ABSTRACT
TCRαβ+ CD4− CD8− double-negative T (DNT) cells are minor populations in peripheral blood, and their roles have mostly been discussed in inflammation and autoimmunity. However, the functions of DNT cells in tumor microenvironment remain to be elucidated. We investigated their characteristics, possible origins and functions in colorectal cancer tissues as well as their corresponding tumor-draining lymph nodes. We found a significant enrichment of DNT cells in tumor tissues compared with their corresponding lymph nodes, especially in tumors with lower T cell infiltration. T cell receptor (TCR) sequence analysis of CD4+ T, CD8+ T and DNT cells indicated that TCR sequences detected in DNT cells were found in CD8+ T cells, but rarely in CD4+ T cells, suggesting that a part of DNT cells was likely to be originated from CD8+ T cells. Through a single-cell transcriptomic analysis of DNT cells, we found that a DNT cell cluster, which showed similar phenotypes to central memory CD8+ T cells with low expression of effector and exhaustion markers, revealed some specific gene expression patterns, including higher GZMK expression. Moreover, in flow cytometry analysis, we found that DNT cells lost production of cytotoxic mediators. These findings imply that DNT cells might function as negative regulators of anti-tumor immune responses in tumor microenvironment.
Introduction
Colorectal cancer (CRC) is one of the most common cancer types with a high mortality rate.Citation1 Currently, treatments for CRC are guided based on tumor stages as well as the microsatellite instability; however, their clinical outcomes show a large variability. Many reports demonstrate that the status of immune cell infiltration and microsatellite instability is proven to be associated with patients’ prognosis and clinical responses to immune checkpoint inhibitors.Citation2,Citation3 CRCs are genetically categorized into two types: one is tumors having extremely high somatic mutations with microsatellite instability caused by deficiency of mismatch repair enzymes or abnormality in certain DNA polymerases, and the other is tumors showing low or absent microsatellite instability and having a modest number of somatic mutations.Citation4 The former type of CRC usually shows high infiltration of immune cells in tumor tissues and better clinical outcome in immune checkpoint inhibitor therapy.Citation3 Tumor immune microenvironment in CRC is not so simple because many types of immune cells are present. Higher CD3+ or CD8+ T cell infiltration has been reported to be associated with better prognosis in CRC patients,Citation2 while infiltration of tumor-associated macrophages, cancer-associated fibroblasts, myeloid-derived suppressor cells and regulatory T cells was correlated with worse clinical outcomes.Citation3 Hence, clarification of the tumor immune microenvironment in individual cancers would help better understand the molecular mechanisms and improve the clinical management of immunotherapy.
Double-negative T cells (DNT cells) are defined as T cells expressing CD3 (or T cell receptor (TCR) αβ) without expression of CD4, CD8 or NK cell markers such as CD56. DNT cells account for 1%–3% of the total peripheral T cells,Citation5 but their functions or cellular origins have not been well elucidated. Some researchers reported that DNT cells originated from the thymus, and others argued that DNT cells are derived from CD4+ or CD8+ T cells.Citation6 Accumulation of DNT cells was recognized in lpr/lpr and gld/gld mice, and in patients with autoimmune lymphoproliferative syndrome, which is caused by an impairment of the Fas/FasL pathway.Citation5–7 In these cases, DNT cells were indicated to have inflammatory and immunomodulatory functions. Several reports indicated possible roles of DNT cells in cancer development. DNT cells might have immunosuppressive effects on CD4+ and antigen-specific CD8+ T cells, and inhibit CD4+ T cell proliferation.Citation8,Citation9 Levels of DNT cells seemed to be positively correlated with tumor metastasis in melanoma.Citation10 However, the immunological functions of DNT cells in tumors have not been well clarified.
In this study, we demonstrate, for the first time, the origin and transcriptomic features of DNT cells in CRC. The results of TCR sequencing and single-cell transcriptome analyses indicate that most of DNT cells in tumors might be derived from CD8+ T cells by losing CD8 expression and may have no or less cytotoxic functions. Our finding might open a new aspect of tumor microenvironment as well as immunotherapy for CRC.
Materials and methods
Patients
CRC patients who received surgical treatment were enrolled after providing written informed consent before their surgery. A total of 42 fresh primary tumor tissues from 41 patients and multiple lymph nodes from 18 patients were collected from individual patients. None of the 41 patients had received any neoadjuvant treatment. The study protocol was approved by the Institutional Review Board of the Japanese Foundation for Cancer Research (2018-GA-1021).
Preparation of single-cell suspensions from tumor tissues
Single-cell suspensions from tumor tissues and lymph nodes were prepared using the method we reported previously.Citation11 Briefly, tissues were dissociated using gentleMACS Dissociator (Miltenyi Biotec, Bergisch Gladbach, Germany) and then treated with collagenase (ThermoFisher Scientific, Waltham, MA, USA) and DNase I (STEMCELL Technologies, Vancouver, Canada) for 60 min at 37°C. After washing with PBS, the cells were passed through a 70 μm nylon cell strainer (Corning Inc., Corning, NY, USA). The passed cells were used for the following flow cytometry and cell sorting analyses.
Flow cytometry
Fluorochrome-conjugated monoclonal antibodies specific to human TCRαβ (clone IP26), CD3 (HIT3a), CD4 (RPA-T4), CD8α (RPA-T8 and SK1), CD56 (HCD56), CCR7 (G043H7), CD45RA (HI100), perforin (B-D48), granzyme B (QA16A02), granzyme K (GM26E7), Fas (DX2) and FasL (NOK-1) were purchased from BioLegend (San Diego, CA, USA). All samples were resuspended in PBS staining buffer containing 0.5% BSA, pre-incubated for 4°C with FcR blocking reagent (Miltenyi Biotec), washed by PBS staining buffer, and then stained with each antibody for 20 min at 4°C. Intracellular staining of perforin, granzyme B and granzyme K was performed according to the manufacturer’s protocol. 7-Aminoactinomycin D (7-AAD) viability staining solution (BioLegend) was used to gate on alive cells. Data were collected on FACSVerse or FACSLyric for the analysis or FACSAriaII for the cell sorting (BD Biosciences, Franklin Lakes, NJ, USA) and then analyzed using FlowJo software (ver.10, BD Biosciences).
Multiplex fluorescence immunohistochemistry
As previously described,Citation12 4-µm sections obtained from formalin-fixed paraffin-embedded tissue blocks were stained for CD3 (1:200, SP7, Abcam, Cambridge, UK), CD4 (1:200, NCL-CD4-368, Novocastra, Milton Keynes, UK), CD8 (1:160, NCL-L-CD8-4B11, Novocastra), CD20 (1:200, L26, Invitrogen, Waltham, MA, USA), CD204 (1:200, KT022, Trans genic, Fukuoka Japan), FOXP3 (1:400, D608R, Cell Signaling Technology, Danvers, MA, USA) and cytokeratin (1:100, AE1/AE3, Dako, Santa Clara, CA, USA) with an Opal IHC kit (AKOYA Biosciences, Menlo Park, CA, USA). Bound primary antibodies were detected using EnVision + System-HRP Labeled Polymer anti-mouse and anti-rabbit (Dako) followed by tyramide signal amplification opal fluorophores (Opal 480, Opal 520, Opal 540, Opal570, Opal 620, Opal 650, Opal 690). The slides were counterstained with DAPI (4′,6-diamidino-2-phenylindole), scanned using the PerkinElmer Vectra Polaris system, and analyzed using the inForm Advanced Image Analysis software (inForm 2.6.0; Akoya Biosciences).
TCR sequencing analysis
TCR sequencing was performed using the methods described previously.Citation13,Citation14 In brief, we used total RNA extracted from 2 × 105 T cells sorted by flow cytometry for cDNA synthesis with a common 5’-RACE adapter using SMART library construction kit (Clontech, Mountain View, CA, USA). TCRα and TCRβ cDNAs were amplified by PCR using a forward primer for the SMART adapter and a reverse primer corresponding to the constant region of TCRα and TCRβ genes. Illumina index sequences with barcode were added using the Nextera XT Index kit (Illumina, San Diego, CA, USA), and then the prepared libraries were sequenced by 300-bp paired-end reads on Illumina MiSeq, using MiSeq Reagent v3 600-cycles kit (Illumina). Sequence data were analyzed using Tcrip software.Citation13
Single-cell transcriptome analysis
Alive CD8+ T cells (TCRαβ+CD56-CD4-CD8α(clone SK1)+) and DNT cells (TCRαβ+CD56-CD4-CD8α(clone SK1)-) were sorted from single-cell suspension obtained from CRC tissues by FACSAriaII (BD Biosciences). To detect surface CD8α protein expression, BD AbSeq antibody-oligos (clone RPA-T8) were conjugated. Single-cell sequence libraries were prepared using the BD Rhapsody Single-Cell Analysis System (BD Biosciences) and were sequenced on Illumina NextSeq (Illumina). Raw sequencing data were processed with Seven Bridges Genomics platform (BD Biosciences). Seurat was used to perform an unsupervised clustering and differential gene expression analysis. Later, genes, which were expressed in less than two cells, were filtered out, and cells with expression of >200 genes and <15% mitochondrial genes were further analyzed. Differentially expressed genes (DEG) were defined as fold changes of >|2.0| and q-values of <1.0 × 10−10 relative to comparator populations. Gene ontology (GO) enrichment analysis for Biological Process was performed using the Panther online tool.Citation15,Citation16 Overrepresentation for each gene set was tested using a Fisher’s exact test and with a false discovery rate threshold of 0.05. Functional module scores were calculated based on the expression levels of top 30 genes which were highly correlated with GZMB (cytotoxicity score) or PDCD1 (exhaustion score), as described previously.Citation17
Statistical analysis
The Mann–Whitney U test was performed to compare differences between two independent groups. The paired t-test was used to compare differences between paired samples.
Results
Higher proportion of DNT cells in CRC tissues than their corresponding lymph nodes
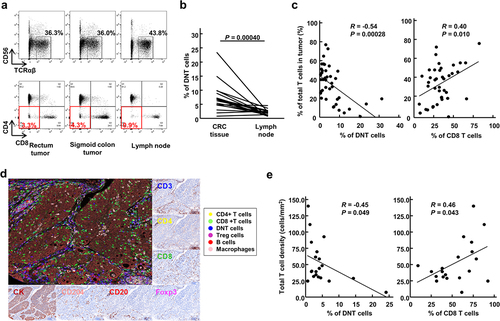
We collected a total of 42 tumor tissues from 41 CRC patients as well as a total of 41 lymph nodes (average 2.3 from each patient) from 18 of the 41 patients. We analyzed proportions of DNT cells, defined as TCRαβ+ cells lacking the expressions of CD4, CD8 and CD56, in these tumor tissues and lymph nodes by flow cytometry (). shows a representative flow cytometry plot of one patient, who had two primary tumors in sigmoid colon and in rectum. The proportions of DNT cells among total TCRαβ+CD56- T cells in both tumor tissues were 3.3% and 4.3%, which were higher than 0.9% in the lymph node. In 16 of the 18 patients, higher levels of DNT cells were detected in tumor tissues than lymph nodes (average 7.4% vs 1.8%, p = .00040; ). We then investigated the relationship between proportions of DNT cells and the extent of T cell infiltration into tumors (defined as proportions of TCRαβ+CD56- cells in total living cells). Tumors with low T cell infiltration showed significantly higher proportions of DNT cells than those with higher T cell infiltration (R = −0.54, p = .00028; ). These results indicated that the proportions of DNT cells were negatively correlated with T cell infiltration in CRC tissues. We also assessed the intra-tumoral immune cell infiltration in 20 randomly selected CRC tissues by multiplex fluorescent immunohistochemistry (mFIHC), and infiltration of DNT cells into tumors were confirmed (). In concordance with flow cytometry data, the proportion of DNT cells detected by mFIHC was inversely correlated with total CD3+ T cell density (R = −0.45, p = .049; ).
Figure 1. Double-negative T (DNT) cells in colorectal cancer (CRC) tissues.
DNT cells derived from CD8+ T cells
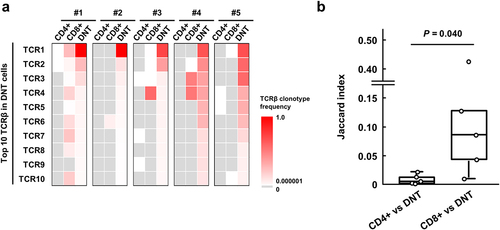
Although various possibilities of the origins of DNT cells have been suggested, none is conclusive. To clarify the origin of DNT cells in CRC tissues, we analyzed TCR clonotypes among CD4+ T, CD8+ T and DNT cells sorted from five CRC tissues, in which we were able to obtain more than 10,000 DNT cells (). The 10 most abundant clonotypes in DNT cells in each tissue are shown in . The sum of the 10 most frequent TCRβ clonotypes in DNT cells in tumor tissues was significantly higher as 80.3% in the average compared to 21.6% and 50.3% in the CD4+ and CD8+ T cells, respectively (p = .0013 and p = .0055), indicating enrichment of certain TCR clonotypes in DNT cells. One or multiple clonotypes of the 10 most common TCRβ clonotypes in DNT cells were also observed commonly in CD8+ T cells, but not in CD4+ T cells in these five CRC tissues we examined. Jaccard index between TCRβ clonotypes in DNT and CD8+ T cells was significantly higher than that between DNT and CD4+ T cells (0.096 vs 0.0090, p = .040, ), suggesting that DNT cells are possibly derived from CD8+ T cells.
Figure 2. Comparison of TCRβ repertoire among CD4+, CD8+ and double-negative T (DNT) cells in colorectal cancer (CRC) tissues.
Single-cell profiling of DNT cells in CRC tissues
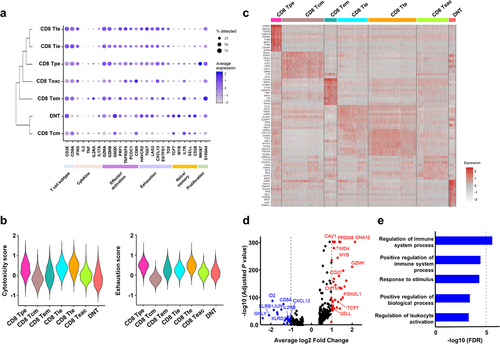
To further characterize DNT cells in CRC tissues, we performed a single-cell transcriptome analysis (). We prepared a total of 25,500 DNT cells and 59,691 CD8+ T cells from tissue samples of four CRC patients and finally obtained RNA sequences from 12,664 cells (). UMAP analysis of RNA expression data of these DNT and CD8+ T cells from the four CRC tissue samples identified unique clustering patterns consisting of seven functional clusters characterized by known gene expression signatures of specific T-cell subtypes (). We found a cluster of “DNT” cells, in which we confirmed mRNA expressions of CD3E, TRAC, TRBC1 and TRBC2, but the absence of CD4 and CD8A mRNA expression as well as CD8α protein expression (). In this cluster, the expression of γδ T cell markers, including TRGC1, TRGC2 and TRDC, and NK cell marker, NCAM1 (CD56), was not detected. Although the cells with no or very low expression of CD8A mRNA or CD8α protein were also present across the other CD8 clusters, suggesting that DNT cells were possibly heterogenous cell populations, these cells were minor populations in each of the CD8 clusters (9.6%-20.8%; and Figure S1 and S2). Therefore, we further analyzed DNT cells in the “DNT” cluster (). DNT cells showed lower expression of effector or activation marker genes, including GZMB, PDCD1 (PD-1) and TNFRSF9 (CD137) as well as exhaustion markers, including LAG3 and CXCL13 than CD8+ T cells, although higher expression levels of naïve or memory marker genes, including IL7R, SELL and TCF7, were detected in the DNT cells (). Through a consensus clustering, we found that the gene expression pattern of DNT cells was most similar to CD8 Tcm (CD8+ central memory T) cell cluster compared to the other effector CD8+ T clusters. Cytotoxicity scores of the DNT cells in this cluster were low as CD8 Tcm compared to effector or exhausted CD8+ T cells, and exhaustion scores were also lower than exhausted CD8+ T cells (). DEG analysis identified 36 up-regulated and 24 down-regulated genes in the DNT cells (). Functional categorization of these up- and down-regulated genes in the DNT cells by GO annotations indicated that the down-regulated genes were significantly enriched in several immune pathways, including positive regulation of immune system process, response to stimulus, and regulation of leukocyte activation (), suggesting that DNT might lose effector or immune activation function. On the other hand, no significant enrichment in any pathways was identified for the up-regulated genes, but several genes involved in the memory function, including TCF7, CD27, MYB and SELL, were highly expressed (). In addition, we identified that GZMK was highly expressed in the DNT cells.
Figure 3. Single-cell transcriptome analysis of CD8+ T and double-negative T (DNT) cells in colorectal cancer (CRC) tissues.
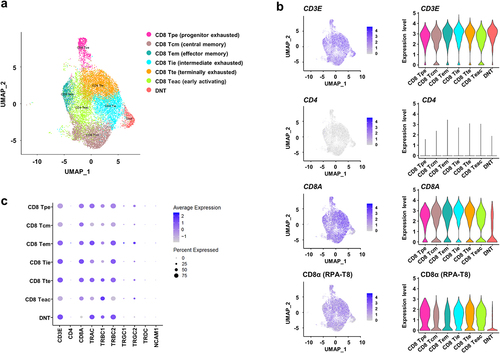
Figure 4. Transcriptional signature of double-negative T (DNT) cells.
Flow cytometry analysis of DNT cells in CRC tissues
Although granzyme K encoded by GZMK is known as a cytolytic molecule as granzymes A and B, GZMK- and TCF7-expression was reported to be a characteristic feature of undifferentiated pre-effector/memory T cells.Citation18 Therefore, we further characterized the DNT cells using flow cytometry (). We first investigated the expression of memory T cell markers, CCR7 and CD45RA, in DNT cells. A CCR7+CD45RA- is known to be characteristic of a central memory phenotype, and CCR7-CD45RA- is considered to be an effector memory phenotype (). We found that DNT cells in tumor tissues showed no naïve phenotype and kept the central memory CCR7+CD45RA- phenotype (). In concordance with single-cell transcriptome analysis, granzyme K expression was detected in average 44.9% (ranging from 15.3% to 72.2%) of the DNT cells using flow cytometry (). To investigate cytotoxic function of DNT cells, we then examined expression of cytolytic molecules in the DNT cells. In the single-cell transcriptome data, the expressions of PRF1 (encoding for perforin) and GZMB (granzyme B) were barely detected in the DNT cells (). We also observed that proportions of cells expressing perforin or granzyme B were much lower in the DNT cell population than CD8+ T cell population by intracellular staining using flow cytometry (). These data supported that DNT cells in tumor tissues were not able to produce cytotoxic mediators and have no or very low cytotoxic function against cancer cells.
Figure 5. Flow cytometry of specific molecules and cytotoxic molecules in double-negative T (DNT) cells.
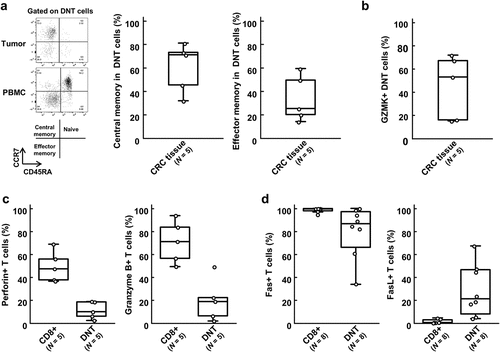
In CD8+ T cells, cytotoxicity also mediates killing of target cells through Fas/FasL-mediated apoptosis.Citation19,Citation20 Since the impairment of this pathway was known to cause accumulation of DNT cells,Citation21 we further investigated Fas and FasL expression levels in the DNT cells. High Fas expression on both CD8+ T and DNT cells was observed. On the other hand, FasL expression was detected only in DNT cells, but not in CD8+ T cells (). Since FasL-expressing cells such as myeloid-derived suppressor cells induced apoptosis of Fas-expressing tumor-infiltrating T cells,Citation22 DNT cells might serve as negative regulators in tumor microenvironment by inducing apoptosis of effector CD8+ T cells via the Fas/FasL pathway.
Discussion
TCRαβ+CD56-CD4-CD8- DNT cells are known to be a minor cell population of T cells, and their functions in tumor microenvironment remain unclear. In this study, we investigated the characteristics, a possible origin(s) and function of DNT cells in CRC tissues, and found a significantly higher proportion of DNT cells in tumor tissues than their draining lymph nodes (). We hypothesized that these DNT cells were likely to be originated from CD8+ T cells possibly through the down-regulation of CD8 expression in activated and clonally expanded CD8+ T cells in tumor microenvironment based on the evidence that DNT cells had clonal TCRs, which were at some extent overlapped with clonotypes of CD8+ T cells in each CRC tissue (). Through the single-cell transcriptome and flow cytometry analyses, we found that these DNT cells showed the central memory-like phenotype, but not the effector phenotype, and would probably have no or very low cytotoxic activity induced by the traditional perforin/granzyme-mediated pathway (). Considering the higher proportion of DNT cells in lower T cell-infiltrated CRC tissues () and a reported suppressive potential to CD8+ T cell activities through the Fas/FasL-mediated pathway in an antigen-specific manner,Citation8 DNT cells might have immune-inhibitory roles in tumor immune microenvironment.
Several reports have suggested that DNT cells in peripheral bloods might have a potent anti-tumor activity against lung cancer cells and leukemia cells in patient-derived xenograft models.Citation23,Citation24 The ex-vivo expanded DNT cells, which may have the selection bias by growth advantages, expressed higher levels of effector/cytolytic molecules, including IFN-γ and TNF-α, and granzyme B. Since allogeneic DNT cells did not induce a host-versus-graft reaction nor cause graft versus host disease, DNT cells are considered as a potential source of novel immunotherapy.Citation25,Citation26 Although the presence of DNT cells in tumor tissues of several cancer types is confirmed, their functions on anti-tumor immunity is still unclear.Citation25,Citation27–30 Immune profiling of infiltrating T cells in hepatocellular carcinoma identified two DNT cell populations; one is γδ DNT cells expressing tissue-resident markers CD69 and CD103 and the other is αβ DNT cells expressing activation markers CD150, CD69, CD137, HLA-DR as well as an immune-inhibitory molecule TIGIT, suggesting the functionally activated DNT cells,Citation30 which were not concordant with our finding (). Another study suggested the defective αβ DNT cells secreting immune suppressive cytokine, IL-10, in mouse glioma and melanoma tissues.Citation31 In our results in , the DNT cells were unlikely to have effector or cytotoxic functions. Due to the limited amount of T cells from the tumor tissues, we were not able to directly examine anti-tumor cytotoxic capacity against autologous cancer cells. Therefore, further analyses using a larger number of DNT cells from large tumor tissues are required to further characterize DNT functions. We identified possible TCRαβ pairs estimated from dominant TCRα and TCRβ sequences in DNT cells (). If these TCRs on the DNT cells are able to bind tumor-specific antigens on HLA molecules on the surface of cancer cells, they may competitively inhibit the antigen-TCR binding in cytotoxic CD8+ T cells. It is also important to identify the target antigens of TCRs that were dominant in DNT cells to further elucidate TCR-dependent or -independent functions of DNT cells.
DEG analysis identified that DNT cells showed the higher expression of GZMK (). Although granzyme K encoded by GZMK is known as a cytolytic molecule in a family of five granzymes in human (granzymes A, B, H, K and M), which is usually expressed in cytotoxic CD8+ T, NK and γδ T cells, co-expression of GZMK and TCF7 was reported to be a characteristic feature of pre-effector/memory T cells.Citation18 Several reports suggested possible immune suppressive function of granzyme K through an interaction with other immune cells, including macrophages. CRC tumors with high infiltration of CD15-high neutrophils contained a population of GZMK-high CD8+ TEM cells, which were related to worse prognosis.Citation32 Santiago L et al. reported that extracellular granzymes, including granzymes A, H and K, promoted CRC development by enhancing gut inflammation through the activation of macrophages.Citation33 Although it was unable to distinguish GZMK expression on CD8+ T or DNT cells, we found significant correlations of GZMK with estimated amounts of M1 and M2 macrophages from RNAseq data in 512 colon and rectal cancers in TCGA database (TCGA-COAD and -READ) (Figure S3), suggesting the possible immune suppressive roles of granzyme K in tumor development possibly through activation of tumor-associated macrophages.
In the current study, we found that DNT cells in tumor microenvironment are possibly heterogeneously present in various T cell subsets and DNT cells showing a central memory phenotype with high expression of GZMK were enriched in primary tumors compared with the corresponding draining lymph nodes. These DNT cells were functionally impaired by lacking of cytotoxic mediators and might be associated with immune desert in tumor microenvironment by inducing Fas-FasL-mediated apoptosis of cytotoxic CD8+ T cells. Although further functional analyses will be required, these findings in the current study might be a new window to elucidate the mechanism of immune dysfunction in tumor microenvironment.
Okamura_DNT_suppleTable1_20240527.pdf
Download PDF (176.1 KB)Okamura DNT_suppleFig.docx
Download MS Word (411.1 KB)Acknowledgments
We thank Junko Sugihara, Kensaku Nemoto and Hiu Ting Chan, Motoyoshi Iwakoshi for technical supports. The super-computing resource was provided by Human Genome Center, the Institute of Medical Science, the University of Tokyo (http://sc.hgc.jp/shirokane.html).
Disclosure statement
KK is a scientific advisor of Cancer Precision Medicine, Inc. YN is a stockholder and a scientific advisor of OncoTherapy Science, Inc.
Data availability statement
All data generated in this manuscript will be provided upon reasonable request to corresponding authors.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/2162402X.2024.2373530
Additional information
Funding
References
- Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394(10207):1467–9. doi:10.1016/S0140-6736(19)32319-0.
- Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313(5795):1960–1964. doi:10.1126/science.1129139.
- Guo L, Wang C, Qiu X, Pu X, Chang P. Colorectal cancer immune infiltrates: significance in patient prognosis and immunotherapeutic efficacy. Front Immunol. 2020;11:1052. doi:10.3389/fimmu.2020.01052.
- Ganesh K, Stadler ZK, Cercek A, Mendelsohn RB, Shia J, Segal NH, Diaz LA. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol. 2019;16(6):361–375. doi:10.1038/s41575-019-0126-x.
- Juvet SC, Zhang L. Double negative regulatory T cells in transplantation and autoimmunity: recent progress and future directions. J Mol Cell Biol. 2012;4(1):48–58. doi:10.1093/jmcb/mjr043.
- Brandt D, Hedrich CM. TCRαβ+ CD3+CD4−CD8− (double negative) T cells in autoimmunity. Autoimmun Rev. 2018;17(4):422–430. doi:10.1016/j.autrev.2018.02.001.
- Li H, Tsokos GC. Double-negative T cells in autoimmune diseases. Curr Opin Rheumatol. 2021;33(2):163–172. doi:10.1097/BOR.0000000000000778.
- Fischer K, Voelkl S, Heymann J, Przybylski GK, Mondal K, Laumer M, Kunz-Schughart L, Schmidt CA, Andreesen R, Mackensen A, et al. Isolation and characterization of human antigen-specific TCRαβ+ CD4−CD8− double-negative regulatory T cells. Blood. 2005;105(7):2828–2835. doi:10.1182/blood-2004-07-2583.
- Voelkl S, Gary R, Mackensen A. Characterization of the immunoregulatory function of human TCR-αβ+ CD4−CD8− double-negative T cells. Eur J Immunol. 2011;41(3):739–748. doi:10.1002/eji.201040982.
- Strippoli S, Fanizzi A, Negri A, Quaresmini D, Nardone A, Armenio A, Sciacovelli AM, Massafra R, De Risi I, De Tullio G, et al. Examining the relationship between circulating CD4− CD8− double-negative T cells and outcomes of immuno-checkpoint inhibitor therapy—looking for biomarkers and therapeutic targets in metastatic melanoma. Cells. 2021;10(2):10. doi:10.3390/cells10020406.
- Okamura K, Nagayama S, Tate T, Chan HT, Kiyotani K, Nakamura Y. Lymphocytes in tumor-draining lymph nodes co-cultured with autologous tumor cells for adoptive cell therapy. J Transl Med. 2022;20(1):241. doi:10.1186/s12967-022-03444-1.
- Ikarashi D, Kitano S, Tsuyukubo T, Takenouchi K, Nakayama T, Onagi H, Sakaguchi A, Yamashita M, Mizugaki H, Maekawa S, et al. Pretreatment tumour immune microenvironment predicts clinical response and prognosis of muscle-invasive bladder cancer in the neoadjuvant chemotherapy setting. Br J Cancer. 2022;126(4):606–614. doi:10.1038/s41416-021-01628-y.
- Fang H, Yamaguchi R, Liu X, Daigo Y, Yew PY, Tanikawa C, Matsuda K, Imoto S, Miyano S, Nakamura Y, et al. Quantitative T cell repertoire analysis by deep cDNA sequencing of T cell receptor α and β chains using next-generation sequencing (NGS). Oncoimmunology. 2014;3(12):e968467. doi:10.4161/21624011.2014.968467.
- Choudhury NJ, Kiyotani K, Yap KL, Campanile A, Antic T, Yew PY, Steinberg G, Park JH, Nakamura Y, O’Donnell PH, et al. Low T-cell receptor diversity, high somatic mutation burden, and high neoantigen load as predictors of clinical outcome in muscle-invasive bladder cancer. Eur Urol Focus. 2016;2(4):445–452. doi:10.1016/j.euf.2015.09.007.
- Thomas PD, Ebert D, Muruganujan A, Mushayahama T, Albou LP, Mi H. PANTHER: making genome-scale phylogenetics accessible to all. Protein Sci. 2022;31(1):8–22. doi:10.1002/pro.4218.
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. Gene Ontol Consortium Nat Genet. 2000;25(1):25–29. doi:10.1038/75556.
- Cheng D, Qiu K, Rao Y, Mao M, Li L, Wang Y, Song Y, Chen J, Yi X, Shao X, et al. Proliferative exhausted CD8+ T cells exacerbate long-lasting anti-tumor effects in human papillomavirus-positive head and neck squamous cell carcinoma. Elife. 2023;12:12. doi:10.7554/eLife.82705.
- Bassez A, Vos H, Van Dyck L, Floris G, Arijs I, Desmedt C, Boeckx B, Vanden Bempt M, Nevelsteen I, Lambein K, et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat Med. 2021;27(5):820–832. doi:10.1038/s41591-021-01323-8.
- Kagi D, Vignaux F, Ledermann B, Burki K, Depraetere V, Nagata S, Hengartner H, Golstein P. Fas and perforin pathways as major mechanisms of T cell-mediated cytotoxicity. Science. 1994;265(5171):528–530. doi:10.1126/science.7518614.
- Lowin B, Hahne M, Mattmann C, Tschopp J. Cytolytic T-cell cytotoxicity is mediated through perforin and Fas lytic pathways. Nature. 1994;370(6491):650–652. doi:10.1038/370650a0.
- Flores-Mendoza G, Rodriguez-Rodriguez N, Rubio RM, Madera-Salcedo IK, Rosetti F, Crispin JC. Fas/FasL signaling regulates CD8 expression during exposure to self-antigens. Front Immunol. 2021;12:635862. doi:10.3389/fimmu.2021.635862.
- Sinha P, Chornoguz O, Clements VK, Artemenko KA, Zubarev RA, Ostrand-Rosenberg S. Myeloid-derived suppressor cells express the death receptor Fas and apoptose in response to T cell–expressed FasL. Blood. 2011;117(20):5381–5390. doi:10.1182/blood-2010-11-321752.
- Merims S, Li X, Joe B, Dokouhaki P, Han M, Childs RW, Wang Z-Y, Gupta V, Minden MD, Zhang L, et al. Anti-leukemia effect of ex vivo expanded DNT cells from AML patients: a potential novel autologous T-cell adoptive immunotherapy. Leukemia. 2011;25(9):1415–1422. doi:10.1038/leu.2011.99.
- Yao J, Ly D, Dervovic D, Fang L, Lee JB, Kang H, Wang Y-H, Pham N-A, Pan H, Tsao M-S, et al. Human double negative T cells target lung cancer via ligand-dependent mechanisms that can be enhanced by IL-15. J Immunother Cancer. 2019;7(1):17. doi:10.1186/s40425-019-0507-2.
- Fang L, Ly D, Wang SS, Lee JB, Kang H, Xu H, Yao J, Tsao M-S, Liu W, Zhang L, et al. Targeting late-stage non-small cell lung cancer with a combination of DNT cellular therapy and PD-1 checkpoint blockade. J Exp Clin Cancer Res. 2019;38(1):123. doi:10.1186/s13046-019-1126-y.
- Vasic D, Lee JB, Leung Y, Khatri I, Na Y, Abate-Daga D, Zhang L. Allogeneic double-negative CAR-T cells inhibit tumor growth without off-tumor toxicities. Sci Immunol. 2022;7(70):eabl3642. doi:10.1126/sciimmunol.abl3642.
- Hall M, Liu H, Malafa M, Centeno B, Hodul PJ, Pimiento J, Pilon-Thomas S, Sarnaik AA. Expansion of tumor-infiltrating lymphocytes (TIL) from human pancreatic tumors. J Immunother Cancer. 2016;4(1):61. doi:10.1186/s40425-016-0164-7.
- Wu Z, Zheng Y, Sheng J, Han Y, Yang Y, Pan H, Yao J. CD3+CD4−CD8− (Double-negative) T cells in inflammation, immune disorders and cancer. Front Immunol. 2022;13:816005. doi:10.3389/fimmu.2022.816005.
- Stankovic B, Bjorhovde HAK, Skarshaug R, Aamodt H, Frafjord A, Muller E, Hammarström C, Beraki K, Bækkevold ES, Woldbæk PR, et al. Immune cell composition in human non-small cell lung cancer. Front Immunol. 2018;9:3101. doi:10.3389/fimmu.2018.03101.
- Di Blasi D, Boldanova T, Mori L, Terracciano L, Heim MH, De Libero G. Unique T-cell populations define immune-inflamed hepatocellular carcinoma. Cell Mol Gastroenterol Hepatol. 2020;9(2):195–218. doi:10.1016/j.jcmgh.2019.08.004.
- Prins RM, Incardona F, Lau R, Lee P, Claus S, Zhang W, Black KL, Wheeler CJ. Characterization of defective CD4−CD8− T cells in murine tumors generated independent of antigen specificity. J Immunol. 2004;172(3):1602–1611. doi:10.4049/jimmunol.172.3.1602.
- Tiberti S, Catozzi C, Croci O, Ballerini M, Cagnina D, Soriani C, Scirgolea C, Gong Z, He J, Macandog AD, et al. GZMKhigh CD8+ T effector memory cells are associated with CD15high neutrophil abundance in non-metastatic colorectal tumors and predict poor clinical outcome. Nat Commun. 2022;13(1):6752. doi:10.1038/s41467-022-34467-3.
- Santiago L, Castro M, Sanz-Pamplona R, Garzon M, Ramirez-Labrada A, Tapia E, Moreno V, Layunta E, Gil-Gómez G, Garrido M, et al. Extracellular granzyme a promotes colorectal cancer development by enhancing gut inflammation. Cell Rep. 2020;32(1):107847. doi:10.1016/j.celrep.2020.107847.
