ABSTRACT
Cancers elicit an immune response by modifying the microenvironment. The immune system plays a pivotal role in cancer recognition and eradication. While the potential clinical value of infiltrating lymphocytes at the tumor site has been assessed in breast cancer, circulating cytokines – the molecules coordinating and fine-tuning immune response – are still poorly characterized.
Using two breast cancer cohorts (MicMa, n = 131, DCTB, n = 28) and the multiplex Luminex platform, we measured the levels of 27 cytokines in the serum of breast cancer patients prior to treatment. We investigated the cytokine levels in relation to clinicopathological characteristics and in perspective of the tumor infiltrating immune cells predicted from the bulk mRNA expression data.
Unsupervised clustering analysis of the serum cytokine levels in the MicMa cohort identified a cluster of pro-inflammatory, pro-angiogenic, and Th2-related cytokines which was associated with poor prognosis. Notably high levels of platelet derived growth factor BB (PDGF) reflected a more aggressive tumor phenotype and larger tumor size. A significant positive correlation between serum levels of interferon gamma-induced protein 10 (IP10) and its mRNA expression at the tumor site suggested that tumor-IP10-production may outflow to the bloodstream. High IP10 serum levels were associated with a worse prognosis. Finally, we found serum levels of both PDGF and IP10 associated with enrichment scores of specific tumor infiltrating immune cells.
Our study suggests that monitoring cytokine circulating levels in breast cancer could be used to characterize breast cancers and the immune composition of their microenvironment through readily available biological material.
Introduction
The immune system plays a key role in cancer recognition and eradication. More than 60 years ago the idea that the immune system recognizes cancer cells and inhibits their growth was introduced.Citation1 Cancers induce a local immune response through the expression of neo-antigens and/or by modifying the microenvironment.Citation2 Characterizing the quality and the quantity of local immune response at the tumor site will improve our understanding of how the microenvironment influences tumor progression and clinical outcome. Cytokines, chemokines, and growth factors are small molecules secreted by stromal, immune, and/or tumor cells, to coordinate and fine tune the immune response.Citation3–Citation5 Cytokine serum levels may represent interesting noninvasive biomarker of tumor-induced immune response, in the detection of cancers or in the monitoring of pathogenesis.
Breast cancer is the most frequent cancer among women worldwide. Classical clinical and pathological markers used to stratify patients are tumor size, estrogen receptor (ER) status, and human epidermal growth factor receptor 2 (HER2) status. The recently clinically approved PAM50 classification divides breast cancers into five subgroups according to gene expression which comprehensively recapitulates the classical pathological markers.Citation6 Lymphocytic infiltration is more abundant in ER negative compared to ER positive breast cancers. In addition, high immune infiltration has been associated with an increased response to neo-adjuvant and adjuvant chemotherapy.Citation7 Various methods have been used to quantify immune infiltration in breast cancer, however, they are all invasive and very few studies have assessed cytokine serum levels as surrogates of immune infiltration.
In a recent study performed in the DCTB cohort, we measured the levels of cytokines in breast tumor interstitial fluid (TIF) and compared it to levels in the interstitial fluid from normal breast specimens (NIF) recovered from the same patient.Citation8 Our results, based on both cytokine measurements and immunohistochemistry (IHC) of immune cells indicated that the presence of a tumor increased the levels of several cytokines in the interstitial fluid. Of particular interest, increased levels of IL5 in the TIF was associated with worse prognosis.Citation8
As tumor interstitial fluid is hardly accessible and to further investigate the relevance of circulating cytokines in breast cancer, we measured serum levels of 27 cytokines in two breast cancer cohorts (MicMa, n = 131 and DCTB, n = 28) using the multiplex Luminex technology. We investigated the relationship between cytokine levels and well-known clinicopathological parameters. We report here findings from both cohorts. Our results indicate that high serum levels of pro-inflammatory and Th2-related cytokines could be associated with a worse prognosis and may be an indicator of more advanced-staged and aggressive cancer. Notably, high serum levels of platelet derived growth factor BB (PDGF) were associated with larger tumors and HER2 positivity. Serum-IP10 levels correlated with its mRNA and protein expression levels within the tumor and with worse prognosis. Finally, we brought serum cytokine levels in perspective of the predicted infiltrating immune cell types at the tumor site to gain insight on how PDGF and IP10 levels may relate to pathogenesis.
Results
Clustering of cytokines in the serum of breast cancer patients
27 cytokine levels were measured in the serum of breast cancer patients using a multiplex bead-based immune assay (Luminex). Two independent cohorts were evaluated: the MicMa (n = 131) collected in Oslo, NorwayCitation9 and the DCTB (n = 28) collected in Denmark.Citation8 Principal component analysis (PCA) demonstrated a separation of the samples from the two cohorts based on the cytokine serum levels, possibly due to batch effects or differences in clinicopathological features (). Observing a strong separation based on PCA, we further analyzed the two cohorts separately as discovery (MicMa) and replication (DCTB).
Figure 1. Clusters of cytokine serum levels in relation to prognosis.
(A) Principal component analysis was performed using the serum levels of the 27 cytokines measured by Luminex. Scatter plot of principal component 1 and 2 are represented, each dot represents a sample projected in the two main principal components (PC1 and PC2); the dots are colored according to the cohort they belong to. (B) Unsupervised clustering of cytokine levels in the MicMa cohort (n = 131) using the R package pheatmap with Euclidean distance and Ward.D linkage. Annotations of the rows of the heatmap indicate histopathological features of the patients: ER and HER2 statuses. Cytokines in columns are annotated in regard to the classical function they exert according to the literature. Cluster were identified using cutree, dendrogram of the unsupervised clustering was colored to identify clusters, in addition solid line boxes highlight the 4 clusters. (C & D) Sample-wise enrichment scores for cluster-4-cytokines was calculated using Gene Set Variation Analysis (GSVA) and the 8 cytokines of cluster 4 (FGF, VEGF, GM-CSF, PDGF, IL17, IL5, IL4, IL15). MicMa and DCTB samples were divided in two groups (high and low score) according to the median for the MicMa and the density distribution of the score plotted in Supplementary Figure 1 for the DCTB. Kaplan-Meier survival curves for the high (red) and low (blue) cluster-4-cytokines scores are depicted for MicMa (C) and DCTB (D). The p-values are from log-rank tests. Hazard Ratio ± 95% confidence interval are shown, for the DCTB cohort the Hazard Ratio was not calculable due to small events rates.
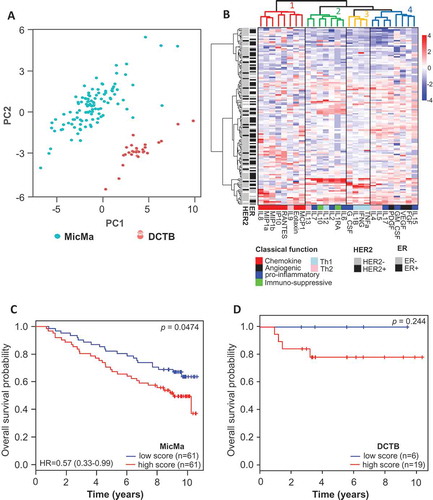
We performed unsupervised clustering based on the cytokine serum levels from MicMa (). The heatmap obtained depicts clusters of cytokines on the x-axis and clusters of patients on the y-axis. Patients’ receptor status (ER and HER2) annotation on the y-axis demonstrate that patients cluster independently of ER and HER2 statuses (). We identified four clusters of cytokines (1: red. 2: green, 3: yellow, 4: blue). Annotation of cytokines according to their classical function (on the bottom of the heatmap), showed that cluster-4-cytokines was composed of cytokines with pro-angiogenic functions and associated with Th2 activity (IL15, VEGF, GM-CSF, PDGF, IL17, IL5, IL4, and FGF). These cytokines are known to act as pro-tumorigenic molecules.Citation10,Citation11 We therefore estimated the variation of cluster-4-cytokines activity over the MicMa cohort in an unsupervised manner using the Gene Set Variation Analysis (GSVA) method.Citation12 We found that breast cancer patients with a cluster-4-cytokines enrichment score above the median had a significantly worse overall survival (). The other clusters enrichment scores, were not associated with survival (Supplementary Figure 1). We also scored the DCTB samples using the GSVA method. For the DCTB, we found more samples with higher cytokine-cluster-4 enrichment scores as depicted in Supplementary Figure 2, we therefore used the bimodal Gaussian distribution of the score and a finite mixture model to define samples with high or low score. We confirmed the tendency of worse prognosis for patients with a higher cytokine-cluster-4 enrichment score ().
This initial analysis of the 27 cytokines highlighted the relevance of measuring circulating cytokines in breast cancer patients’ serum.
Assessment of individual cytokines
We next investigated the relationship between each cytokine and clinical parameters (see Supplementary Table 1–3, for summary of the statistics according to ER status HER2 status and tumor size). In both MicMa and the DCTB cohorts IL17, a cytokine mainly produced by Th17 cells,Citation13 was the only cytokine with significantly different serum levels according to hormone receptor status (ER or PR); showing higher expression in the serum of breast cancer patients with an ER negative (,)) or PR negative tumor (Supplementary Figure 3A,B).
Figure 2. Serum cytokine levels and clinicopathological parameters.
(A & B) Boxplots represent the average serum levels of IL17 (log pg/mL) in ER positive (ER pos, gray) and ER negative (ER neg, white) tumors in the MicMa (A) and DCTB (B) cohorts. IL17 levels are significantly higher in ER negative samples. (C & D) Average PDGF serum levels (log pg/mL) are visualized using boxplot in regard to HER2 status in the MicMa (C) and the DCTB (D) cohorts. White boxes, HER2 negative samples (HER2 neg), pink boxes HER2 positive samples (HER2 pos). (E & F) Boxplots represent the average serum levels of PDGF (log pg/mL) in small (< 2cm, white) or larger (> 2cm, blue) tumors in the MicMa (E) and DCTB (F) cohorts. PDGF levels are significantly higher in the serum of patient with bigger tumors. The size 2cm was chosen as a cutoff to reflect the TNM staging of the American Cancer Society. Mann-Whitney test p-values are denoted in the bottom right of each boxplot.
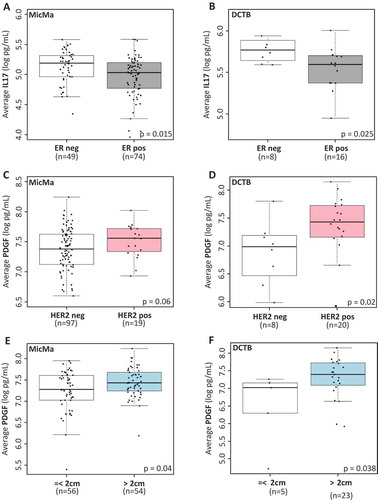
Platelet derived growth factor BB (PDGF) is a growth factor mainly produced by activated platelets, macrophages, endothelial, smooth, and tumor cells.Citation14 We found higher serum levels of PDGF associated with HER2 positive tumors ( and ) and with larger tumor size ( and ). PDGF is part of the cluster-4-cytokine identified in ; and high levels may therefore reflect the presence of a more aggressive breast cancer type. Indeed, patients with the highest serum levels of PDGF (highest tertile) had a trend for worse prognosis (Supplementary Figure 4A,B; MicMa: p = 0.06, DCTB: p = 0.24, log-rank tests).
In one cohort only: significantly lower levels of G-CSF were associated with HER2 positivity in the MicMa study, while high levels of IL2, IL17, FGF, and GM-CSF were detected among HER2 positive DCTB patients (Supplementary Table 2). Furthermore, higher levels of IP10 in MicMa and of IL2, IL8, and IL9 in DCTB, were associated with larger tumors (Supplementary Table 3).
We assessed cytokine serum levels according to the PAM50 molecular subtypes in the MicMa study;Citation15 two cytokines were significantly associated with the PAM50 subtypes (Kruskal-Wallis test < 0.05). High PDGF serum levels were associated with the HER2-enriched subtype (Supplementary Figure 5A). This confirmed an association between PDGF and HER2-related breast cancer phenotype. High IL8 serum levels were associated with the Normal-like subtype (Supplementary Figure 5B).
Together, these analyses suggest that serum levels of PDGF and IL17 may reflect important clinical and molecular parameters of the breast tumor.
Serum cytokines levels and their intra-tumor mRNA expression levels
To investigate whether the levels of serum cytokines may be affected or may derive from tumor-produced cytokines, we retrieved the tumor expression data for the MicMa (GSE19783),Citation16 while for the DCTB, we generated mRNA expression profiles from fresh frozen tumors. The DCTB data set of mRNA expression is now available in ArrayExpress with accession number E-MTAB-7201. 98 MicMa and 25 DCTB samples were assessed, for both serum cytokine levels and mRNA profiling. Focusing on each cytokine serum level and its corresponding mRNA expression at the tumor site we found that IP10 serum level positively significantly correlated with its gene expression (CXCL10) at the primary tumor in both cohorts; MicMa (Pearson r = 0.33, p = 0.001) and DCTB (Pearson r = 0.67, p = 0.0003) ( and ), Supplementary Table 4). IP10 (CXCL10) is a pro-inflammatory cytokine which may promote breast cancer cell proliferation.Citation17 Survival analyses revealed that high serum levels of IP10 (> median) were significantly associated with worse outcome in the MicMa cohort (, p = 0.004). Although not significant, the same trend was observed in the smaller DCTB cohort (). We further validated this finding in the METABRIC cohort,Citation18 a large and well documented breast cancer cohort in terms of mRNA expression and survival data. In accordance to what was observed in the MicMa cohort, we found in the METABRIC that high mRNA levels (higher than the median) of CXCL10 significantly associated with worse survival ().
Figure 3. Cytokines serum levels and corresponding tumor mRNA expression.
(A & B) Correlation analysis for IP10 serum levels and CXCL10 mRNA expression from the bulk tumor. Dots are colored according to ER status. (C & D) Patients were divided into two groups according to the median expression of IP10. Kaplan-Meier survival curves for the high (red) and low (blue) levels of IP10 are depicted for MicMa (C) and DCTB (D). (E) Patients of the METABRIC cohort were divided into two groups according to the median mRNA levels of CXCL10. Kaplan-Meier survival curves for the high (red) and low (blue) CXCL10 are depicted. The p-values are from log-rank tests. Hazard Ratio ± 95% confidence interval are shown.
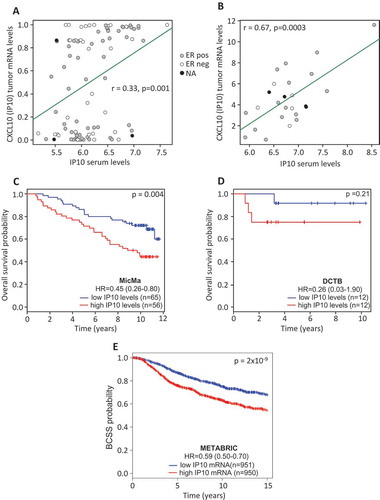
Our data suggest a continuum between the characteristics of the tumor microenvironment (production of IP10 at the primary tumor site) and of its macroenvironment (leakage of tumor-produced IP10 into the bloodstream). In addition, high IP10 serum levels may indicate worse prognosis.
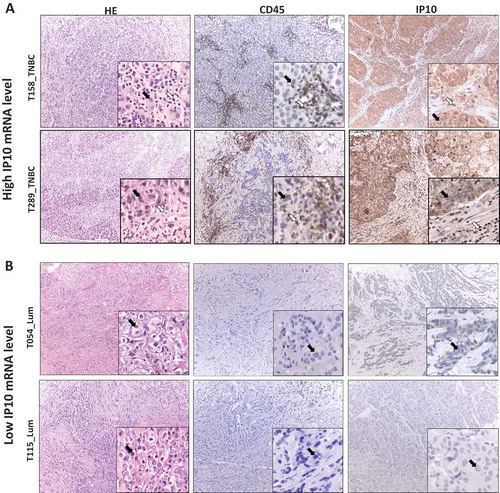
We further investigated whether high CXCL10 mRNA expression was associated with high IP10 protein production and which cells were likely to produce this cytokine using immunohistochemistry (IHC). Using the DCTB cohort, for which we had formalin fixed paraffin embedded (FFPE) staining available, we selected two samples with high CXCL10 mRNA expression (highest quartile, ) and two with low (lowest quartile, ). Samples with high CXCL10 mRNA also showed high IP10 protein expression. As previously described, we found IP10 expressed by both cancer cells and tumor-infiltrating lymphocytes, with slightly higher expression levels in tumor cells. Also, higher IP10 and/or CXCL10 was found in tumors with high proportion of T-cell lymphocytes which agrees with our recent data.Citation8
Figure 4. IP10 protein expression in breast tumors associates with high and low IP10 mRNA.
The representative IHC images of tissue sections from two pairs of breast tumors exhibiting high (Panel A, T289 and T158) and low (Panel B, T115 and T054) mRNA levels of IP10. Left sections: hematoxylin and eosin (H&E) staining for histopathological topography examination of the selected FFPE breast tissue samples. Middle sections: IHC staining with anti-CD45 specific antibodies for evaluating tumor infiltrating lymphocytes. Right sections: IHC staining with of IP10 antibodies (see Materials and Methods for details). Black arrows show the tumor cells. White arrows show the TILs. The magnification is x10 for the whole section and x40 for the insertions.
Our results demonstrate the association between IP10 and worse prognosis and emphasize the relevance of measuring this cytokine in the serum of breast cancer patients.
Tumor immune infiltration and circulating cytokine levels
To further investigate the possible mechanisms by which cytokine serum levels may be related to pathogenesis, we analyzed cytokine profiles in perspective of the tumor infiltrating immune cells. Using expression data and the algorithm, xCell, we inferred the extent of immune cell infiltration in the tumor of the patients in both cohorts. xCell uses the single sample gene set enrichment analysis (ssGSEA) methodCitation19 to predict the relative enrichment of 64 cell types by using gene expression values from bulk tumor samples. Here, we focused on the 26 most common immune cell types (Supplementary Table 5).
Immunohistochemistry (IHC) has been previously performed on the DCTB cohort to estimate the type of leukocyte infiltration present in the tumors, notably, cytotoxic T-lymphocytes (anti-CD8+ antibodies), and tumor-associated macrophages (TAMs) (anti-CD68 antibodies) were assessedCitation8. We therefore examined the relationship between intensity of CD8 and CD68 staining and xCell scores for immune cell types expressing these surface markers. We found a positive correlation between IHC staining and xCell scores (Supplementary Figure 6A and 6B) which indicates that xCell scoring could be a good surrogate to investigate immune cell infiltration.
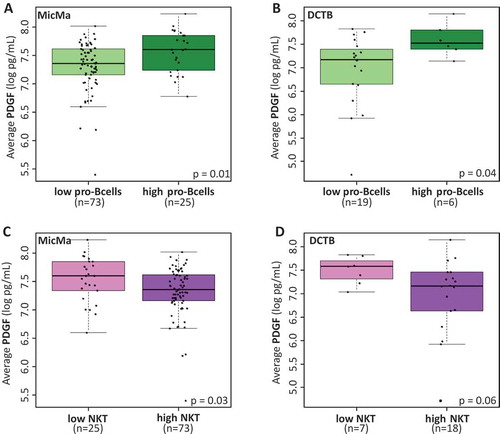
As our results pointed to PDGF, IL17 and IP10 as relevant cytokines to measure in the serum, in respect to the tumor phenotype, we sought for association between infiltrating immune cell types at the tumor site and the serum levels of these two cytokines. We found that patients with the highest levels of pro-B cells (highest quartile) at their tumor site also had significantly higher levels of PDGF in matched serum ( and ). Furthermore, samples with low natural killer T (NKT) scores (lowest quartile) had higher serum levels of PDGF ( and ). In summary, high levels of PDGF in the serum, which we found associated with more advanced and aggressive tumors may also relate to lower levels of NKT and higher levels of pro-B cells at the tumor site.
Figure 5. Cytokine levels and infiltrating immune cells.
(A & B) Boxplots represent the average serum levels of PDGF (pg/mL) in respect to low or high (highest quartile) scores for pro-B cells infiltration at the tumor site in the MicMa (A) and DCTB (B) cohorts. PDGF levels are significantly higher in the serum of patient with higher pro-B cells scores. (C & D) Boxplots of serum PDGF levels according to low (lowest quartile) and high scores for natural killer T cells (NKT) infiltration at the tumor site in the MicMa (C) and DCTB (D) cohorts. Mann-Whitney test p-values are denoted in the bottom right of each boxplot.
IP10 serum levels on the other hand, were found positively correlated with type M1 macrophages (MicMa, Pearson r = 0.24, p = 0.05; DCTB, Pearson r = 0.57, p = 0.003) (Supplementary Figure 7). Raw Pearson r values between the measured cytokines serum levels and inferred immune cell infiltration are denoted in Supplementary Table 6 when p value < 0.1. Altogether, these analyses provide clues to understand the mechanisms by which the serum cytokine levels associate with clinicopathological parameters.
Discussion
The immune system plays a pivotal role in cancer recognition and eradication.Citation20 Experimental and clinical evidence suggests that cancers elicit an immune response through the expression of neo-antigens or by modifying the tumor microenvironment. Recent studies have assessed the relevance of immune infiltration in regard to risk of relapse, clinical subtypes or response to therapy in breast cancers.Citation21–Citation24 Here, we focused on circulating cytokine levels and found that PDGF, IL17, and IP10 serum levels may be used to characterize breast cancers and the immune composition of their microenvironment.
Our previous studies of integrated molecular profiles of invasive breast tumors and ductal carcinoma in situ (DCIS) obtained by PARADIGM revealed differential vascular and interleukin signaling associated with different tumor phenotype. The interleukin signaling profiles observed in invasive cancers were absent or weakly expressed in healthy tissue, but already prominent in DCIS. The most prominent difference associated with mammographic density in healthy breast tissue was that of STAT4 signaling.Citation25 We have also shown that the density of immune cells that infiltrate a breast tumor tissue is highly dependent on tumor subtype.Citation26 The immune cell types found in the vicinity of growing tumor cells is very wide and complex, the predominant type of infiltrating leukocyte, or their location within the growing tumor influence pathogenesis.Citation27 However, there are no good clinical guidelines for the use of information on immune infiltration into the treatment of breast cancer, although in a number of studies the presence of such cells have been linked to a favorable response. Citation28,Citation29
Cytokines, the molecules orchestrating the immune response, are still poorly characterized in breast cancer. We previously highlighted the relevance of measuring cytokines in breast cancer; we found several cytokines levels elevated in the tumor interstitial fluids when compared to the interstitial fluid of normal tissue.Citation8 In the current study, we measured 27 cytokines in the serum of breast cancer patients from two cohorts (MicMa, n = 131) and DCTB (n = 28). We assessed cytokine serum levels in perspective of clinicopathological features. We extracted the significant results in the two cohorts MicMa (discovery) and DCTB (validation). Even though, we use two independent cohorts in this study, the low number of samples in the DCTB cohort may have hindered the validation of results discovered in the MicMa cohort. We believe that the smaller sample size in the DCTB cohort was the reason why the significant survival results found in the MicMa were not fully validated in the DCTB cohort. In addition, ER, PR, and HER2 definitions were different in both cohorts; for example, the MicMa tumors were considered ER positive if more than 10% of the tumor cells were positively immunostained, while in DCTB which is a “newer” cohort, the current St Gallen 2015 (> 1%) criteria was used.Citation30 This may lead to differences in classification of the samples and may also have interfered with validation of some results. Finally, even though the number of tests was relatively small (27 cytokines) we did not correct our p-values in each cohort according to multiple testing and instead focused on the significant results found in both cohorts. The MicMa and the DCTB cohort display different clinicopathological features as depicted in Supplementary Table 7, which may affect our ability to validate some findings. Despite, all the differences inherent to these two cohorts, we were able to find serum cytokine levels significantly associated with clinicopathological features in both cohorts. While our study clearly needs further validation, these are important data and resources which demonstrate that measurements in breast cancer patients’ serums provide relevant information regarding primary tumor characteristics. In short, our results would benefit from additional independent validations, but give sufficient ground for more targeted cytokine measurement and emphasize the potential of using readily available serum samples in breast cancer research and clinical practice.
We found PDGF levels significantly higher in the serum of patients with larger tumors or with a positive HER2 status. These results suggest that high serum levels of PDGF may reflect a more aggressive and advanced tumor phenotype. Matsumoto et al., showed that higher levels of PDGF-bb were associated with poor prognosis in esophageal cancers,Citation31 while Eide et al found higher PDGF-bb serum levels in lung cancer compared to COPD patients.Citation32 In the MicMa cohort higher levels of PDGF were associated with worse prognosis, the same tendency although not significant, was observed in the DCTB cohort. The PDGF family of growth factors play a role in lymphangiogenesis.Citation33 Even though we did not find a correlation between PDGF serum levels and its tumor mRNA expression, we previously reported that the tumor interstitial fluid levels of PDGF were higher than in the normal interstitial fluid,Citation8 which suggest that the tumor microenvironment may contribute to the production of PDGF.
Using mRNA expression data to predict for different immune cell type infiltration, we found a possible association between PDGF serum levels and pro-B cells infiltration. Both B cells and PDGF have been shown to be involved in lymph-angiogenesis.Citation34 The data we report here are correlative and from small cohorts, and further study will be necessary to investigate the possible interrelationship between pro-B cells, PDFG and lymph-angiogenesis. As proposed also by others Citation35 our study comes to suggest that PDGF acts as a pro-tumorigenic cytokine and its inhibition may enhance the efficacy of chemotherapy.
In this study, we found higher levels of IL17 in the serum of patients with an ER negative tumor when compared to the serum ER positive patients. IL17 is a pleiotropic cytokine mainly secreted by Th17 cells, which may exert pro- or anti- tumorigenic effects.Citation13 Elevated levels of IL17 have been found in the microenvironment of ER negative breast tumors and associated with a worse prognosis.Citation36,Citation37 Our study reinforces the association between IL17 levels and ER status and further suggest that the higher levels of IL17 and therefore Th17 cell activity observed in the microenvironment of ER negative breast cancers can also be measured in the serum.
We previously reported higher IP10 levels on the tumor interstitial fluid of breast cancer patients. IP10 is widely expressed by both cancer cells and tumor-infiltrating lymphocytes. Our recently published analysis of paired normal and tumor biopsies demonstrated that the expression of IP10 is mainly restricted to the ductal epithelial cells. A paired analysis of matched Tumor and Normal interstitial fluid demonstrated that IP10 is significantly elevated in tumor samples. Our data provided an evidence for the secretion of more IP10 from breast tumor tissues than from non-malignant breast tissues.Citation8
Here we report a positive correlation between IP10 tumor-derived mRNA expression and circulating serum levels. IP10 act as a chemo-attractant for monocytesCitation38,Citation39 which may explain the herein reported positive correlation between IP10 and type M1 Macrophages. IP10 has been reported as a pro- tumorigenic chemokine. Increased expression of IP10 and its receptor CXCR3 has been observed in many cancers, including malignant melanoma,Citation40 ovarian carcinoma,Citation41 multiple myeloma,Citation42 B-cell lymphoma,Citation43 and basal cell carcinoma.Citation44 The role of IP10 and its receptor in breast cancer progression has been demonstrated using breast cancer cell lines.Citation17 Our results suggest that high IP10 serum levels correlate with high IP10 in the tumor microenvironment and may be a measure of worse prognosis.
The tumor microenvironment may accumulate high concentration of cytokines and other molecules such as metabolites which may affect systemically the concentrations of the same molecules or others. Apart for IP10, we report here modest/no correlations between serum level of cytokines and their mRNA expression at the tumor site. Systemically the expression of the cytokines measured here may be affected by many factors, the primary breast tumor is one of them, but other such as lifestyle, environmental cues or administration of anti-inflammatory/painkiller drugs should be considered in further studies to further assess the continuum between the tumor micro- and macro-environment.
In conclusion patients with advanced stage disease appear to experience a simultaneous immunostimulation and immunosuppression which may lead to disturbed cytokines production.Citation45 Breast cancer tumors are highly heterogeneous. However, there are common clinical characteristics in patients with more advanced tumor phenotype. We report that PDGF and IP10 serum levels as paraneoplastic signals may reflect the presence of larger tumor and a worse prognosis. We further suggest that cytokine serum levels may be associated with specific immune cell types at the tumor site. Our results emphasize the value of measuring serum cytokine levels in breast cancer.
Materials and methods
Patient material
MicMA cohort
Operable early breast cancer patients were included in the Oslo1 micrometastasis observational study between 1995 and 1998.Citation46 Related to the current results, sera were collected at the time of surgery. A subset of the patient’s fresh primary tumors was collected for detailed molecular analyses, a cohort called MicMa. Only patients within the MicMa subset were included in the current analysis. The study is approved by the Regional Ethics Committee. Written informed consent was obtained from all participants, the study was conducted in accordance with the Declaration of Helsinki. Sera from 137 MicMa samples were available, six patients were excluded from the analysis due to administration of neo-adjuvant therapy. For the remaining 131 patients, their characteristics are shown in Supplementary Table 7. Immunostaining was performed using mouse antibody against estrogen receptor and progesterone receptor (clones 6F11 and 1A6, respectively; Novocastra, Cat. Nos. NCL-L-ER-6F11 and NCL-L-PGR), c-erbB-2 (clone CB11; BioGenex, Cat. No. AM134). Automated immunostaining systems were used, Ventana Medical Systems, Inc (Tucson, AZ). Immunopositivity was recorded if more than 10% (ER, PR, c-erbB-2) of the tumor cells were immunostained as previously described.Citation47 The gene expression data was generated using Agilent whole genome 4x44K oligo array (Agilent Technologies, Cat. No. G4112F) as previously describedCitation16 and is available at GEO with accession number GSE19783.
DCTB cohort
The DCTB study was conducted in compliance with the Helsinki II Declaration, written informed consent was obtained from all participants. The project was approved by the Copenhagen and Frederiksberg regional division of the Danish National Committee on Biomedical Research Ethics (KF 01–069/03). 79 breast cancer patients were included in this cohort as previously described.Citation8 In our recent publication using this cohort, tumor interstitial fluid was extracted from small surgically resected breast tumor pieces, matched sera from 28 women were obtained prior surgery. Characteristics of these 28 patients are shown in Supplementary Table 7. Samples were considered negative for ER when the percentage of nuclear immunoreactivity within the invasive cancer cells was < 1%. Samples with HER2/centromere 17 ratio > 2 were considered HER2 positive.
mRNA expression profiling
Total RNA was isolated using the TRIzol reagent (Thermo Fisher Scientific, Cat. No. 15596026) following the manufacturer’s instructions. NanoDrop spectrophotometeric analysis (Thermo Fisher Scientific, Waltham, MA, USA) assessed RNA concentration and purity. RNA quality and integrity were assessed by the 2100 Bioanalyzer instrument (Agilent Technologies, Santa Clara, CA, USA). mRNA expression was measured using SurePrint G3 Human GE 8x60K one-color microarrays from Agilent (Agilent Technologies, Cat. No. G4851A) according to the manufacturer’s protocol and using 100 ng of RNA as input for amplification.
Scanning was performed with Agilent Scanner G2565A, and signals were extracted using Feature Extraction v.10.7.3.1 (Agilent Technologies). Non-uniform spots were excluded and missing data were imputed using local least squares imputation (LLSimpute from the R package “pcaMethods”).Citation48 Arrays were log2-transformed and quantile-normalized. mRNA expression data have been submitted to ArrayExpress database under accession number E-MTAB-7201. A total of 96 samples were measured for gene expression.
Cytokine profiling
A total of 27 molecules including interleukins, chemokines, growth factors, interferon (IFN), and tumor necrosis factor (TNF), were measured in a 27-plex commercially available cytokine panel (Bio-Rad Laboratories, Cat. No. M500KCAF0Y) and were analyzed with the Luminex xMAP 200 platform (Luminex Corporation, Austin, TX, USA). The assays included a series of known concentrations to generate standard curves. All samples were analyzed in duplicates, the mean values of the duplicates were used for the further analysis. The results obtained were collected and processed with Bio-Plex Manager 6.0 (Bio-Rad Laboratories). Details of the cytokines are given in Supplementary Table 8. Natural log transformed cytokines levels (pg/mL) for the MicMa and the DCTB cohorts are given in Supplementary Table 9, summary statistics are given in Supplementary Table 10. IL15 and IL1RA serum levels were often very low or undetectable; many samples display a zero value for these two cytokines, therefore interpretation of the association of these serum cytokine levels with any other feature must be interpreted cautiously.
Immunohistochemistry (IHC)
IHC analysis was performed as described previously.Citation8 Briefly, FFPE blocks were prepared from 2 to 3 various parts of the tissue piece. Immune cells were assessed using an antibody recognizing CD45 (DAKO, mouse monoclonal antibody (clone 2B11+ PD7/26); dilution: 1:400) and IP10 with an anti-IP10 (Abcam, rabbit polyclonal (ab9807); dilution: 1:100). Standardization of the dilution, incubation, and development times appropriate for each antibody allowed an accurate comparison of expression levels in all cases. Positive and negative control slides were analyzed in parallel, with the latter incubated with PBS instead of primary antibodies.
Statistical analysis
All analyses were performed in the R Citation49 version 3.3.2. After natural log transformation, cytokine levels were analyzed in relation to categorical clinical parameters using Mann-Whitney-U (MWU) or Kruskal-Wallis tests. Unless otherwise stated, results were considered statistically significant, if the two-sided p-value < 0.05. Hierarchical clustering with Euclidean distance and Ward.D linkage was performed on the log transformed cytokine levels using the R package pheatmap (version 1.08)Citation50 clusters were identified using the cutree function. Pearson correlations were visualized using R package corrplot (version 0.84). Survival analysis using Kaplan-Meier estimates for overall survival were compared by the log-rank test. Hazard ratios and their 95% confidence intervals were estimated using the survival R package based on the log rank test. Cytokine-cluster4 scores were calculated using the GSVA Bioconductor package.Citation12 Enrichment score for immune cell type at the tumor site was calculated using xCell R package with default parameters.Citation19 xCell relies on single-sample GSEA (ssGSEA) and newly generated gene signatures for 64 cell types to digitally dissect the tumor microenvironment. In our study, we focused on 27 immune cell types, the xCell scores from these 26 immune cell types can be found in Supplementary Table 5.
Abbreviations
| DCIS | = | ductal carcinoma in situ |
| ER | = | estrogen receptor |
| HER2 | = | Human epidermal growth factor receptor 2 |
| IHC | = | immunohistochemistry |
| NIF | = | normal interstitial fluid |
| PAM50 | = | Prediction Analysis of Microarray 50 |
| PCA | = | Principal component analysis |
| PR | = | progesterone receptor |
| ssGSEA | = | single sample gene set enrichment analysis |
| TAMs | = | tumor-associated macrophages |
| TIF | = | tumor interstitial fluid |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Download Zip (415.5 KB)Acknowledgments
Sera from the MicMa cohorts were collected with the support of the grants from the Norwegian Cancer Society (D99061, PI: Bjørn Naume), the Norwegian Research Council (155218/300) and the SalusAnsvar Award to ALBD. DCTB project was supported by grants from The Eurocan Platform, which has received funding from the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement No. 260791. The Danish Cancer Society, the “Race Against Breast Cancer” foundation, the John and Birthe Meyer Foundation, the Danish National Research Foundation (Centre of Excellence: CARD, DNRF125). Cytokine profiling was performed with a grant from Strategiske Ahus midler, grant 266972. Shakila Jabeen was financed by the South Eastern Norway Health Authority (grant 272904 to Vessela Kristensen). J.E is funded by Cancerfonden (CAN 2015/739), Sweden.
Supplemental Material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Burnet FM. The concept of immunological surveillance. Prog Exp Tumor Res. 1970;13:1–27.
- Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–998. doi:10.1038/ni1102-991.
- Parrish-Novak J, Dillon SR, Nelson A, Hammond A, Sprecher C, Gross JA, Johnston J, Madden K, Xu W, West J, et al. Interleukin 21 and its receptor are involved in NK cell expansion and regulation of lymphocyte function. Nature. 2000;408:57–63. doi:10.1038/35040504.
- Tan JT, Dudl E, LeRoy E, Murray R, Sprent J, Weinberg KI, Surh CD. IL-7 is critical for homeostatic proliferation and survival of naive T cells. Proc Natl Acad Sci U S A. 2001;98:8732–8737. Epub 2001 Jul 10.. doi:10.1073/pnas.161126098.
- Zou L, Barnett B, Safah H, Larussa VF, Evdemon-Hogan M, Mottram P, Wei S, David O, Curiel TJ, Zou W. Bone marrow is a reservoir for CD4+CD25+ regulatory T cells that traffic through CXCL12/CXCR4 signals. Cancer Res. 2004;64:8451–8455. doi:10.1158/0008-5472.CAN-04-1987.
- Js P, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi:10.1200/JCO.2008.18.1370.
- Pruneri G, Vingiani A, Denkert C. Tumor infiltrating lymphocytes in early breast cancer. Breast. 2018;37:207–214. doi:10.1016/j.breast.2017.03.010.
- Espinoza JA, Jabeen S, Batra R, Papaleo E, Haakensen V, Timmermans Wielenga V, Møller Talman M-L, Brunner N, Børresen-Dale A-L, Gromov P, et al. Cytokine profiling of tumor interstitial fluid of the breast and its relationship with lymphocyte infiltration and clinicopathological characteristics. Oncoimmunology. 2016;5:e1248015. doi:10.1080/2162402X.2016.1248015.
- Naume B, Zhao X, Synnestvedt M, Borgen E, Russnes HG, Lingjaerde OC, Strømberg M, Wiedswang G, Kvalheim G, Kåresen R, et al. Presence of bone marrow micrometastasis is associated with different recurrence risk within molecular subtypes of breast cancer. Mol Oncol. 2007;1:160–171. doi:10.1016/j.molonc.2007.03.004.
- Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi:10.1038/nature07205.
- West NR, McCuaig S, Franchini F, Powrie F. Emerging cytokine networks in colorectal cancer. Nat Rev Immunol. 2015;15:615–629. doi:10.1038/nri3896.
- Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi:10.1186/1471-2105-14-7.
- Chen K, Kolls JK. Interluekin-17A (IL17A). Gene. 2017;614:8–14. doi:10.1016/j.gene.2017.01.016.
- Papadopoulos N, Lennartsson J. The PDGF/PDGFR pathway as a drug target. Mol Aspects Med. 2018;62:75-88. doi: 10.1016/j.mam.2017.11.007. Epub 2017 Nov 15.
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi:10.1038/35021093.
- Enerly E, Steinfeld I, Kleivi K, Leivonen SK, Aure MR, Russnes HG, Rønneberg JA, Johnsen H, Navon R, Rødland E, et al. miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS One. 2011;6:e16915. doi:10.1371/journal.pone.0016915.
- Datta D, Flaxenburg JA, Laxmanan S, Geehan C, Grimm M, Waaga-Gasser AM, Briscoe DM, Pal S. Ras-induced modulation of CXCL10 and its receptor splice variant CXCR3-B in MDA-MB-435 and MCF-7 cells: relevance for the development of human breast cancer. Cancer Res. 2006;66:9509–9518. doi:10.1158/0008-5472.CAN-05-4345.
- Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi:10.1038/nature10983.
- Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi:10.1186/s13059-017-1349-1.
- Blankenstein T, Coulie PG, Gilboa E, Jaffee EM. The determinants of tumour immunogenicity. Nat Rev Cancer. 2012;12:307–313. doi:10.1038/nrc3246.
- Ali HR, Chlon L, Pharoah PD, Markowetz F, Caldas C, Ladanyi M. Patterns of immune infiltration in breast cancer and their clinical implications: a gene-expression-based retrospective study. PLoS Med. 2016;13:e1002194. doi:10.1371/journal.pmed.1002194.
- Dannenfelser R, Nome M, Tahiri A, Ursini-Siegel J, Vollan HKM, Haakensen VD, Helland Å, Naume B, Caldas C, Børresen-Dale A-L, et al. Data-driven analysis of immune infiltrate in a large cohort of breast cancer and its association with disease progression, ER activity, and genomic complexity. Oncotarget. 2017;8:57121–57133. doi:10.18632/oncotarget.19078.
- Desmedt C, Salgado R, Fornili M, Pruneri G, Van Den Eynden G, Zoppoli G, Rothé F, Buisseret L, Garaud S, Willard-Gallo K, et al. Immune infiltration in invasive lobular breast cancer. J Natl Cancer Inst. 2018. doi:10.1093/jnci/djx268.
- Loi S, Sirtaine N, Piette F, Salgado R, Viale G, Van Eenoo F, Rouas G, Francis P, Crown JPA, Hitre E, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J Clin Oncol. 2013;31:860–867. doi:10.1200/JCO.2011.41.0902.
- Kristensen VN, Vaske CJ, Ursini-Siegel J, Van Loo P, Nordgard SH, Sachidanandam R, Sorlie T, Warnberg F, Haakensen VD, Helland A, et al. Integrated molecular profiles of invasive breast tumors and ductal carcinoma in situ (DCIS) reveal differential vascular and interleukin signaling. Proc Natl Acad Sci U S A. 2012;109:2802–2807. doi:10.1073/pnas.1108781108.
- Quigley D, Silwal-Pandit L, Dannenfelser R, Langerod A, Vollan HK, Vaske C, Siegel JU, Troyanskaya O, Chin S-F, Caldas C, et al. Lymphocyte invasion in IC10/basal-like breast tumors is associated with wild-type TP53. Mol Cancer Res. 2015;13:493–501. doi:10.1158/1541-7786.MCR-14-0387.
- Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi:10.1038/nrc3245.
- Ignatiadis M, Singhal SK, Desmedt C, Haibe-Kains B, Criscitiello C, Andre F, Loi S, Piccart M, Michiels S, Sotiriou C. Gene modules and response to neoadjuvant chemotherapy in breast cancer subtypes: a pooled analysis. J Clin Oncol. 2012;30:1996–2004. doi:10.1200/JCO.2011.39.5624.
- Ono M, Tsuda H, Shimizu C, Yamamoto S, Shibata T, Yamamoto H, Hirata T, Yonemori K, Ando M, Tamura K, et al. Tumor-infiltrating lymphocytes are correlated with response to neoadjuvant chemotherapy in triple-negative breast cancer. Breast Cancer Res Treat. 2012;132:793–805. doi:10.1007/s10549-011-1554-7.
- Esposito A, Criscitiello C, Curigliano G. Highlights from the 14(th) St Gallen international breast cancer conference 2015 in vienna: dealing with classification, prognostication, and prediction refinement to personalize the treatment of patients with early breast cancer. Ecancermedicalscience. 2015;9:518. doi:10.3332/ecancer.2015.518.
- Matsumoto S, Yamada Y, Narikiyo M, Ueno M, Tamaki H, Miki K, Wakatsuki KOHEI, Enomoto KOJI, Yokotani TOMOYO, Nakajima YOSHIYUKI. Prognostic significance of platelet-derived growth factor-BB expression in human esophageal squamous cell carcinomas. Anticancer Res. 2007;27:2409–2414.
- Eide HA, Halvorsen AR, Sandhu V, Fane A, Berg J, Haakensen VD, Kure EH, Brustugun OT, Kiserud CE, Kyte JA, et al. Non-small cell lung cancer is characterised by a distinct inflammatory signature in serum compared with chronic obstructive pulmonary disease. Clin Transl Immunol. 2016;5:e109. doi:10.1038/cti.2016.65.
- Vincent L, Rafii S. Vascular frontiers without borders: multifaceted roles of platelet-derived growth factor (PDGF) in supporting postnatal angiogenesis and lymphangiogenesis. Cancer Cell. 2004;6:307–309. doi:10.1016/j.ccr.2004.09.024.
- Dubey LK, Karempudi P, Luther SA, Ludewig B, Harris NL. Interactions between fibroblastic reticular cells and B cells promote mesenteric lymph node lymphangiogenesis. Nat Commun. 2017;8:367. doi:10.1038/s41467-017-00504-9.
- Pietras K, Rubin K, Sjoblom T, Buchdunger E, Sjoquist M, Heldin CH, Östman A. Inhibition of PDGF receptor signaling in tumor stroma enhances antitumor effect of chemotherapy. Cancer Res. 2002;62:5476–5484.
- Chen WC, Lai YH, Chen HY, Guo HR, Su IJ, Chen HH. Interleukin-17-producing cell infiltration in the breast cancer tumour microenvironment is a poor prognostic factor. Histopathology. 2013;63:225–233. doi:10.1111/his.12156.
- Cochaud S, Giustiniani J, Thomas C, Laprevotte E, Garbar C, Savoye AM, Curé H, Mascaux C, Alberici G, Bonnefoy N, et al. IL-17A is produced by breast cancer TILs and promotes chemoresistance and proliferation through ERK1/2. Sci Rep. 2013;3:3456. doi:10.1038/srep03456.
- Roberts WK, Blachere NE, Frank MO, Dousmanis A, Ransohoff RM, Darnell RB. A destructive feedback loop mediated by CXCL10 in central nervous system inflammatory disease. Ann Neurol. 2015;78:619–629. doi:10.1002/ana.24494.
- Vazirinejad R, Ahmadi Z, Kazemi Arababadi M, Hassanshahi G, Kennedy D. The biological functions, structure and sources of CXCL10 and its outstanding part in the pathophysiology of multiple sclerosis. Neuroimmunomodulation. 2014;21:322–330. doi:10.1159/000357780.
- Monteagudo C, Martin JM, Jorda E, Llombart-Bosch A. CXCR3 chemokine receptor immunoreactivity in primary cutaneous malignant melanoma: correlation with clinicopathological prognostic factors. J Clin Pathol. 2007;60:596–599. doi:10.1136/jcp.2005.032144.
- Furuya M, Suyama T, Usui H, Kasuya Y, Nishiyama M, Tanaka N, Ishiwata I, Nagai Y, Shozu M, Kimura S. Up-regulation of CXC chemokines and their receptors: implications for proinflammatory microenvironments of ovarian carcinomas and endometriosis. Hum Pathol. 2007;38:1676–1687. doi:10.1016/j.humpath.2007.03.023.
- Pellegrino A, Antonaci F, Russo F, Merchionne F, Ribatti D, Vacca A, Vaccaa A, Dammaccoa F. CXCR3-binding chemokines in multiple myeloma. Cancer Lett. 2004;207:221–227. doi:10.1016/j.canlet.2003.10.036.
- Jones D, Benjamin RJ, Shahsafaei A, Dorfman DM. The chemokine receptor CXCR3 is expressed in a subset of B-cell lymphomas and is a marker of B-cell chronic lymphocytic leukemia. Blood. 2000;95:627–632.
- Lo BK, Yu M, Zloty D, Cowan B, Shapiro J, McElwee KJ. CXCR3/ligands are significantly involved in the tumorigenesis of basal cell carcinomas. Am J Pathol. 2010;176:2435–2446. doi:10.2353/ajpath.2010.081059.
- Lippitz BE. Cytokine patterns in patients with cancer: a systematic review. Lancet Oncol. 2013;14:e218–e228. doi:10.1016/S1470-2045(12)70582-X.
- Naume B, Wiedswang G, Borgen E, Kvalheim G, Karesen R, Qvist H, Janbu J, Harbitz T, Nesland JM. The prognostic value of isolated tumor cells in bone marrow in breast cancer patients: evaluation of morphological categories and the number of clinically significant cells. Clin Cancer Res. 2004;10:3091–3097. doi:10.1158/1078-0432.CCR-04-1002.
- Wiedswang G, Borgen E, Karesen R, Kvalheim G, Nesland JM, Qvist H, Schlichting E, Sauer T, Janbu J, Harbitz T, et al. Detection of isolated tumor cells in bone marrow is an independent prognostic factor in breast cancer. J Clin Oncol. 2003;21:3469–3478. doi:10.1200/JCO.2003.02.009.
- Stacklies W, Redestig H, Scholz M, Walther D, Selbig J. pcaMethods–a bioconductor package providing PCA methods for incomplete data. Bioinformatics. 2007;23:1164–1167. doi:10.1093/bioinformatics/btm069.
- R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2018.
- Kolde R pheatmap: pretty Heatmaps. R package version 1.0.8. 2015.
