ABSTRACT
Clusterin is a glycoprotein able to mediate different physiological functions such as control of complement activation, promotion of unfolded protein clearance and modulation of cell survival. Clusterin is overexpressed in many types of cancers and a large body of evidence suggests that it promotes carcinogenesis and tumor progression. We have previously described a novel clusterin glycoform present in human semen, but not in serum, highly enriched in terminal fucose motifs. Here we show that human luminal breast cancer (LBC) clusterin also bears terminal fucosylated glycans, conferring clusterin the ability to interact with DC-SIGN, a C-type lectin receptor expressed by myeloid cells. This clusterin glycosylation pattern was absent or diminished in non-involved juxtatumoral tissue, suggesting that fucosylated clusterin might represent a cancer associated glycoform. We also found that DC-SIGN is expressed by luminal breast cancer intratumoral macrophages. Moreover, experiments performed in vitro using semen fucosylated clusterin and monocyte derived macrophages showed that the interaction of semen clusterin with DC-SIGN promoted a proangiogenic profile, characterized by a high production of VEGF, IL-8 and TNF-α. Our results reveal an unexpected complexity on the structure and function of secretory clusterin produced by tumors and suggest that fucosylated clusterin produced by luminal breast cancer cells might play a role in tumor progression by promoting the release of pro-angiogenic factors by intratumoral macrophages.
Introduction
Clusterin is a multifunctional glycoprotein present in almost all tissues and body fluids. It was shown to be involved in a number of physiological and pathological processes including apoptosis modulation, protein homeostasis, Alzheimer´s disease and cancer progression.Citation1
Clusterin expression is dysregulated in many types of tumors, including colon, gastric, prostate, ovarian, bladder, lung and breast cancer.Citation2 Clusterin expression usually correlates with bad prognostic, disease recurrence or resistance to therapy.Citation3–Citation6 Intriguingly, the nature of these associations is still controversial. While the expression of clusterin in the cytosol of tumor cells inhibits apoptosis promoting tumor progression and resistance to chemotherapy, nuclear clusterin seems to induce cell death.Citation7 In addition, clusterin is also secreted to the extracellular milieu, however, its role on the tumor microenvironment remains poorly understood.
Clusterin is a highly glycosylated protein with 30 percent of its molecular mass contributed by glycans.Citation8,Citation9 We have previously described a novel clusterin glycoform in semen.Citation10 Semen clusterin bears highly fucosylated LewisX and LewisY type glycans.Citation10 In contrast, serum clusterin is decorated by non-fucosylated terminal sialylated glycans.Citation8,Citation10 Lewis type fucosylated glycan motifs enable semen clusterin to interact with DC-SIGN, a C type lectin receptor expressed by myeloid cells, preventing HIV recognition by dendritic cells.Citation10 Moreover, we have also reported that the interaction of semen clusterin with DC-SIGN promotes antigen capture by dendritic cells (DCs) and DC differentiation into a tolerogenic profile characterized by an increased ability to promote the expansion of FOXP3+ regulatory T cells.Citation11
A key feature of cancer is the acquisition of glycan motifs different than those expressed by healthy tissues, a phenomenon often referred as “aberrant glycosylation”.Citation12,Citation13 The expression of fucosylated Lewis type motifs is one of the most common alterations in the glycosylation pattern of cancer cells.Citation14 The neo-expression of Sialyl LewisX motifs renders neoplastic cells able to bind endothelium selectins promoting metastasis. Intriguing, many tumors express non-sialylated Lewis type glycans, which are unable to bind selectins. The contribution of this glycosylation pattern to tumor growth has not been clearly characterized.Citation15
In the present study we show that human breast tumors express fucosylated clusterin. Interestingly, fucosylated clusterin was absent or diminished in juxtatumor tissues suggesting that it might represent a cancer associated glycoform. Moreover, we here report the expression of DC-SIGN by intratumoral macrophages, which represent the most prominent myeloid cell population infiltrating breast tumors.Citation16 Using an in vitro model, we show that fucosylated clusterin turns macrophages into a pro-angiogenic profile, suggesting a novel pathway linking clusterin and tumor growth.
Results
Fucosylated clusterin expression in breast cancer
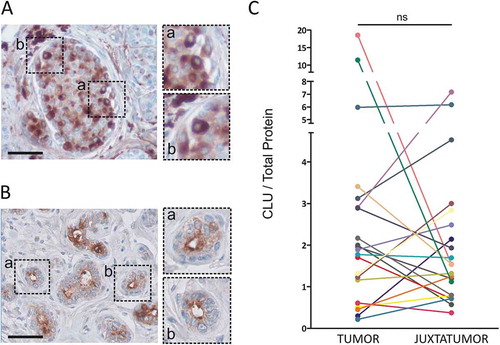
We first analyzed the expression of clusterin in luminal breast cancer samples. The patient cohort characteristics are described in . For each patient, a fragment of tumor and pathologically non-involved juxtatumoral tissue were obtained. The pattern of clusterin expression was analyzed by immunohistochemistry (,b)). As described previously, clusterin is expressed in the cytoplasm of breast cancer cells ()) and shows an apical expression pattern in normal epithelial cells ()). Clusterin expression was quantified by ELISA in disaggregated tumor samples and their juxtatumoral counterparts. As shown in ), no statistically significant differences were found in the mean total amount of clusterin between tumoral and juxtatumoral tissues (n = 21).
Table 1. Patient sample characteristics.
Figure 1. Clusterin expression in luminal breast cancer samples. A and B. Clusterin expression was analyzed by immunohistochemistry in tumoral (a) and juxtatumoral (b) tissues. Representative pictures are shown (n = 3, bars: 50 µm). C. Clusterin expression was quantified in tumor samples and their juxtatumoral counterparts by ELISA. Results are expressed as the amount of clusterin/total protein (µg of clusterin/mg of total protein, n = 21, ns = no statistically significant differences, p = .31).
We and others have previously described a novel fucosylated glycoform of clusterin present in semen. In contrast to serum clusterin, semen clusterin bears abundant LewisX and LewisY type fucosylated motifs.Citation10,Citation17 Because changes in the expression status of glycan structures represent a common feature of cancer, and considering that fucosylation of tumor glycoproteins appears to be associated with increased tumorigenicity, invasiveness and metastatic ability,Citation14,Citation15 we speculated that breast carcinoma might express fucosylated clusterin. To test this hypothesis, we developed two different ELISA assays based on the ability of the lectins Ulex europaeus and Lotus tetragonolobus to bind terminal fucose motifs. The Ulex europaeus agglutinin-1 (UEA-1) binds to LewisY as well as to other terminal fucosylated motifs.Citation18 The Lotus tetragonolobus lectin binds to LewisX and LewisY motifs.Citation19 The characteristics of the ELISA assay are illustrated in ). As shown in ) (blue bars), using Ulex europaeus agglutinin-1, fucosylated semen clusterin, but not serum clusterin or recombinant clusterin, was detected through this assay. As expected, α-L-fucose completely abrogated semen clusterin detection (red bars). Similar results were obtained using Lotus tetragonolobus (not shown). Then, we analyzed the expression of fucosylated clusterin by breast tumoral and juxtatumoral samples. Results in ,d) show that tumor samples express higher amounts of terminal fucosylated clusterin compared with juxtatumoral samples.
Figure 2. Fucosylated clusterin (fCLU) is expressed in luminal breast cancer and interacts with DC-SIGN. a. Scheme of the ELISA assay used to detect fucosylated clusterin. Clusterin is captured by a mAb directed to clusterin and the presence of fucosylated clusterin (fCLU) is revealed by using either biotinilated Ulex europaeus agglutinin-I lectin (UEA-I) or Lotus tetragonolobus lectin (LT). b. Fucosylated semen clusterin, but not serum or recombinant clusterin (blue bars), was detected by the assay described in (a). The addition of α-L-fucose prevents the recognition of fucosylated semen clusterin (red bars). Results are expressed as the mean ± SD of 4 independent experiments. c and d. Fucosylated clusterin expression in breast tumor and juxtatumoral samples (n = 14–21) was analyzed using Lotus tetragonolobus lectin (c) or Ulex europaeus agglutinin-I (d). Results are expressed as the amount of fucosylated clusterin relative to the amount of total clusterin (left panels) or total protein (right panels) (***p < .001, **p < .01, *p < .05). E. The ability of clusterin from tumor samples and juxtatumoral samples to bind to DC-SIGN was evaluated. Upper panel: the presence of clusterin was evaluated using a mAb directed to clusterin. Lower panel: the membrane was stripped and revealed using DC-SIGN-huFc and HRP conjugated anti-huIgG. Of note, total clusterin but not total protein was used to normalize the amount of sample loaded into the gel for each tumor and juxtatumor sample pair. Accordingly, no loading control was analyzed. Panel F shows the ratio between DC-SIGN-huFC binding and total CLU. The fold change between tumor and juxtatumor CLU ratios are shown.
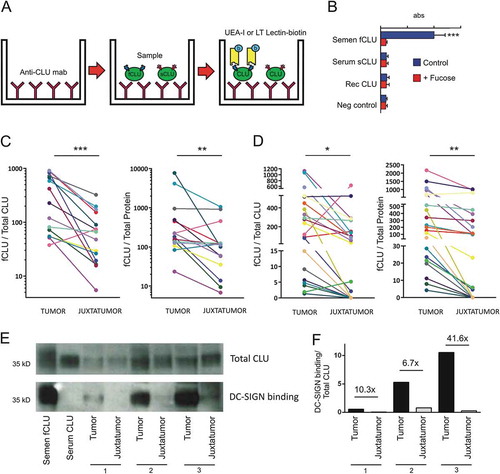
We have previously reported that the binding of seminal plasma clusterin to DC-SIGN depends on terminal fucosylated LewisX and LewisY motifs.Citation10 The interaction of fucosylated ligands with DC-SIGN was shown to promote the acquisition of a tolerogenic profile by DCs.Citation20–Citation23 We hypothesized that fucosylated clusterin from breast tumors might facilitate tumor escape from immune destruction by interacting with DC-SIGN expressed by infiltrating myeloid cells. To test this hypothesis, we first analyzed whether fucosylated clusterin from luminal breast tumors actually interacted with DC-SIGN. In these experiments, 3 pairs of tumor and juxtatumoral samples were analyzed in western blot like assays. In this assay, the binding of clusterin to DC-SIGN is evaluated by adding soluble DC-SIGN-huFc chimera instead of a primary antibody. For each pair, a similar amount of total clusterin was lo added into the gel, as shown on the upper panel of ) using anti-clusterin antibodies. The lower panel of ) shows that tumor clusterin, but not clusterin from juxtatumoral tissues, effectively binds DC-SIGN. ) shows the ratio between DC-SIGN-huFC binding and total CLU. The fold change between tumor and juxtatumor CLU ratios are shown. Overall, these observations indicate that clusterin present in breast tumor samples bears terminal fucosylated motifs and binds to DC-SIGN.
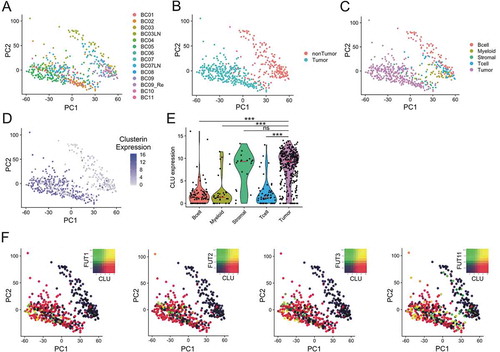
The synthesis of fucosylated Lewis type motifs is regulated by the action of different fucosyltransferases (FUTs) in the Golgy complex. To study the expression of fucosyltransferases and clusterin we analyze a cohort of breast cancer patients previously caracherized by Chung et al, using single cell transcriptomics.Citation24 This study provides the transcriptome analysis of 515 single cells from tumor samples of 11 patients with different breast cancer subtypes. Unsupervised principal component analysis (PCA) shows extensive intratumoral heterogeneity ()). In ,c), tumor and different lineages of non-tumor cells where identified using gene expression signatures previously described.Citation24 As shown on ,e), clusterin is predominantly expressed by tumor and stromal cells, but not by tumor infiltrating immune cells. We then analyzed the expression of FUTs that might be involved in clusterin fucosylation. The expression of different FUTs in breast cancer has already been reported.Citation25–Citation27 The expression of FUT1, FUT2 and FUT3 was detected in tumor cells while FUT11 expression was observed in tumor and non-tumor cells. As shown in ), clusterin and FUTs are coexpressed in a proportion of the cells. These cells are potential sources of fucosylated clusterin in breast cancer tissues.
Figure 3. Single-cell analysis in breast cancer. Unsupervised principal component analysis (PCA) on the transcriptome of single-cell profiles (dots) colored by donor (a), cell tumor state (b) and cell type (c) showing the separation between tumor and non-tumor cells. D. Clusterin expression is dominant tumor cells. Unsupervised PCA of single-cell profiles with coloring of clusterin expression. E. Violin plots comparing clusterin expression based on cell types. Red lines show median expression for each group. P-values computed are pairwise Wilcoxon rank sum test comparing clusterin expression in tumor cells and other groups (*** = p < .001). F. Co-expression of Fucosyltransferases I, II, III and XI (FUT1, FUT2, FUT3 and FUT11) with clusterin in tumor cells is shown by PCA of single-cell profiles, showing clusterin expre ssion (red), FUT expression (green) and co-expression (yellow) or non-expression (black) of clusterin and FUTs.
Analysis of DC-SIGN expression in breast cancer
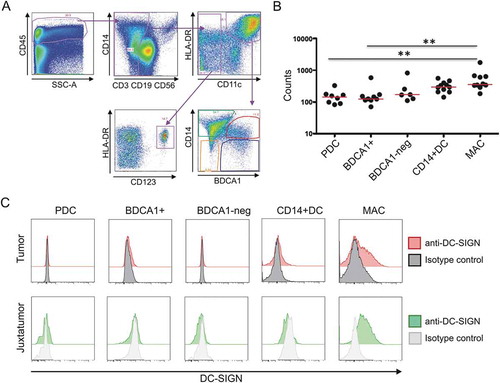
We next analyzed DC-SIGN expression by the different populations of DC and macrophages found in breast tumor samples using flow cytometry. The gating strategy is illustrated in ). As previously described,Citation28 we identified four major DC populations: CD11c− CD123+ plasmacytoid pre-DCs, CD11c+ BDCA1+ CD14− DCs, CD11c+ BDCA1− CD14− DCs, and CD11c+ BDCA1+ CD14+ inflammatory DCs. The most prominent intratumoral myeloid cell population, however, was shown to be represented by macrophages, defined as CD11c+ BDCA1− CD14+ cells. We analyzed DC-SIGN expression on the surface of these populations by flow cytometry. As shown on ), DC-SIGN was only expressed on the membrane of macrophages from tumoral as well as juxtatumoral tissues, but not on the DC populations analyzed. Next, we analyzed DC-SIGN mRNA expression by RNA sequencing on each sorted population. Among all the cell populations analyzed on tumor samples, macrophages showed the highest DC-SIGN mRNA expression ()).
Figure 4. DC-SIGN expression by tumoral macrophages. a. Gating strategy used to identify the different populations of DCs and macrophages in tumoral and juxtatumoral samples. CD45+, lineage negative (CD3, CD19, CD56) and HLA-DR+ cells were gated and four different populations of DCs were identified: CD11c- CD123+ plasmacytoid pre-DCs (PDC), CD11c+ BDCA1+ CD14- DCs (BDCA1+), CD11c+ BDCA1- CD14- DCs (BDCA1-), and CD11c+ BDCA1+ CD14+ inflammatory DCs (CD14+ DC). Macrophages were identified as CD11c+ BDCA1- CD14+ cells (MAC). b. The four populations of tumoral DCs and tumoral macrophages were sorted, and the expression of DC-SIGN mRNA was determined by RNA-seq (n = 6–10). c. Flow cytometry analysis of cell surface DC-SIGN expression by the different populations of DCs and macrophages from tumoral and juxtatumoral samples. A representative experiment (n = 5) is shown.
Fucosylated clusterin modulates the differentiation profile of macrophages
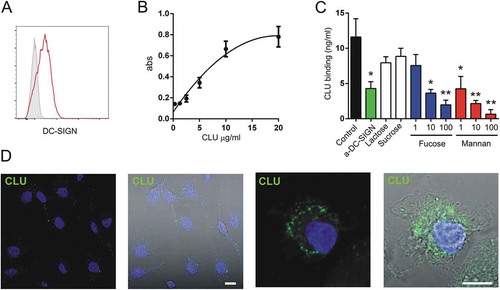
Then, we analyzed whether fucosylated clusterin might be able to switch the differentiation of macrophages into a functional profile able to promote tumor growth. To this aim we developed an in vitro assay using fucosylated clusterin isolated from human semen and human monocyte derived macrophages. As shown in ,b), monocyte-derived macrophages express DC-SIGN and bind fucosylated clusterin in a concentration-dependent mode, reaching a plateau at concentrations of 20 µg/ml. Clusterin binding was prevented by the DC-SIGN ligands mannan or α-L-fucose as well as by treatment of macrophages with a blocking mAb directed to DC-SIGN ()). Upon binding, fucosylated clusterin was rapidly internalized by macrophages, as revealed by confocal microscopy ()).
Figure 5. Fucosylated clusterin interacts with macrophages. a. Expression of DC-SIGN by monocyte-derived macrophages. A representative experiment (n = 7) is shown. b. Macrophages were incubated for 30 min at 4°C with different concentrations of fucosylated clusterin isolated from semen. Cells were then washed, lysed and the binding of clusterin was revealed by ELISA. Results are expressed as the mean ± SD of 4 independent experiments. c. Macrophages were incubated for 30 min at 4°C with fucosylated clusterin (10 µg/ml), in the absence or presence of a neutralizing mAb directed to DC-SIGN (5 µg/ml), lactose (100 µg/ml), sucrose (100 µg/ml), mannan (1–100 µg/ml), or α-L-fucose (1–100 µg/ml). Cells were then washed, lysed and the binding of clusterin was revealed by ELISA. Results are expressed as the mean ± SD of 3 experiments (**p < .01, *p < .05). d. Macrophages were incubated for 15 min at 37°C with fucosylated clusterin (10 µg/ml) and clusterin endocytosis (green) was analyzed by confocal microscopy (n = 5, bars:10µm).
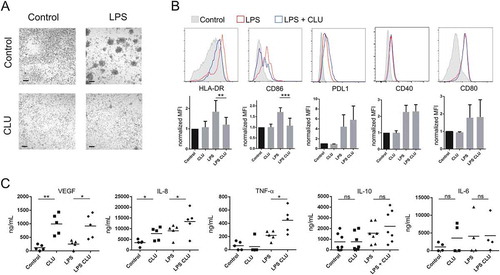
Having shown that fucosylated clusterin effectively interacts with macrophages via DC-SIGN, we then analyzed its influence on the phenotype and functional profile of macrophages. In these experiments, monocytes were cultured for 5 days with M-CSF, in the absence or presence of semen fucosylated clusterin. Then, the cells were cultured for additional 24 hours with or without LPS and the phenotype and the pattern of cytokine production were analyzed. As shown in ), fucosylated clusterin inhibited the classical clustering of LPS stimulated macrophages. As shown in ), fucosylated clusterin-treated macrophages failed to upregulate the expression of HLA-DR and CD86 induced by LPS but not the expression of CD40, CD80 and PDL-1 ()). Of note, fucosylated clusterin significantly increased the production of the angiogenic factors VEGF and IL-8, evaluated in both, untreated and LPS-treated macrophages. Moreover, it significantly increased the production of TNF-α by LPS-activated macrophages, without modifying the production of IL-10 and IL-6 ()). IL-12 and IL-1 were undetectable in all the conditions assessed (not shown). We conclude that fucosylated clusterin inhibits the expression of cell surface molecules involved in antigen presentation and promotes the differentiation of macrophages into a pro-angiogenic profile.
Figure 6. Fucosylated clusterin promotes the differentiation of macrophages into a proangiogenic profile. a-c. Macrophages were differentiated from monocytes cultured with M-CSF for 5 days, in the absence or presence of semen fucosylated clusterin. Then, cells were stimulated, or not, with LPS (10 ng/ml), and the pattern of cell clustering (a), the expression of HLA-DR, CD86, PDL-1, CD40 and CD80 (b), and the production of VEGF, IL-8, TNF-α, IL-10, and IL-6 (c) were evaluated by optical microscopy, flow cytometry or ELISA. Representative pictures (bars = 100µm) and histograms (n = 3–9) are shown in a and b. On b, the control condition mean fluorescent intensities (MFI) of each independent experiment were normalized as 1. The mean of 5–9 independent experiments is shown. The mean of 5–7 independent experiments is shown in c (**p < .01, *p < .05).
Discussion
Since its discovery by Fritz and coworkers in 1983, clusterin has shown to be involved in a variety of physiological and pathological processes such as cell death regulation, lipid transport, sperm motility, unfolded protein clearance, Alzheimer’s disease and cancer.Citation1,Citation2,Citation29 An altered expression of clusterin in breast cancer was first described by Redondo et al. in a large cohort of cancer patients, showing a relationship between clusterin overexpression and advanced disease.Citation30 It is now accepted that clusterin expression is associated with a poor prognosis and disease recurrence in breast cancer as well as in many other tumor types such as gastric, bladder, cervical, prostate, lung and colorectal cancer.Citation2,Citation31–Citation33 However, discrepant results have been published in lung and prostate cancer regarding the value of clusterin expression as a predictive marker of a better response to therapy.Citation34–Citation37 A possible explanation for these controversial observations could be related to the existence of two different clusterin isoforms, an anti-apoptotic form usually called secretory clusterin and a nuclear form with pro-apoptotic activity. These different isoforms might result from distinct transcripts generated by alternative transcription initiation, possibly driven by two distinct promoters.Citation7
We have recently characterized a novel clusterin glycoform present in semen that express abundant fucosylated LewisX and LewisY motifs.Citation10 Importantly, semen fucosylated clusterin, but not serum sialylated clusterin, binds to DC-SIGN, a C-type lectin receptor expressed by dendritic cells at the female genital mucosa, inhibiting its interaction with HIV-1.Citation10 In a similar way than other seminal plasma glycoproteins, a multivalent presentation of Lewisx/y sequences appears to be required for the interaction between clusterin and DC-SIGN.Citation17 Moreover, in agreement with previous reports analyzing DC-SIGN interaction with fucosylated ligands, we showed that fucosylated semen clusterin promotes the acquisition of a tolerogenic profile by conventional dendritic cells, suggesting a novel mechanism through which semen might promote tolerance to male antigens, thereby contributing to successful pregnancy.Citation11 These results are in line with a large body of evidence sustaining that glycan functional groups are able to elicit tolerogenic effects that protect human gametes and offspring in utero from immune rejection. Interestingly, microorganisms and tumors seem to mimic this strategy in order to evade the immune system.Citation38,Citation39
Our present results demonstrate the expression of fucosylated clusterin in breast tumor samples and its ability to interact with DC-SIGN. Fucosylated clusterin was absent or diminished in juxtatumor tissues, suggesting that it might be considered as a cancer associated glycoform. Indeed, aberrant fucosylation of proteins is frequently found in cancer cells.Citation14,Citation15 In spite that we did not analyze whether breast cancer clusterin and seminal plasma clusterin bear identical fucosylated motifs, we demonstrate that clusterin from both sources express terminal fucosylated motifs able to bind DC-SIGN. In fact, our observations suggest that these motifs are responsible for the recognition of tumor clusterin by DC-SIGN. However, due to the low amount of clusterin recovered from tumor samples, we could not assay whether removal of fucose motifs by fucosidases prevent the ability of tumor clusterin to interact with DC-SIGN, as we have previously shown for semen clusterin.Citation10
We speculated that the ability of tumor fucosylated clusterin to interact with DC-SIGN might induce the acquisition of a tolerogenic profile by tumor DCs, promoting tumor escape from the immune response. However, contrasting with observations made in other human tumors, we found that infiltrating DCs in breast cancer do not express DC-SIGN. By contrast, intratumoral macrophages, which represent the predominant myeloid cell population infiltrating breast tumors, as well as juxtatumoral macrophages express DC-SIGN. Using semen clusterin as a source of fucosylated clusterin and monocyte-derived macrophages, we found that clusterin not only inhibited the ability of LPS to stimulate the up-regulation of T cell activating molecules by macrophages, such as MHC class II and CD86, but also promoted the differentiation of macrophages into profile characterized by a high production of VEGF, IL-8 and TNF-α. A weakness of our study is that we were unable to isolate enough amounts of breast cancer clusterin to perform functional studies using tumor clusterin instead of semen clusterin. Further studies are needed to characterize the effect of tumor clusterin on the phenotype and function of tumor infiltrating DC-SIGN+ macrophages.
Interestingly, the presence of fucosylated clusterin in tumors does not appear to be restricted to breast cancer. In fact, we also detected the expression of fucosylated clusterin in tumor samples from patients with cervical cancer (unpublished results), suggesting that it might represent a common marker for different tumors. There is a large body of evidence indicating that lectin-glycan interactions modulate tumor angiogenesis, invasion and metastasis as well as tumor escape from the immune response.Citation13,Citation40 Here we propose that tumor fucosylated clusterin might be considered as a novel player in the tumor microenvironment able to promote tumor progression by interacting with tumor infiltrating DC-SIGN+ macrophages.
Materials and methods
Patients and samples
Breast tumor and juxtatumoral samples from previously untreated cancer patients undergoing surgery were obtained from the Institut Curie Hospital (Paris, France) in accordance with Institut Curie ethical guidelines and the Declaration of Helsinki. Patient characteristics are summarized in . Samples were mechanically homogenized in a phosphate-buffered saline (PBS) containing an EDTA-free protease inhibitor cocktail (Roche) and stored at −80°C.
ELISA for the detection of fucosylated clusterin
An ELISA plate was treated overnight at 4°C with a PBS-diluted IgG anti-clusterin mAb (R&D mAb 29372, 5µg/ml). The plate was then blocked during 1 h using PBS containing 3% BSA (blocking buffer) (Sigma) at room temperature, and washed 3 times using PBS containing 0.05% Tween. The samples were diluted using blocking buffer, and incubated for 2 h at room temperature in pre-treated plates. After washing with PBS containing 0.05% Tween, the biotinylated Ulex europaeus-1 (50µg/ml) or Lotus tetragonolobus (10µg/ml) lectin diluted in blocking buffer were added, incubated for 30 min at room temperature and washed 3 times. The presence of fucosylated clusterin was revealed using streptavidin-HRP and TMB ELISA substrate.
Total clusterin was detected by ELISA using a conventional ELISA kit (Human Clusterin DuoSet ELISA, R&D Systems).
DC-SIGN binding
Western blot using DC-SIGN-huFc chimera (R&D 161-DC-050) was carried out as described.Citation10 Samples were analyzed by SDS-PAGE (4–12% polyacrylamide gel under reducing conditions), transferred onto a nitrocellulose blot, treated with DC-SIGN-huFc chimera (1 µg/ml in a buffer containing 20 mM Tris, 150 mM NaCl, 1 mM CaCl2, and 2 mM MgCl2 for 4 h at 4°C), and stained with secondary antibodies. Alternatively, membranes were treated with polyclonal goat IgG anti-clusterin (R&D AF2937) and stained with secondary antibodies.
Binding of fucosylated clusterin to macrophages
Peripheral blood mononuclear cells were isolated from blood of healthy donors by density gradient centrifugation using Ficoll-Hypaque (GE Healthcare). Monocytes (>90% purity) were obtained using CD14 microbeads (Miltenyi Biotec). To obtain macrophages, monocytes were cultured for 5 days in RPMI 1640 medium containing 10% fetal bovine serum (Gibco) and recombinant M-CSF (100 ng/ml)(Miltenyi Biotec). Macrophages were incubated with fucosylated clusterin (10 µg/ml), isolated from human seminal plasma as described before,Citation10 for 30 min at 4°C, extensively washed and lysed. The amount of clusterin in cell lysates was measured by ELISA. In some experiments, the binding of fucosylated clusterin to macrophages was assessed in the presence of mannan (Sigma), a blocking mAb directed to DC-SIGN (NIH AIDS reagent program, cat no 6884, 5 µg/ml) or α-L-fucose (Sigma).
Confocal microscopy
Macrophages were placed on poly-L-lysine–coated glass coverslips (12 mm) for 30 min at room temperature. Clusterin binding and endocytosis was carried out as follows: the cells were incubated for 60 min at 4°C with fucosyalted clusterin (10 µg/ml). The cells were washed and the coverslips were covered with 100 ml of complete medium and incubated at 37°C for 15 min to allow endocytosis. The cells were washed with PBS and fixed in 4% paraformaldehyde for 15 min on ice. Then, cells were treated with 0.1 mM glycine in PBS and incubated with mAbs directed to clusterin for 1 h, washed and stained with conjugated secondary antibodies (Jackson Immuno Research, USA). Coverslips were mounted on glass slides using Fluoromount G. Immunofluorescence images were acquired with a Zeiss LSM 800 confocal microscope. The row images are available and can be requested to the corresponding author.
Fucosylated clusterin treatment of macrophages
Monocytes were isolated by positive selection using magnetic beads (Miltenyi Biotec), and cultured in RPMI 1640 medium containing 10% of inactivated-fetal bovine serum (Gibco) and recombinant M-CSF (100 ng/ml) (Miltenyi Biotec), in the presence or absence (control) of fucosylated clusterin (20 µg/ml) isolated from human seminal plasma. After 5 days, the cells were stimulated with LPS (10 ng/ml) for 24h at 37ºC. Then, cells and supernatants were harvested and analyzed by flow cytometry and ELISA, respectively.
Flow cytometry
Tumor samples were cut into small fragments, digested with 0.1 mg/ml of Liberase TL (Roche) in the presence of 0.1 mg/mL DNase (Roche) for 20 min, before the addition of 10 mM EDTA. Cells were then filtered through a 40 mm cell strainer (BD Falcon) and washed. Staining was performed using the following antibodies: anti-CD45 APC-Cy7 (BD), anti-CD14 Qdot 605 (Thermo), anti-CD3 Alexa700 (Biolegend), anti-CD19 Alexa700 (Biolegend), anti-CD56 Alexa700 (Biolegend), anti-HLA-DR BV711 (Biolegend), anti-CD11c PC5 (Beckman Coulter), anti-BDCA-1 PE (BD), anti-CD123 PCy7 (BD) and anti-C-SIGN FITC (BD). Cells were analyzed by a LSRFORTESSA X-20 instrument (BD Biosciences). Data was analyzed with FlowJo (Tree Star). The FACS files are available and can be requested to the corresponding author.
RNA-seq data
We used the RNA-seq data obtained and previously published by Michea et al.Citation28 The data have been deposited in the NCBI Sequence Read Archive (SRA) with the accession code PRJNA380940.
Single cell analysis
Breast cancer SMART-Seq2 data was downloaded from the NCBI Gene Expression Omnibus database (accession number : GSE75688). SRA files were converted to FASTQ files with SRA Toolkit (version 2.3.2-4). RNA AU: The funding information provided has been checked against the Open Funder Registry and we failed to find a match. Please check and resupply the funding details. genome sequences (hg19) using the 2-pass mode of STAR (version 2.5.3.a)Citation41 (parameters: quantMode GeneCounts, twopassMode Basic, alignSJDBoverhangMin 1, bamRemoveDuplicatesType UniqueIdentical, winAnchorMultimapNmax 1000, outFilterMultimapNmax 1000, outFilterScoreMinOverLread 0.33, outFilterMatchNminOverLread 0.33, outFilterMismatchNoverLmax 0.04, outMultimapperOrder Random, sjdbOverhang 99). Raw count tables were obtained with Subread tool FeatureCounts (version 1.5.1)Citation42 and formatted as a matrix with a python script (version 3.6.2). Then, ENSEMBL IDs in the matrix were changed to gene symbols using biotools online converter (https://www.biotools.fr/human/ensembl_symbol_converter).
Metadata from the authors and the raw matrix were used to create a SingleCellExperiment (version 1.4.1) R object. Quality control, normalization and filtering steps were performed in R environment (version 3.5.2). From the initial 563 cells profiles, 36 were removed because of missing information about their cell type in the metadata. Cells were filtered by their library size and their number of genes (respectively 7 and 2 cells were removed) using 3*Median Absolute Deviation (MAD) method from Scater (version 1.10.1).Citation43 Counts were normalized using deconvolution strategy, which divide the counts for each cell by the size factor for that cell. This was performed using Scater (version 1.10.1) which perform the normalization and Scran (version 1.10.2)Citation44 which compute size factors. Only genes with at least 1 count by cell were kept considering their size factors. Finally, a PCA on quality control metrics was performed to remove outlier cells profiles that have substantially different quality control metrics from the others (8 removed) using Scater (version 1.10.1). Expression scaling, Principal Component Analysis and data visualization were done using R packages Seurat 3.0Citation45 on the 510 remaining single-cell profiles.
Statistics
Statistical comparisons were performed by using paired Student’s t-test or ANOVA. The p values <.001(***), <0.01 (**) and <0.05 (*) were considered statistically significant. The single cell analysis was performed using pairwise Wilcoxon rank sum test.
Disclosure of potential conflicts of interest
The authors declare no commercial or financial conflicts of interest.
Acknowledgments
We particularly thank Sophie Viel (INSERM U932), Philemon Sirven (Clinical Immunology Laboratory) and Anne Vincent-Salomon (Center of Clinical Investigations CICBT507 IGR/Curie and Department of Pathology) for providing clinical samples and associated clinical data and for organizing the logistics of these samples. We thank to the Pathex and Flow Cytometry platforms from Institute Curie. Thanks to all the participating patients.
Additional information
Funding
References
- Rohne P, Prochnow H, Koch-Brandt C. The CLU-files: disentanglement of a mystery. Biomol Concepts. 2016;7:1–15. doi:10.1515/bmc-2015-0026.
- Tellez T, Garcia-Aranda M, Redondo M. The role of clusterin in carcinogenesis and its potential utility as therapeutic target. Curr Med Chem. 2016;23:4297–4308.
- Garcia-Aranda M, Tellez T, Munoz M, Redondo M. Clusterin inhibition mediates sensitivity to chemotherapy and radiotherapy in human cancer. Anticancer Drugs. 2017;28:702–716. doi:10.1097/CAD.0000000000000507.
- Miyake H, Gleave M, Kamidono S, Hara I. Overexpression of clusterin in transitional cell carcinoma of the bladder is related to disease progression and recurrence. Urology. 2002;59:150–154.
- Miyake H, Hara S, Arakawa S, Kamidono S, Hara I. Over expression of clusterin is an independent prognostic factor for nonpapillary renal cell carcinoma. J Urol. 2002;167:703–706.
- Steinberg J, Oyasu R, Lang S, Sintich S, Rademaker A, Lee C, Kozlowski JM, Sensibar JA. Intracellular levels of SGP-2 (Clusterin) correlate with tumor grade in prostate cancer. Clin Cancer Res. 1997;3:1707–1711.
- Rizzi F, Coletta M, Bettuzzi S. Chapter 2: clusterin (CLU): from one gene and two transcripts to many proteins. Adv Cancer Res. 2009;104:9–23. doi:10.1016/S0065-230X(09)04002-0.
- Kapron JT, Hilliard GM, Lakins JN, Tenniswood MP, West KA, Carr SA, Crabb JW. Identification and characterization of glycosylation sites in human serum clusterin. Protein Sci. 1997;6:2120–2133. doi:10.1002/pro.5560061007.
- Jones SE, Jomary C. Clusterin. Int J Biochem Cell Biol. 2002;34:427–431. doi:10.1016/S1357-2725(01)00155-8.
- Sabatte J, Faigle W, Ceballos A, Morelle W, Rodríguez Rodrígues C, Remes Lenicov F, Thépaut M, Fieschi F, Malchiodi E, Fernández M, et al. Semen clusterin is a novel DC-SIGN ligand. J Immunol. 2011;187:5299–5309. doi:10.4049/jimmunol.1101889.
- Merlotti A, Dantas E, Remes Lenicov F, Ceballos A, Jancic C, Varese A, Rubione J, Stover S, Geffner J, Sabatté J. Fucosylated clusterin in semen promotes the uptake of stress-damaged proteins by dendritic cells via DC-SIGN. Hum Reprod. 2015;30:1545–1556. doi:10.1093/humrep/dev113.
- Croci DO, Cerliani JP, Pinto NA, Morosi LG, Rabinovich GA. Regulatory role of glycans in the control of hypoxia-driven angiogenesis and sensitivity to anti-angiogenic treatment. Glycobiology. 2014;24:1283–1290. doi:10.1093/glycob/cwu083.
- Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev. 2015;15:540–555. doi:10.1038/nrc3982.
- Blanas A, Sahasrabudhe NM, Rodríguez E, van Kooyk Y, van Vliet SJ. Fucosylated antigens in cancer: an alliance toward tumor progression, metastasis, and resistance to chemotherapy. Front Oncol. 2018;8:39. doi:10.3389/fonc.2018.00039.
- Miyoshi E, Moriwaki K, Nakagawa T. Biological function of fucosylation in cancer biology. J Biochem. 2008;143:725–729. doi:10.1093/jb/mvn011.
- Williams CB, Yeh ES, Soloff AC. Tumor-associated macrophages: unwitting accomplices in breast cancer malignancy. NPJ Breast Cancer. 2016;2. doi:10.1038/npjbcancer.2015.25.
- Clark GF, Grassi P, Pang P-C, Panico M, Lafrenz D, Drobnis EZ, Baldwin MR, Morris HR, Haslam SM, Schedin-Weiss S, et al. Tumor biomarker glycoproteins in the seminal plasma of healthy human males are endogenous ligands for DC-SIGN. Mol Cell Proteomics. 2012;11:M111 008730. doi:10.1074/mcp.M111.008730.
- Baldus SE, Thiele J, Park YO, Hanisch FG, Bara J, Fischer R. Characterization of the binding specificity of Anguilla anguilla agglutinin (AAA) in comparison to Ulex europaeus agglutinin I (UEA-I). Glycoconj J. 1996;13:585–590.
- Yan L, Wilkins PP, Alvarez-Manilla G, Do SI, Smith DF, Cummings RD. Immobilized Lotus tetragonolobus agglutinin binds oligosaccharides containing the Le(x) determinant. Glycoconj J. 1997;14:45–55.
- Gringhuis SI, Den Dunnen J, Litjens M, van der Vlist M, Geijtenbeek TB. Carbohydrate-specific signaling through the DC-SIGN signalosome tailors immunity to Mycobacterium tuberculosis, HIV-1 and Helicobacter pylori. Nat Immunol. 2009;10:1081–1088. doi:10.1038/ni.1778.
- van Gisbergen KP, Aarnoudse CA, Meijer GA, Geijtenbeek TB, van Kooyk Y. Dendritic cells recognize tumor-specific glycosylation of carcinoembryonic antigen on colorectal cancer cells through dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin. Cancer Res. 2005;65:5935–5944. doi:10.1158/0008-5472.CAN-04-4140.
- van Gisbergen KP, Sanchez-Hernandez M, Geijtenbeek TB, van Kooyk Y. Neutrophils mediate immune modulation of dendritic cells through glycosylation-dependent interactions between Mac-1 and DC-SIGN. J Exp Med. 2005;201:1281–1292. doi:10.1084/jem.20041276.
- Conde P, Rodriguez M, van der Touw W, Jimenez A, Burns M, Miller J, Brahmachary M, Chen H-M, Boros P, Rausell-Palamos F, et al. DC-SIGN(+) macrophages control the induction of transplantation tolerance. Immunity. 2015;42:1143–1158. doi:10.1016/j.immuni.2015.05.009.
- Chung W, Eum HH, Lee H-O, Lee K-M, Lee H-B, Kim K-T, Ryu HS, Kim S, Lee JE, Park YH, et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat Commun. 2017;8:15081. doi:10.1038/ncomms15081.
- Carrascal MA, Silva M, Ramalho JS, Pen C, Martins M, Pascoal C, Amaral C, Serrano I, Oliveira MJ, Sackstein R, et al. Inhibition of fucosylation in human invasive ductal carcinoma reduces E-selectin ligand expression, cell proliferation, and ERK1/2 and p38 MAPK activation. Mol Oncol. 2018;12:579–593. doi:10.1002/1878-0261.12163.
- Do Nascimento JC, Ferreira SA, Vasconcelos JL, Da Silva-Filho JL, Barbosa BT, Bezerra MF, Rocha CRC, Beltrão EIC. Fut3 role in breast invasive ductal carcinoma: investigating its gene promoter and protein expression. Exp Mol Pathol. 2015;99:409–415. doi:10.1016/j.yexmp.2015.08.015.
- Li N, Liu Y, Miao Y, Zhao L, Zhou H, Jia L. MicroRNA-106b targets FUT6 to promote cell migration, invasion, and proliferation in human breast cancer. IUBMB Life. 2016;68:764–775. doi:10.1002/iub.1541.
- Michea P, Noël F, Zakine E, Czerwinska U, Sirven P, Abouzid O, Goudot C, Scholer-Dahirel A, Vincent-Salomon A, Reyal F, et al. Adjustment of dendritic cells to the breast-cancer microenvironment is subset specific. Nat Immunol. 2018;19:885–897. doi:10.1038/s41590-018-0145-8.
- Fritz IB, Burdzy K, Setchell B, Blaschuk O. Ram rete testis fluid contains a protein (clusterin) which influences cell-cell interactions in vitro. Biol Reprod. 1983;28:1173–1188. doi:10.1095/biolreprod28.5.1173.
- Redondo M, Villar E, Torres-Munoz J, Tellez T, Morell M, Petito CK. Overexpression of clusterin in human breast carcinoma. Am J Pathol. 2000;157:393–399. doi:10.1016/S0002-9440(10)64552-X.
- Yom CK, Woo HY, Min SY, Kang SY, Kim HS. Clusterin overexpression and relapse-free survival in breast cancer. Anticancer Res. 2009;29:3909–3912.
- Niu ZH, Wang Y, Chun B, Li CX, Wu L. Secretory clusterin (sCLU) overexpression is associated with resistance to preoperative neoadjuvant chemotherapy in primary breast cancer. Eur Rev Med Pharmacol Sci. 2013;17:1337–1344.
- Kruger S, Ola V, Fischer D, Feller AC, Friedrich M. Prognostic significance of clusterin immunoreactivity in breast cancer. Neoplasma. 2007;54:46–50.
- Rizzi F, Bettuzzi S. The clusterin paradigm in prostate and breast carcinogenesis. Endocr Relat Cancer. 2010;17:R1–17. doi:10.1677/ERC-09-0140.
- Li H, Liu S, Zhu X, Yang S, Xiang J, Chen H. Clusterin immunoexpression and its clinical significance in patients with non-small cell lung cancer. Lung. 2010;188:423–431. doi:10.1007/s00408-010-9248-1.
- Panico F, Casali C, Rossi G, Rizzi F, Morandi U, Bettuzzi S, Davalli P, Corbetta L, Storelli ES, Corti A, et al. Prognostic role of clusterin in resected adenocarcinomas of the lung. Lung Cancer. 2013;79:294–299. doi:10.1016/j.lungcan.2012.11.024.
- Panico F, Rizzi F, Fabbri LM, Bettuzzi S, Luppi F. Clusterin (CLU) and lung cancer. Adv Cancer Res. 2009;105:63–76. doi:10.1016/S0065-230X(09)05004-0.
- Clark GF, Oehninger S, Patankar MS, Koistinen R, Dell A, Morris HR, Koistinen H, Seppälä M. A role for glycoconjugates in human development: the human feto-embryonic defence system hypothesis. Hum Reprod. 1996;11:467–473. doi:10.1093/humrep/11.3.467.
- Clark GF. The role of glycans in immune evasion: the human fetoembryonic defence system hypothesis revisited. Mol Hum Reprod. 2014;20:185–199. doi:10.1093/molehr/gat064.
- Rabinovich GA, Croci DO. Regulatory circuits mediated by lectin-glycan interactions in autoimmunity and cancer. Immunity. 2012;36:322–335. doi:10.1016/j.immuni.2012.03.004.
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi:10.1093/bioinformatics/bts635.
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi:10.1093/bioinformatics/btt656.
- McCarthy DJ, Campbell KR, Lun ATL, Wills QF. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–1186. doi:10.1093/bioinformatics/btw777.
- Lun AT, Bach K, Marioni JC. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016;17:75. doi:10.1186/s13059-016-0947-7.
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi:10.1038/nbt.4096.
