ABSTRACT
Deregulation or loss of the human leukocyte antigen class I (HLA-I) molecules on tumor cells leading to inhibition of CD8+ T cell recognition is an important tumor immune escape strategy, which could be caused by a posttranscriptional control of molecules in the HLA-I pathway mediated by RNA-binding proteins (RBPs). So far, there exists only limited information about the interaction of RBPs with HLA-I-associated molecules, but own work demonstrated a binding of the heterogeneous ribonucleoprotein C (hnRNP C) to the 3’ untranslated region (UTR) of the TAP-associated glycoprotein tapasin (tpn). In this study, in silico analysis of pan-cancer TCGA datasets revealed that hnRNP C is higher expressed in tumor specimens compared to corresponding normal tissues, which is negatively correlated to tpn expression, T cell infiltration and the overall survival of tumor patients. Functional analysis demonstrated an upregulation of tpn expression upon siRNA-mediated downregulation of hnRNP C, which is accompanied by an increased HLA-I surface expression. Thus, hnRNP C has been identified to target tpn and its inhibition could improve the HLA-I surface expression on melanoma cells suggesting its use as a possible biomarker for T-cell-based tumor immunotherapies.
Introduction
During the last two decades, tumor immunotherapy has rapidly developed, but still has a limited efficacy in solid tumors including melanoma.Citation1 Although immune checkpoint inhibitors (ICPi) represent an important pillar of the current immunotherapeutic options and combination therapies, which have dramatically changed the outcome of melanoma patients, long-term success of this treatment is only reported for <20% of patients.Citation2–4 Therefore, novel immunotherapeutic targets and better insights into the mechanisms associated with melanoma development and immune evasion are urgently required.
A few tumor immunotherapies, including cancer vaccines, ICP blockade and chimeric antigen receptor (CAR) T cells, are currently FDA approved or implemented in clinical studies, and the different tumor immunotherapeutic strategies have some disadvantages and not every patient will respond to this treatment option.Citation5 The characterization of the underlying mechanisms of tumor immune escape is the cornerstone for the selection of the appropriate immunotherapeutic approach. In addition to the expression of inhibitory ICP, the loss or downregulation of molecules of the human leukocyte antigen class I (HLA-I) antigen processing and presentation pathway is a major immune evasion process, since their deficiencies block the presentation of tumor-associated antigens to CD8+ cytotoxic T lymphocytes (CTL).Citation6 A number of studies demonstrated an impaired expression of different components of the HLA-I antigen processing and presentation machinery (APM) in multiple tumor types at distinct tumor stages.Citation7,Citation8 However, the underlying mechanisms of the absent and/or reduced function of HLA-I APM components are not uniform and significantly vary including the changes at the genetic, transcriptional, posttranscriptional, posttranslational and epigenetic levels.Citation9,Citation10 In this context, the posttranscriptional regulation of the expression of HLA-I APM components mediated by microRNAs (miRNAs) and RNA-binding proteins (RBPs) has recently rekindled interest.Citation11–13
RBPs are proteins binding to the RNA sequences including in cells thereby participating in the formation of ribonucleoprotein (RNP) complexes.Citation14,Citation15 Due to their wide range of regulatory functions, RBPs play an important role not only in physiological process but also in the development of many diseases.Citation16,Citation17 There exists evidence that RBPs also have crucial functions in tumors based on their deregulation in different tumor specimens compared to normal tissues.Citation18 RBPs have been shown to modulate tumor metastasis and epithelial mesenchymal transition (EMT) progression. For example, PCBP1 regulates breast cancer invasiveness,Citation19 heterogeneous ribonucleoprotein A1 ((hnRNP) A1 affects hepatocellular carcinoma migrationCitation20 and MEX3A promotes angiogenesis in colorectal cancer.Citation21 Nevertheless, there exists only limited information about the interaction of RBPs with HLA-I-associated molecules. So far, only MEX3B and Syncrip have been identified to target HLA-A.Citation13,Citation22 In addition, hnRNP R has been shown as a positive regulator of both classical HLA-I and the non-classical HLA-G antigens, thereby modulating the NK cell cytotoxicity.Citation23 Other HLA-I APM components, such as tapasin (tpn), have not yet been investigated regarding their association with RBPs. Tpn is an important component of the HLA-I APM that coordinates with the transporter associated with antigen processing (TAP) proteins the peptide loading onto HLA-I molecules, which is a key determinant for successful immune responses.Citation24 New studies reveal a selector function of the tpn-ERp57 complex by regulating the peptide kinetic stability, peptide abundance and peptide proofreading,Citation25,Citation26 suggesting an important role of tpn for peptide loading and editing processes on HLA-I molecules. Own previous mass spectrometric data revealed that the heterogeneous nuclear ribonucleoprotein C1/C2 (hnRNP C), an RBP with heterogeneous nuclear RNA (hnRNA) and associated with pre-mRNAs in the nucleus thereby influencing the pre-mRNA processing,Citation27 targets the 3’ untranslated region (UTR) of tpn. Based on its wide range of functions, in particular the modulation of growth properties, recent studies have linked the expression of this RBP to the development of many tumors,Citation28–30 including lung cancer,Citation31 gastric cancerCitation32 and hepatocellular carcinoma.Citation33 In melanoma, hnRNP C increases the tumor cell migration and invasion via the interaction with several proteins.Citation34–36 However, the role of hnRNP C on the expression and function of immune modulatory molecules remains to be explored.
In this study, we identified that hnRNP C can bind to tpn, leading to an inhibition of its expression, while knockdown of hnRNP C can upregulate tpn expression, thereby enhancing HLA-I surface expression and CD8+ T cell recognition. These experimental data were confirmed by bioinformatics analyses of the Cancer Genome Atlas (TCGA) and might have important implications for the implementation of T cell-based immunotherapies.
Methods and materials
Cell culture and transfection
The human metastatic melanoma cell lines Buf1379 and Buf1402 were kindly provided by Soldano Ferrone (Department of Surgery, Massachusetts General Hospital, Harvard Medical School, Boston, MA, USA). Both melanoma cell lines were cultured in Roswell Park Memorial Institute 1640 medium (RPMI1640, Invitrogen, Carlsbad, CA, USA) at 37°C in 5% CO2 humidified air supplemented with 10% fetal calf serum (FCS) (PAN, Aidenbach, Germany), 3% L-glutamine (Lonza, Basel, Switzerland) and 3% pen/strep (Sigma-Aldrich, Missouri, USA).
For transfection, 2.5 × 105 melanoma cells/well were seeded into six-well plates. After 16–20 h, cells were transfected with siRNA (si-hnRNP C, 2 ng/ml, EHU133931, Sigma-Aldrich, St. Louis, MO, USA) or the negative control (NC, EHUEGFP, Sigma-Aldrich) using lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. Furthermore, untreated parental melanoma cells served as additional control. The cells were collected 48 h after transfection or approx. 64 h after seeding for subsequent RNA, protein and flow cytometric analyses.
RNA isolation and real-time quantitative reverse-transcription PCR (RT-qPCR)
Total RNA was isolated using the TRIzol reagent (Invitrogen) following the manufacturer’s instruction and then subjected to cDNA synthesis (Thermo Scientific, Rockford, IL, USA). For RT-qPCR, the SYBR qPCR master mix (Vazyme, Nanjing, PRC) was used with the primers listed in Table S1, and the data were normalized to the housekeeping genes including β-actin, delta-amino levulinate synthase (ALSA1) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and mRNA levels were normalized to parental cells.
Protein extraction and Western blot analysis
Total cellular protein was isolated with a lysis buffer (Thermo Scientific) containing protease and phosphatase inhibitors (Thermo Scientific) followed by the quantification of proteins with the Pierce BCA Protein Assay kit (Thermo Scientific). About 25 µg protein/sample was separated in Bolt™ 4 to 12% mini protein gels (Invitrogen) and transferred onto membranes through Blot2 Transfer Stacks (Invitrogen). Subsequently, the membranes were blocked with milk for 1 h at room temperature and incubated with the following primary antibodies (Abs) overnight at 4°C: anti-HNRNPC (1:1000, PA5-24221 Thermo Scientific), anti-tpn (1:1000, ab13518, Abcam, Cambridge, UK), anti-β-actin (1:1000, ab6276, Abcam) and anti-HLA-I heavy chain (HC) (1:750; HC-10), which was kindly provided by Prof. Dr Soldano Ferrone (Harvard University, Boston, USA). This was followed by staining of the membranes with a horseradish peroxidase-conjugated goat anti-mouse/rabbit Ab (Cell Signaling Technology, Inc., Danvers, USA) as a secondary Ab for 1 h at room temperature. A LAS-3000 imaging system (Fujifilm, Tokyo, Japan) was employed to image the chemiluminescent blots. The signal intensity was determined by densitometric analysis using ImageJ (NIH, Bethesda, Maryland, USA) and normalized to the β-actin staining.
Flow cytometry
For determination of the HLA-I surface expression by flow cytometry, 1 × 105–1 × 106 cells were incubated with an anti-HLA-ABC (Beckman, Brea, California, USA) or anti-HLA-BC (Invitrogen) mAb, respectively, for 15 min at room temperature, then washed with PBS and measured on a Navios 3L10C (Beckman Coulter GmbH, Krefeld, Germany) or a LSRFortessa (BD Biosciences, Heidelberg, Germany) flow cytometer. The data were evaluated with the Kaluza (Beckman Coulter) or FACS Diva (BD Biosciences) analysis software and calculated as mean specific fluorescence intensity (MFI).
Gene set enrichment analysis (GSEA)
To explore the hnRNP C-mediated regulation of gene expression, the RNA-seq dataset from 67 metastatic melanoma cases from the TCGA-SKCM dataset was analyzed using the GSEA software (UC SanDiego, Broad Institute).Citation37 The molecular signature database (MSigDB) was used to determine which gene sets correlated with the hnRNP C expression.
Bioinformatics and statistical analysis
The probability of melanoma patients’ overall survival (OS) in dependence of hnRNP C expression was determined by the Kaplan–Meier estimation employing 459 cases from the “SKCM Cancer” and “Bhardwaj” and 44 cases of a metastatic melanoma patient’s dataset from R2 database (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi). In addition, using 67 samples of distantly metastasized melanoma from the TCGA-SKCM dataset, the link between immune infiltration and the expression levels of HnRNP Chigh and hnRNP Clow was determined by CIBERSORT. For the correlation of the hnRNP C expression, in pan-cancer with tpn as well as MHC-I expression, GAPIA, ENCORI and the GSEA web tools were employed. The data were presented as mean ± standard deviation (SD) and analyzed by GraphPad Prism (GraphPad Software, LLC, San Diego, USA), ImageJ software and Microsoft Office (Microsoft Corporation, Redmond, WA, USA). Paired and/or unpaired t-tests were performed. p < 0.05 (*), p < 0.01 (**) or p < 0.001 (***) were statistically significant.
Results
Clinical relevance of hnRNP C expression regarding the survival of pan-cancer patients
Since hnRNP C overexpression was reported in several cancer types including melanoma, the prognostic relevance of hnRNP C expression in melanoma patients was investigated using the datasets “R2: Tumor Melanoma – Bhardwaj − 44 – MAS5.0 – u133p2” and “R2: Tumor Melanoma – TCGA − 470 – rsem – tcgars” from the R2 genomic analysis web tool. In both datasets, lower hnRNP C mRNA transcript levels were correlated with a higher OS probability (), suggesting an association of lower hnRNP C expression with a better OS of melanoma patients. These data were in line with a majority of cancer types also demonstrating a correlation of increased patients’ survival with reduced hnRNP C expression ( and Figure S1). In contrast, the hnRNP C expression levels were not linked to the patients’ survival in gastric and ovarian cancers (Figure S2), while the expression of hnRNPC positively correlated with the patients’ survival in lung and breast carcinoma (Figure S3). Comparison of the expression of hnRNP C in different tumor types to that of corresponding normal tissues using the TCGA dataset analyzed with the GAPIA web tool demonstrated higher hnRNP C transcript levels in most cancer types than in the corresponding non-neoplastic tissues suggesting that hnRNP C expression has a possible tumor-promoting effect (). Similar results were also obtained in vitro with higher hnRNP C expression in Buf1379 and Buf1402 melanoma cell lines than in melanocytes (Figure S4). In contrast, for acute myeloid leukemia (LAML), subsequent correlation analyses suggest that hnRNP C has no tumor-promoting effect in this disease.
Figure 1. Correlation of increased patients’ overall survival with reduced hnRNP C expression in different cancer types including melanoma.
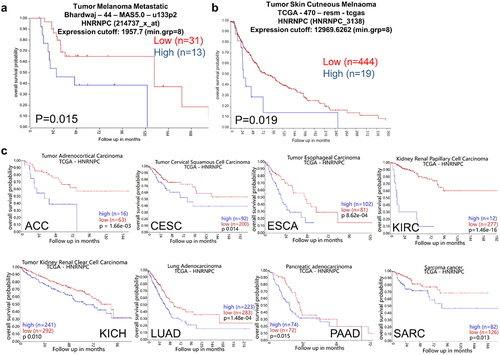
Figure 2. Higher hnRNP C expression levels in tumor samples compared to paired tumor adjacent samples in pan-cancer.
Correlation between hnRNP C, tpn and HLA-I expression in pan-cancer than adjacent normal tissue
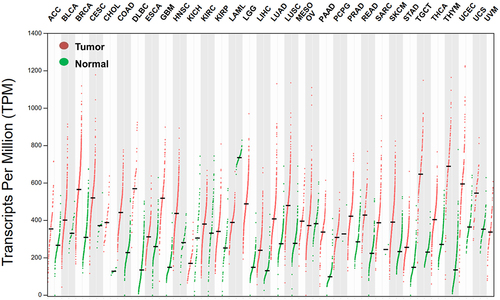
Since hnRNP C has been shown to bind to the 3’UTR of tpn, the link between hnRNP C mRNA levels and tpn expression was analyzed in the TCGA dataset using the ENCORI web tool. A negative correlation of both genes was found in 13 different cancer types including melanoma with a high correlation coefficient (R value) (), suggesting that the binding of hnRNP C to the 3’UTR of tpn inhibits its expression. In contrast, there exists no correlation between hnRNP C and tpn in some tumors, such as breast cancer (Figure S5A), while, e.g., in pancreatic cancer, a positive correlation was detected (Figure S5B). To further verify the relationship between hnRNP C and MHC class I APM components, GSEA was performed using 67 metastatic melanoma cases from the TCGA dataset. This bioinformatics analysis in combination with the Molecular Signatures Database demonstrated a negative correlation of hnRNP C mRNA levels and the MHC class I complex “GOBP-PEPTIDE-ANTIGEN-ASSEMBLY-WITH-MHC-PROTEIN-COMPLEX” (Figure S6, Table S2), thereby strengthening the evidence of an inverse correlation of hnRNP C and tpn.
Figure 3. Inverse correlation of hnRNP C and tpn expression in different cancer types.
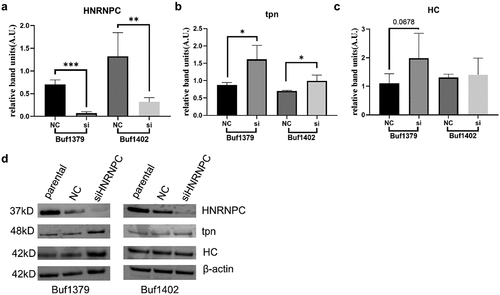
Upregulation of tpn expression, but not of other HLA-I APM components by knockdown of hnRNP C
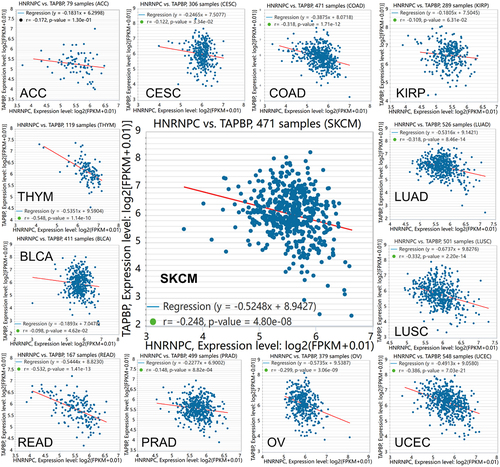
To gain functional insights into the hnRNP C-mediated tpn regulation, hnRNP C was silenced in the two metastatic melanoma cell lines Buf1379 and Buf1402. Successful knockdown of hnRNP C () significantly increased the tpn mRNA expression levels (). To investigate whether hnRNP C also influenced other HLA-I APM molecules, the mRNA expression levels of TAP1, TAP2, β2-microglobulin (β2m) and HLA-I HC were determined by q-PCR in hnRNP Clow (hnRNP C knockdown) vs. hnRNP Chigh (NC, negative controls) cells. As shown in , the mRNA levels of TAP1, TAP2, and β2m of the hnRNP Chigh and hnRNP Clow transfectants were comparable, while the HLA-B and HLA-C mRNA expression levels were slightly altered upon siRNA-mediated silencing of hnRNP C and HLA-A expression was only affected in hnRNP Clow Buf1402 cells. Since interferon-γ (IFN-γ) is master regulator and most potent inducer of major HLA-I APM components,Citation38 the effect of hnRNP C silencing on IFN signaling components was also investigated. As shown in Figure S7, an upregulation of JAK1, STAT2 and STAT1 was detected after hnRNP C knockdown in Buf1379 and Buf1402 cells. Next to mRNA levels, the tpn protein was 2-fold upregulated after hnRNP C knockdown () in both melanoma cells compared to the NC group. Although the non-targeting siRNA also slightly influenced the hnRNP C protein expression in both cell lines, tpn protein expression was significantly altered in hnRNP Clow Buf1402 and Buf1379, which is accompanied by a slight upregulation of the HLA-I HC as determined by staining of the Western blot with the anti-HC10 mAb ().
Figure 4. Upregulation of tpn mRNA levels by knockdown of hnRNP C.
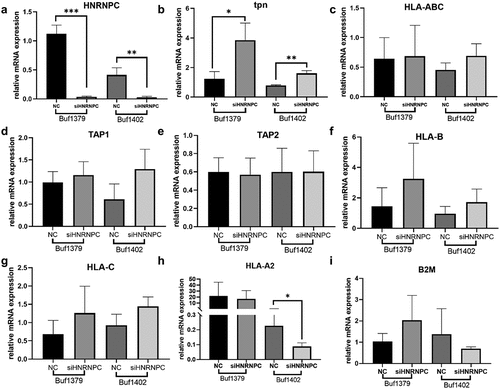
Figure 5. Upregulation of tpn protein levels by knockdown of hnRNP C.
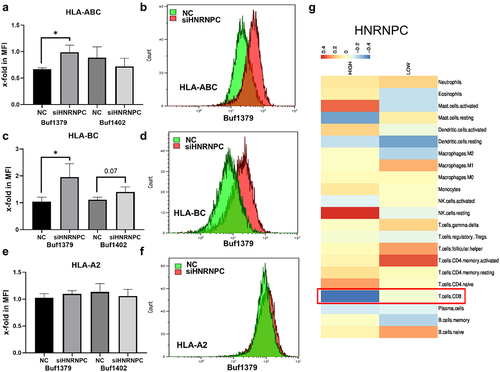
Inverse correlation of hnRNP C and activation of HLA-I antigens and tumor immune cell infiltration
Silencing hnRNP C can upregulate the expression of tpn mRNA and protein level, which is also inversely correlated with HLA-I pathway-associated molecules, as determined by flow cytometry. After hnRNP C knockdown, HLA-ABC and HLA-BC are upregulated on the cell surface of Buf1379 cells (). While only HLA-BC was slightly upregulated in Buf1402 cells (), which correlated with the Western blot results of the HLA-I and for HLA-A, in contrast, hnRNP C silencing did not affect HLA-A2 surface expression in both melanoma cell lines (). Several studies revealed that proteasomal subunitsCitation39 contribute to the development of CD8+ T cells and HLA-I-mediated activation of CD8+ T cells accompanied by IFN-γ production,Citation40 suggesting that hnRNP C inhibits HLA-I pathway components in melanoma and consequently the CD8+ T cell activation. In addition, in breast and lung cancer, the loss of HLA-I and associated molecules significantly correlated with a reduced number of tumor infiltrating T lymphocytes,Citation41 while high HLA-I expression levels were associated with an increased immune cell infiltration.Citation42 Since the HLA-I pathway inhibition by hnRNP C overexpression might affect the immune cell infiltration in melanoma cells, the correlation between hnRNP C expression and immune cell infiltration was investigated using 67 metastatic melanoma cases from the TCGA-SKCM dataset, which were divided into hnRNP Chigh and hnRNP Clow expressors. It is well established that CD8+ T cells recognize antigens presented by HLA-I,Citation43 and HLA-I expression affects CD8+ T cell activation and is associated with the level of T cell infiltration. As shown in , the frequency of CD8+ T cell infiltration was inversely correlated with the hnRNP Chigh group, suggesting that hnRNP C overexpression might impair the CD8+ T cell-mediated tumor recognition due to downregulation of the HLA-I pathway.
Figure 6. Effect of siRNA-mediated downregulation of hnRNP C on HLA-I cell surface expression and immune cell infiltration.
Discussion
Our study focused on the role of hnRNP C in the regulation of tpn mRNA, a component of the HLA class I pathway. In silico analysis as well as experimental approaches showed that hnRNP C binds to the tpn 3’ UTR and down-regulation of hnRNP C significantly increases the expression of tpn thereby improving the expression of HLA-I on the surface of melanoma cells. HnRNP C, a member of the subfamily of ubiquitously expressed hnRNPs, was reported to be highly expressed in most tumor entities, but the underlying molecular mechanisms of hnRNP C function in tumors have not yet been well defined.Citation44 So far, high levels of hnRNP C expression in tumors strongly correlated with the tumor phenotype, like increased proliferation and migration,Citation31,Citation45 but its effect on immunomodulatory molecules has not yet been addressed. Own studies and published data demonstrated that the loss or downregulation of HLA-I molecules in tumors is one major tumor immune escape mechanism.Citation46–48 This abnormality can be caused by gene mutations or loss of heterogeneity of, e.g., HLA-I HC and β2m.Citation49 In addition, epigenetic, transcriptional and/or post-transcriptional regulation can control the HLA-I APM component expression.Citation10,Citation50 These different causes gave rise to the same result: impaired HLA class I surface expression and T cell-mediated immune evasion. So far, some information is available about the impact of miRNAs on the HLA-I molecules,Citation11,Citation47 while the effects of RBPs on these molecules are with few exceptions mainly unexplored.
During the last years, hnRNP C has been reported to regulate the growth properties of tumors,Citation44,Citation51 which was associated with a worse survival of patients.Citation33,Citation52 These data are in accordance with recent studies indicating that lower hnRNP C expression levels were associated with a better survival for patients with different cancer types. In melanoma patients, two TCGA datasets revealed that hnRNP C expression is associated with the patients’ outcome. In addition, hnRNP C transcript levels were higher in most cancer types when compared to paired normal tissues, which confirmed earlier studiesCitation53,Citation54 suggesting that hnRNP C might be an accelerator in these tumors. Furthermore, our study demonstrated a strong negative correlation of hnRNP C with tpn in many cancer types, suggesting a functional association of both molecules. This is in line with the fact that hnRNP C can bind to the tpn 3’UTR.Citation55 Subsequently, additional in silico analyses were performed, which confirmed our results. HnRNP C is negatively linked to the expression of HLA-I, which might be due to the binding of hnRNP C to the tpn 3’ UTR.
In eukaryotes, genes are also subjected to processing, translocation and stabilization prior to translation. The transcriptional process and post-transcriptional co-regulation are strictly controlled by RBPs, which bind to the 3‘UTR of respective mRNAs,Citation56 thereby mainly leading to a reduced mRNA expression accompanied by a diminished protein expression.Citation57,Citation58 Consistent with these findings, hnRNP C impaired tpn expression at the mRNA and protein levels, which was reverted by siRNA-mediated hnRNP C knockdown. In addition, other molecules of this pathway, such as HLA-B and HLA-C, and/or the HLA-I HC mRNA levels were also upregulated upon hnRNP C knockdown. One possible explanation might be that hnRNP C also binds to the HLA-I 3’ UTR accompanied by a reduced HLA-I surface expression, which could be reverted after hnRNP C knockdown. Abnormal expression of hnRNP C in many malignant tumors negatively correlated with patients’ prognosis,Citation28–33 indicating its value as prognostic marker and therapeutic target. Interestingly, Wu and coauthors have summarized the implementation of small-molecule inhibitors targeting RBPs, which makes hnRNP C a possible therapeutic target, but altering its expression or inhibiting its function using small-molecule inhibitors,Citation59 thereby improving HLA-I expression. Thus, hnRNP C not only has the potential as an important molecular biomarker for disease outcome as well as a possible therapeutic target but also as a small inhibitory molecule, which might be used as a single treatment or in combination with immunotherapies to improve the patients’ outcome.
Author contributions
Conceptualization: Y.W., B.S. Methodology Y.W., B.S. Investigation: Y.W., B.S. Visualization: Y.W. Funding acquisition: B.S. Writing: Y.W., B.S.
Abbreviations
| Ab | = | Antibody |
| ALSA1 | = | Delta-amino levulinate synthase |
| APM | = | Antigen processing machinery |
| CAR | = | Chimeric antigen receptor |
| CTL | = | Cytotoxic T lymphocyte |
| GAPDH | = | Glyceraldehyde-3-phosphate dehydrogenase |
| GSEA | = | Gene set enrichment analysis |
| HC | = | Heavy chain |
| HLA | = | Human leukocyte antigen |
| hnRNA | = | Heterogeneous nuclear RNA |
| hnRNP C | = | Heterogeneous nuclear ribonucleoproteins C1/C2 |
| hnRNP | = | Heterogeneous ribonucleoprotein family |
| ICP | = | Immune checkpoint |
| ICPi | = | Immune checkpoint inhibitor |
| MFI | = | Mean fluorescence intensity |
| MHC | = | Major histocompatibility complex |
| miRNA | = | microRNA |
| OS | = | Overall survival |
| RBP | = | RNA-binding protein |
| RNP | = | Ribonucleoprotein |
| SD | = | Standard deviation |
| TAP | = | Transporter associated with antigen processing |
| TCGA | = | The Cancer Genome Atlas |
| tpn | = | Tapasin |
| UTR | = | Untranslated region |
Supplemental Material
Download MS Word (2.2 MB)Acknowledgments
We would like to thank Maria Heise for her excellent secretarial work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All the data are available in the manuscript or the supplementary materials.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/2162402X.2024.2370928
Additional information
Funding
References
- Su D, Kluger H, Olino K. Educational review: clinical application of immune checkpoint blockade for the treatment of melanoma. Ann Surg Oncol. 2023. doi:10.1245/s10434-023-14587-w. [ published Online First: 20231121].
- Sun Q, Hong Z, Zhang C, Wang L, Han Z, Ma D. Immune checkpoint therapy for solid tumours: clinical dilemmas and future trends. Signal Transduct Target Ther. 2023;8(1):320. doi:10.1038/s41392-023-01522-4. [ published Online First: 20230828].
- Hieken TJ, Kreidieh F, Aedo-Lopez V, Block MS, McArthur GA, Amaria RN. Neoadjuvant immunotherapy in melanoma: the paradigm shift. Am Soc Clin Oncol Educ Book. 2023;43(43):e390614. doi:10.1200/EDBK_390614.
- Haslam A, Prasad V. Estimation of the percentage of US patients with cancer who are eligible for and respond to checkpoint inhibitor immunotherapy drugs. JAMA Netw Open. 2019;2(5):e192535. doi:10.1001/jamanetworkopen.2019.2535. [ published Online First: 20190503].
- Jang A, Lichterman JN, Zhong JY, Shoag JE, Garcia JA, Zhang T, Barata PC. Immune approaches beyond traditional immune checkpoint inhibitors for advanced renal cell carcinoma. Hum Vaccin Immunother. 2023;19(3):2276629. doi:10.1080/21645515.2023.2276629. [ published Online First: 20231110].
- Wu X, Li T, Jiang R, Yang X, Guo H, Yang R. Targeting MHC-I molecules for cancer: function, mechanism, and therapeutic prospects. Mol Cancer. 2023;22(1):194. doi:10.1186/s12943-023-01899-4. [ published Online First: 20231202].
- Garrido F, Aptsiauri N. Cancer immune escape: MHC expression in primary tumours versus metastases. Immunol. 2019;158(4):255–11. doi:10.1111/imm.13114. [ published Online First: 20191001].
- Dhatchinamoorthy K, Colbert JD, Rock KL. Cancer immune evasion through loss of MHC class I antigen presentation. Front Immunol. 2021;12:636568. doi:10.3389/fimmu.2021.636568. [ published Online First: 20210309].
- Rene C, Lozano C, Eliaou JF. Expression of classical HLA class I molecules: regulation and clinical impacts: Julia Bodmer award review 2015. HLA. 2016;87(5):338–349. doi:10.1111/tan.12787. [ published Online First: 20160406].
- Campoli M, Ferrone S. HLA antigen changes in malignant cells: epigenetic mechanisms and biologic significance. Oncogene. 2008;27(45):5869–5885. doi:10.1038/onc.2008.273.
- Lazaridou MF, Gonschorek E, Massa C, Friedrich M, Handke D, Mueller A, Jasinski-Bergner S, Dummer R, Koelblinger P, Seliger B, et al. Identification of miR-200a-5p targeting the peptide transporter TAP1 and its association with the clinical outcome of melanoma patients. OncoImmunol. 2020;9(1):1774323. doi:10.1080/2162402X.2020.1774323.
- Mari L, Hoefnagel SJM, Zito D, van de Meent M, van Endert P, Calpe S, Sancho Serra MDC, Heemskerk MHM, van Laarhoven HWM, Hulshof MCCM, et al. microRNA 125a regulates MHC-I expression on esophageal adenocarcinoma cells, associated with suppression of antitumor immune response and poor outcomes of patients. Gastroenterol. 2018;155(3):784–798. doi:10.1053/j.gastro.2018.06.030. [ published Online First: 20180607].
- Huang L, Malu S, McKenzie JA, Andrews MC, Talukder AH, Tieu T, Karpinets T, Haymaker C, Forget MA, Williams LJ, et al. The RNA-binding protein MEX3B mediates resistance to cancer immunotherapy by downregulating HLA-A expression. Clin Cancer Res. 2018;24(14):3366–3376. doi:10.1158/1078-0432.CCR-17-2483. [ published Online First: 20180301].
- Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8(6):479–490. doi:10.1038/nrm2178.
- Glisovic T, Bachorik JL, Yong J. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582(14):1977–1986. doi:10.1016/j.febslet.2008.03.004. [ published Online First: 20080313].
- Ariyachet C, Solis NV, Liu Y, Prasadarao NV, Filler SG, McBride AE. SR-like RNA-binding protein Slr1 affects Candida albicans filamentation and virulence. Infect Immun. 2013;81(4):1267–1276. doi:10.1128/IAI.00864-12. [ published Online First: 20130204].
- Brochu C, Cabrita MA, Melanson BD, Hamill JD, Lau R, Pratt MAC, McKay BC. NF-κB-dependent role for cold-inducible RNA binding protein in regulating interleukin 1β. PLOS ONE. 2013;8(2):e57426. doi:10.1371/journal.pone.0057426. [ published Online First: 20130221].
- Wang ZL, Li B, Luo YX. Comprehensive genomic characterization of RNA-Binding proteins across human cancers. Cell Rep. 2018;22(1):286–298. doi:10.1016/j.celrep.2017.12.035.
- Streitfeld WS, Dalton AC, Howley BV, Howe PH. PCBP1 regulates LIFR through FAM3C to maintain breast cancer stem cell self-renewal and invasiveness. Cancer Biol Ther. 2023;24(1):2271638. doi:10.1080/15384047.2023.2271638. [ published Online First: 20231106].
- Jung M, Ji E, Kang H, Kim C, Ryu S, Han S, Cha S, Lee EK. The microRNA-195-5p/hnRNP A1 axis contributes to the progression of hepatocellular carcinoma by regulating the migration of cancer cells. Biochem Biophys Res Commun. 2023;686:149183. doi:10.1016/j.bbrc.2023.149183. [ published Online First: 20231030].
- Lu Y, Bi T, Zhou S. MEX3A promotes angiogenesis in colorectal cancer via glycolysis. Libyan J Med. 2023;18(1):2202446. doi:10.1080/19932820.2023.2202446.
- Kulkarni S, Ramsuran V, Rucevic M, Singh S, Lied A, Kulkarni V, O’hUigin C, Le Gall S, Carrington M. Posttranscriptional regulation of HLA-A protein expression by alternative polyadenylation signals involving the RNA-binding protein syncrip. J Immunol. 2017;199(11):3892–3899. doi:10.4049/jimmunol.1700697. [ published Online First: 20171020].
- Reches A, Nachmani D, Berhani O, Duev-Cohen A, Shreibman D, Ophir Y, Seliger B, Mandelboim O. HNRNPR regulates the expression of classical and nonclassical MHC class I proteins. J Immunol. 2016;196(12):4967–4976. doi:10.4049/jimmunol.1501550. [ published Online First: 20160518].
- van Hateren A, Elliott T. Visualising tapasin- and TAPBPR-assisted editing of major histocompatibility complex class-I immunopeptidomes. Curr Opin Immunol. 2023;83:102340. doi:10.1016/j.coi.2023.102340. [ published Online First: 20230526].
- Muller IK, Winter C, Thomas C, Spaapen RM, Trowitzsch S, Tampé R. Structure of an MHC I–tapasin–ERp57 editing complex defines chaperone promiscuity. Nat Commun. 2022;13(1):5383. doi:10.1038/s41467-022-32841-9. [ published Online First: 20220914].
- Boulanger DSM, Douglas LR, Duriez PJ, Kang Y, Dalchau N, James E, Elliott T. Tapasin-mediated editing of the MHC I immunopeptidome is epitope specific and dependent on peptide off-rate, abundance, and level of tapasin expression. Front Immunol. 2022;13:956603. doi:10.3389/fimmu.2022.956603. [ published Online First: 20221031].
- Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17(7):909–915. doi:10.1038/nsmb.1838. [ published Online First: 20100704].
- Zheng J, Wu S, Tang M, Xi S, Wang Y, Ren J, Luo H, Hu P, Sun L, Du Y, et al. USP39 promotes hepatocellular carcinogenesis through regulating alternative splicing in cooperation with SRSF6/HNRNPC. Cell Death Dis. 2023;14(10):670. doi:10.1038/s41419-023-06210-3. [ published Online First: 20231011].
- Zhou Y, Xue X, Luo J, Li P, Xiao Z, Zhang W, Zhou J, Li P, Zhao J, Ge H, et al. Circular RNA circ-FIRRE interacts with HNRNPC to promote esophageal squamous cell carcinoma progression by stabilizing GLI2 mRNA. Cancer Sci. 2023;114(9):3608–3622. doi:10.1111/cas.15899. [ published Online First: 20230707].
- Cheng Y, Li L, Wei X, Xu F, Huang X, Qi F, Zhang Y, Li X. HNRNPC suppresses tumor immune microenvironment by activating treg cells promoting the progression of prostate cancer. Cancer Sci. 2023;114(5):1830–1845. doi:10.1111/cas.15745. [ published Online First: 20230213].
- Cai Y, Lyu T, Li H, Liu C, Xie K, Xu L, Li W, Liu H, Zhu J, Lyu Y, et al. LncRNA CEBPA-DT promotes liver cancer metastasis through DDR2/β-catenin activation via interacting with hnRNPC. J Exp Clin Cancer Res. 2022;41(1):335. doi:10.1186/s13046-022-02544-6. [ published Online First: 20221206].
- He Q, Yang C, Xiang Z, Huang G, Wu H, Chen T, Dou R, Song J, Han L, Song T, et al. LINC00924-induced fatty acid metabolic reprogramming facilitates gastric cancer peritoneal metastasis via hnRNPC-regulated alternative splicing of Mnk2. Cell Death Dis. 2022;13(11):987. doi:10.1038/s41419-022-05436-x. [ published Online First: 20221123].
- Liu D, Luo X, Xie M, Zhang T, Chen X, Zhang B, Sun M, Wang Y, Feng Y, Ji X, et al. HNRNPC downregulation inhibits IL-6/STAT3-mediated HCC metastasis by decreasing HIF1A expression. Cancer Sci. 2022;113(10):3347–3361. doi:10.1111/cas.15494. [ published Online First: 20220812].
- Pan C, Wu Q, Feng N. A systematic pan-cancer study demonstrates the oncogenic function of heterogeneous nuclear ribonucleoprotein C. Aging (Albany NY). 2022;14(6):2880–2901. doi:10.18632/aging.203981. [ published Online First: 20220328].
- Cholewa BD, Pellitteri-Hahn MC, Scarlett CO, Ahmad N. Large-scale label-free comparative proteomics analysis of polo-like Kinase 1 inhibition via the small-molecule inhibitor BI 6727 (Volasertib) in BRAF V600E mutant melanoma cells. J Proteome Res. 2014;13(11):5041–5050. doi:10.1021/pr5002516. [ published Online First: 20140609].
- Hwang SJ, Seol HJ, Park YM, Kim KH, Gorospe M, Nam DH, Kim HH. MicroRNA-146a suppresses metastatic activity in brain metastasis. Molecules And Cells. 2012;34(3):329–334. doi:10.1007/s10059-012-0171-6. [ published Online First: 20120903].
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi:10.1038/ng1180.
- Hazini A, Fisher K, Seymour L. Deregulation of HLA-I in cancer and its central importance for immunotherapy. J Immunother Cancer. 2021;9(8):e002899. doi:10.1136/jitc-2021-002899.
- Matsuda-Lennikov M, Ohigashi I, Takahama Y. Tissue-specific proteasomes in generation of MHC class I peptides and CD8+ T cells. Curr Opin Immunol. 2022;77:102217. doi:10.1016/j.coi.2022.102217. [ published Online First: 20220608].
- Mani R, Abdelaziz MH, Michelon A, Suzuki Y. Human MHC class I molecule, HLA-A2.1, mediates activation of CD8+ T cell IFN-γ production and the T cell-dependent protection against reactivation of cerebral toxoplasma infection. Front Immunol. 2022;13:1005059. doi:10.3389/fimmu.2022.1005059. [ published Online First: 20221013].
- Perea F, Sanchez-Palencia A, Gomez-Morales M, Bernal M, Concha Á, García MM, González-Ramírez AR, Kerick M, Martin J, Garrido F, et al. HLA class I loss and PD-L1 expression in lung cancer: impact on T-cell infiltration and immune escape. Oncotarget. 2018;9(3):4120–4133. doi:10.18632/oncotarget.23469. [ published Online First: 20171219].
- Han SH, Kim M, Chung YR, Woo JW, Choi HY, Park SY. Expression of HLA class I is associated with immune cell infiltration and patient outcome in breast cancer. Sci Rep. 2022;12(1):20367. doi:10.1038/s41598-022-24890-3. [ published Online First: 20221127].
- Wang Y, Jasinski-Bergner S, Wickenhauser C, Seliger B. Cancer immunology: immune escape of tumors—expression and regulation of HLA class I molecules and its role in immunotherapies. Advan Anatom Pathol. 2023;30(3):148–159. doi:10.1097/PAP.0000000000000389. [ published Online First: 20221215].
- Wu Y, Zhao W, Liu Y, Tan X, Li X, Zou Q, Xiao Z, Xu H, Wang Y, Yang X, et al. Function of HNRNPC in breast cancer cells by controlling the dsRNA-induced interferon response. Embo J. 2018;37(23). doi:10.15252/embj.201899017. [ published Online First: 20180829].
- Shi S, Wu T, Ma Z, Zhang X, Xu K, Tian Q, Gao L, Yin X, Xu S, Yang S, et al. Serum-derived extracellular vesicles promote the growth and metastasis of non-small cell lung cancer by delivering the m6A methylation regulator HNRNPC through the regulation of DLGAP5. J Cancer Res Clin Oncol. 2023;149(8):4639–4651. doi:10.1007/s00432-022-04375-6. [ published Online First: 20220929].
- Bao Y, Qiao Y, Choi JE, Zhang Y, Mannan R, Cheng C, He T, Zheng Y, Yu J, Gondal M, et al. Targeting the lipid kinase PIKfyve upregulates surface expression of MHC class I to augment cancer immunotherapy. Proc Natl Acad Sci USA. 2023;120(49):e2314416120. doi:10.1073/pnas.2314416120. [ published Online First: 20231127].
- Lazaridou MF, Massa C, Handke D, Mueller A, Friedrich M, Subbarayan K, Tretbar S, Dummer R, Koelblinger P, Seliger B, et al. Identification of microRNAs targeting the transporter associated with antigen processing TAP1 in melanoma. J Clin Med. 2020;9(9):2690. doi:10.3390/jcm9092690. [ published Online First: 20200820].
- Friedrich M, Vaxevanis CK, Biehl K, Mueller A, Seliger B. Targeting the coding sequence: opposing roles in regulating classical and non-classical MHC class I molecules by miR-16 and miR-744. J Immunother Cancer. 2020;8(1):e000396. doi:10.1136/jitc-2019-000396.
- Matsui M, Kawano M, Matsushita S. Introduction of a point mutation into an HLA class I single-chain trimer induces enhancement of CTL priming and antitumor immunity. Mol Ther Methods Clin Dev. 2014;1:14027. doi:10.1038/mtm.2014.27. [ published Online First: 20140702].
- Jongsma MLM, Guarda G, Spaapen RM. The regulatory network behind MHC class I expression. Mol Immunol. 2019;113:16–21. doi:10.1016/j.molimm.2017.12.005. [ published Online First: 20171208].
- Wu Z, Zuo X, Zhang W, Li Y, Gui R, Leng J, Shen H, Pan B, Fan L, Li J, et al. m6A-modified circTET2 interacting with HNRNPC regulates fatty acid oxidation to promote the proliferation of chronic lymphocytic leukemia. Adv Sci. 2023;10(34):e2304895. doi:10.1002/advs.202304895. [ published Online First: 20231011].
- Gu Z, Yang Y, Ma Q, Wang H, Zhao S, Qi Y, Li Y. HNRNPC, a predictor of prognosis and immunotherapy response based on bioinformatics analysis, is related to proliferation and invasion of NSCLC cells. Respir Res. 2022;23(1):362. doi:10.1186/s12931-022-02227-y. [ published Online First: 20221219].
- Xia N, Yang N, Shan Q. HNRNPC regulates RhoA to induce DNA damage repair and cancer-associated fibroblast activation causing radiation resistance in pancreatic cancer. J Cell Mol Med. 2022;26(8):2322–2336. doi:10.1111/jcmm.17254. [ published Online First: 20220311].
- Huang GZ, Wu QQ, Zheng ZN, Shao TR, Chen YC, Zeng WS, Lv XZ. M6A-related bioinformatics analysis reveals that HNRNPC facilitates progression of OSCC via EMT. Aging (Albany NY). 2020;12(12):11667–11684. doi:10.18632/aging.103333. [ published Online First: 20200611].
- Wang Y, Lazaridou MF, Massa C. 910 Overexpression of miR-155–5p can upregulate antigen processing and presentation pathway via targeting tapasin. J Immunother Cancer. 2021;9(Suppl 2):A956–A56. doi:10.1136/jitc-2021-SITC2021.910.
- Kattan FG, Koukouraki P, Anagnostopoulos AK, Tsangaris GT, Doxakis E. RNA binding protein AUF1/HNRNPD regulates nuclear export, stability and translation of SNCA transcripts. Open Biol. 2023;13(11):230158. doi:10.1098/rsob.230158. [ published Online First: 20231122].
- Chen Z, Pi H, Zheng W, Guo X, Shi C, Wang Z, Zhang J, Qu X, Liu L, Shen H, et al. The 3′ non-coding sequence negatively regulates PD-L1 expression, and its regulators are systematically identified in pan-cancer. Genes (Basel). 2023;14(8):1620. doi:10.3390/genes14081620. [ published Online First: 20230813].
- Yang YC, Lin YW, Lee WJ, Lai FR, Ho KH, Chu CY, Hua KT, Chen JQ, Tung MC, Hsiao M, et al. The RNA-binding protein KSRP aggravates malignant progression of clear cell renal cell carcinoma through transcriptional inhibition and post-transcriptional destabilization of the NEDD4L ubiquitin ligase. J Biomed Sci. 2023;30(1):68. doi:10.1186/s12929-023-00949-9. [ published Online First: 20230814].
- Wu P. Inhibition of RNA-binding proteins with small molecules. Nat Rev Chem. 2020;4(9):441–458. doi:10.1038/s41570-020-0201-4. [ published Online First: 20200715].
