ABSTRACT
The oncoantigen nucleophosmin-anaplastic lymphoma kinase (NPM-ALK) induces cellular and humoral immune responses in patients with NPM-ALK-positive anaplastic large cell lymphoma (ALCL). We characterize the NPM-ALK-specific T-cell responses in a cohort of pediatric and adolescent ALCL-patients in remission without Human Leucocyte Antigen (HLA)-preselection.
First, we assessed NPM-ALK-reactive T-cell responses and their HLA-class I restriction in patients by using dendritic cells (DCs) transfected with in vitro transcribed (IVT) NPM-ALK-RNA for CD8 (n = 20) or CD3 (n = 9) T-cell stimulation. NPM-ALK-specific T-cells were detected in twelve of 29 patients (nine of 20 with CD8-selected and three of nine with CD3-selected cells). Recognition of NPM-ALK was restricted by HLA-C alleles in six of eight, and by HLA-B alleles in four of eight analyzed patients. No NPM-ALK-reactivity was detected in 20 healthy individuals.
Second, in order to define possible immunogenic NPM-ALK-epitope regions, DCs pulsed with pools of overlapping long NPM-ALK-peptides were used to stimulate T-cells in further 22 patients and ten controls. Responsive T-cells were detected in 15 patients and in five controls. A peptide pool located in the middle of the kinase domain induced ALK-reactive T-cells in 14 of 15 responsive patients. We could narrow to single peptides between p327-p370 of NPM-ALK in four patients.
In conclusion, using IVT-RNA, 40% of NPM-ALK-positive ALCL-patients in remission had detectable NPM-ALK-specific T-cell responses which were mainly restricted by HLA-B and -C alleles. Peptide stimulation of T-cells revealed responses in almost 70% of patients and allowed describing an immunogenic region located in the ALK-kinase domain.
Introduction
Anaplastic lymphoma kinase (ALK)-positive anaplastic large cell lymphoma (ALCL) is characterized by a specific translocation involving the ALK gene on chromosome 2 leading to the expression of constitutively active ALK-fusion proteins which are heavily involved in lymphoma pathogenesis.Citation1 Almost 90% of ALK-positive ALCLs in children and adolescents express the oncogenic nucleophosmin (NPM)-ALK fusion protein resulting from translocation t(2;5)(p23;q35).Citation2
The relapse rate in children and adolescents with ALK-positive ALCL reaches 30% with current standard chemotherapies, indicating that new therapeutic approaches need to be explored.Citation3,Citation4 Even though remission can be induced in a high proportion of relapsing patients by targeted agents like ALK-inhibitors and Brentuximab Vedotin,Citation5–Citation6 cure of the disease has not been reported by these drugs. The high efficacy of consolidation of relapsed patients by allogeneic blood stem cell transplantation hints at a possible involvement of the immune system in the control of ALCL.Citation7–Citation9
The limitation of ALK expression to immune-privileged sites and its central role in the development and maintenance of ALCL suggest that it has the potential to serve as an attractive target for immunotherapy.Citation10,Citation11 Previous studies on the immunogenicity of ALK revealed that patients with ALK-positive ALCL mount ALK-specific humoral and cellular immune responses. More than 90% of patients have detectable anti-ALK antibodies in their blood. B-cell Antibody-titer inversely correlates with the risk of relapse.Citation12,Citation13 Applying a reverse immunological approach, ALK-specific CD8+ and CD4+ T-cells have been detected in HLA-preselected ALCL-patients using ALK-derived peptides.Citation14–Citation17 Regarding ALK-specific CD8+ T-cell response, ALK-derived peptides p280-89 and p376–85 predicted to bind to the prevalent HLA-class I allele A*02:01 induced peptide-reactive T-cell responses in HLA-A*02:01 positive patients and healthy controls. In healthy donors these CD8+ T-cells predominantly exhibited a naïve phenotype, whereas effector and memory CD8+ T-cells were detected in ALK-positive ALCL-patients.Citation15 Using a mouse model of vaccination with truncated ALK cDNA, CD8+ T-cells were shown to mediate protection against ALK-positive ALCL, even when vaccination was used in a therapeutic setting of established lymphoma.Citation18 This study demonstrated the therapeutic potential of ALK vaccination in vivo.
As a prerequisite for the development of an NPM-ALK-directed immunotherapy, we aimed to characterize the CD8+ T-cell responses in a large cohort of NPMALK-positive ALCL-patients involving the complete HLA-type of each patient and by ensuring endogenous processing and presentation of NPM-ALK peptide to T-cells. The two specific goals of our study were to define the HLA-class I restriction of NPM-ALK-reactive CD8+ T-cells in responsive patients and the identification of immunogenic NPM-ALK-regions recognized by responding T-cells.
To achieve the first goal, we developed a test system using autologous dendritic cells (DCs) as antigen-presenting cells (APCs) and in vitro transcribed RNA (IVT-RNA) encoding full length NPM-ALK as the antigenic format, thereby ensuring endogenous processing of peptides for presentation.Citation19 COS-7 cells, co-transfected with each patient´s individual HLA-class I alleles and NPM-ALK-encoding plasmids, allowed to identify the HLA-class I restriction of the NPM-ALK-specific T-cells in responding patients. We previously reported the applicability of this test system in five ALCL-patients in remission after chemotherapy. NPM-ALK-reactive CD8+ T-cells were detected in three of them and the response was restricted by HLA-C alleles.Citation19 These first patients were selected on the basis of an initial high antibody titer as a possible surrogate marker for a strong anti-ALK immune response. Now, we report the results using this approach in a large cohort of 29 patients in order to define the proportion of responding patients and their restricting HLA-class I alleles as well as to correlate the T-cell response to the ALK-antibody titer and clinical characteristics.
To address the second question, we selected overlapping long peptides as antigen format to stimulate and detect NPM-ALK-specific T-cell responses. The long peptides ensured peptide processing for presentation by HLA-molecules on APCs.Citation20,Citation21 These peptides were covering the NPM-ALK fusion region, the entire kinase domain and the ALK-antibody binding region. The peptide selection was based upon the location of known antigenic sites and possible immunogenic regions.Citation15–Citation19,Citation22 Detection of the potential immunogenic epitope region of NPM-ALK was performed on 22 additional patients. Both peptide-pulsed DCs and IVT-RNA-transfected DCs were used as target cells to confirm a peptide-induced response.
Results
NPM-ALK-reactive T-cell response against antigen IVT-RNA
To enrich the T-cell responses directed against the NPM-ALK oncoprotein, NPM-ALK IVT-RNA-based in vitro T-cell stimulation was performed. Due to the limiting amounts of patient materials, and in order to increase the potency of the T-cell stimulation assays, we applied a microculture-based approach.Citation19 Peripheral blood lymphocytes obtained from altogether 29 NPM-ALK-positive paediatric and adolescent ALCL-patients including the five patients reported earlierCitation19 who were in clinical remission for 1–15 years and from 20 healthy donors were analyzed by this approach for their anti-NPM-ALK T-cell responses. From 20 patients, purified CD8+ T-cells were stimulated with autologous RNA-transfected DCs and tested for recognition of NPM-ALK. In nine patients CD3-selected T-cells were applied in order to get a first hint for a possible CD4+ T-cell response in addition to CD8+ T-cells reactive against NPM-ALK (). Responder T-cells were tested after three stimulations for recognition of autologous DCs transfected with IVT-RNA encoding NPM-ALK in an IFN-γ ELISPOT assay.
Table 1. NPM-ALK-specific T-cell responses in NPM-ALK-positive ALCL-patients analyzed against in vitro transcribed RNA.
In nine of 20 patients, microculture responders CD8+ T-cells were able to recognize autologous DCs transfected with NPM-ALK IVT-RNA (). In responding patients, NPM-ALK-reactivity was observed in 1-3 microcultures out of 6-8 stimulated microcultures. IFN-γ spot numbers in positive microcultures ranged from 3- to 47-fold above the background reactivity (). Microcultures with the strongest NPM-ALK-reactivity were observed in patient R2. NPM-ALK-reactive CD8+ T-cells were not detected in the microcultures generated from the 15 healthy individuals.
Figure 1. CD8+ T-cell responses after in vitro stimulation with in vitro transcribed -RNA encoding NPM-ALK in ALCL-patients.Purified T-cells (either CD8+ or CD3+) from ALCL-patients in remission were stimulated with irradiated autologous DCs transfected with NPM-ALK IVT-RNA. After two weekly re-stimulations, day 19 responder T-cells were tested in an IFN-γ ELISPOT assay for recognition of autologous DCs untransfected or transfected with IVT-RNA encoding NPM-ALK or – as a control – HCMV pp65 (3-6 × 103/well). Data are shown for representative microculture (mc) responder populations of patients R2, R4, R8, R10, R13, R15, in whom CD8+ blood-derived T-cells had been stimulated (a), and of patients R20 and R21, in whom stimulations had been performed on CD3+ blood T cells. The NPM-ALK-responder populations were partially inhibited with the pan HLA-class I-specific antibody W6/32 (b). Bars represent the means of duplicates ± standard deviation.
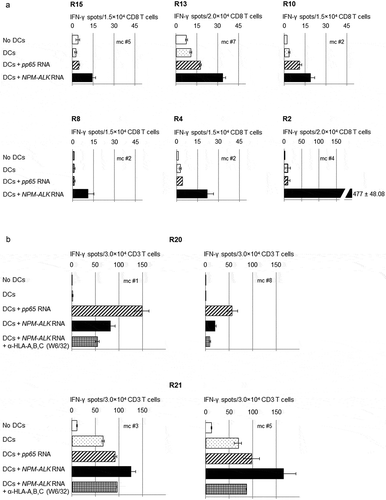
NPM-ALK-reactive T-cells were enriched and detected in three of nine ALCL-patients analyzed starting from CD3+ blood T lymphocytes (). In responding patients, the observed number of NPM-ALK-reactive microcultures varied from two to six out of eight stimulated microcultures. IFN-γ spot numbers in positive microcultures ranged from 2- to 50-fold above the background reactivity (). Microcultures with the strongest NPM-ALK-reactivity were found in patient R20. NPM-ALK-reactive CD3+ T-cells were not detected in the microcultures generated from the five healthy individuals. As a control for the chosen in vitro stimulation conditions and for the functionality of IVT-RNA, anti-CMV-pp65-reactivity was confirmed in all CMV-seropositive ALCL-patients and controls tested (Supplementary Table 1).
To differentiate between NPM-ALK-specific CD4+ and CD8+ T-cell responses, in the patients with detectable NPM-ALK-responsive CD3+ T-cells, the T-cell receptor/HLA-class I interaction was blocked by a pan-HLA-class I-reactive antibody in two analyzed patients R20 and R21 (). The partial inhibition of T-cell reactivity against NPM-ALK indicates the presence of both HLA-class I and -class II restricted T-cell responses against NPM-ALK in CD3-stimulated patients.
The RNA-based T-cell stimulation approach without HLA-preselection, using autologous DC as APCs, allowed for the detection of NPM-ALK-specific CD8+ T-cells in the peripheral blood of around 40% of well characterized and uniformly treated ALCL-patients in first remission but not in healthy controls. NPM-ALK-reactive T-cells were present in NPM-ALK-positive ALCL-patients even 15 years after diagnosis. No significant differences were observed between patients with or without detectable CD8+ T-cell response regarding median age at diagnosis, the median time from diagnosis to analysis, gender, anti-ALK antibody titer at diagnosis and time of analysis, minimal disseminated disease, clinical risk features or histological subtype (Supplementary Table 2A).
HLA-class I restriction of anti-NPM-ALK CD8+ T-cell responses
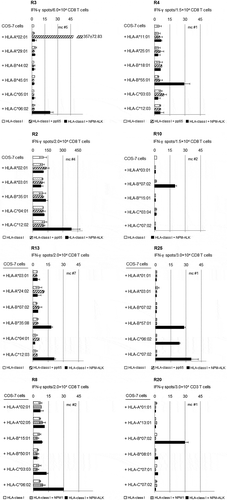
To assess the HLA-class I alleles involved in the presentation of immunogenic antigens to CD8+ T-cells, the microculture-responders of ALCL-patients were tested for their HLA-restriction using COS-7 cells as targets that were co-transfected with plasmids encoding NPM-ALK and the patients’ individual HLA-class I alleles. Restricting HLA-class I alleles were identified in eight out of twelve NPM-ALK-reactive patients. In six of them, recognition of NPM-ALK was restricted by HLA-C alleles and in four of them the response against NPM-ALK was restricted by HLA-B alleles (). In two NPM-ALK-reactive patients, both HLA-B and -C alleles were contributing to the recognition of COS-7 transfectants ( and ). As a control, HLA-class I alleles restricting the recognition of HCMV pp65, were determined in selected patients and healthy donors (Supplementary Table 1). For comparison, the HLA-class I alleles of all patients and healthy donors are shown in the Supplementary Table 3. To rule out that T-cells were directed against NPM1 alone, responder T-cells were tested for the recognition of COS-7/NPM1/HLA transfectants. No NPM1-reactivity was detected in microcultures of the six analyzed patients (data not shown).
Figure 2. Identification of HLA-class I restriction molecules for the CD8+ T-cell response against NPM-ALK in ALCL-patients.
On day 19, microculture (mc) responder populations obtained from indicated patients after RNA stimulation of blood-derived T-cells were tested in parallel IFN-γ ELISPOT assays for the recognition of COS-7 cells (2.0 × 104/well) co-transfected with plasmids encoding the respective patient´s HLA-class I alleles and NPM-ALK. Bars represent the means of duplicates ± standard deviation. (A part of this figure is reproduced with permission from reference 20.)
NPM-ALK-reactive T-cell response against NPM-ALK-derived overlapping long peptides
In order to identify relevant immunogenic NPM-ALK-regions as well as target CD8+ and CD4+ T-cell responses, we tested T-cells from 22 further ALK-positive ALCL-patients and ten healthy controls for their reactivity against a panel of NPM-ALK-derived synthetic long peptides. We chose 25 long peptides (contiguous 30mers overlapping by 16 amino acids) for CD3+ T-cell stimulation as they are taken up by APCs and processed into shorter peptides that can be presented by a broad range of HLA-class I and ‐class II molecules. Similar to the RNA approach, this method also opens up the possibility to assess T-cell responses in ALCL-patients taking into account the entire individual HLA-types. The overlapping peptides covered the NPM-ALK fusion region, the ALK-kinase domain and the juxta-kinase part of ALK. With the exception of a long peptide covering the fusion region, six overlapping peptides were combined in each pool (pools A – D) (Supplementary Table 4 and Supplementary Figure 1). CD3+ T-cells were stimulated in separate microcultures with autologous DCs loaded with one peptide pool. After two re-stimulations, responder lymphocytes were analyzed with an IFN-γ ELISPOT assay for the recognition of DCs pulsed with the fusion peptide, peptide pools as well as all 25 NPM-ALK peptides or later with individual peptides of a positive pool.
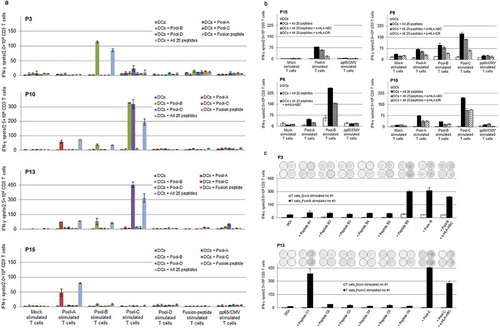
NPM-ALK peptide-reactive T-cells were detected in peripheral blood samples of 15 of the 22 ALCL-patients, who were in remission for up to eight years after treatment (). In responding patients’ responder CD3+ T-cells were able to recognize autologous DCs pulsed with at least one peptide pool. T-cell reactivity against a particular peptide-pool was observed in all the microcultures stimulated with the same peptide pool. IFN-γ spot numbers in positive microcultures ranged from 2- to 65-fold above the background reactivity. Microculture-responders with the strongest peptide pool-reactivity were those from patient P10.
Table 2. NPM-ALK-specific T-cell responses in NPM-ALK-positive ALCL-patients analyzed with NPM-ALK-derived long peptides.
The T-cell responses were directed against one or more peptide pools. In patients, responses were seen against pools A, B and C, but not against pool D and not against the fusion peptide ( and ). In 14 of the 15 responding patients, we identified peptide pool B as consistent inducer of T-cell responses. T-cell responses directed against pool A and pool C peptides were detected in five and six patients, respectively ( and ). In two pool C reactive patients, responder T-cells from pool C stimulated microcultures were able to recognize autologous DCs pulsed with peptide pool C as well as pool B. This indicates that the recognized T-cell epitope is located in the overlapping part of peptide pool B and pool C. In seven of the 15 reactive patients, more than one peptide pool induced a T-cell response ( and ). There were no significant differences observed between patients with or without a detectable CD3+ T-cell response regarding biological and clinical risk features (Supplementary Table 2B).
Figure 3. Detection of anti-ALK CD3+ T-cells in ALCL-patients after stimulation with NPM-ALK derived overlapping long peptides.
Purified CD3+ T-cells were stimulated with autologous DCs loaded with NPM-ALK derived peptide-pools (supplementary Figure 1 and supplementary Table 3). After two re-stimulations, on day 19, responder T-cells (2–3 × 104/well) were tested in an IFN-γ ELISPOT assay for the recognition of DCs (4–6 × 103/well) pulsed with peptides. Representative anti-NPM-ALK-reactive T-cell responses directed against peptide pool A, pool B and or pool C. Shown are example patients with one or more than one peptide pool stimulating anti-NPM-ALK T-cell responses (a). CD3+ T-cell responses against all 25 peptides were partially blocked by using either the anti-HLA-ABC antibody (W6/32) or the anti-HLA-DR antibody (L243) (b). After one more re-stimulation of reactive microculture (mc) under similar culture conditions (day 26), by testing the individual peptides of positive pools in IFN-γ ELISpot tests in two patients, single immunogenic B6 and C1 peptides were identified from the T-cell reactive pool B and pool C, respectively (c). Bars represent the means of duplicates ± standard deviation. Representative images of ELISPOT wells are shown in .
NPM-ALK-peptide pool-reactive CD3+ T-cells were also detected in microcultures generated from five of ten healthy individuals. In peptide pool-reactive donors, the observed number of reactive microcultures varied from one to two out of four stimulated microcultures. IFN-γ spot numbers in positive microcultures ranged from 2- to 10-fold above the background reactivity. T-cell responses against pool B peptides were detected in all of the reactive individuals. T-cells directed against pool A, pool C and pool D each were detected once in different individuals (supplementary Table 5 and supplementary Figure 2a). As a control for the chosen conditions for the peptide stimulation, anti-CMV-pp65-reactivity was confirmed in all CMV-seropositive ALCL-patients (data not shown).
To further characterize the NPM-ALK-directed CD3+ responses and to differentiate between CD4+ and CD8+ T-cell responses, blocking experiments were performed with pan-HLA-class I- and -class II-blocking antibodies. In all 13 analyzed NPM-ALK peptide-reactive patients, T-cell receptor/peptide-HLA-class I interactions were partially blocked by an anti-HLA-ABC antibody, confirming the presence of CD8+ T-cell responses against NPM-ALK-derived peptides (). In seven of nine responding patients analyzed, the HLA-DR–specific antibody had a partial blocking effect on CD3+ T-cell responses, indicating recognition of the NPM-ALK-derived peptides by CD4+ T-cells in the HLA-class II context (). Similar effects of HLA-class I and HLA-DR blocking were observed in healthy individuals with NPM-ALK responses (Supplementary Figure 2b).
To identify single peptides from reactive peptide pools, pool-reactive microcultures were re-stimulated once more under similar culture conditions and responder T cells were tested in an IFN-γ ELISPOT assay for the recognition of DCs pulsed with individual peptides of a positive pool. Single peptides from reactive peptide pools were tested in four patients with sufficient responder and target cells available in order to narrow down the immunogenic NPM-ALK-regions. In three pool B reactive patients, detectable CD3+ T-cell responses were directed against peptide B6 – p327-356 of NPM-ALK (Figure 3C). In one pool C responding patient, peptide C1 – p341-370 was specifically recognized by the reactive T-cells (Figure 3C).
In addition, reactivity with peptide pools was dissected in four reactive healthy controls (three for pool B and one for pool B and pool D). In pool B responding healthy controls, detectable CD3+ T-cell responses were directed against peptide B2 – p271-300 and B6 – p327-356 (Supplementary Figure 2C). In the pool D reactive donors, responder T-cells specifically recognized peptide D2 – p439-468 and peptide D3 – p453-482 (Supplementary Figure 2C).
In order to quantify the enrichment of single peptide-specific T-cells in a pool-stimulated microcultures, we performed an IFN-γ secretion assay. After single peptide identification in one pool B and pool D reactive healthy control, responding microculture T-cells from two different time points (day 14 – after 2 stimulations and day 26 – after 4 stimulations) were tested for IFN-γ secretion. After the two additional stimulations, significant increases of IFN-γ-secreting CD3+ T-cells were observed in pool B (12.5-fold) and pool D (14-fold) reactive microcultures (Supplementary Figure 2D).
Thus, the overlapping long peptide based T-cell stimulation induced NPM-ALK-specific CD8 + T-cell as well as CD4 + T-cell responses and enabled the identification of immunogenic ALK-regions in NPM-ALK+ ALCL-patients irrespective of individual HLA-type.
Confirmation of T-cell specificity and proof of endogenous processing and presentation of antigens
To confirm the specificity of T-cell responses generated against NPM-ALK induced by stimulation with IVT-RNA, responder T-cells from reactive microcultures were tested in additional IFN-γ ELISpot assays for the recognition of targets (autologous DCs or CD3− CD14− PBMCs, in case of sufficient material availability) pulsed with the NPM-ALK-derived peptide pools.
T-cells generated with RNA-transfected DCs were able to recognize targets loaded with NPM-ALK-derived peptide pools in all five analyzed patients. Recognition of peptide pool-loaded targets was partially blocked by an antibody against pan-HLA-class I in the CD3+ T-cell stimulated patients R20 and R25 and completely abrogated by the same antibody in the CD8+ T cell-selected patients R2 and R3 (supplementary Figure 3A). Responder T-cells from three ALCL-patients and four healthy donors without detectable NPM-ALK-reactive T-cell responses after RNA stimulations were also unable to recognize target cells loaded with NPM-ALK-derived peptide pools (data not shown).
To confirm the specificity of T cells responding to long peptides, peptide pool-reactive T-cells were tested for recognition of targets endogenously expressing NPM-ALK. Reactive microcultures from two ALCL-patients and two healthy donors with responder T-cells stimulated with NPM-ALK-derived overlapping long peptides were able to recognize targets transfected with NPM-ALK encoding IVT-RNA (Supplementary Figure 3B). In two ALCL-patients and three healthy individuals, who had no reactivity against NPM-ALK-derived peptides, stimulated T-cells failed to target DCs or CD3− CD14− PBMCs transfected with NPM-ALK IVT-RNA (data not shown).
These results confirm that NPM-ALK reactive T-cells, which could be stimulated by either synthetic long peptide-pulsed DCs or IVT-RNA-transfected DCs, recognize the respective other antigenic format on target cells.
Discussion
Understanding the characteristics of a spontaneous autologous T-cell response against a specific tumor-associated antigen facilitates the development of immunotherapeutic approaches to augment antigen-specific immune responses. We report the characterization of the spontaneous NPM-ALK-specific T-cell response in a large cohort of uniformly treated children and adolescents with NPM-ALK-positive ALCL in first remission, involving the complete HLA-repertoire of each patient and by ensuring endogenous peptide processing for presentation.
RNA-transfected DCs are efficient stimulators of antigen-specific T-cell responses and antitumor immunity in humans.Citation23–Citation25 The use of IVT-RNA a priori ensures endogenous processing of all possible immunogenic peptides and incorporation of all potential epitopes in the context of the complete individual HLA-repertoire. Applying an RNA-based in vitro T-cell stimulation approach, detection of tumor antigen-specific T-cell responses were reported in small cohorts of various cancer patients.Citation26–Citation30 However, there are no reports about a comparable examination of a tumor antigen-specific autologous spontaneous CD8+ T-cell response in a large cohort of cancer patients in remission.
In the recent past, autologous DCs transfected with RNA were used in several vaccination studies in different cancers. For example, total tumor-RNA transfected DC-vaccines were used for the induction of T-cell responses in renal cell carcinoma, chronic lymphocytic leukemia and melanoma.Citation31–Citation33 In addition, IVT-RNA encoding tumor-associated antigens were used to detect anti-tumor responses in prostate cancer, acute myeloid leukaemia and melanoma patients.Citation34–Citation37 Regardless of whether specific responses were boosted by vaccination or not, the percentages of patients with detectable antigen-specific T-cell response using RNA as antigen format for detection in those studies were comparable to our observation of spontaneous anti-NPM-ALK T-cell response in ALCL-patients.
HLA-B- and -C-restricted, but not HLA-A-restricted anti-NPM-ALK T-cell responses were detected after NPM-ALK RNA stimulation in eight NPM-ALK-reactive ALCL-patients. The possible explanation for the lack of HLA-A2-restricted T-cell responses that have been observed beforeCitation15,Citation16 may be that the HLA-A binding-epitopes are poorly or not processed from IVT-RNA-encoded protein by the predominant immunoproteasomes expressed in mature DCs.Citation38–Citation40 Inefficient processing of HLA-A2-restricted peptides has also been reported in other tumor models. For example, melanoma-associated proteins MAGE-A3 and Melan-A/Mart-1 contain HLA-A2-restricted peptides that are poorly or not processed by the immunoproteasomes of professional APCs.Citation41–Citation43 Previous reports on the recognition of synthetic HLA-A2-binding short (9mer) peptides by CD8+ T-cells can be explained by the fact that exogenous short peptides (8-10mers) do not require intracellular processing by APCs, as they can bind directly to HLA-class I molecules on the cell surfaceCitation44 or can be internalized and directly bind to MHC-class I molecules in the endoplasmic reticulum.Citation45,Citation46
In our second T-cell stimulation approach, NPM-ALK-derived overlapping long peptides were used as an antigen format for a more potent T-cell stimulation.Citation47,Citation48 It has been shown that compared to protein, overlapping long peptides were rapidly and much more efficiently processed by DCs, resulting in an increased antigen presentation to T-cells.Citation49,Citation50 The NPM-ALK peptides were selected based on immunogenic regions assumed according to published data covering the fusion site, the entire kinase domain and ALK-antibody binding region.Citation15–Citation19,Citation22 After stimulation with long peptides, NPM-ALK peptide-reactive T-cells were detected in peripheral blood of 15 out of 22 analyzed ALCL-patients in remission.
Compared to RNA stimulation, a higher percentage of ALCL-patients exhibited T-cell responses against NPM-ALK after peptide stimulation (68% vs. 41%). Furthermore, the cohort of patients analyzed with peptide stimulation included fewer patients with a high anti-ALK antibody titer which may be a surrogate marker for an NPM-ALK-specific immune response.Citation12,Citation13 Therefore, the observed difference between both antigen formats seems quite remarkable. This finding is consistent with the observations that long peptides are cross-presented through varied intracellular routes and are much more efficiently processed by DCs, resulting in an increased antigen presentation to T-cells, compared to the whole protein translated from IVT-RNA.Citation49,Citation50
As recognized in previous studies,Citation14,Citation15 NPM-ALK-reactive T-cells were detected in healthy individuals, however, only after stimulation with long peptides in our studies. A potential explanation for this discrepancy may again be that the presented peptides are less efficiently processed in RNA-transfected DCs.Citation38–Citation40 Recognition of NPM-ALK by peptide-stimulated T-cells in healthy individuals indicates the presence of T-cells specific for NPM-ALK, which is in the observed range of about one T-cell in 105 - 106 naïve T-cells for a given antigen.Citation51,Citation52
Inhibition experiments with HLA-class I- and -class II-specific antibodies indicated that we detected both, CD8+ and CD4+ T-cells against NPM-ALK when we stimulated CD3+ T lymphocytes. This is in line with reports in the literatureCitation17 and supports the presumption that the immune response against NPM-ALK involves CD4+ T-cells.
T-cell responses after stimulation with overlapping peptides were directed against the ALK-kinase domain (covered by peptide pool A, B and C). This region was the main part of ALK included in a DNA-vaccination with truncated ALK (p157-458) which induced a protective CD8 + T-cell response in a mouse lymphoma model.Citation18 The more prominent recognition of pool B peptides (covering p257-356) observed in our study indicates that this region may indeed represent a dominant immunogenic site of ALK. This is in line with observations reported before on CD8+ and CD4+ T-cell responses against ALK in a small number of ALCL-patients when short peptides were applied as antigen format for lymphocyte stimulation (peptides were comprising p280-289 and p278-301, respectively).Citation15,Citation17
Some of the NPM-ALK protein regions found by us to contain T-cell epitopes [kinase domain (peptide B6; p327-356) and juxta-kinase domain (peptide D2 and D3; p439-482)] that have been described as ALK antibody-binding sites as well.Citation53 Recent reports on the humoral response against the cytoplasmic domain of ALK suggest that antibodies bind to proximal and distal regions with a higher affinity than to other parts of the cytoplasmic domain.Citation53,Citation54 Inclusion of these potential antibody-binding regions in further analysis of ALK-specific spontaneous T-cell responses in ALCL-patients might help identifying further immunodominant T-cell epitope(s) suitable for ALCL immunotherapy.
NPM-ALK-reactive T-cells generated by stimulation with one of the antigen formats recognized both formats. These results confirm that the T-cells are specific for NPM-ALK and also the endogenous processing and the presentation of NPM-ALK-peptides on the cell surface.
The assay was selected and adapted for its suitability to characterize different aspects of the NPM-ALK-specific T-cell response in ALCL-patients. Using T-cell stimulation with IVT-RNA-transfected DC assured presentation of endogenously processed peptides from the whole protein and allowed for determination of the presenting HLA alleles; overlapping long NPM-ALK derived peptides were chosen to define immunogenic regions of NPM-ALK. The necessary cell number to reach these goals required a relatively large amount of blood from the patients. Therefore, the patients were selected in good clinical condition in first complete remission after chemotherapy and were at least 14 years of age at the time of analysis. It is, therefore, not unexpected that there was no correlation between the detection of a NPM-ALK-specific T-cell response with clinical or biological characteristics of the patients. Analysis of a possible prognostic impact of an NPM-ALK-specific T-cell response as shown for the humoral anti-ALK response, or a longitudinal analysis of the T-cell response during therapy necessitates the development of a different assay.Citation12,Citation13
In summary, our analysis of the spontaneous anti-NPM-ALK T-cell response in a large cohort of ALCL-patients shows that T-cell responses against NPM-ALK can be detected in a high proportion of patients. NPM-ALK-specific CD8+ T-cell responses were restricted mainly by HLA-B and -C alleles. Further, the ALK-kinase region contains immunodominant peptides inducing a T-cell response in humans. The proximal and distal part of ALK should be included in future immunogenicity testing. Next steps are the characterization of the NPM-ALK-epitopes inducing a CD4+ T-cell response and their restricting HLA-class II alleles. These data not only confirm the presence of a spontaneous autologous immune response but also set a basis for the development of a peptide-based vaccination able to augment protective CD8+ and CD4+ T-cell anti-tumor responses. The monitoring of NPM-ALK-specific immune responses during therapy will provide new insights into the interplay between ALCL and the immune system.
Material and methods
Patients and healthy donors
The ALCL-patients had been included in the studies NHL-BFM95 or ALCL99 and were treated with comparable BFM-type front-line therapy (Supplementary Figure 4). Leucocyte fractions from 30 Cytomegalovirus (CMV)-seropositive healthy blood donors, received from the Institute of Transfusion Medicine and Clinical Immunology of the University Hospital Giessen and Marburg, Giessen, Germany, were also analyzed.
The patient selection criteria and technical procedures for blood collection have been described before.Citation19 In short, the patients had to be in clinical remission without relapse after first-line therapy – 14 years or older at the time of analysis without concurring illness or immunosuppressive therapy. After written informed consent, 100 ml peripheral blood was collected into citrate phosphate dextrose (CPD)-containing bags using a modified umbilical cord blood collection system (Maco Pharma International GmbH, Langen, Germany). The study was approved by the Ethical committee of the medical faculty of the Justus-Liebig-University of Giessen (number 193/11).
To detect anti-NPM-ALK T-cell responses and identify their HLA-restriction alleles by ensuring endogenous peptide processing from the whole NPM-ALK protein, T-cells from 29 NPM-ALK-positive ALCL-patients and 20 controls were stimulated with autologous DCs transfected with IVT-RNA encoding full-length NPM-ALK. The results of five of these patients and five controls have been reported earlier.Citation19 Twenty of the patients were analyzed for CD8+ and nine for CD3+ T-cell responses ().
In the second approach addressing the question of the NPM-ALK antigenic region, CD3+ T-cells of 22 ALK-positive ALCL-patients were stimulated with autologous DCs pulsed with overlapping long peptides derived from the NPM-ALK oncoprotein (). Additionally, ten healthy donors were analyzed for NPM-ALK-reactive T-cells using this method.
Generation of HLA-class I alleles and NPM-ALK-cDNA expression plasmids
The HLA-class I alleles of patients and healthy donors were identified by high-resolution (4-digit) HLA-class I genotyping. HLA-class I allele plasmids, not yet available in the laboratory (most HLA-class I alleles had previously been cloned in other projects), were cloned into the pcDNA3.1/V5-His TOPO TA expression vector system as described before.Citation19 The pcDNA3-NPM-ALK plasmid, encoding the NPM-ALK full-length fusion protein, was obtained from S.W. Morris.Citation55 The construction of plasmid pcDNA3.1.pp65 TOPO was described previously.Citation20 The NPM-1 expression plasmid pcDNA3.1.NPM1 TOPO TA was generated by sub-cloning the entire NPM1-coding sequence from the GFP-NPM WT plasmidCitation56 (Addgene, Cambridge, MA/USA).
In vitro transcription of antigen-encoding RNA
pcDNA3-NPM-ALK was linearized with the restriction enzyme Xho I, and pcDNA3.1.pp65 with the restriction enzyme Apa I (both from New England Biolabs, Frankfurt, Germany). In vitro transcription and the polyadenylation of the resulting IVT-RNA were performed using the MESSAGE mMACHINE T7 Ultra Kit (Life Technologies, Darmstadt, Germany) according to the manufacturer’s guidelines.
Synthesis of NPM-ALK-derived long peptides
Twenty-five NPM-ALK-derived synthetic long peptides were tested for their capacity to stimulate and detect NPM-ALK-specific T-cell responses. To enable efficient cross-presentation, the selected length of the long peptides was 30 amino acids (aa) with an overlap of 16 aa to include most of the possible CD4+ and CD8+ T-cell epitopes. These peptides were custom synthesized by JPT (JPT Peptide Technologies, Berlin, Germany). The purity of the peptides was above 80%, and their composition was confirmed by mass spectrometry analysis. The lyophilized peptides were dissolved in 50 µl DMSO per peptide. After solubilization, a 5 mM stock solution was prepared using sterile deionized water. The final DMSO concentration did not exceed >1% when dissolved in culture media. Overlapping long peptides used in this study were pooled as described in supplementary data (Supplementary Table 4). The final concentration of NPM-ALK peptide-pools was 1 µM for each peptide.
PepTivator® CMV-pp65 (from Miltenyi Biotec, Bergisch Gladbach, Germany) consists of 15mer peptides with 11 aa overlap, covering the complete sequence of phosphoprotein 65 (pp65) of human CMV. It was used as experimental control to detect pp65-specific T-cells in all patients and healthy donors. CMV (pp65) PepTivator® was used at a final concentration of 0.6 nM of each peptide.
T-cell isolation and generation of antigen-presenting cells
CPD-anti-coagulated blood from patients and leukocyte fractions from healthy individuals were processed on the day of sample collection. Peripheral blood mononuclear cells (PBMCs) were isolated using Ficoll/Hypaque 1.077 g/ml (Axis-Shield PoC AS, Oslo, Norway) density gradient centrifugation. CD8+ or CD3+ T-cells were purified from PBMCs using CD8- or CD3-microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany), respectively. The T-cell fractions were frozen in Cryo-safe I (C.C. Pro GmbH, Oberdorla, Germany) medium for later stimulation and ex-vivo testing.
DCs were generated from microbead-sorted CD14+ cells by using GM-CSF and IL-4 followed by a combination of proinflammatory cytokines and maturation status of the DCs was determined as previously described.Citation19,Citation57 These mature DCs were irradiated and transfected with antigen-coding IVT-RNA using the nucleofection system (Lonza GmBH, Cologne, Germany). The nucleofection efficiency was measured by flow cytometry as described before.Citation19 The RNA-transfected DCs were then used as APCs in the subsequent in vitro stimulation of T-cells. In addition, portions of the transfected DCs were frozen in Cryo-safe I medium for later re-stimulation and testing.
In vitro stimulation of T-cells with DCs transfected with NPM-ALKRNA
For in vitro stimulation and detection of NPM-ALK-specific T-cells, we used the T-cell stimulation protocol as described before.Citation19 Briefly, PBMC-derived CD8+ or CD3+ T-cells were plated at 1 to 2 × 105 per well in a 96-well U-bottom plate in AIM-Vstim consisting of AIM-V (Life Technologies, Darmstadt, Germany) supplemented with 5% human serum (HS) (Biochrom, Berlin, Germany) and 20 IU/ml interleukin (IL)-2 (Novartis Pharma, Nürnberg, Germany). T-cells were stimulated with autologous DCs transfected with NPM-ALK IVT-RNA, pp65 IVT-RNA, or a mock control. From each patient’s blood-donation 6–8 NPM-ALK-stimulation microcultures and from each healthy blood-donation 8–16 NPM-ALK-stimulation microcultures were initiated. All responder T-cells were re-stimulated on days 7 and 14 under the same culture conditions using the DCs transfected with antigen-encoding IVT-RNA. Responder T-cells were tested on day 19 in an interferon-gamma (IFN-γ) ELISPOT assay for recognition of respective antigens.
In case of NPM-ALK-reactivity, if sufficient responder and target materials were available, reactive T-cells were tested in another IFN-γ ELISpot assay for the recognition of targets (DCs or CD3− CD14− PBMCs) pulsed with the NPM-ALK-derived peptide pools.
In vitro stimulation of CD3 + T-cells with DCs loaded with overlapping long peptides
To detect NPM-ALK-reactive T-cells, microbeads-purified CD3+ T-cells were plated at 2 to 5 × 105 per well in a 96-well or 48-well flat-bottom plate in AIM-Vstim culture medium. CD3+ T-cells were stimulated with autologous DCs loaded with NPM-ALK-derived overlapping long peptide-pools (with 1 µM final peptide concentration) in a 10:1 ratio (effector: target). As stimulation controls, we included mock stimulation and stimulation with the CMV-pp65 peptide pool (PepTivator®) as standard negative and positive control, respectively. From each donation, 2–4 NPM-ALK stimulation microcultures were initiated. After two re-stimulations under the same culture conditions, day 19 responder lymphocytes were analyzed in an IFN-γ ELISPOT assay for the recognition of DCs pulsed with the peptide pools.
In case of NPM-ALK peptide pool-reactivity, reactive microcultures were restimulated once more under similar culture conditions depending on the availability of stimulator cells. Responder T-cells were tested for recognition of targets (DCs or CD3− CD14− PBMCs) loaded with individual peptides of a positive pool or transfected with IVT-RNA encoding NPM-ALK oncoprotein.
IFN-γ ELISPOT assay
To determine the NPM-ALK-specific T-cell responses, various IFN-γ ELISPOT (20 hours) assays were performed, as previously described.Citation19
Recognition of NPM-ALK antigen: To test the T-cells generated against NPM-ALK IVT-RNA, DCs (5.0–10.0 × 103 cells/well) transfected with IVT-RNA-encoding NPM-ALK or control antigens were used as target cells. In case of peptide-based stimulation, responder lymphocytes were tested for the recognition of DCs (3.0–6.0 × 103) pulsed with the peptide pools or individual peptides of a positive pool (1 µM for each peptide). For HLA-blocking experiments, stimulator cells were pre-incubated with the anti-HLA-class I (clone: W6/32) or anti-HLA-DR (clone: L243) monoclonal antibody (both from BIOZOL Diagnostica Vertrieb GmbH, Eching, Germany) at a concentration of 20 µg/ml for 2 hours at 37°C, and then cultured with effector cells. The target cells were re-suspended in AIM-V/5% HS and plated in ELISPOT plates as indicated.
Recognition of COS-7/NPM-ALK transfectants: To assess both, antigen reactivity and HLA-class I restriction for antigen presentation, COS-7 cells (German Collection of Microorganisms and Cell Cultures, DSMZ) co-transfected with plasmids encoding the respective patient’s/donor’s HLA-class I alleles and NPM-ALK, NPM1 or pp65 were used as APCs at a density of 2.0 × 104 cells/well. COS-7 cells were transfected using LipofectaminTM 2000 (Invitrogen, Karlsruhe, Germany), according to the manufacturer’s guideline.
Confirmation of Antigen-specific T-cell responses: To confirm the antigen-specificity, frozen T-cells from RNA-stimulated microcultures were retested for the recognition of NPM-ALK derived peptide pulsed targets and peptide-stimulated microcultures were retested for the recognition of RNA transfected cells. The targets were either DCs (3.0–5.0 × 103 cells/well) or CD3− CD14− PBMCs (1.5–3.0 × 104 cells/well) (depending on the availability of material) and pulsed with the NPM-ALK-derived peptide pools (1 µM for each peptide) or transfected with NPM-ALK IVT-RNA. The target cells were re-suspended in AIM-V/5% HS and plated in ELISPOT plates as indicated.
Responder T-cells (0.5–6.0 × 104/well) were added 2-hours after peptide-pulsing, 4-hours after RNA-transfection or 24-hours after plasmid DNA-transfection. After 20-hours of co-culture, the cells were discarded and the ELISPOT plates were developed according to the manufacturer’s instructions (Mabtech, Nacka Strand, Sweden). IFN-γ spots were primarily counted with a computer-assisted video-imaging analysis system Bioreader® 4000 (from BIO-SYS GmbH, Karben, Germany) and if required then further validated by advanced ImmunoSpot Series 5 Versa Analyzer (C.T.L. Europe, Bonn, Germany). Responses in a given microculture were defined as ‘‘positive’’ when the mean number of IFN-γ spot-forming T-cells against the test antigen was at least two-fold higher than the background reactivity (baseline response against either antigen-negative target cells only or targets expressing an irrelevant antigen).
IFN-γ secretion assay
The IFN-γ secretion assay technique allows identifying antigen-specific IFN-γ secreting T-cells in a mixed population of cells.Citation58 NPM-ALK-peptide specific IFN-γ-secreting T-cells were detected by using the IFN-γ secretion assay kit according to the manufacturer’s instructions (Miltenyi Biotec, Bergisch Gladbach, Germany). Responder T-cells, frozen before, from peptide pool-reactive donation were thawed and cultured overnight in AIM-V medium supplemented with 5% human serum and 5 IU/ml IL-2. Then, the T-cells were seeded in a 48-well plate (1 – 2x10Citation59/well) in AIM-Vstim and stimulated at a 10:1 ratio with autologous DCs pulsed either with the peptide pools or with individual peptides of a positive-pool. After 12 hours incubation in the presence or absence of antigen, T-cells from each 48-well were processed according to the manufacturer’s guidelines and resuspended in the flow cytometry-buffer. IFN-γ secreting cells were immediately analyzed by the FACS Calibur system (BD).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Authors’ contributions
VKS, SW, AR, TW, CDW and WW designed the project. VKS, SW, SS and SST performed the experiments. SW, SR, AR, and WW collected patient blood samples and provided the clinical data. VL and TW contributed to the experimental setup and ELISPOT analysis. HH performed HLA-typing. WW, CDW, and AR coordinated the study. VKS, CDW and WW analyzed the data and VKS and WW wrote the manuscript. All authors approved the final version of the manuscript.
Supplemental Material
Download MS Word (57.1 KB)Supplemental Material
Download MS Word (4.2 MB)Acknowledgments
The authors thank the patients and donors who participated in the study. We thank Jutta Schieferstein, Claudia Keller and Susanne Suesslin for expert technical assistance, Ulrike Meyer for data management and the physicians at the NHL-BFM study center for visiting patients and collecting blood samples. We also thank to Dr. Martin Zimmermann, Hannover, Germany, for the statistical analyses.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Chiarle R, Voena C, Ambrogio C, Piva R, Inghirami G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat Rev Cancer. 2008;8:11–23. doi:10.1038/nrc2291.
- Damm-Welk C, Klapper W, Oschlies I, Gesk S, Rottgers S, Bradtke J, Siebert R, Reiter A, Woessmann W. Distribution of NPM1-ALK and X-ALK fusion transcripts in paediatric anaplastic large cell lymphoma: a molecular-histological correlation. Br J Haematol. 2009;146:306–309. doi:10.1111/j.1365-2141.2009.07754.x.
- Brugieres L, Le Deley MC, Rosolen A, Williams D, Horibe K, Wrobel G, Mann G, Zsiros J, Uyttebroeck A, Marky I, et al. Impact of the methotrexate administration dose on the need for intrathecal treatment in children and adolescents with anaplastic large-cell lymphoma: results of a randomized trial of the EICNHL Group. J Clin Oncol. 2009;27:897–903. doi:10.1200/JCO.2008.18.1487.
- Alexander S, Kraveka JM, Weitzman S, Lowe E, Smith L, Lynch JC, Chang M, Kinney MC, Perkins SL, Laver J, et al. Advanced stage anaplastic large cell lymphoma in children and adolescents: results of ANHL0131, a randomized phase III trial of APO versus a modified regimen with vinblastine: a report from the children’s oncology group. Pediatr Blood Cancer. 2014;61:2236–2242. doi:10.1002/pbc.25187.
- Gambacorti Passerini C, Farina F, Stasia A, Redaelli S, Ceccon M, Mologni L, Messa C, Guerra L, Giudici G, Sala E, et al. Crizotinib in advanced, chemoresistant anaplastic lymphoma kinase-positive lymphoma patients. J Natl Cancer Inst. 2014;106:djt378. doi:10.1093/jnci/djt378.
- Pro B, Advani R, Brice P, Bartlett NL, Rosenblatt JD, Illidge T, Matous J, Ramchandren R, Fanale M, Connors JM, et al. Five-year results of brentuximab vedotin in patients with relapsed or refractory systemic anaplastic large cell lymphoma. Blood. 2017;130:2709–2717. doi:10.1182/blood-2017-05-780049.
- Woessmann W, Peters C, Lenhard M, Burkhardt B, Sykora KW, Dilloo D, Kremens B, Lang P, Fuhrer M, Kuhne T, et al. Allogeneic haematopoietic stem cell transplantation in relapsed or refractory anaplastic large cell lymphoma of children and adolescents–a Berlin-Frankfurt-Munster group report. Br J Haematol. 2006;133:176–182. doi:10.1111/j.1365-2141.2006.06004.x.
- Strullu M, Thomas C, Le Deley MC, Chevance A, Kanold J, Bertrand Y, Jubert C, Dalle JH, Paillard C, Baruchel A, et al. Hematopoietic stem cell transplantation in relapsed ALK+ anaplastic large cell lymphoma in children and adolescents: a study on behalf of the SFCE and SFGM-TC. Bone Marrow Transplant. 2015;50:795–801. doi:10.1038/bmt.2015.57.
- Fukano R, Mori T, Kobayashi R, Mitsui T, Fujita N, Iwasaki F, Suzumiya J, Chin M, Goto H, Takahashi Y, et al. Haematopoietic stem cell transplantation for relapsed or refractory anaplastic large cell lymphoma: a study of children and adolescents in Japan. Br J Haematol. 2015;168:557–563. doi:10.1111/bjh.13167.
- Werner MT, Zhao C, Zhang Q, Wasik MA. Nucleophosmin-anaplastic lymphoma kinase: the ultimate oncogene and therapeutic target. Blood. 2017;129:823–831. doi:10.1182/blood-2016-05-717793.
- Stadler S, Singh VK, Knorr F, Damm-Welk C, Woessmann W. Immune Response against ALK in Children with ALK-Positive Anaplastic Large Cell Lymphoma. Cancers (Basel). 2018;10:114. doi:10.3390/cancers10040114.
- Ait-Tahar K, Damm-Welk C, Burkhardt B, Zimmermann M, Klapper W, Reiter A, Pulford K, Woessmann W. Correlation of the autoantibody response to the ALK oncoantigen in pediatric anaplastic lymphoma kinase-positive anaplastic large cell lymphoma with tumor dissemination and relapse risk. Blood. 2010;115:3314–3319. doi:10.1182/blood-2009-11-251892.
- Mussolin L, Damm-Welk C, Pillon M, Zimmermann M, Franceschetto G, Pulford K, Reiter A, Rosolen A, Woessmann W. Use of minimal disseminated disease and immunity to NPM-ALK antigen to stratify ALK-positive ALCL patients with different prognosis. Leukemia. 2013;27:416–422. doi:10.1038/leu.2012.205.
- Passoni L, Scardino A, Bertazzoli C, Gallo B, Coluccia AM, Lemonnier FA, Kosmatopoulos K, Gambacorti-Passerini C. ALK as a novel lymphoma-associated tumor antigen: identification of 2 HLA-A2.1-restricted CD8+ T-cell epitopes. Blood. 2002;99:2100–2106. doi:10.1182/blood.V99.6.2100.
- Passoni L, Gallo B, Biganzoli E, Stefanoni R, Massimino M, Di Nicola M, Gianni AM, Gambacorti-Passerini C. In vivo T-cell immune response against anaplastic lymphoma kinase in patients with anaplastic large cell lymphomas. Haematologica. 2006;91:48–55.
- Ait-Tahar K, Cerundolo V, Banham AH, Hatton C, Blanchard T, Kusec R, Becker M, Smith GL, Pulford K. B and CTL responses to the ALK protein in patients with ALK-positive ALCL. Int J Cancer. 2006;118:688–695. doi:10.1002/ijc.21410.
- Ait-Tahar K, Barnardo MC, Pulford K. CD4 T-helper responses to the anaplastic lymphoma kinase (ALK) protein in patients with ALK-positive anaplastic large-cell lymphoma. Cancer Res. 2007;67:1898–1901. doi:10.1158/0008-5472.CAN-06-4427.
- Chiarle R, Martinengo C, Mastini C, Ambrogio C, D’Escamard V, Forni G, Inghirami G. The anaplastic lymphoma kinase is an effective oncoantigen for lymphoma vaccination. Nat Med. 2008;14:676–680. doi:10.1038/nm1769.
- Singh VK, Werner S, Hackstein H, Lennerz V, Reiter A, Wolfel T, Damm-Welk C, Woessmann W. Analysis of nucleophosmin-anaplastic lymphoma kinase (NPM-ALK)-reactive CD8(+) T cell responses in children with NPM-ALK(+) anaplastic large cell lymphoma. Clin Exp Immunol. 2016;186:96–105. doi:10.1111/cei.12842.
- Melief CJ. van der Burg SH. Immunotherapy of established (pre)malignant disease by synthetic long peptide vaccines. Nat Rev Cancer. 2008;8:351–360. doi:10.1038/nrc2373.
- Tomita Y, Nishimura Y. Long peptide-based cancer immunotherapy targeting tumor antigen-specific CD4(+) and CD8(+) T cells. Oncoimmunology. 2013;2:e25801. doi:10.4161/onci.25801.
- Pulford K, Lamant L, Morris SW, Butler LH, Wood KM, Stroud D, Delsol G, Mason DY. Detection of anaplastic lymphoma kinase (ALK) and nucleolar protein nucleophosmin (NPM)-ALK proteins in normal and neoplastic cells with the monoclonal antibody ALK1. Blood. 1997;89:1394–1404.
- Nair S, Boczkowski D. RNA-transfected dendritic cells. Expert Rev Vaccines. 2002;1:507–513. doi:10.1586/14760584.1.4.507.
- Weiner DB. RNA-based vaccination: sending a strong message. Mol Ther. 2013;21:506–508. doi:10.1038/mt.2013.26.
- McNamara MA, Nair SK, Holl EK. RNA-Based Vaccines in Cancer Immunotherapy. J Immunol Res. 2015;2015:794528. doi:10.1155/2015/794528.
- Nair SK, Boczkowski D, Morse M, Cumming RI, Lyerly HK, Gilboa E. Induction of primary carcinoembryonic antigen (CEA)-specific cytotoxic T lymphocytes in vitro using human dendritic cells transfected with RNA. Nat Biotechnol. 1998;16:364–369. doi:10.1038/nbt0498-364.
- Heiser A, Dahm P, Yancey DR, Maurice MA, Boczkowski D, Nair SK, Gilboa E, Vieweg J. Human dendritic cells transfected with RNA encoding prostate-specific antigen stimulate prostate-specific CTL responses in vitro. J Immunol. 2000;164:5508–5514. doi:10.4049/jimmunol.164.10.5508.
- Morandi F, Chiesa S, Bocca P, Millo E, Salis A, Solari M, Pistoia V, Prigione I. Tumor mRNA-transfected dendritic cells stimulate the generation of CTL that recognize neuroblastoma-associated antigens and kill tumor cells: immunotherapeutic implications. Neoplasia. 2006;8:833–842. doi:10.1593/neo.06415.
- Knights AJ, Nuber N, Thomson CW, de la Rosa O, Jager E, Tiercy JM, van Den Broek M, Pascolo S, Knuth A, Zippelius A. Modified tumour antigen-encoding mRNA facilitates the analysis of naturally occurring and vaccine-induced CD4 and CD8 T cells in cancer patients. Cancer Immunol Immunother. 2009;58:325–338. doi:10.1007/s00262-008-0556-8.
- Gholamin M, Moaven O, Farshchian M, Mahmoudi M, Sankian M, Memar B, Forghani MN, Malekzadeh R, Rajabi-Mashhadi MT, Abbaszadegan MR. Induction of cytotoxic T lymphocytes primed with tumor RNA-loaded dendritic cells in esophageal squamous cell carcinoma: preliminary step for DC vaccine design. BMC Cancer. 2010;10:261. doi:10.1186/1471-2407-10-261.
- Su Z, Dannull J, Heiser A, Yancey D, Pruitt S, Madden J, Coleman D, Niedzwiecki D, Gilboa E, Vieweg J. Immunological and clinical responses in metastatic renal cancer patients vaccinated with tumor RNA-transfected dendritic cells. Cancer Res. 2003;63:2127–2133.
- Muller MR, Tsakou G, Grunebach F, Schmidt SM, Brossart P. Induction of chronic lymphocytic leukemia (CLL)-specific CD4- and CD8-mediated T-cell responses using RNA-transfected dendritic cells. Blood. 2004;103:1763–1769. doi:10.1182/blood-2003-06-2097.
- Kyte JA, Aamdal S, Dueland S, Saeboe-Larsen S, Inderberg EM, Madsbu UE, Skovlund E, Gaudernack G, Kvalheim G. Immune response and long-term clinical outcome in advanced melanoma patients vaccinated with tumor-mRNA-transfected dendritic cells. Oncoimmunology. 2016;5:e1232237. doi:10.1080/2162402X.2016.1232237.
- Su Z, Dannull J, Yang BK, Dahm P, Coleman D, Yancey D, Sichi S, Niedzwiecki D, Boczkowski D, Gilboa E, et al. Telomerase mRNA-transfected dendritic cells stimulate antigen-specific CD8+ and CD4+ T cell responses in patients with metastatic prostate cancer. J Immunol. 2005;174:3798–3807. doi:10.4049/jimmunol.174.6.3798.
- Van Tendeloo VF, Van de Velde A, Van Driessche A, Cools N, Anguille S, Ladell K, Gostick E, Vermeulen K, Pieters K, Nijs G, et al. Induction of complete and molecular remissions in acute myeloid leukemia by Wilms’ tumor 1 antigen-targeted dendritic cell vaccination. Proc Natl Acad Sci U S A. 2010;107:13824–13829. doi:10.1073/pnas.1008051107.
- Van Nuffel AM, Benteyn D, Wilgenhof S, Pierret L, Corthals J, Heirman C, van der Bruggen P, Coulie PG, Neyns B, Thielemans K, et al. Dendritic cells loaded with mRNA encoding full-length tumor antigens prime CD4+ and CD8+ T cells in melanoma patients. Mol Ther. 2012;20:1063–1074. doi:10.1038/mt.2012.11.
- Benteyn D, Van Nuffel AM, Wilgenhof S, Corthals J, Heirman C, Neyns B, Thielemans K, Bonehill A. Characterization of CD8+ T-cell responses in the peripheral blood and skin injection sites of melanoma patients treated with mRNA electroporated autologous dendritic cells (TriMixDC-MEL). Biomed Res Int. 2013;2013:976383. doi:10.1155/2013/976383.
- Morel S, Levy F, Burlet-Schiltz O, Brasseur F, Probst-Kepper M, Peitrequin AL, Monsarrat B, Van Velthoven R, Cerottini JC, Boon T, et al. Processing of some antigens by the standard proteasome but not by the immunoproteasome results in poor presentation by dendritic cells. Immunity. 2000;12:107–117. doi:10.1016/S1074-7613(00)80163-6.
- Schultz ES, Chapiro J, Lurquin C, Claverol S, Burlet-Schiltz O, Warnier G, Russo V, Morel S, Levy F, Boon T, et al. The production of a new MAGE-3 peptide presented to cytolytic T lymphocytes by HLA-B40 requires the immunoproteasome. J Exp Med. 2002;195:391–399. doi:10.1084/jem.20011974.
- Anderson KS, Zeng W, Sasada T, Choi J, Riemer AB, Su M, Drakoulakos D, Kang YJ, Brusic V, Wu C, et al. Impaired tumor antigen processing by immunoproteasome-expressing CD40-activated B cells and dendritic cells. Cancer Immunol Immunother. 2011;60:857–867. doi:10.1007/s00262-011-0995-5.
- van der Bruggen P, Bastin J, Gajewski T, Coulie PG, Boel P, De Smet C, Traversari C, Townsend A, Boon T. A peptide encoded by human gene MAGE-3 and presented by HLA-A2 induces cytolytic T lymphocytes that recognize tumor cells expressing MAGE-3. Eur J Immunol. 1994;24:3038–3043. doi:10.1002/eji.1830241218.
- Valmori D, Gileadi U, Servis C, Dunbar PR, Cerottini JC, Romero P, Cerundolo V, Levy F. Modulation of proteasomal activity required for the generation of a cytotoxic T lymphocyte-defined peptide derived from the tumor antigen MAGE-3. J Exp Med. 1999;189:895–906. doi:10.1084/jem.189.6.895.
- Chapatte L, Ayyoub M, Morel S, Peitrequin AL, Levy N, Servis C, Van Den Eynde BJ, Valmori D, Levy F. Processing of tumor-associated antigen by the proteasomes of dendritic cells controls in vivo T-cell responses. Cancer Res. 2006;66:5461–5468. doi:10.1158/0008-5472.CAN-05-4310.
- Rock KL, Rothstein L, Benacerraf B. Analysis of the association of peptides of optimal length to class I molecules on the surface of cells. Proc Natl Acad Sci U S A. 1992;89:8918–8922. doi:10.1073/pnas.89.19.8918.
- Pm D, Jw Y, Porgador A, Rn G, Jr B. Direct delivery of exogenous MHC class I molecule-binding oligopeptides to the endoplasmic reticulum of viable cells. Proc Natl Acad Sci U S A. 1997;94:8064–8069. doi:10.1073/pnas.94.15.8064.
- Luft T, Rizkalla M, Tai TY, Chen Q, MacFarlan RI, Davis ID, Maraskovsky E, Cebon J. Exogenous peptides presented by transporter associated with antigen processing (TAP)-deficient and TAP-competent cells: intracellular loading and kinetics of presentation. J Immunol. 2001;167:2529–2537. doi:10.4049/jimmunol.167.5.2529.
- Welters MJ, Kenter GG, Piersma SJ, Vloon AP, Lowik MJ, Berends-van der Meer DM, Jw D, Ar V, Ar W, Oostendorp J, et al. Induction of tumor-specific CD4+ and CD8+ T-cell immunity in cervical cancer patients by a human papillomavirus type 16 E6 and E7 long peptides vaccine. Clin Cancer Res. 2008;14:178–187. doi:10.1158/1078-0432.CCR-07-1880.
- Leffers N, Vermeij R, Bn H, Ur S, Wolf R, Ie H, Ag VDZ, Kj M, Sh VDB, Daemen T, et al. Long-term clinical and immunological effects of p53-SLP(R) vaccine in patients with ovarian cancer. Int J Cancer. 2012;130:105–112. doi:10.1002/ijc.25980.
- Rosalia RA, Quakkelaar ED, Redeker A, Khan S, Camps M, Drijfhout JW, Silva AL, Jiskoot W, van Hall T, van Veelen PA, et al. Dendritic cells process synthetic long peptides better than whole protein, improving antigen presentation and T-cell activation. Eur J Immunol. 2013;43:2554–2565. doi:10.1002/eji.201343324.
- Zhang H, Hong H, Li D, Ma S, Di Y, Stoten A, Haig N, Di Gleria K, Yu Z, Xu XN, et al. Comparing pooled peptides with intact protein for accessing cross-presentation pathways for protective CD8+ and CD4+ T cells. J Biol Chem. 2009;284:9184–9191. doi:10.1074/jbc.M809456200.
- Jenkins MK, Moon JJ. The role of naive T cell precursor frequency and recruitment in dictating immune response magnitude. J Immunol. 2012;188:4135–4140. doi:10.4049/jimmunol.1102661.
- Yu W, Jiang N, Ebert PJ, Kidd BA, Muller S, Lund PJ, Juang J, Adachi K, Tse T, Birnbaum ME, et al. Clonal deletion prunes but does not eliminate self-specific alphabeta CD8(+) T Lymphocytes. Immunity. 2015;42:929–941. doi:10.1016/j.immuni.2015.05.001.
- Knorr F, Weber S, Singh VK, Pulford K, Reiter A, Woessmann W, Damm-Welk C. Epitope mapping of anti-ALK antibodies in children with anaplastic large cell lymphoma. Clin Immunol. 2018;195:77–81. doi:10.1016/j.clim.2018.07.008.
- Awad MM, Mastini C, Blasco RB, Mologni L, Voena C, Mussolin L, Mach SL, Adeni AE, Lydon CA, Sholl LM, et al. Epitope mapping of spontaneous autoantibodies to anaplastic lymphoma kinase (ALK) in non-small cell lung cancer. Oncotarget. 2017;8:92265–92274. doi:10.18632/oncotarget.21182.
- Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, Look AT. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science. 1994;263:1281–1284. doi:10.1126/science.8122112.
- Wang W, Budhu A, Forgues M, Wang XW. Temporal and spatial control of nucleophosmin by the Ran-Crm1 complex in centrosome duplication. Nat Cell Biol. 2005;7:823–830. doi:10.1038/ncb1282.
- Dauer M, Obermaier B, Herten J, Haerle C, Pohl K, Rothenfusser S, Schnurr M, Endres S, Eigler A. Mature dendritic cells derived from human monocytes within 48 hours: a novel strategy for dendritic cell differentiation from blood precursors. J Immunol. 2003;170:4069–4076. doi:10.4049/jimmunol.170.8.4069.
- Desombere I, Meuleman P, Rigole H, Willems A, Irsch J, Leroux-Roels G. The interferon gamma secretion assay: a reliable tool to study interferon gamma production at the single cell level. J Immunol Methods. 2004;286:167–185. doi:10.1016/j.jim.2004.01.001.
- Mosse YP, Voss SD, Lim MS, Rolland D, Minard CG, Fox E, Adamson P, Wilner K, Blaney SM, Weigel BJ. Targeting ALK with crizotinib in pediatric anaplastic large cell lymphoma and inflammatory myofibroblastic tumor: a children’s oncology group study. J Clin Oncol. 2017;35:3215–3221. doi:10.1200/JCO.2017.73.4830.
