obtained by cultivating cells with Earle's Balanced Salt Solution (EBSS) (Ref# 24010–043). Hypoxia experiments were carried out using C-Shuttle Glove Box coupled hypoxia chamber (BioSpherix).
Reagents and antibodies
BrHpp and Zoledronate (ZOL) were obtained from Innate Pharma (Marseilles, France). and Novartis (United Kingdom) respectively. Recombinant human (Rh) IL-2 was obtained from BD Biosciences (San Jose, CA, USA) and TAPI-1 from Peptides International (Louisville, KY, USA). The mAbs used for functional experiments and cytometry are listed in Supplementary Table 3.
Vγ9Vδ2 T cells expansion
Effector γδ T cells were established and maintained as previously described.Citation44 Peripheral Blood Mononuclear Cells (PBMCs) from healthy donors were provided by the local Blood Bank (EFS) and isolated by density gradient centrifugation. PBMCs were stimulated with ZOL (1μM) or BrHpp (3 μM) and rhIL-2 (200IU/ml) at Day 0. From Day 5, rhIL-2 was renewed every two days and cells were kept at 1.5 × 106 /ml for 15 days. The purity of γδ T cells was determined to be greater than 80%.
HEKShBTN3A
BTN3A Knock-down HEK293FT cells (sh#284; clone#30) were cultured and transfected with BTN3A1, BTN3A2, BTN3A3 mutated cDNA-containing plasmids, as described.Citation23
CRISPR plasmid construction
CRISPR plasmid construction was adapted from the protocol previously described.Citation45 pSpCas9(BB)-2 A-GFP (PX458) was a gift from Feng Zhang (Addgene plasmid # 48138). PX458 plasmid was linearized using BbsI (37°C, one hour digestion). sgRNA targeting the sequence 5′-GAGTGAGCAGCTGGACCAAGAGG-3′ within the signal peptide of BTN3A2 (Third exon) was devised manually. The guide oligonucleotides (oligont) targeting BTN3A2 were purchased from Invitrogen:
Top strand oligont was: 5′-CACCGAGTGAGCAGCTGGACCAAG-3′ and Bottom strand oligont was: 5′- AAACCTTGGTCCAGCTGCTCACTC-3′. The guide oligont for the top and bottom strand contain overhangs for ligation into the pair of BbsI sites inPX458 (in bold case). The sgRNA oligont were annealed in a thermocycler by using the following parameters: 95°C for 5 min; ramp down to 85°C at 2°C/s, then ramp down to 25°C at 0.1°C/s. sgRNA oligont were cloned into PX458 plasmid by incubating them with PX458 and T4 ligase (New England Biolabs) during one hour at 37°C. Then, 20 uL of E. Coli DH5-α strain (from Invitrogen) were transformed with 2 uL ligation product and immediately seeded on LB-agar ampicillin plates (50 ug/ml). Primary screening of clones with the correct insertion was performed with a BbsI/AgeI double digestion of colonies' minipreps. Confirmation of positive clones carrying the correct insertion was assessed by Sanger sequencing (GATC Biotech).
BxPC-3 transfection and clonal isolation
BxPC-3 cells were seeded the day before transfection at 70% confluency. BxPC-3 cells were then transfected with FuGENE®HD (Promega®) at a 4:1 ratio (Reagent/DNA ratio). 48 hours after transfection, the BxPC-3 clones which expressed the highest GFP levels were sorted and isolated by FACS (BD FACSAria™ II). Afterwards, 250 GFP+ cells were cultured in ClonaCell™-TCS Medium (StemCell™ Technologies) (Hydroximethylcellulose-based semisolid medium) with1% Penicillin/Streptomycin, 20% RPMI 1640 with 10% FBS in order to allow the formation of monoclonal colonies. Thirty days later, BxPC-3 colonies were manually picked from the semisolid medium plate then expanded and viably frozen to allow for functional experiments.
Genomic PCR and sequence analysis of BxPC-3 CRISPR clones
Primers flanking the CRISPR plasmid putative cut site on BTN3A2 were manually devised and purchased from Invitrogen®. BTN3A2 Forward primer: 5′-TCCCACAC CTTCTGGTATCTCT-3′; BTN3A2 Reverse primer: 5′-TGTGTCTTACTCAAGGGCCTCA-3′. The PCR reaction was performed with Phusion Hot Start II DNA Polymerase (ThermoFisher™ Scientific). The PCR reaction was performed in a thermocycler by using the following parameters: 1. – Initial denaturation at 98.0°C during 30s, 2.-Denaturation at 98.0°C during 20s, 3. – Primer annealing at 67°C during 20s, 4. – Extension at 72°C during 30°C and 5. – Final extension at 72°C during 5 minutes. 35 amplification cycles were performed on each PCR reaction.
Once obtained, the PCR products were purified with Nucleospin® Gel and PCR Clean-up (MACHEREY-NAGEL GmbH & Co). DNA concentration of purified PCR products was measured with Nanodrop® ND-1000 (ThermoFisher™ Scientific). Sanger sequencing of purified PCR products was subsequently performed (GATC Biotech). Quality of sequencing results was visually assessed with FinchTV (Geospiza™). DNA sequences were aligned one to each other with the multiple sequence alignment tool MUSCLE (EMBL-EBI). If two or more sequences were observed from the CRISPR plasmid cut site for one purified PCR product, each allelic sequence was then analyzed cloning the PCR pool into pCR®-Blunt plasmid (ThermoFisher™ Scientific).
Purified PCR products cloning into pCR®-Blunt plasmid and allelic sequencing
pCR®-Blunt plasmid was linearized performing a digestion with StuI during two hours at 37°C. Then, the digestion product was run on a 2% agarose gel. The 3.5 Kb band was extracted from the gel using Nucleospin® Gel and PCR Clean-up (MACHEREY-NAGEL GmbH & Co). Purified PCR products were ligated into linearized pCR®-Blunt plasmid incubating them in the presence of T4 ligase at room temperature during two hours. Afterwards, 20 µl of E. Coli DH5-α strain (from Invitrogen) were transformed with 2 µl ligation product, overgrowth during one hour in S.O.C. medium (Invitrogen®) and seeded on LB-agar Kanamycin plates (50 ug/ml). The day after, colonies were picked and cultured in 2 ml LB kanamycin overnight. Then, plasmid DNA was isolated from minipreps with Nucleospin® Plasmid (MACHEREY-NAGEL GmbH & Co). Plasmids concentration was measured with Nanodrop® ND-1000 (ThermoFisher™ Scientific). Sanger sequencing of purified PCR products was subsequently performed (GATC Biotech). Quality of sequencing results was visually assessed with FinchTV (Geospiza™). DNA sequences were aligned one to each other with the multiple sequence alignment tool MUSCLE (EMBL-EBI).
Generation of anti-human BTN3A mAbs and BTN3A-Fc
BTN3A1-Fc, BTN3A2-Fc and BTN3A3-Fc and anti-BTN3A mAbs clones 20.1, 103.2 and 108.5 were generated as previously described.Citation26 Clones 20.1 and 103.2 were further labeled for cytometry using Alexa Fluor® 647 Protein Labeling Kit (Life Technologies).
Determination of BTN3A isoforms expression by quantitative RT-PCR (qRT-PCR) in Wild Type PDAC cell lines
RNA from cells was prepared using Trizol (Invitrogen, Cergy Pontoise, France) according to the manufacturer's instructions. RNA concentration was determined by absorption and RNA integrity was checked on RNA Nano chips (Agilent, Santa Clara, CA). Reverse transcription (RT) reactions were performed on 1 µg of total RNA using Go Script (Promega, Madison, WI) according to the manufacturer's protocol.
qPCR reactions were run in duplicate on two independent cDNA preparations. qPCR was performed in Stratagene MX3005P machine (Agilent, Santa Clara, CA) using TaqMan® Universal Master Mix II, with UNG (Applied biosystem (Invitrogen), Cergy Pontoise, France). The crossing point (Cp), defined as the point at which the fluorescence rises appreciably above the background fluorescence, was determined for each transcript. The 2ΔΔCp method was used to analyze the relative gene expression. The Peptidylprolyl Isomerase A (PPIA) gene (ref 4331182) was chosen as control. Three BTN3A isoforms are measured: BTN3A1 (Hs01063368_m1), BTN3A2 (Hs00389328_m1) and BTN3A3 (Hs00757230_m1).
Transcriptomic analysis
Transcriptomic analysis was performed by using the software PROGgeneV2. This tool allows us to analyze TCGA public databases according to different survival measurements as overall survival.Citation46 The pancreatic cancer TCGA database pools 170 patients and clusters by high and low expression of selected gene expression (compared to the global median expression) in order to create Kaplan Meier overall survival curves compared using LogRank Test.
Western blot
Ten cm-culture dishes were placed on ice, washed in PBS and cells were dissociated and lysed in 250 μl of ice-cold HNTG buffer (50 mM HEPES pH 7, 50 mM NaF, 1 mM EGTA, 150 mM NaCl, 1% Triton X-100, 10% glycerol, and 1.5 mM MgCl2) in the presence of protease inhibitors (Roche Applied Science) and 100 μM Na3VO4. Protein quantification in all cell lysates was performed according to the manufacturer (Biorad quantification kit). Proteins were resolved by SDS-PAGE 10%, followed by western blotting. Primary antibodies (anti-BTN3A 20.1 mAb, anti-HIF1α (BD Biosciences), anti-γ tubulin, anti-Grb2 (SantaCruz technology)) were detected with peroxidase-conjugated anti-mouse IgG1 and anti-rabbit IgG antibody (Jackson Laboratory) and immunoreactive bands were confirmed using enhanced chemiluminescent reagents (Pierce). Quantification of the density of immunoreactive bands was performed using the ImageJ program.
Flow cytometry
2 × 105 PBMC were washed in PBS (Cambrex Bio Science) and incubated at 4°C for 20 min with specified mAb. Following incubation and washing, samples were analyzed on LSRFortessa or FACS Canto II (Becton Dickinson) using DIVA software (BD bioscience, Mountain View, CA). For analysis of CD107 expression, γδ T cells were incubated at 37°C with target cells in the presence of anti-CD107a/b and Golgi stop with or without BrHpp, anti-BTN3A 20.1 mAb or anti-BTN3A 108.5 mAb in normoxia or hypoxia. When specified, the target cell was pretreated O/N with zoledronate (ZOL) 25 µM then washed before co-culture with γδ T cells. After 4 hours, cells were collected, washed in PBS and analyzed by flow cytometry. To study cytokine production, cells were further permeabilized with Permwash (BD bioscience) to allow intracellular staining with labeled antibodies
Cytotoxic activity analysis
1 × 106 target cells were incubated with 20 μCi of 51Cr (Perkin-Elmer) for 60 minutes and mixed with effector cells in a Effector: Target (E:T) ratio of 30:1; 10:1; 1:1. After 4 hours of incubation at 37°C, 50 μl supernatant of each sample was transferred in LUMA plates and radioactivity was determined by a gamma counter. The percentage of specific lysis was calculated using the formula [(experimental – spontaneous release / total –spontaneous release) × 100] and expressed as the mean of triplicate.
Enzyme linked immunoSorbent assay (ELISA)
“Pan-BTN3A” and “BTN3A1-specific” sandwich ELISAs were conceived by Dynabio®. Were used as capture antibodies: 1/anti-BTN3A mAb clone that recognizes the 3 BTN3A-Fc recombinant proteins i.e BTN3A1-Fc, BTN3A2-Fc and BTN3A3-Fc for “Pan-BTN3A” ELISA and 2/anti-BTN3A mAb clone that only recognizes BTN3A1-Fc for “BTN3A1-specific” ELISA. Anti-BTN3A 103.2 mAb that recognizes the 3 BTN3A isoforms was biotinylated for detection of BTN3A isoforms. The efficiency of biotinylation was validated comparing to detection mAb used for Pancreatitis-Associated Protein (PAP) ELISA test (Dynabio®) and using recombinant BTN3A-Fc proteins. After blockade of the plate, supernatants of pancreatic cell lines, plasmas of patients or BTN3A1-Fc, BTN3A2-Fc and BTN3A3-Fc recombinant proteins used as standards were added. After repeated washes, biotinylated detection-anti-BTN3A mAb were added. Revelation was achieved with avidin-HRP. The optical density of each well was determined using a microplate reader set to 450 nm. The concentration of each BTN3A isoform was assessed following the standard curve obtained with BTN3A1-Fc protein.
Statistical analysis
Results are expressed as median ± interquartile range. Statistical analysis was performed using two-tailed paired t-test, Wilcoxon test, Mann–Whitney t test and Spearman correlation. P values < 0.05 were considered significant. Survival curves were compared using LogRank Test. Analyses were performed using GraphPad Prism program.
TMA statistical analysis
To identify differentially expressed biomarkers, ANOVA analysis was performed. Since TMA comprises 2 groups of PDAC patients staged N0 and N1, ANOVA analysis was followed by post hoc analysis with Tukey-Kramer test. Then, percentage of staining was compared between patients N0 and patients N1 using Mann-Whitney Test. The analysis was performed using NCSS software (Kaysville, Utah).
Study approval
Written informed consent was obtained from patients included in this study, in accordance with the Declaration of Helsinki. The study was approved by the local institutional review boards of the Institut-Paoli-Calmettes.
Competing financial interests
DO is founder of Imcheck Therapeutics.
Author contributions
AB designed research, performed experimental work, analyzed and interpreted data and wrote the manuscript. CL performed experimental work and analyzed data and contributed to draft the manuscript. EF, JLB, CC, ASC performed experimental work and analyzed data. ND designed research and provided PDX-derived cell lines. SC and MM contributed to the design of the project research. VS, EV, GM and MG provided patients samples and analyzed data. JCD designed, performed and analyzed ELISA test. DO and JLI designed research, contributed to the analysis and interpretation of data and to draft the manuscript.
supp_data.pdf
Download PDF (501 KB)Acknowledgments
We thank the CRCM animal core, cytometry and immunomonitoring facilities for their technical assistance. We acknowledge S.Just-Landi, S.Pastor, C.Bontemps, C. Kozaczyk and C.Pasero for technical support. We thank C.Allasia for TMA statistical analysis. We thank S.Hannify for commenting and editing the manuscript
This study was supported by research grant from Fondation ARC to AB and CC, Fondation pour la Recherche Médicale to EF, Ligue contre le Cancer to JLB and Institut National du Cancer to ASC, ANR GDStress, Fondation pour la Recherche Médicale (FRMDEQ20140329534), Instituto de Salud Carlos III (PI13/02192, co-funded by FEDER-European Union). DO is senior member of Institut Universitaire de France. CIBEREHD is funded by the Instituto de Salud Carlos III.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. [https://doi.org/10.3322/caac.21332]. PMID:26742998
- Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–23. [https://doi.org/10.1056/NEJMoa1003466]. PMID:20525992
- Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–54. [https://doi.org/10.1056/NEJMoa1200690]. PMID:22658127
- Farren MR, Mace TA, Geyer S, Mikhail S, Wu C, Ciombor K, Tahiri S, Ahn D, Noonan AM, Villalona-Calero M, et al. Systemic immune activity predicts overall survival in treatment-naïve patients with metastatic pancreatic cancer. Clin Cancer Res Off J Am Assoc Cancer Res. 2016;22:2565–74. [https://doi.org/10.1158/1078-0432.CCR-15-1732]
- Diana A, Wang LM, D'Costa Z, Allen P, Azad A, Silva MA, Soonawalla Z, Liu S, McKenna WG, Muschel RJ, et al. Prognostic value, localization and correlation of PD-1/PD-L1, CD8 and FOXP3 with the desmoplastic stroma in pancreatic ductal adenocarcinoma. Oncotarget. 2016;7(27):40992–1004. [https://doi.org/10.18632/oncotarget.10038]. PMID:27329602
- Daley D, Zambirinis CP, Seifert L, Akkad N, Mohan N, Werba G, Barilla R, Torres-Hernandez A, Hundeyin M, Mani VR, et al. γδ T cells support pancreatic oncogenesis by restraining αβ T cell activation. Cell. 2016;166:1485–1499.e15. [https://doi.org/10.1016/j.cell.2016.07.046]. PMID:27569912
- Chang Q, Jurisica I, Do T, Hedley DW. Hypoxia predicts aggressive growth and spontaneous metastasis formation from orthotopically grown primary xenografts of human pancreatic cancer. Cancer Res. 2011;71:3110–20. [https://doi.org/10.1158/0008-5472.CAN-10-4049]. PMID:21343390
- Hasmim M, Messai Y, Ziani L, Thiery J, Bouhris JH, Noman MZ, Chouaib S. Critical role of tumor microenvironment in shaping NK cell functions: implication of hypoxic stress. Front Immunol. 2015;6:482. [https://doi.org/10.3389/fimmu.2015.00482]. PMID:26441986
- Noman MZ, Janji B, Kaminska B, Van Moer K, Pierson S, Przanowski P, Buart S, Berchem G, Romero P, Mami-Chouaib F, et al. Blocking hypoxia-induced autophagy in tumors restores cytotoxic T-cell activity and promotes regression. Cancer Res. 2011;71:5976–86. [https://doi.org/10.1158/0008-5472.CAN-11-1094]. PMID:21810913
- Noman MZ, Buart S, Van Pelt J, Richon C, Hasmim M, Leleu N, Suchorska WM, Jalil A, Lecluse Y, El Hage F, et al. The cooperative induction of hypoxia-inducible factor-1 alpha and STAT3 during hypoxia induced an impairment of tumor susceptibility to CTL-mediated cell lysis. J Immunol. 2009;182:3510–21. [https://doi.org/10.4049/jimmunol.0800854]. PMID:19265129
- Kitayama J, Atomi Y, Nagawa H, Kuroda A, Mutoh T, Minami M, Juji T. Functional analysis of TCR gamma delta+ T cells in tumour-infiltrating lymphocytes (TIL) of human pancreatic cancer. Clin Exp Immunol. 1993;93:442–7. [https://doi.org/10.1111/j.1365-2249.1993.tb08198.x]. PMID:8370173
- Helm O, Mennrich R, Petrick D, Goebel L, Freitag-Wolf S, Röder C, Kalthoff H, Röcken C, Sipos B, Kabelitz D, et al. Comparative characterization of stroma cells and ductal epithelium in chronic pancreatitis and pancreatic ductal adenocarcinoma. PLoS ONE. 2014;9(5):e94357. [https://doi.org/10.1371/journal.pone.0094357]. eCollection 2014.
- Di Marco Barros R, Roberts NA, Dart RJ, Vantourout P, Jandke A, Nussbaumer O, Deban L, Cipolat S, Hart R, Iannitto ML, et al. Epithelia use butyrophilin-like molecules to shape organ-specific γδ t cell compartments. Cell. 2016;167:203–218.e17. [https://doi.org/10.1016/j.cell.2016.08.030]. PMID:27641500
- Nicol AJ, Tokuyama H, Mattarollo SR, Hagi T, Suzuki K, Yokokawa K, Nieda M. Clinical evaluation of autologous gamma delta T cell-based immunotherapy for metastatic solid tumours. Br J Cancer. 2011;105:778–86. [https://doi.org/10.1038/bjc.2011.293]. PMID:21847128
- Corvaisier M, Moreau-Aubry A, Diez E, Bennouna J, Mosnier JF, Scotet E, Bonneville M, Jotereau F. V gamma 9 V delta 2 T cell response to colon carcinoma cells. J Immunol. 2005;175:5481–8. [https://doi.org/10.4049/jimmunol.175.8.5481]. PMID:16210656
- Cordova A, Toia F, La Mendola C, Orlando V, Meraviglia S, Rinaldi G, Todaro M, Cicero G, Zichichi L, Donni PL, et al. Characterization of human γδ T lymphocytes infiltrating primary malignant melanomas. PloS One. 2012;7:e49878. [https://doi.org/10.1371/journal.pone.0049878]. PMID:23189169
- Gober H-J, Kistowska M, Angman L, Jenö P, Mori L, De Libero G. Human T cell receptor gammadelta cells recognize endogenous mevalonate metabolites in tumor cells. J Exp Med. 2003;197:163–8. [https://doi.org/10.1084/jem.20021500]. PMID:12538656
- Li J, Herold MJ, Kimmel B, Müller I, Rincon-Orozco B, Kunzmann V, Herrmann T. Reduced expression of the mevalonate pathway enzyme farnesyl pyrophosphate synthase unveils recognition of tumor cells by Vgamma9Vdelta2 T cells. J Immunol. 2009;182:8118–24. [https://doi.org/10.4049/jimmunol.0900101]. PMID:19494338
- Benzaïd I, Mönkkönen H, Stresing V, Bonnelye E, Green J, Mönkkönen J, Touraine JL, Clézardin P. High phosphoantigen levels in bisphosphonate-treated human breast tumors promote Vgamma9Vdelta2 T-cell chemotaxis and cytotoxicity in vivo. Cancer Res. 2011;71:4562–72. [https://doi.org/10.1158/0008-5472.CAN-10-3862]. PMID:21646473
- Märten A, von Lilienfeld-Toal M, Büchler MW, Schmidt J. Soluble MIC is elevated in the serum of patients with pancreatic carcinoma diminishing gammadelta T cell cytotoxicity. Int J Cancer J Int Cancer. 2006;119:2359–65. [https://doi.org/10.1002/ijc.22186]
- McAllister F, Bailey JM, Alsina J, Nirschl CJ, Sharma R, Fan H, Rattigan Y, Roeser JC, Lankapalli RH, Zhang H, et al. Oncogenic Kras activates a hematopoietic-to-epithelial IL-17 signaling axis in preinvasive pancreatic neoplasia. Cancer Cell. 2014;25:621–37. [https://doi.org/10.1016/j.ccr.2014.03.014]. PMID:24823639
- Kabelitz D, Wesch D, Pitters E, Zöller M. Characterization of tumor reactivity of human V gamma 9 V delta 2 gamma delta T cells in vitro and in SCID mice in vivo. J Immunol. 2004;173:6767–76. [https://doi.org/10.4049/jimmunol.173.11.6767]. PMID:15557170
- Harly C, Guillaume Y, Nedellec S, Peigné CM, Mönkkönen H, Mönkkönen J, Li J, Kuball J, Adams EJ, Netzer S, et al. Key implication of CD277/butyrophilin-3 (BTN3A) in cellular stress sensing by a major human γδ T-cell subset. Blood. 2012;120:2269–79. [https://doi.org/10.1182/blood-2012-05-430470]. PMID:22767497
- Rhodes DA, Chen HC, Price AJ, Keeble AH, Davey MS, James LC, Eberl M, Trowsdale J. Activation of Human γδ T Cells by Cytosolic Interactions of BTN3A1 with Soluble Phosphoantigens and the Cytoskeletal Adaptor Periplakin. J Immunol. 2015;194(5):2390–8. [https://doi.org/10.4049/jimmunol.1401064]. PMID:25637025
- Benyamine A, Le Roy A, Mamessier E, Gertner-Dardenne J, Castanier C, Orlanducci F, Pouyet L, Goubard A, Collette Y, Vey N, et al. BTN3A molecules considerably improve Vγ9Vδ2 T cells-based immunotherapy in acute myeloid leukemia. Oncoimmunology. 2016;5:e1146843. [https://doi.org/10.1080/2162402X.2016.1146843]. PMID:27853633
- Compte E, Pontarotti P, Collette Y, Lopez M, Olive D. Frontline: Characterization of BT3 molecules belonging to the B7 family expressed on immune cells. Eur J Immunol. 2004;34:2089–99. [https://doi.org/10.1002/eji.200425227]. PMID:15259006
- Le Page C, Marineau A, Bonza PK, Rahimi K, Cyr L, Labouba I, Madore J, Delvoye N, Mes-Masson AM, Provencher DM, et al. BTN3A2 expression in epithelial ovarian cancer is associated with higher tumor infiltrating T cells and a better prognosis. PloS One. 2012;7:e38541. [https://doi.org/10.1371/journal.pone.0038541]. PMID:22685580
- Cubillos-Ruiz JR, Martinez D, Scarlett UK, Rutkowski MR, Nesbeth YC, Camposeco-Jacobs AL, Conejo-Garcia JR. CD277 is a negative co-stimulatory molecule universally expressed by ovarian cancer microenvironmental cells. Oncotarget. 2010;1:329–38. [https://doi.org/10.18632/oncotarget.165]. PMID:21113407
- Zhu M, Yan C, Ren C, Huang X, Zhu X, Gu H, Wang M, Wang S, Gao Y, Ji Y, et al. Exome array analysis identifies variants in SPOCD1 and BTN3A2 that affect risk for gastric cancer. Gastroenterology. 2017;152(8):2011–2021. [https://doi.org/10.1053/j.gastro.2017.02.017]. [Epub 2017 Feb 27]
- Deer EL, González-Hernández J, Coursen JD, Shea JE, Ngatia J, Scaife CL, Firpo MA, Mulvihill SJ. Phenotype and genotype of pancreatic cancer cell lines. Pancreas. 2010;39:425–35. [https://doi.org/10.1097/MPA.0b013e3181c15963]. PMID:20418756
- Elsässer HP, Lehr U, Agricola B, Kern HF. Structural analysis of a new highly metastatic cell line PaTu 8902 from a primary human pancreatic adenocarcinoma. Virchows Arch B Cell Pathol Incl Mol Pathol. 1993;64:201–7. [https://doi.org/10.1007/BF02915113]. PMID:8287116
- Elsässer HP, Lehr U, Agricola B, Kern HF. Establishment and characterisation of two cell lines with different grade of differentiation derived from one primary human pancreatic adenocarcinoma. Virchows Arch B Cell Pathol Incl Mol Pathol. 1992;61:295–306. [https://doi.org/10.1007/BF02890431]. PMID:1348891
- Duconseil P, Gilabert M, Gayet O, Loncle C, Moutardier V, Turrini O, Calvo E, Ewald J, Giovannini M, Gasmi M, et al. Transcriptomic analysis predicts survival and sensitivity to anticancer drugs of patients with a pancreatic adenocarcinoma. Am J Pathol. 2015;185(4):1022–32. [https://doi.org/10.1016/j.ajpath.2014.11.029]. PMID:25765988
- Zocchi MR, Costa D, Venè R, Tosetti F, Ferrari N, Minghelli S, Benelli R, Scabini S, Romairone E, Catellani S, et al. Zoledronate can induce colorectal cancer microenvironment expressing BTN3A1 to stimulate effector γδ T cells with antitumor activity. Oncoimmunology. 2017;6:e1278099. [https://doi.org/10.1080/2162402X.2016.1278099]. PMID:28405500
- Kilcollins AM, Li J, Hsiao C-HC, Wiemer AJ. HMBPP analog prodrugs bypass energy-dependent uptake to promote efficient BTN3A1-mediated malignant cell lysis by Vγ9Vδ2 T lymphocyte effectors. J Immunol Baltim Md 1950. 2016;197:419–28.
- Sandstrom A, Peigné CM, Léger A, Crooks JE, Konczak F, Gesnel MC, Breathnach R, Bonneville M, Scotet E, Adams EJ. The intracellular B30.2 domain of butyrophilin 3A1 binds phosphoantigens to mediate activation of human Vγ9Vδ2 T cells. Immunity. 2014;40:490–500. PMID:24703779
- Wang H, Henry O, Distefano MD, Wang YC, Räikkönen J, Mönkkönen J, Tanaka Y, Morita CT. Butyrophilin 3A1 plays an essential role in prenyl pyrophosphate stimulation of human Vγ2Vδ2 T cells. J Immunol. 2013;191:1029–42. PMID:23833237
- Messal N, Mamessier E, Sylvain A, Celis-Gutierrez J, Thibult M-L, Chetaille B, Firaguay G, Pastor S, Guillaume Y, Wang Q, et al. Differential role for CD277 as a co-regulator of the immune signal in T and NK cells. Eur J Immunol. 2011;41:3443–54. PMID:21918970
- Palakodeti A, Sandstrom A, Sundaresan L, Harly C, Nedellec S, Olive D, Scotet E, Bonneville M, Adams EJ. The molecular basis for modulation of human Vγ9Vδ2 T cell responses by CD277/butyrophilin-3 (BTN3A)-specific antibodies. J Biol Chem. 2012;287:32780–90. PMID:22846996
- Liu Z, Guo B, Lopez RD. Expression of intercellular adhesion molecule (ICAM)-1 or ICAM-2 is critical in determining sensitivity of pancreatic cancer cells to cytolysis by human gammadelta-T cells: implications in the design of gammadelta-T-cell-based immunotherapies for pancreatic cancer. J Gastroenterol Hepatol. 2009;24:900–11. PMID:19175829
- Guillaumond F, Leca J, Olivares O, Lavaut MN, Vidal N, Berthezène P, Dusetti NJ, Loncle C, Calvo E, Turrini O, et al. Strengthened glycolysis under hypoxia supports tumor symbiosis and hexosamine biosynthesis in pancreatic adenocarcinoma. Proc Natl Acad Sci U S A. 2013;110:3919–24. PMID:23407165
- Charpin C, Tavassoli F, Secq V, Giusiano S, Villeret J, Garcia S, Birnbaum D, Bonnier P, Lavaut MN, Boubli L, et al. Validation of an immunohistochemical signature predictive of 8-year outcome for patients with breast carcinoma. Int J Cancer J Int Cancer. 2012;131:E236–243.
- Giusiano S, Secq V, Carcopino X, Carpentier S, Andrac L, Lavaut MN, Allasia C, Bonnier P, Iovanna JL, Garcia S, et al. Immunohistochemical profiling of node negative breast carcinomas allows prediction of metastatic risk. Int J Oncol. 2010;36:889–98. PMID:20198333
- Gertner J, Wiedemann A, Poupot M, Fournié JJ. Human gammadelta T lymphocytes strip and kill tumor cells simultaneously. Immunol Lett. 2007;110:42–53. [https://doi.org/10.1016/j.imlet.2007.03.002]. PMID:17451812
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308. [https://doi.org/10.1038/nprot.2013.143]. PMID:24157548
- Goswami CP, Nakshatri H. PROGgene: gene expression based survival analysis web application for multiple cancers. J Clin Bioinforma. 2013;3:22.[https://doi.org/10.1186/2043-9113-3-22]. PMID:24165311
