ABSTRACT
Colorectal cancer (CRC) is the third most prevalent cancer worldwide with a high mortality rate (20–30%), especially due to metastasis to adjacent organs. Clinical responses to chemotherapy, radiation, targeted and immunotherapies are limited to a subset of patients making metastatic CRC (mCRC) difficult to treat. To understand the therapeutic modulation of immune response in mCRC, we have used a genetically engineered mouse model (GEMM), “KPN”, which resembles the human ‘CMS4’-like subtype. We show here that transforming growth factor (TGF-β1), secreted by KPN organoids, increases cancer cell proliferation, and inhibits splenocyte activation in vitro. TGF-β1 also inhibits activation of naive but not pre-activated T cells, suggesting differential effects on specific immune cells. In vivo, the inhibition of TGF-β inflames the KPN tumors, causing infiltration of T cells, monocytes and monocytic intermediates, while reducing neutrophils and epithelial cells. Co-inhibition of TGF-β and PD-L1 signaling further enhances cytotoxic CD8+T cells and upregulates innate immune response and interferon gene signatures. However, simultaneous upregulation of cancer-related metabolic genes correlated with limited control of tumor burden and/or progression despite combination treatment. Our study illustrates the importance of using GEMMs to predict better immunotherapies for mCRC.
Introduction
Colorectal carcinoma (CRC) is one of the most prevalent cancers worldwide and has a mortality rate of 20–30%. Metastasis of tumor to adjacent organs is the major cause of death and almost 20% of the patients diagnosed with CRC have metastasis.Citation1,Citation2 The current options for targeted therapies are very limited such as anti-Vascular Endothelial Growth Factor (VEGF), Anti-Epidermal Growth Factor Receptor (EGFR) and anti-Programmed Cell Death-1 (PD1) but only a fraction of patients respond to the therapies. Many treated individuals either do not respond to these therapies (innate resistance) or show recurrence or develop resistance to treatments (adaptive resistance).Citation3,Citation4 Patient stratification is limited to screening of PD-L1 expression for immunotherapy, and no other clinical biomarkers have been identified to differentiate responders from non-responders.Citation5–7 Hence, there is a need to identify combinatorial therapies to the right subset of patients with CRC.
CRC can be differentiated into four Consensus Molecular Subtypes (CMS) based on microsatellite instability (MSI) and chromosomal instability (CIN). Driver mutations in genes like APC, TP53, SMAD4, KRAS, and PIK3CA initiate tumor development and chromosomal instability, making the tumor invasive and metastatic.Citation8 In CRC, mutations in KRAS (40–50%), TP53 (43%), and NOTCH (16.5–25%) genes are common,Citation7,Citation9,Citation10 and 23% tumors are CMS4 (mesenchymal) subtype.Citation11,Citation12 CMS4 tumors are characterized by SMAD4 mutations, TGF-β upregulation, presence of endothelial cells and cancer-associated fibroblasts (CAFs), making these tumors highly metastatic with poor survival.Citation11,Citation13 Hence, it is critical to use for appropriate preclinical models resembling human CRC subtypes to test immune therapies that may improve treatment of CMS4 mCRC.
The KPN mouse model (villinCreERKrasG12D/+Trp53fl/fl Rosa26N1icd/+) represents the CMS4 CRC.Citation14 These genes are under the control of cre-recombinase driven by a tamoxifen-inducible villin promoter expressed in intestinal epithelial cells. Mutation in the Kras affects the RAS/MAPK pathway, resulting in uncontrolled cell proliferationCitation15 Deletion of Trp53 results in genetic instability.Citation16 Constitutively active truncated Notch1 intracellular domain “N1icd” drives KPN toward an immunosuppressive TGF-β-driven tumor microenvironment (TME).Citation14 The combination generates tumors in the intestine with metastases in the liver and adjoining organs.Citation14 Late-stage liver metastasis is modeled using the intra-splenic transplantation. Organoids derived from KPN liver metastases are injected as single cells into the spleen, rapidly generating metastasis in the liver. Thus, these intra-splenic KPN mice provide a model to understand therapy-driven immune regulation in mCRC.
The TGF-β cytokine superfamily includes ligands TGF-β1,2 and 3.Citation12 Upon activation, the inactive TGF-β1 ligand binds to the serine/threonine kinase receptors, TGF-βRI (ALK5) and TGF-βRII. This binding activates or represses target genes associated with either tumor suppression or progression.Citation12 Tumorigenic effects of TGF-β include promoting epithelial to mesenchymal transition (EMT), angiogenesis, immune evasion, myofibroblast generation, and metastasis.Citation12,Citation17 TGF-β also inhibits natural killer (NK), CD8+T cells, M1-like macrophages, and N1 neutrophils and promotes the activity of T regulatory cells (Treg), M2-like macrophages, and N2 neutrophils near the TME, resulting in disease aggravationCitation18,Citation19 Blocking circulating TGF-β inhibits immune suppression and tumor progression. ALK5 inhibitor (ALK5i) or anti-TGF-β antibody inhibit TGF-β signaling pathway by either inhibiting phosphorylation of TGF-βRI kinase domain, or by blocking TGF-β ligand to receptor interactionsCitation14,Citation20,Citation21
Programmed cell death ligand 1(PD-L1/CD274), expressed by tumor cells and antigen presenting cells (APCs), is the ligand for Programmed Cell Death 1(PD-1/CD279), on T cells. The inhibitory interaction between PD1 with PD-L1 enables the tumor cells to escape host immune surveillance by suppressing antigen recognition, lymphocyte infiltration, and effector functions.Citation22 Antibody targeting of PD-L1 inhibits PD1/PD-L1 interactions, potentially allowing T cells to proliferate, perform their effector functions, and reestablish the anti-tumor immune response.Citation23
Though there have been significant advancements in precision/targeted therapy for cancer, limited information is available about the optimum treatment of CMS4 subtype of tumors with high TGF-β and activated Notch signaling. Since KRAS, TRP53, and NOTCH1 gene mutations play a critical role in mCRC in humans, the human-CMS4 like CRC tumor characteristics in KPN makes it an interesting model to evaluate therapy driven anti-tumor immune response. We have used this model to answer (a) whether targeting TGF-β alongside with immune checkpoint blockade would help in modulating immune response and curing liver metastasis of KPN tumors and (b) what could be the underlying mechanisms of resistance if the combination therapy is not efficacious. To test this hypothesis, in vitro and in vivo experiments were conducted using KPN organoids and with the help of techniques such as flow cytometry, immunohistochemistry and TempOseq-based RNA sequencing we have investigated the mechanisms modulating the immune response and potential resistance mechanisms in mCRC after co-inhibition of TGF-β and PD-L1 signaling pathways. Our study highlights the importance of using GEM models like KPN to predict novel immune therapy combinations that may have a higher chance of translatability to the clinic.
Results
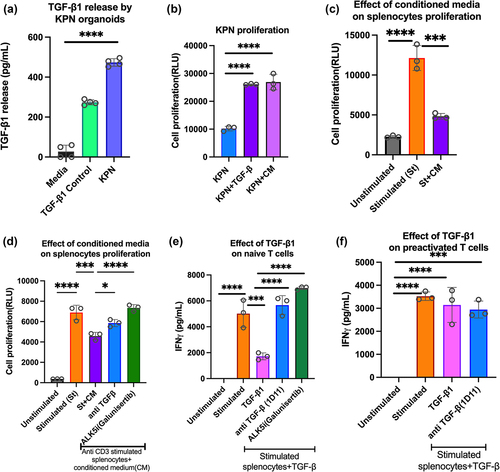
KPN organoids secrete TGF-β1, enhancing tumour proliferation and inhibiting splenocyte activation
TGF-β signaling is characteristic of the CMS4 CRC, and Notch drives epithelial TGF-β2 expression in KPN tumors.Citation14 Since TGF-β1 is the most abundant isoform,Citation17 we tested its role in KPN organoids, and in inhibiting T cell activation. In vitro, cell culture supernatant of KPN organoids contained TGF-β1 (), and conditioned media (CM) from organoid cultures increased KPN proliferation to a similar extent as addition of recombinant TGF-β1 (). To evaluate TGF-β1-mediated immune cell suppression, anti-CD3-stimulated splenocytes were cultured with TGF-β1 or CM. Reductions in cell proliferation and IFN-γ release were observed with TGF-β1 and CM, and restored with ALK5i (Galunisertib), and anti-TGF-β antibody (1D11) (, FigS1A-S1B). Because TGF-β1 drives both pro and antitumorigenic responses, we tested its effects on naïve and preactivated T cells. IFN-γ release and proliferation of activated splenocytes (naïve T cells) was inhibited by TGF-β1, and inhibition was blocked by anti-TGF-β antibody or ALK5i ( FigS1C). In contrast, inhibition was not observed when splenocytes were pre-stimulated, rested, and restimulated with added TGF-β1 (, FigS1D). Thus, CM from KPN tumors has tumor proliferative and immunosuppressive properties like recombinant TGF-β1, and TGF-β1 inhibits activation of naïve but not pre-stimulated T cells.
Figure 1. Effect of TGF-β1 on organoids and immune cells.
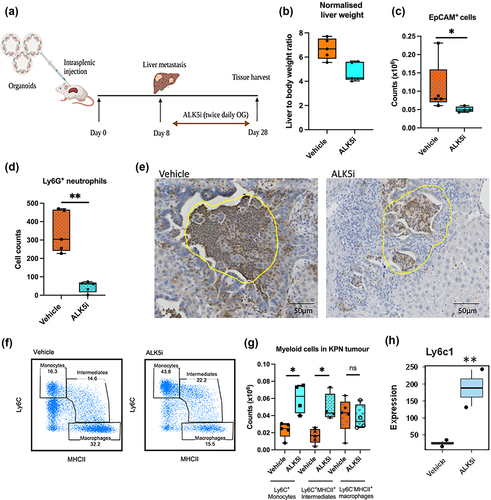
TGF-β inhibitor enhances Ly6C+MHCII+ monocytic infiltrate into KPN liver metastases
To understand immune responses in KPN metastatic tumors treated with ALK5i, liver metastases were generated by intra-splenic transplantation of KPN organoids. A week after transplantation, treatment with ALK5i or vehicle was performed twice daily for 3 weeks (). Livers were then weighed and sectioned. Treatment with ALK5i reduced the liver: body weight ratio, suggesting a decrease in liver metastases (). Liver metastases were digested and stained to identify EpCAM+ epithelial cells and infiltrating immune cells. Flow cytometry showed that ALK5i treatment significantly reduces the number of EpCAM+ cells (). Immunohistochemistry (IHC) showed reduction numbers of Ly6G+ neutrophils (). A significant increase in numbers and frequency of Ly6C+ monocytes and Ly6C+MHCII+ monocytic intermediates occurred after ALK5i treatment (, FigS 2A, S2B, S2C). There was no change in numbers of Ly6C−MHCII+ macrophages (). Further, RNA expression analysis by TempO-Seq also showed upregulation of Ly6c1 after ALK5i treatment (). More CD64+ cells and Ly6C+MHCII+ intermediates were also observed in liver tissue of mice with low KPN metastatic tumor burden (Fig-S2D). These data indicate that KPN tumors have an inflammatory TME which is modulated after ALK5i treatment.
Figure 2. TGF-β1 inhibitor reduces neutrophils, epithelial cells and enhances inflammatory monocytes in KPN metastases in vivo.
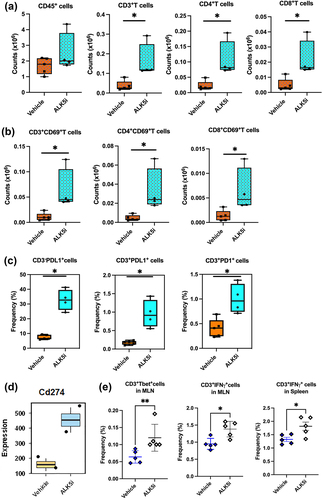
TGF-β inhibitor enhances infiltration of lymphocytes into KPN liver metastases
Since lymphoid cells play a critical role in anti-tumor responses, we also evaluated changes in T-lymphocytes after ALK5i treatment (Fig-S3). Increases in the numbers of tumor infiltrating CD4+T and CD8+T cells were observed after treatment (). Total CD45+cells did not change in frequency or numbers. The T lymphocytes were also activated, with higher CD69 expression after ALK5i treatment (). Since T cells were activated, we evaluated their expression of immune checkpoint proteins-PD1 and PD-L1. ALK5i significantly increased expression of PD-L1 on both CD3− and CD3+ cells. Interestingly, a higher proportion of CD3− cells were PD-L1+ than CD3+ cells (). TempO-Seq analysis of liver metastases also showed an increase in Cd274 (PD-L1) gene expression (). The tSNE plot shows an increase in the frequency of CD3+, CD4+ and CD8+T cells, CD69, PD1 and PD-L1 expression on T cells, and a decrease in the frequency of epithelial cells after treatment, indicating enhanced anti-tumor immune responses (Fig S4A and S4B). We also observed an increase in the frequencies of CD3+T-bet+, CD3+IFN-γ+ cells in the mesenteric lymph nodes (MLN) and spleens from KPN mice after ALK5i treatment (), indicating enhanced systemic Th1 immune responses in KPN tumor-bearing mice.
Figure 3. TGF-β1 inhibitor enhances lymphocyte infiltration and PD-1/PD-L1 expression into KPN metastases.
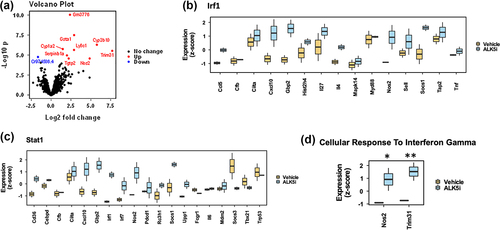
TGF-β inhibition upregulates interferon-associated genes in KPN liver metastases
To understand the effects of ALK5i on immune associated pathways, we analyzed formalin fixed paraffin embedded (FFPE) liver sections from metastatic tissue by whole mouse transcriptome RNA expression analysis using the TempO-Seq platform. Volcano plot shows significantly upregulated and downregulated genes between vehicle and ALK5i treatment groups (). Differential gene expression (DEG) analysis show that the genes associated with the closely related interferon regulatory factor (IRF)-1, and STAT1 pathways are enriched and most activated upstream regulators (TRUSST database) (). Gene ontology (GO) analysis also showed an increase in the expression of genes: Nos2Citation24 and Trim31,Citation25 in the Cellular_Response_to_Interferon_Gamma pathway, that are associated with modulation of tumor associated immune responses (). Thus, ALK5i enhances IRF and STAT1 gene expression, and enhances anti-tumor immune responses.
Figure 4. TGF-β1 inhibitor enhances expression of IRF1 associated genes.
Co-inhibition of TGF-β and PD-L1 boosts infiltration of CD8+ T cells in KPN liver metastases
The PD1/PD-L1 axis is often associated with T cell exhaustion, leading to immune evasion and tumor progression.Citation23 Since ALK5i treatment of KPN metastases led to high PD1/PD-L1 expression but did not completely inhibit tumor growth, we hypothesized that dual inhibition of TGF-β and PD1/PD-L1 would further reduce KPN metastasis. KPN metastases were therefore treated with combination of ALK5i and anti-PD-L1 antibody, or with the individual treatments, or appropriate vehicle/isotype controls (). Samples were analyzed at endpoint by flow cytometry. Frequencies of CD3+T cells are shown in . No changes were observed in the counts of CD45+ leukocytes (). ALK5i-treated animals showed a significant increase in counts of CD3+T cells and CD4+T cells over vehicle-treated mice. The combination treatment also caused a significant increase in the frequency of CD3+T cells over the vehicle. No changes were observed in CD3+ or CD4+T cell frequency or counts after anti-PD-L1 treatment (). Unlike CD4, CD8+T cells did not significantly increase in number after ALK5i treatment. However, Anti-PD-L1 single and combination treatments showed significant increases in CD8+T cells in the KPN tumor ().
Figure 5. Inhibition of TGF-β and PD-L1 boosts infiltration of CD8+T cells.
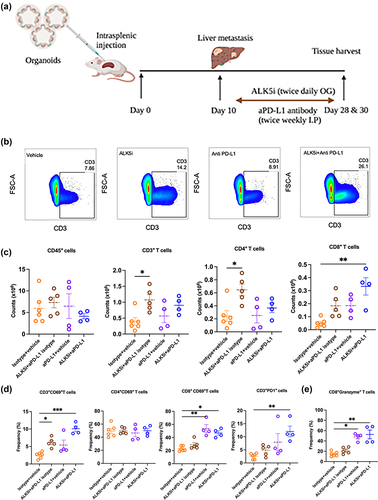
We also evaluated activation of these infiltrating T cells, by assessing their CD69 expression. ALK5i alone and in combination with anti-PD-L1 caused a significant increase in the frequency of CD69+CD3+T cells and CD69+CD8+T cells, indicating CD8+T cell activation () The combination treatment also elevated PD1 expression on CD3+T cells (). Intracellular cytokine staining showed higher frequency of granzymeB+CD8+T cells after anti-PD-L1 single, and combination treatment with ALK5i (). This increase in the frequency of CD8+PD1+, CD8+CD69+ and granzymeB+CD8+T cells was also observed in livers of mice with low KPN metastatic tumor burden (Fig-S5A-5D). Overall, co-inhibition of TGF-β and PD1/PD-L1 signaling increases infiltration of granzymeB-expressing cytolytic CD8+T cells into KPN metastatic tumors.
Co-inhibition of TGF-β and PD-L1 upregulates innate immune responses in the KPN liver metastases
To investigate mechanisms modulating the immune response in KPN tumors after combination treatment, we performed RNA sequencing of metastases from FFPE liver sections by TempO-Seq. We observed differential expression of genes (DEG) between vehicle, single and combination treatment groups (Fig-S6A-C, supplementary table S1). Since DEG were highest between vehicle and combination groups, pathway analysis was used to compare between them. This showed an increase in genes regulating and activating innate_immune_response, cell_surface_receptor_signalling, Fc_receptor_signalling, and metabolic_processes and cellular response to interferon gamma (, S6D-F). Thus, co-inhibition of TGF-β and PD-L1 pathways boosts the interferon gene expression in KPN metastases ().
Figure 6. Co-inhibition of TGF-β and PD-L1 upregulates IRF-1 and cancer associated metabolic pathway genes.
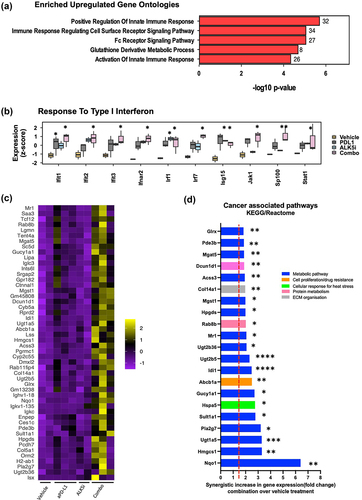
Upregulation of cancer associated metabolic pathway genes associates with tumour resistance after dual treatment
Though we observed increases in both infiltration of CD8+T lymphocytes and IFN gene expression, there was no observed reduction of met counts, liver to body weight ratio, or in the proportion of epithelial cells after combination treatment (data not shown). Thus, we investigated potential mechanisms underlying this tumor resistance. We evaluated genes that were upregulated after combination treatment () and identified the KEGG and Reactome pathways relating to the top 50 upregulated genes. The majority of the genes synergistically upregulated by inhibition of both PD-L1 and TGF-β were associated with metabolic pathways (Nqo1, Hmgsc1, Sult1a1, Pla2g7, Ugt1A5, Ugt2B5, Abcb1a), that enhance cancer cell proliferation, invasion, and metastasis (Fig-6D). This upregulation of cancer-related metabolic genes correlated with limited control of tumor burden, despite the combination treatment. Our data therefore indicate that the combination of ALK5i with anti-PD-L1 potentially induces tumor resistance, and correlates with upregulation of cancer-associated metabolic pathways in the KPN tumor.
Discussion
mCRC has a 5-year overall survival of 14% and almost 50% of patients undergoing surgery develop metastasis within these 5 years.Citation5,Citation26 Hence, there is significant unmet clinical need. The KPN model provides an investigational tool to understand mechanisms modulating therapy-induced immune responses, to help develop better immunotherapy combinations to treat CMS4 type mCRC.
Jackstadt et al. has reported comparable TGF-β1 expression in both primary tumors and organoids derived from KPN mice.Citation14 TGF-β1 secreted by KPN organoids has a direct effect on KPN tumors, enhancing proliferation, and inhibiting IFN-γ release by activated T cells. Immunosuppressive effect of TGF-β was primarily on naive and not pre-activated T cells. Similarly, TGF-β inhibits proliferation of naïve T cells but not antigen experienced cells in diabetes.Citation27 However, a contrary report showed TGF-β1 attenuated effector function of tumor antigen-specific human memory CD8+T cells.Citation28 The effects of TGF-β on naïve and preactivated T cells could be different due to downregulation of TGF-βRII expression on activated T cells.Citation29 Thus, the timing of TGF-β inhibition and the mode of T cell stimulation play critical roles in determining its pleiotropic effects.
Since KPN tumors resemble the CMS4-CRC with high TGF-β, we evaluated the effect of ALK5i on myeloid cells in these tumors. High numbers of Ly6C+ monocytes and Ly6C+MHCII+ monocytic intermediates reveal an inflammatory TME. This increase in monocytic intermediates is like that observed in the inflamed intestine.Citation30 TGF-β is also indispensable for monocyte to macrophage differentiation in the intestine and could also play this role in tumors.Citation31 Neutrophils contribute to inflammation and metastasis, and a decrease in Ly6G+ neutrophil after ALK5i treatment modulates inflammatory TME.
ALK5i treatment enhanced T cell infiltration into tumors, increasing numbers of activated CD69+CD4+ and CD8+T cells. In addition, fewer EpCAM+ epithelial cells indicated a reduction in tumor cellularity. TempO-seq analysis confirmed upregulation of IRF-associated genes in the tumor and enhanced systemic Th1 immune responses. Therefore, inhibiting TGF-β in KPN tumors enhances the anti-tumor immune response.
Despite the ALK5i-mediated enhancement in infiltrating T cells and inflammatory myeloid cells, treatment was not highly effective. The T-lymphocytes showed high expression of exhaustion markers PD1 and PD-L1. Exhaustion of T cells could enable tumor resistance, so we treated tumors with a combination of ALK5i and anti-PD-L1. This significantly increased the activated CD8+T cells, and enhanced expression of granzymeB+CD8+T cells. Our data indicate that combination treatment of KPN tumors with ALK5i and anti PD-L1 antibody skews toward Type 1 interferon and interferon gamma mediated immune response by innate cells and CD8+CTLs. These cells are often associated with improved survival of colorectal and other cancersCitation32.
TempO-Seq analysis showed upregulation of innate immune response and IRF pathway genes (IRF1, IRF7, IFNAR2, JAK1, STAT1), yet no reduction was observed in the metastatic tumors. Thus, the combination of ALK5i and anti-PD-L1 was insufficient to eliminate metastatic KPN tumors. An increase in the gene expression of Nos2 was observed in the combination treatment group when compared to vehicle. Nos2 has a dichotomous role in modulating tumor immune response as it is induced by interferon gamma and is often associated with tumorigenesis and poor prognosis in cancer.Citation33,Citation34 In addition to Nos2, an increase in the expression of cancer-associated metabolic genes (Nqo1, Hmgsc1, Pla2g7, Sult1a1, Ugt1A5, Ugt2B5, Abcb1a) indicates reprogramming of tumor metabolic pathways, potentially leading to the observed tumor resistance.Citation35–42 Abcb1 encodes the ABC transporter protein, which confers multidrug resistance in cancer, fosters cancer stem cell-like properties, and facilitates epithelial–mesenchymal transition.Citation43 Interestingly, inhibiting ABCB1 overcomes acquired resistance of non-small cell lung cancer cells to MET inhibitors.Citation41
The metabolic genes, Nqo1, Hmgcs1, Pla2g7, and Sult1a1, encode enzymes such as cytosolic reductase, mevalonate, phospholipase, and sulfotransferase. Like Abcb1, upregulation of these genes is closely linked with the advancement of cancer and its metastatic spread.Citation35,Citation37,Citation39,Citation42 Some of these metabolic genes modulate anti-tumor immune responses. For instance, Sult1a1 contributes to immune exclusion, which manifests as a negative correlation with crucial immune cell populations including CD8+ cells, CD4+ cells, macrophages, and neutrophils.Citation42 Furthermore, therapeutic targeting of NQO1 and HMGCS1 can augment T-cell responses by fostering pyroptosis, an inflammatory form of cell death, and ultimately enhancing anti-tumor immunity.Citation44,Citation45 Recent findings suggest that PLA2G7high macrophages create an immunosuppressive microenvironment in the tumor and hinder CD8 T-cell activation, and its inhibition enhances efficacy of anti-PD1 treatment.Citation46 These findings highlight the therapeutic potential of modulating metabolic pathways to boost the immune response against cancer.
Another colon cancer metastasis mouse model, AKPT, showed tumor regression after inhibition of TGF-β and PD-1/PD-L1.Citation19 Enhanced Notch 1 activation and neutrophil infiltration differentiates the KPN model used here from other CRC models, including AKPT, and these factors may influence the therapeutic response. In a phase II trial for CMS4 mCRC, patients received bintrafusp alfa, a dual PD-L1 antibody/TGFβ trap alongside radiation. Though changes in IFN-γ signature was observed, efficacy was low.Citation47 This correlates with our observation of combination treatment in the KPN tumors. Therefore, aggressive mCRC will require additional treatment along with TGF-β and PD-L1 inhibitors. GEMMs like KPN, resembling human CRC,Citation48 will continue to be helpful in evaluating pre-clinical efficacy and immune modulatory mechanisms in hard-to-treat metastatic colorectal cancer.
Materials and methods
KPN organoid culture
KPN liver metastasis organoids were generated as previously described.Citation14 Organoids were cultured in Base Medium including Supplements (BMS), with 50 ng/mL Human EGF and 100 ng/mL murine noggin (Peprotech, USA). Organoids were mixed with Growth Factor Reduced Matrigel (Corning, USA) before seeding. Organoids were frozen in Recovery cell culture freezing medium (ThermoFisher, USA) and thawed as required.
For in vitro assays, organoids (BVKPN RKAC13.1e) were trypsinized with TrypLE and washed with 1 mL FACS buffer (PBS +2%FBS +1 mM EDTA). For flow cytometry, cells were stained with fixable viability dye (FVD) (ThermoFisher, USA) and anti-mouse EpCAM (clone G8.8, Biolegend, USA). Supernatant from KPN organoid culture at 72 h was used for TGF-β1 estimation using ELISA (R&D systems, USA). For proliferation assay, KPN cells were washed and seeded in BMS medium. Recombinant mouse TGF-β1 (0.5 ng/mL, R&D systems, USA) or conditioned medium (CM), from organoids cultured for 72 h, was added and proliferation measured after 72 h using cell titer glo (Promega, USA).
Splenocyte proliferation assay
Splenocytes were harvested from C57BL/6 mice and frozen in RPMI with 90%FBS (Gibco, USA) and 10% DMSO (Sigma, USA). On the day of the assay, frozen cells were revived in RPMI media containing 10% FBS and seeded in a 96 well plate as 1–2×105 million cells/well. The cells were stimulated (St) using plate-coated anti-CD3 (0.5 μg/mL) in the presence of either TGF-β1 (0.5 ng/mL), or 50 μL of CM. Some wells were supplemented with anti-TGF-β1 antibody (10 μg/mL, Bioxcell, USA) or ALK5i (Galunisertib,10 μM, Tocris, U.K). After 72 h, proliferation was measured by cell titer glo (Promega, USA) and cytokine release by IFN-γ ELISA (R&D systems, USA).
T cell re-stimulation assay
To assess the effects on previously activated T cells, frozen splenocytes seeded in anti-CD3 coated (0.5 μg/mL) plates. After 24 h, cells were washed and rested overnight. Cells were re-seeded for anti-CD3 re-stimulation and treated as described above.
Intra-splenic model of metastatic cancer and treatments
Mouse experimental work was carried out in accordance with UK Home Office regulations (Project Licenses 70/8646 and 70/9112), adhered to ARRIVE guidelines, and were subject to ethical review at the University of Glasgow. Injection of liver metastatic KPN organoids were performed as previously describedCitation14 on C57BL/6 mice, 6–7 weeks old (Charles River, UK) under specific pathogen-free conditions. Mice received a single intra-splenic injection of 5 × 105 BVKPN RKAC3.2f cells in 50 µL of phosphate buffer saline (PBS).
ALK5i treatment of KPN metastatic tumour bearing mice
One week after intra-splenic injection mice received either vehicle (n = 5) or ALK5i treatment (n = 5). 50 mg/kg of ALK5i (AstraZeneca AZ12601011) was dosed twice daily by oral gavage in 100 µL of 0.5% Hydroxypropyl Methylcellulose (HPMC) and 0.1% Tween-80 (vehicle). After 3 weeks the mice were euthanized and tissues were collected for analysis.
ALK5i and anti-PD-L1 treatment
One week after intrasplenic injection, mice received (a) rat IgG2b isotype antibody (Bioxcell BE0090) and vehicle (0.5% HPMC/0.1% Tween-80) (n = 6), (b) ALK5i and isotype antibody (n = 6), (c) anti-PD-L1 antibody (Bioxcell BE0101) and vehicle (n = 6), or (d) combination of ALK5i and anti-PD-L1 antibody (n = 6). Anti-PD-L1 and isotype antibody were injected at 10 mg/kg intraperitoneal (IP) twice per week. After 3 weeks, mice were euthanized and tissues were collected for analysis.
Tumour digestion flow cytometry
Macroscopic tumor metastases were dissected from liver tissue, minced (~0.5 mm), and then digested for 30 minutes using 1.5 mg/mL Collagenase V, 0.1 mg/mL of Hyaluronidase (Sigma, USA) and 100 µg/mL DNase I (Roche, Switzerland) in RPMI complete medium. Samples were filtered and the enzyme was neutralized. 1 × 106 cells from each sample were used for Fc blocking (Biolegend, USA) at 4°C followed by staining for a lymphoid panel: viability dye (Thermo, USA), anti-mouse CD45 (30-F11), CD3 (17A2), CD4 (RM4–5), CD8 (53–6.7), CD69 (H1.2F3), PD-1 (29F.1A12), PD-L1 (10F.9G2) and EpCAM (G8.8). For the myeloid panel: viability dye, anti-mouse CD45 (30-F11), B220 (RA3-6B2), CD64 (X54–5/7.1), Ly6C (HK1.4) and MHC-II (M5/114.15.2). All antibodies were from Biolegend, USA. Cells were washed and resuspended in FACS buffer and acquired using BD Fortessa/LSRII/Aria.
Spleen and lymph node digestion
Spleen and mLN were harvested, chopped into fine pieces, and digested using collagenase D (1 mg/mL, Sigma, USA) in complete RPMI, for 25 min at 37°C. Cells were filtered and washed with FACS buffer. RBC lysis was carried out for spleen cells using ACK lysis buffer (Sigma, USA).
Transcription factor staining
2×106 cells were Fc blocked (Biolegend) at 4°C for 20 min, washed, and stained with viability dye (Thermo, USA), anti-mouse CD45 and CD3 (Biolegend, USA). The cells were washed and fixed with fixation/permeabilization reagent (Thermo, USA) for 30 min at RT, washed with permeabilization buffer (Thermo), and stained with anti-mouse T-Bet (4B10, Biolegend, USA).
Intracellular staining
2×106 cells were resuspended in complete RPMI with cell stimulation cocktail (Thermo, USA). After washing and Fc blocking, they were stained with viability dye (Thermo, USA), anti-mouse CD45 (30-F11), CD3 (17A2), CD4 (RM4–5), CD8 (53–6.7), CD69 (H1.2F3), (Biolegend, USA). Cells were washed, fixed and permeabilized (Thermo, USA), and were stained with anti-mouse IFN-γ (XMG1.2) and granzyme B(QA16A02) (Biolegend, USA). Cells were washed and resuspended in permeabilization buffer before acquisition using BD Fortessa or LSRII flow cytometers.
Analysis
Flowjo v10.8.1 was used for the analysis of flow cytometry data.
Immunohistochemistry (IHC)
4 µm FFPE sections of liver with metastases were H&E stained, using standard methods. For Ly6G detection, FFPE sections were deparaffinized and hydrated using xylene, graded alcohol and rinsed with distilled water. Endogenous peroxide was quenched using Bloxall (Vector labs). 0.01 M citrate buffer pH 6.0 was used for antigen retrieval. Anti-mouse Ly6G (Bioxcell BE0075–1), ImmPRESS reagent (Vector Lab), rewashed, and Impact DAB chromogen solution were used to visualize Ly6G+ cells. Sections were counter stained with Haematoxylin. Metastatic areas (~4×105µm2) were analyzed to count Ly6G+ cells using Qupath v0.3.2.
RNA sequencing by TempO-seq
Tumour areas from FFPE liver sections were excised for targeted sequencing-based RNA expression analysis using Tempo-Seq. Metastatic regions from liver sections were pooled (2–3 mice/group) for sequencing. Differential gene expression was calculated using DESEQ2 (1.38.3). Data were explored using SearchlightCitation49(V2.0,0) specifying DE workflow for Vehicle vs PDL1, Vehicle vs ALK5i, and Vehicle vs Combination, and one MDE workflow combining the 3DE. Significance threshold is Wald test with BH correction, adjusted p < 0.05. Over-representation analysis used GO mouse biological process database and TRUSST mouse database for upstream regulator analysis.
Synergistic response of combination treatment over vehicle was calculated using formula: {expected = mean (ALK5i-Vehicle) + (aPDL1-Vehicle), observed = mean (combination), synergy = ratio of observed/expected}, and confirmed using Bliss analysis (EAB=EA+EB-EA*EB), where EA=ALK5i and EB=anti PD-L1 and EAB is the additive effect. For a gene to be synergistic, counts in combination treatment group > EAB.
Statistical analysis
In vitro data were analyzed as mean±SD and statistical analyses was performed using One-way ANOVA and Tukey’s multiple comparison test. Statistical analysis between two in vivo groups was done using an unpaired Mann Whitney test. Data between four treatment groups were analyzed using Kruskal Wallis with Dunn’s multiple comparison test. Analysis was performed with GraphPad Prism software and p-value *<0.05 was considered significant. Sample sizes are included in figure legends.
Author contributions
RN, TL, AA, CB performed experiments. RN, TL, and SM conceived and planned elements of the study. RN analyzed the data and wrote the manuscript. JC analyzed the RNA sequencing data. SM, OS, RJBN, RJ, TL, AA, and JC reviewed the manuscript and provided critical comments.
Supplemental Material
Download Zip (18.4 MB)Acknowledgments
We thank Biocon limited, Bengaluru, India for supporting this programme of research. We thank Professor William Cushley and Dr. Seth Coffelt for helpful discussions; Rachel Ridgway for her timely support with in vivo experiments; and Diane Vaughan, Colin Nixon, and Fiona McMonagle for their support with the University of Glasgow Flow Core Facility and immunohistochemistry.
Disclosure statement
RN was employed with Biocon Limited, Bengaluru, India during this work.
Data availability statement
The data generated in this study are available upon request from the author.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/2162402X.2024.2330194.
Additional information
Funding
References
- Riihimaki M, Hemminki A, Sundquist J, Hemminki K. Patterns of metastasis in colon and rectal cancer. Sci Rep. 2016;6(1):6. doi:10.1038/srep29765.
- Pretzsch E, Bösch F, Neumann J, Ganschow P, Bazhin A, Guba M, Werner J, Angele M. Mechanisms of metastasis in colorectal cancer and metastatic organotropism: hematogenous versus peritoneal spread. J Oncol. 2019 Sep;19:1–13. doi:10.1155/2019/7407190.
- Hammond WA, Swaika A, Mody K. Pharmacologic resistance in colorectal cancer: A review. Ther Adv Med Oncol. 2016 Jan;8(1):57–84. doi:10.1177/1758834015614530.
- Koyama S, Akbay EA, Li YY, Herter-Sprie GS, Buczkowski KA, Richards WG, Gandhi L, Redig AJ, Rodig SJ, Asahina H, et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun. 2016 Feb;17(7):10501. doi:10.1038/ncomms10501.
- Kim SY, Kim TW. Current challenges in the implementation of precision oncology for the management of metastatic colorectal cancer. ESMO Open. 2020 Mar;55(2):e000634. doi:10.1136/esmoopen-2019-000634.
- Knickelbein K, Zhang L. Mutant KRAS as a critical determinant of the therapeutic response of colorectal cancer. Genes Dis. 2015 Mar;2(1):4–12. doi:10.1016/j.gendis.2014.10.002.
- Meng M, Zhong K, Jiang T, Liu Z, Kwan HY, Su T. The current understanding on the impact of KRAS on colorectal cancer. Biomed Pharmacother. 2021 Aug;140:111717. doi:10.1016/j.biopha.2021.111717.
- Schmitt M, Greten FR. The inflammatory pathogenesis of colorectal cancer. Nat Rev Immunol. 2021 Oct;21(10):653–667. doi:10.1038/s41577-021-00534-x.
- Dos Santos W, Sobanski T, de Carvalho AC, Evangelista AF, Matsushita M, Berardinelli GN, de Oliveira MA, Reis RM, Guimarães DP. Mutation profiling of cancer drivers in Brazilian colorectal cancer. Sci Rep. 2019;9(1): doi:10.1038/s41598-019-49611-1.
- Philipovskiy A, Ghafouri R, Dwivedi AK, Alvarado L, McCallum R, Maegawa F, Konstantinidis IT, Hakim N, Shurmur S, Awasthi S, et al. Association between tumor mutation profile and clinical outcomes among hispanic-latino patients with metastatic colorectal cancer. Front Oncol. 2022;11:11. doi:10.3389/fonc.2021.772225.
- Dienstmann R, Vermeulen L, Guinney J, Kopetz S, Tejpar S, Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer. 2017 Feb;17(2):79–92. doi:10.1038/nrc.2016.126.
- Batlle E, Massagué J. Transforming growth factor-β signaling in immunity and cancer. Immunity. 2019 Apr 16;50(4):924–940. doi:10.1016/j.immuni.2019.03.024.
- Marisa L, Blum Y, Taieb J, Ayadi M, Pilati C, Le Malicot K, Lepage C, Salazar R, Aust D, Duval A, et al. Intratumor CMS heterogeneity impacts patient prognosis in localized colon cancer. Clin Cancer Res. 2021;27(17):4768–4780. doi:10.1158/1078-0432.CCR-21-0529.
- Jackstadt R, van Hooff SR, Leach JD, Cortes-Lavaud X, Lohuis JO, Ridgway RA, Wouters VM, Roper J, Kendall TJ, Roxburgh CS, et al. Epithelial NOTCH signaling rewires the tumor microenvironment of colorectal cancer to drive poor-prognosis subtypes and metastasis. Cancer Cell. 2019 Sep 16;36(3):319–336.e7. doi:10.1016/j.ccell.2019.08.003.
- Hamarsheh S, Groß O, Brummer T, Zeiser R. Immune modulatory effects of oncogenic KRAS in cancer. Nat Commun. 2020;11(1). doi:10.1038/s41467-020-19288-6.
- Kennedy MC, Lowe SW. Mutant p53: it’s not all one and the same. Cell Death Differ. 2022 May;2929(5):983–987. doi:10.1038/s41418-022-00989-y.
- Kubiczkova L, Sedlarikova L, Hajek R, Sevcikova S. TGF-β – an excellent servant but a bad master. J Transl Med. 2012 Sep 3;10(1):183. doi:10.1186/1479-5876-10-183.
- Calon A, Espinet E, Palomo-Ponce S, Tauriello DVF, Iglesias M, Céspedes MV, Sevillano M, Nadal C, Jung P, Zhang X-F, et al. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22(5):571–584. doi:10.1016/j.ccr.2012.08.013.
- Tauriello DVF, Palomo-Ponce S, Stork D, Berenguer-Llergo A, Badia-Ramentol J, Iglesias M, Sevillano M, Ibiza S, Cañellas A, Hernando-Momblona X, et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 2018;554(7693):538–543. doi:10.1038/nature25492.
- Spender LC, Ferguson GJ, Hughes GD, Davies BR, Goldberg FW, Herrera B, Taylor RG, Strathearn LS, Sansom OJ, Barry ST, et al. Preclinical evaluation of AZ12601011 and AZ12799734, inhibitors of transforming growth factor β superfamily type 1 receptors. Mol Pharmacol. 2019;95(2):222–234. doi:10.1124/mol.118.112946.
- Yingling JM, McMillen WT, Yan L, Huang H, Sawyer JS, Graff J, Clawson DK, Britt KS, Anderson BD, Beight DW, et al. Preclinical assessment of galunisertib (LY2157299 monohydrate), a first-in-class transforming growth factor-β receptor type I inhibitor. Oncotarget. 2017 Dec 31;9(6):6659–6677. doi:10.18632/oncotarget.23795.
- Patsoukis N, Wang Q, Strauss L, Boussiotis VA. Revisiting the PD-1 pathway. Sci Adv. 2020 Sep 18;6(38):eabd2712. doi:10.1126/sciadv.abd2712.
- Saleh R, Taha RZ, Toor SM, Sasidharan Nair V, Murshed K, Khawar M, Al-Dhaheri M, Petkar MA, Abu Nada M, Elkord E, et al. Expression of immune checkpoints and T cell exhaustion markers in early and advanced stages of colorectal cancer. Cancer Immunol Immun. 2020;69(10):1989–1999. doi:10.1007/s00262-020-02593-w.
- Klug F, Prakash H, Huber PE, Seibel T, Bender N, Halama N, Pfirschke C, Voss R, Timke C, Umansky L, et al. Low-dose irradiation programs macrophage differentiation to an iNOS+/M1 phenotype that orchestrates effective T cell immunotherapy. Cancer Cell. 2013;24(5):589–602. doi:10.1016/j.ccr.2013.09.014.
- Guo Y, Li Q, Zhao G, Zhang J, Yuan H, Feng T, Ou D, Gu R, Li S, Li K, et al. Loss of TRIM31 promotes breast cancer progression through regulating K48- and K63-linked ubiquitination of p53. Cell Death Disease. 2021;12(10): doi:10.1038/s41419-021-04208-3.
- Bhullar DS, Barriuso J, Mullamitha S, Saunders MP, O’Dwyer ST, Aziz O. Biomarker concordance between primary colorectal cancer and its metastases. EBioMedicine. 2019;40:363–374. doi:10.1016/j.ebiom.2019.01.050.
- Filippi CM, Juedes AE, Oldham JE, Ling E, Togher L, Peng Y, Flavell RA, von Herrath MG. Transforming growth factor-β suppresses the activation of CD8+ T-Cells when naive but promotes their survival and function Once antigen experienced. Diabetes. 2008 Oct;57(10):2684–2692. doi:10.2337/db08-0609.
- Ahmadzadeh M, Rosenberg SA. TGF-β1 attenuates the acquisition and expression of effector function by tumor Antigen-Specific Human Memory CD8 T cells. J Immunol. 2005;174(9):5215–5223. doi:10.4049/jimmunol.174.9.5215.
- Cottrez F, Groux H. Regulation of TGF-β response during T cell activation is modulated by IL-10. J Immunol. 2001 Jul 15;167(2):773–8. doi:10.4049/jimmunol.167.2.773.
- Bain CC, Bravo-Blas A, Scott CL, Gomez Perdiguero E, Geissmann F, Henri S, Malissen B, Osborne LC, Artis D, Mowat AM, et al. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat Immunol. 2014 Oct;15(10):929–937. doi:10.1038/ni.2967.
- Schridde A, Bain CC, Mayer JU, Montgomery J, Pollet E, Denecke B, Milling SWF, Jenkins SJ, Dalod M, Henri S, et al. Tissue-specific differentiation of colonic macrophages requires TGFβ receptor-mediated signaling. Mucosal Immunol. 2017;10(6):1387–1399. doi:10.1038/mi.2016.142.
- Prizment AE, Vierkant RA, Smyrk TC, Tillmans LS, Nelson HH, Lynch CF, Pengo T, Thibodeau SN, Church TR, Cerhan JR, et al. Cytotoxic t cells and granzyme b associated with improved colorectal cancer survival in a prospective cohort of older women. Cancer Epidem Biomar. 2017;26(4):622–631. doi:10.1158/1055-9965.EPI-16-0641.
- de Oliveira GA, Cheng RYS, Ridnour LA, Basudhar D, Somasundaram V, McVicar DW, Monteiro HP, Wink DA. Inducible nitric oxide synthase in the carcinogenesis of gastrointestinal cancers. Antioxid Redox Signal. 2017 Jun 20;26(18):1059–1077. doi:10.1089/ars.2016.6850.
- Cheng RYS, Ridnour LA, Wink AL, Gonzalez AL, Femino EL, Rittscher H, Somasundaram V, Heinz WF, Coutinho L, Rangel MC, et al. Interferon-gamma is quintessential for NOS2 and COX2 expression in ER- breast tumors that lead to poor outcome. Cell Death Disease. 2023 May 11;14(5):319. doi:10.1038/s41419-023-05834-9.
- Yang Y, Zhu G, Dong B, Piao J, Chen L, Lin Z. The NQO1/PKLR axis promotes lymph node metastasis and breast cancer progression by modulating glycolytic reprogramming. Cancer Lett. 2019;453:170–183. doi:10.1016/j.canlet.2019.03.054.
- Oh ET, Kim JW, Kim JM, Kim SJ, Lee JS, Hong SS, Goodwin J, Ruthenborg RJ, Jung MG, Lee H-J, et al. NQO1 inhibits proteasome-mediated degradation of HIF-1α. Nat Commun. 2016;7(1): doi:10.1038/ncomms13593.
- Walsh CA, Akrap N, Garre E, Magnusson Y, Harrison H, Andersson D, Jonasson E, Rafnsdottir S, Choudhry H, Buffa F, et al. The mevalonate precursor enzyme HMGCS1 is a novel marker and key mediator of cancer stem cell enrichment in luminal and basal models of breast cancer. PLoS One. 2020;15(7):e0236187. doi:10.1371/journal.pone.0236187.
- Pandyra AA, Mullen PJ, Goard CA, Ericson E, Sharma P, Kalkat M, Yu R, Pong JT, Brown KR, Hart T, et al. Genome-wide RNAi analysis reveals that simultaneous inhibition of specific mevalonate pathway genes potentiates tumor cell death. Oncotarget. 2015;6(29):26909–26921. doi:10.18632/oncotarget.4817.
- Vainio P, Lehtinen L, Mirtti T, Hilvo M, Seppänen-Laakso T, Virtanen J, Sankila A, Nordling S, Lundin J, Rannikko A, et al. Phospholipase PLA2G7, associated with aggressive prostate cancer, promotes prostate cancer cell migration and invasion and is inhibited by statins. Oncotarget. 2011;2(12):1176–1190. doi:10.18632/oncotarget.397.
- Allain EP, Rouleau M, Lévesque E, Guillemette C. Emerging roles for UDP-glucuronosyltransferases in drug resistance and cancer progression. Br J Cancer. 2020 Apr;122122(9):1277–1287. doi:10.1038/s41416-019-0722-0.
- Sugano T, Seike M, Noro R, Soeno C, Chiba M, Zou F, Nakamichi S, Nishijima N, Matsumoto M, Miyanaga A, et al. Inhibition of ABCB1 overcomes cancer stem cell–like properties and acquired resistance to MET inhibitors in non–small cell lung cancer. Mol Cancer Ther. 2015;14(11):2433–2440. doi:10.1158/1535-7163.MCT-15-0050.
- Li Y, Ding Q, Xiong Z, Wen H, Feng C. Overexpression of steroid sulfotransferase genes is associated with worsened prognosis and with immune exclusion in clear cell-renal cell carcinoma. Aging. 2019;11(20):9209–9219. doi:10.18632/aging.102392.
- Vaidyanathan A, Sawers L, Gannon AL, Chakravarty P, Scott AL, Bray SE, Ferguson MJ, Smith G. ABCB1 (MDR1) induction defines a common resistance mechanism in paclitaxel- and olaparib-resistant ovarian cancer cells. Br J Cancer. 2016;115(4):431–441. doi:10.1038/bjc.2016.203.
- Li X, Liu Z, Zhang A, Han C, Shen A, Jiang L, Boothman DA, Qiao J, Wang Y, Huang X, et al. NQO1 targeting prodrug triggers innate sensing to overcome checkpoint blockade resistance. Nat Commun. 2019;10(1): doi:10.1038/s41467-019-11238-1.
- Zhou W, Liu H, Yuan Z, Zundell J, Towers M, Lin J, Lombardi S, Nie H, Murphy B, Yang T, et al. Targeting the mevalonate pathway suppresses ARID1A-inactivated cancers by promoting pyroptosis. Cancer Cell. 2023;41(4):740–756.e10. doi:10.1016/j.ccell.2023.03.002.
- Zhang F, Liu W, Meng F, Jiang Q, Tang W, Liu Z, Lin X, Xue R, Zhang S, Dong L. Inhibiting PLA2G7 reverses the immunosuppressive function of intratumoral macrophages and augments immunotherapy response in hepatocellular carcinoma. J Immunother Cancer. 2024 Jan 25;12(1):e008094. doi:10.1136/jitc-2023-008094.
- Mehrvarz Sarshekeh A, Lam M, Zorrilla IR, Holliday EB, Das P, Kee BK, Overman MJ, Parseghian CM, Shen JPYC, Tam A, et al. Consensus molecular subtype (CMS) as a novel integral biomarker in colorectal cancer: a phase II trial of bintrafusp alfa in CMS4 metastatic CRC. J Clin Oncol. 2020;38(15_suppl):4084–4084. doi:10.1200/JCO.2020.38.15_suppl.4084.
- Amirkhah R, Gilroy K, Malla SB, Lannagan TRM, Byrne RM, Fisher NC, Corry SM, Mohamed N-E, Naderi-Meshkin H, Mills ML, et al. MmCMS: mouse models’ consensus molecular subtypes of colorectal cancer. Br J Cancer. 2023;128(7):1333–1343. doi:10.1038/s41416-023-02157-6.
- Cole JJ, Faydaci BA, McGuinness D, Shaw R, Maciewicz RA, Robertson NA, Goodyear CS. Searchlight: automated bulk RNA-seq exploration and visualisation using dynamically generated R scripts. BMC Bioinf. 2021;22(1): doi:10.1186/s12859-021-04321-2.
