ABSTRACT
Functional roles of SIGLEC15 in hepatocellular carcinoma (HCC) were not clear, which was recently found to be an immune inhibitor with similar structure of inhibitory B7 family members. SIGLEC15 expression in HCC was explored in public databases and further examined by PCR analysis. SIGLEC15 and PD-L1 expression patterns were examined in HCC samples through immunohistochemistry. SIGLEC15 expression was knocked-down or over-expressed in HCC cell lines, and CCK8 tests were used to examine cell proliferative ability in vitro. Influences of SIGLEC15 expression on tumor growth were examined in immune deficient and immunocompetent mice respectively. Co-culture system of HCC cell lines and Jurkat cells, flow cytometry analysis of tumor infiltrated immune cells and further sequencing analyses were performed to investigate how SIGLEC15 could affect T cells in vitro and in vivo. We found SIGLEC15 was increased in HCC tumor tissues and was negatively correlated with PD-L1 in HCC samples. In vitro and in vivo models demonstrated inhibition of SIGLEC15 did not directly influence tumor proliferation. However, SIGLEC15 could promoted HCC immune evasion in immune competent mouse models. Knock-out of Siglec15 could inhibit tumor growth and reinvigorate CD8+ T cell cytotoxicity. Anti-SIGLEC15 treatment could effectively inhibit tumor growth in mouse models with or without mononuclear phagocyte deletion. Bulk and single-cell RNA sequencing data of treated mouse tumors demonstrated SIGLEC15 could interfere CD8+ T cell viability and induce cell apoptosis. In all, SIGLEC15 was negatively correlated with PD-L1 in HCC and mainly promote HCC immune evasion through inhibition of CD8+ T cell viability and cytotoxicity.
GRAPHICAL ABSTRACT
Introduction
Hepatocellular carcinoma (HCC) is one of the leading causes for cancer-associated death worldwide.Citation1,Citation2 Fifty percent of patients afflicted by HCC experience early recurrence after surgery, and the overall response to chemotherapy is not satisfied.Citation3 Although immunotherapy targeting PD-1/PD-L1 or CTLA-4 axis has achieved success in some patients, the objective response rate for HCC ranges from 15–20%, which has led to the exploration of combining therapy or additional targets in order to increase immunotherapy efficacy in patients with limited results.Citation4–8
PD1 and CTLA-4 are both B7 family members, which played vital roles in immune regulation, and a series of studies tried to screen other B7 family members as potential immune regulators, which could be targets to ‘tip-off’ immune exhaustion in cancer treatment.Citation9–12 SIGLEC15, expressed by macrophages, has recently been reported to share a similar genetic sequence and protein structure with inhibitory B7 family members and could suppress CD8+ T cell function in melanoma, which was formerly reported to be involved in bone resorption, immune defenses and inflammation.Citation13–16 Liver cancer otherwise has been considered to have a suppressive immune microenvironment with unique immune cell populations, metabolic changes and intrinsic oncogenic signals.Citation17–21 The expression patterns and immunoregulatory roles of SIGLEC15 in HCC have not been elucidated, and in our investigation, we found that SIGLEC15 was negatively correlated with PD-L1 expression in HCC samples, relating an immune-barren microenvironment. Tumor-derived SIGLEC15 did not directly impact tumor growth; rather, it could lead to tumor immune evasion through induction of cytotoxic CD8+ T cell apoptosis, which could be redirected and reduced by anti-SIGLEC15 treatment. The preclinical results of our study provided rationale for precision immunotherapy in treatment of advanced HCC.
Methods
Reverse transcript quantitative polymerase chain reaction (RT – qPCR) analysis
Total RNA of transfected cell lines was extracted with TRI reagent (Sigma) according to manufactured protocols, and then chloroform and isopropanol were respectively used to filter procured total RNA through centrifugation at 10,000 rpm for 10 min. Tumor tissue was frozen in liquid nitrogen and then put into 1.5 ml tubes with grinding beads, which were filled with 1 ml TRI reagent (Sigma); afterward, tissues were grand at a frequency of 60 Hz for 30s to get total RNA before centrifugation. Master Mix (TAKARA) was used to generate complementary DNA for PCR analysis according to the protocols, and finally, SYBR GREEN (TAKARA) kit was used for RT – qPCR analysis of the quantitated cDNA solution. Primers for targeted genes were generated by GenePharma (China) (Table S4).
Western-blot analysis (WB)
Cell samples were washed with iced PBS for 3 times, and then NP40, containing proteinase inhibitors, was used for cell lysis. Yielded cell lysate was then loaded for centrifugation at 10,000 rpm for 10 min at 4°C to get rid of the cell debris. Protein buffer was firstly examined by BCA kit to estimate the protein concentration of each sample, after which cell lysates were added into each cell of prepared gels for electrophoresis (300 mA, 30 min). Afterwards, separated proteins in gel were transferred onto PVDF membranes (45 µm, Millipore) through wet transfer method (120 V, 1.5 h). The whole membrane was then blocked with QuickBlock Western solution (Beyotime P0252) for 15 min. Membranes were trimmed according to the respective molecular weights of targeted genes, and then trimmed membrane lanes were for each primary antibody incubation (4°C, overnight; SIGLEC15 1:1000; PD-L1 1:1000) (Table S2). Before further incubation with secondary antibodies (room temperature, 30 min), membrane lanes were washed twice with TBST. Immobilon Western HRP Substrate (Millipore) was used for exposure detection.
Cell lines and genetic modification
The cell lines of L02 (RRID: CVCL_6926), HepG2 (RRID: CVCL_0027), Hep3B (RRID: CVCL_0326), PLC (RRID: CVCL_0485), HCCLM3 (RRID: CVCL_6832), Huh7 (RRID: CVCL_0036), HCC7721 (RRID: CVCL_0534), MHCC97H (RRID: CVCL_4972), MHCC97L (RRID: CVCL_4973) and Hep1–6 (RRID: CVCL_0327) used in our investigation were provided by the Liver Cancer Institute of Zhongshan Hospital, affiliated with Fudan University, Shanghai, China. The Jurkat cell line (RRID: CVCL_0065) was purchased from GeneChem (China). Lentiviruses targeting human SIGLEC15 were generated by GeneChem (China); plasmids for human SIGLEC15 over-expression and Cas9/gRNAs, targeting mouse Siglec15, were generated by GenoMediTech (China) (Table S4).
Cell viability analysis
To examine the growth ability of transfected tumor cells, 5 × 103 cells of tested cell line were seeded in each well of a 96-well plate, which was then cultivated under condition of 37°C and 5% CO2 for 48 h. Then solution of Cell Counting Kit-8 (BOSTER, AR1160) were diluted and added into cells of a 96-well plate according to the manufactured protocols. Light absorbance (OD450 minus OD600) was measured after 2 h incubation at 37°C, which was for further analysis of viability difference between groups, and 3 time points were examined for each transfected cell line.
Construction of orthotopic and percutaneous mouse models
BALB/c nude (immunodeficient, RRID: IMSR_JAX:002019) and C57BL (immunocompetent, RRID: IMSR_ORNL:C57BL) male mice of 6–8 weeks old were used for tumor model construction in our investigation. 5 mice, fed with ordinary diet, were raised in one cage in specific pathogen free (SPF) environment. Huh7 (RRID: CVCL_0036, 1 × 106), MHCC97H (RRID: CVCL_4972, 1 × 106) and Hep1–6 (RRID: CVCL_0327, 2 × 105) cells were injected percutaneously into the right flank of BALB/c nude or C57BL mice to generate percutaneous tumor models. For construction of orthotopic liver tumor models, sliced Hep1–6 tumor tissue (1 mm3) was surgically implanted into mouse liver after anesthesia.
To evaluate the treatment efficacy of anti-SIGLEC15 immunotherapy, eight days after percutaneous cell injection, SIGLEC15-blocking antibody (200 µg each) was intraperitoneally injected into each mice twice a week. Anti-SIGLEC15 antibody was generated by SanYou Biotech (China) according to related patterns.Citation13 A caliper was used to measure the sizes of percutaneous tumors.
Extraction of tumor-infiltrated lymphocytes
Percoll solution (GE Health) was used to separate lymphocytes from tumor tissues according to the published protocols. Briefly, tumors were trimmed into 2 mm3 pieces in dish before digestion (10 ml of solution with 0.1% DNase I and 0.1% collagenase IV).Citation22,Citation23 The mixed tissue solution was then incubated at 37°C for 30 min with a speed of 120 rounds/min for proper tissue digestion. Then, the mixed solution was applied to a gentle MACS dis-associator for further decomposition, which was set to the according grinding mode. After decomposition process, tubes with tissue solution were put on ice, and 10 ml of 2% FACS solution was added into each tube to neutralize tissue digestion enzymes; afterward, the mixed solution was filtered through a 70 µm mesh to get rid of the tissue debris. Centrifugation at 1500 rpm was performed to obtain cell pellets in filtered tissue solution, which was then further separated by Percoll solution at gradient dilutions of 30% and 70%. Cell pellets between 30% and 70% Percoll interface were mononuclear lymphocytes and could be collected and washed with cold PBS before further analysis.
Cell staining process and flow cytometry analysis
Extracted lymphocytes were allocated into 1.5 ml tubes (106 each) in 100 µl staining buffer (BioLegend). TruStain FcXtrade (BioLegend) was applied to block Fc receptors for 5 min at 4°C, and then antibodies for cell surface markers were added for staining process (4°C, 30 min). For intracellular staining, cell pellets, after surface marker staining process, were washed twice with cold staining buffer and then centrifuged at 1500 rpm; eBioscience Foxp3/Transcription Factor Staining Buffer Set (Invitrogen, Thermo Fisher Scientific) was used for cell pellets fixation and permeabilization (1 h, room temperature) according to the manufactured protocols. Final cell pellets were then washed twice with fixation solution and suspended in 500 µl staining buffer (BioLegend) for flow cytometry analysis on the BD FACS Aria III platform.
Co-culture system of tumor and Jurkat cell lines
To investigate the influences of SIGLEC15 expression on T cells, we constructed a co-culture system of tumor and Jurkat cells in vitro. Briefly, tumor cells were seeded into a 6-well plate for cultivation (37°C, 5% CO2), and after 80% coverage of each cell was achieved, 1 × 105 Jurkat cells were added into the cell medium for co-culture of 48 h. Then floating cells in medium were collected through centrifugation at 1000 rpm. Collected cell pellets were stained for CD3 and GZMB expression for further flow cytometry analysis.
Magnetic sorting of immune cells
Anti-CD8a magnetic beads (StemCell) were used for enrichment of tumor-infiltrated CD8+ T cells, and anti-CD45 magnetic beads (StemCell) were used to enrich CD45+ lymphocytes for further single-cell sequencing analysis according to the manufactured protocols. Briefly, extracted lymphocytes were washed with cell medium and adapted to certain cell concentration. Then antibody cocktail in the kit was added into the cell medium according to the protocols, and cell solution was further mixed and incubated at room temperature for 5 min before adding the magnetic beads for another round of mixture and incubation for 3 min at room temperature. Finally, the tube, containing cell pellets, was placed into a magnet for incubation of 5 min. After pouring the unwanted cell pellets out, the enriched targeting cells were remained and could be reconstituted into certain concentration for further usage.
Single cell sequencing (scRNA-seq) and bulk sequencing analysis
Tumor cell suspension was incubated with anti-CD45 magnetic beads (StemCell) for lymphocyte enrichment. After that, CD45+ and CD45- cells were mixed at a ratio of 1:1 for further sequencing analysis on a 10× Genomics Platform with cell viability over 90%. A total of 17,100 cells were yielded in sequencing, and after filtering (percent.mt < 5%, nFeature < 2500 & >200), 11241 cells (IgG: 5765 cells, anti-SIGLEC15: 5482 cells) were remained for further analysis. Cell clusters were classified as lymphocytes and non-lymphocytes according to Cd45 expression and annotated according to specific markers (Table S5). Magnetic enriched CD8a positive T cells from mouse tumor models were collected and prepared for total RNA extraction, which was further examined in quality control process and then loaded onto sequencing platform (DNB-seq). The yielded data were applied to following data filtration, and mapped genetic expressions were finally used in downstream analysis.
Monocytes depletion
To investigate whether tumor-derived SIGLEC15 expression was sufficient to induce immune evasion in tumor microenvironment, anionic clophosomes (BioLegend) were used to deplete mononuclear phagocytes in mouse model through percutaneous injection at a dose of 100 µg per mouse. To consistently eradicate mononuclear phagocytes, anionic clophosome was injected twice a week. Clearance of mononuclear phagocytes in mouse tumors was confirmed by flow cytometry analysis through staining and gating CD11b and F4/80 positive cells.
Tumor tissue microarray (TMA)
Patient tumor samples were fixed in formaldehyde for 48 h and then embedded in wax. Afterwards, tissues were loaded and pinned onto glass plates with two points representing one sample. The aligned tissue array was then stored at 4°C for usage. A cohort of 221 HCC patients, who were treated in Zhongshan Hospital affiliated with Fudan University, Shanghai, China, were analyzed (Table S1).
Immunohistochemistry analysis (IHC)
Slides were incubated in Tris-EDTA buffer at 100°C for epitope retrieval. Then, intrinsic peroxidase activity was blocked for 15 min with 3% hydrogen peroxide at room temperature. Blocking buffer containing 5% bovine serum albumin was used for blockade for 30 min. Afterward, slides were incubated with primary antibodies at specific dilution ratios (anti-SIGLEC15, 1:400, NOVUS, Cat#NBP2–41162; anti-PD-L1, 1:30, Abcam, Cat#ab205921, RRID: AB_2687878) overnight in 4°C environment, followed by incubation with anti-rabbit or commonly used secondary antibodies for 60 min at room temperature. Diaminobenzidine chromogen substrate was used to detect antibody staining. Hematoxylin was used to counterstain the slides for 5 min after three washes with deionized water. Staining scores of each marker were analyzed by two independent physicians, according to staining areas and staining intensity, which were rated from point 0 to 4.
Bioinformatic analysis
HCC Sequencing data from the Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC) databases were used for expression analysis.Citation24–26 edgeR package (edgeR, RRID:SCR_012802) was used to examine differentially expressed genes between groups.Citation27,Citation28 The ggplot2 and Seurat packages were used for graphic demonstration.Citation29 Monocle package was used for trajectory analysis.Citation30–32
Ethical statement
All samples from patients, used in the investigation, were obtained with informed consent, which was approved by the local ethical committee of Zhongshan Hospital Ethical Review Board.
Statistics
Expression differences between two groups were examined with student t tests. Survival differences between groups were examined with the log-rank test. p values under .05 was considered significant.
Results
SIGLEC15 was negatively correlated with PD-L1 in HCC samples, relating an immune-barren microenvironment and unfavorable prognosis
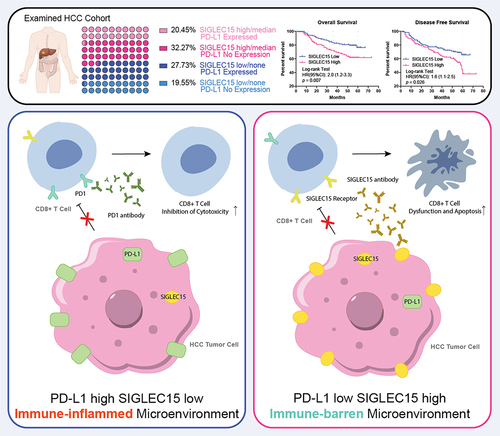
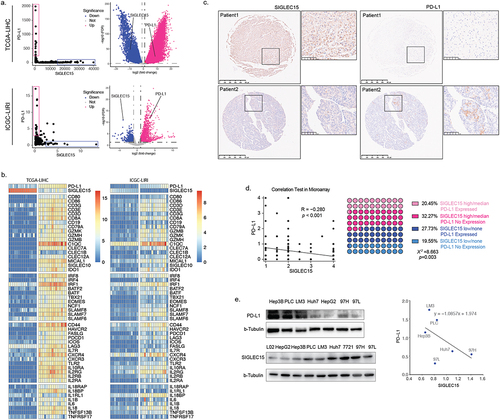
We examined SIGLEC15 expression in databases and found that SIGLEC15 mRNA expression was increased in tumor samples, which was further confirmed by PCR analysis of paired HCC samples (). Further IHC analysis of 221 HCC patient tumor tissues (characteristics shown in Table S1) showed that SIGLEC15 protein expression in tumors was related to worse overall survival (OS) and disease-free survival (DFS) of patients (). We examined the correlation between SIGLEC15 expression and immune microenvironment characteristics through bioinformatic analysis, and we found mRNA expression of PD-L1 and SIGLEC15 in public HCC datasets (TCGA-LIHC, ICGC-LIRI) demonstrated polarized patterns: SIGLEC15-high samples turned to have lower expression of PD-L1, while PD-L1-high samples demonstrated fewer mRNA counts of SIGLEC15, with different genetic profiles between sample groups (). Examination of differentially expressed genes between SIGLEC15-high and PD-L1-high samples demonstrated that immune-related cell and functional signatures were highly expressed in PD-L1 high samples, while SIGLEC15-high samples showed an immune-barren status in contrast (). We additionally examined IHC expression of PD-L1 in HCC patient cohort, in which 32.27% samples showed no expression of PD-L1 and high expression of SIGLEC15, while 27.73% samples were PD-L1 positive with low or none expression of SIGLEC15 (). Further examination of HCC cell lines also showed negatively correlated expression patterns between SIGLEC15 and PD-L1, and SIGLEC15 was relatively higher in HCC cell lines with aggressive traits (). In summary, SIGLEC15 was examined to negatively correlate with PD-L1 expression in HCC tumor samples, and SIGLEC15-high samples shared an immune-barren microenvironment with shorter overall survival and disease-free survival.
Figure 1. SIGLEC15 was highly expressed in HCC tumor samples and was related to deleterious survival of patients.
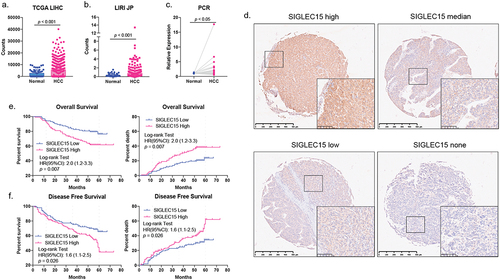
Figure 2. SIGLEC15 expression was negatively correlated with PD-L1 expression in HCC samples, relating to a barren immune microenvironment in tumor.
SIGLEC15 expressed by HCC tumor cells mainly promote tumor growth through evasion of cytotoxic CD8+ T cell killing
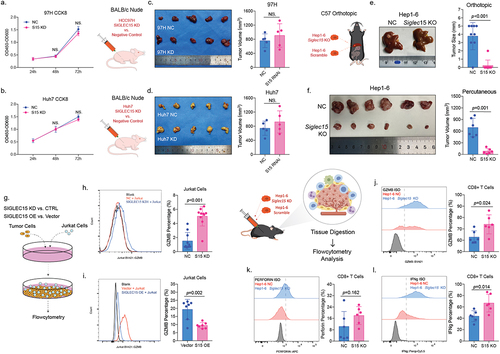
To explore whether SIGLEC15 expression could directly influence tumor growth, we knocked down SIGLEC15 in cell lines of MHCC97H and Huh7 (Supplemental Figure S1a-c), and found decrease of SIGLEC15 expression in MHCC97H and Huh7 cells did not affect cell growth in vitro through CCK8 examination (). Also, in vivo analysis showed that knockdown of SIGLEC15 in tumor cells did not affect tumor growth in immune deficient mice (BALB/c nude) (). However, in immunocompetent C57 mice, knockout of Siglec15 in Hep1–6 HCC tumor cells could significantly inhibit tumor growth in orthotopic and percutaneous tumor models (). Since mature and functional T cell populations are deficient in BALB/c nude mice, we wondered whether SIGLEC15 influenced cytotoxic T cells to promote tumor growth and constructed a co-culture system of HCC and T cell lines (). We found SIGLEC15 knock-down in Huh7 cells could increase GZMB expression in Jurkat cells (CD3+) after co-culture of 48 h, while overexpression of SIGLEC15 could decrease GZMB expression in Jurkat cells accordingly (). We then examined tumor-infiltrating CD8+ T cells in mouse model and it demonstrated that knockout of Siglec15 in Hep1–6 cells could restore CD8+ T cell cytotoxicity with increased expression of GZMB and IFNg in tumor microenvironment (). In conclusion, SIGLEC15, expressed by tumor cells in HCC, mainly promoted malignancy growth through induction of CD8+ T cell anergy in tumor microenvironment.
Figure 3. SIGLEC15, expressed by tumor cells, mainly promoted HCC growth through immune evasion.
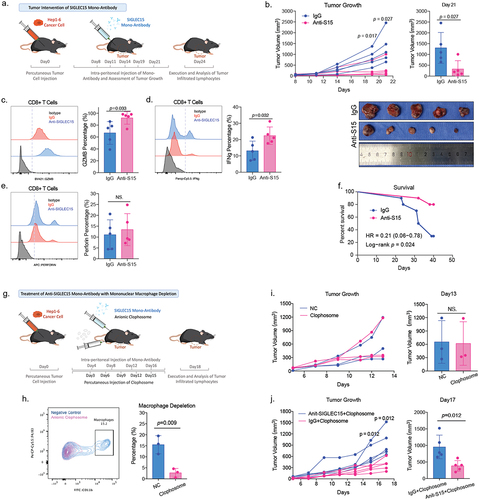
Anti-SIGLEC15 treatment in HCC mouse model could reinvigorate CD8+ T cell cytotoxicity and inhibit tumor growth
We further applied SIGLEC15 antibody in subcutaneous HCC mouse models to investigate whether targeting SIGLEC15 in tumor could inhibit HCC growth in preclinical models (). We found anti-SIGLEC15 therapy could sufficiently reduce tumor volumes and prolong survival of tumor-bearing mice (). Analysis of tumor-infiltrating CD8+ T cells indicated anti-SIGLEC15 treatment significantly reinvigorated cytotoxic functions of CD8+ T cells in tumor, increasing GZMB and IFNg expression accordingly (). Former study demonstrated SIGLEC15, expressed by tumor associated macrophages, could dampen functions of CD8+ T cells in melanoma, and we depleted mononuclear phagocytes, basically macrophages (F4/80+ and CD11b+), in mouse model to testify whether tumor-derived SIGLEC15 was crucial in immune evasion of HCC (). After depletion of mononuclear phagocytes, it showed anti-SIGLEC15 treatment still significantly inhibited tumor growth in comparison to IgG group, while simply depleting mononuclear phagocytes did not influence tumor growth in vivo (). So, tumor-derived SIGLEC15 expression was sufficient to induce immune evasion in HCC, and anti-SIGLEC15 treatment in preclinical HCC mouse models could reinvigorate cytotoxic CD8+ T cells to inhibit tumor growth.
Figure 4. Anti-SIGLEC15 immunotherapy could reduce tumor burden and promote CD8+ T cell cytotoxic functions in mouse models.
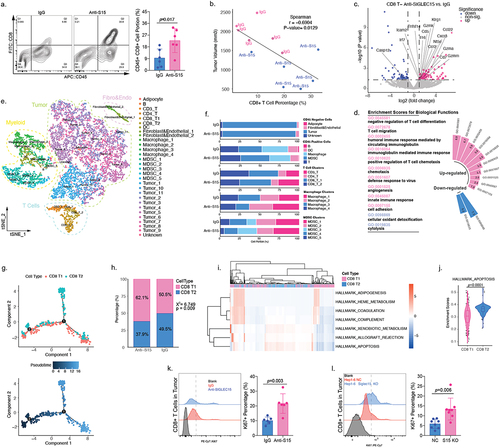
SIGLEC15 could induce infiltrated CD8+ T cell apoptosis in HCC tumor microenvironment
We examined CD8+ T cell portions between anti-SIGLEC15 and IgG treatment groups, and we found anti-SIGLEC15 treatment could increase CD8+ T cell infiltration in HCC tumor model (). It turned out that CD8+ T cell infiltrating levels were negatively correlated with tumor burden in mouse models (). To understand how SIGLEC15 affected CD8+ T cells in HCC tumor microenvironment, we additionally collected magnetically enriched CD8+ T cells in tumor tissues for sequencing analysis. Differentially expressed genes between groups (anti-SIGLEC15 vs. IgG) were enriched for further annotation, and it demonstrated that CD8+ T cells were highly activated and mobile after anti-SIGLEC15 treatment, while apoptotic signatures were down-regulated, such as Casp12 (). We additionally performed scRNA-seq analysis of mouse tumor samples (anti-SIGLEC15 vs. IgG), and finally 11,247 cells were yielded after cell filtration for downstream analysis (); anti-SIGLEC15 treatment mainly influenced T cell and myeloid cell cluster portions between groups (). We wondered how CD8+ T cells changed after deletion of SIGLEC15 signals and performed trajectory analysis accordingly. It appeared CD8 T2 cluster took a single arm in CD8+ T cell developmental trajectories; also, anti-SIGLEC15 could alter CD8+ T cell trajectories by increasing CD8 T1 and reducing CD8 T2 cell portions in the process (). Analysis of signaling changes between CD8+ T cell clusters demonstrated that the apoptotic signaling was significantly lower in CD8 T1 cluster (). We further examined cell viability of tumor infiltrated CD8+ T cells through examination of Ki67 expression and found Siglec15 knockout or anti-SIGLEC15 treatment could both increase CD8+ T cell viability in tumor (). In summary, SIGLEC15 could induce CD8+ T cell apoptosis to promote immune evasion in HCC, and anti-SIGLEC15 treatment could modify CD8+ T cell developmental trajectories in HCC tumor model, increasing CD8+ T cell viability, promoting CD8+ T cell tumor infiltration and reinvigorate CD8+ T cell cytotoxicity at the same time.
Figure 5. SIGLEC15 could induce CD8+ T cell apoptosis in HCC tumor microenvironment.
Discussion
In our investigation, we found that SIGLEC15, expressed by HCC tumor cells, mainly promoted malignancy growth through immune evasion, inhibiting cytotoxicity and viability of CD8+ T cells in tumor microenvironment; SIGLEC15 expression levels in HCC tumor cells did not directly influence tumor cell viability. However, in a study concerning anaplastic thyroid carcinoma, knockout of Siglec15 could reduce expression of STAT1, STAT3 and VEGF in tumor, leading to tumor cell apoptosis and reduced tumor volume.Citation33 Former studies demonstrated that SIGLEC15 were found to be expressed in macrophages and dendritic cells, which mainly regulate cell functions through downstream regulation of DAP12 and DAP10,Citation34 and in differentiated myeloid cells, such as osteoclasts, SIGLEC15 was examined to function with DAP12 in maintenance of osteoclast physiological functions.Citation35 The explicit roles of SIGLEC15 in tumor may vary according to different tissue origins, and our results demonstrated that in HCC, SIGLEC15 expression in tumor cell mainly promote tumor progression through immune evasion.
Studies showed that in melanoma and pancreatic adenocarcinoma tumor microenvironment, SIGLEC15 associated macrophages were polarized to induce CD8+ T cell dysfunction;Citation13,Citation36 we wonder if SIGLEC15-associated macrophages took the primal immune editing roles in CD8+ T cell dysfunction and delete mononuclear phagocytes in anti-SIGLEC15 treated mouse models accordingly. Our results showed anti-SIGLEC15 treatment still significantly inhibited tumor growth even after deletion of mononuclear phagocytes, emphasizing the crucial immunoregulatory function of SIGLEC15 in HCC tumor cells.
PD-L1 expression is an important indicator of immunotherapy efficacy across cancer types,Citation37 and a series of studies demonstrated that combination therapy could increase treatment responses through up-regulation of PD-L1 in tumor microenvironment.Citation38 In our investigation, SIGLEC15 expression was found negatively correlated with PD-L1 in HCC cell lines and tumor samples. SIGLEC15 mRNA was also found negatively correlated with PD-L1 in public melanoma sequencing data, and in a study concerning non-small-cell lung carcinoma (NSCLC), SIGLEC15 expression was higher in adenocarcinoma samples, while on the other hand, PD-L1 positive rate was higher in samples of squamous cell carcinoma.Citation13,Citation39 In various solid tumors, high expression of SIGLEC15 in malignancy or macrophages was all related to poor survival of patients, providing rationale for SIGLEC15-targeting therapy in cancer patients,Citation36,Citation40 and preliminary results from an ongoing clinical trial indicated that anti-SIGLEC15 therapy achieved prolonged stabilization of disease in 54% (20/37) of recruited cancer patients, and in 20% (2/10) anti-PD1-refractory NSCLC patients, objective responses were observed.Citation15 In our examined HCC patient cohort, 27% (61/221) samples demonstrated expression of PD-L1 and low/none expression of SIGLEC15, while 32% (71/221) patients showed high expression of SIGLEC15 and no expression of PD-L1, who may be potential candidates for anti-SIGLEC15 immunotherapy.
There are several limitations of our study. First, the examination of SIGLEC15 expression in HCC tumor samples was only performed in a small cohort of Chinese patients from a single center, which may not represent the overall characteristics of patients. Second, our results mainly relied on cell and mouse models, which may not replicate actual tumor microenvironment that develop over a long period of time in human.
Conclusion
SIGLEC15 was negatively correlated with PD-L1 expression in HCC and could promote tumor immune evasion through induction of CD8+ T cell apoptosis, relating to an immune barren tumor microenvironment.
Author contribution
ZC, MCH, QHY, LG and HL designed the investigation, and ZC, MCY, BZ, LJ, QY, SL, BHZ, JLY, WTZ and XQL conducted experiments of the study. ZC, MCY and BZ drafted the manuscript, and JZ, JF, YFX, YSX, MCH, QHY, LG and HL helped to revise and make adaptations. MCH, QHY, LG and HL administered the project. All authors are responsible for the content of the investigation.
Supplemental Tables.xlsx
Download MS Excel (24.7 KB)Suppl Figure1.png
Download PNG Image (379.1 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Sequencing data for the manuscript were deposited in the Gene Expression Omnibus database under the accession number of GSE218411. Public datasets of LIHC and LIRI projects were downloaded from TCGA and ICGC databases respectively. There was no original code generated in the manuscript.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/2162402X.2024.2376264
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–12. doi:10.3322/caac.21492.
- Singal AG, Lampertico P, Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: new trends. J Hepatol. 2020;72(2):250–261. doi:10.1016/j.jhep.2019.08.025.
- Jindal A, Thadi A, Shailubhai K. Hepatocellular carcinoma: etiology and current and future drugs. J Clin Exp Hepatol. 2019;9(2):221–232. doi:10.1016/j.jceh.2019.01.004.
- Cheng AL, Hsu C, Chan SL, Choo SP, Kudo M. Challenges of combination therapy with immune checkpoint inhibitors for hepatocellular carcinoma. J Hepatol. 2020;72(2):307–319. doi:10.1016/j.jhep.2019.09.025.
- Meric-Bernstam F, Larkin J, Tabernero J, Bonini C. Enhancing anti-tumour efficacy with immunotherapy combinations. Lancet. 2021;397(10278):1010–1022. doi:10.1016/S0140-6736(20)32598-8.
- Upadhaya S, Neftelino ST, Hodge JP, Oliva C, Campbell JR, Yu JX. Combinations take centre stage in PD1/PDL1 inhibitor clinical trials. Nat Rev Drug Discov. 2021;20(3):168–169. doi:10.1038/d41573-020-00204-y.
- El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, Kim TY, Choo S-P, Trojan J, Welling TH. et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389(10088):2492–2502. doi:10.1016/S0140-6736(17)31046-2.
- Yau T, Kang YK, Kim TY, El-Khoueiry AB, Santoro A, Sangro B, Melero I, Kudo M, Hou M-M, Matilla A. et al. Efficacy and safety of nivolumab plus ipilimumab in patients with advanced hepatocellular carcinoma previously treated with Sorafenib: the CheckMate 040 randomized clinical trial. JAMA Oncol. 2020;6(11):e204564. doi:10.1001/jamaoncol.2020.4564.
- Edner NM, Carlesso G, Rush JS, Walker LSK. Targeting co-stimulatory molecules in autoimmune disease. Nat Rev Drug Discov. 2020;19(12):860–883. doi:10.1038/s41573-020-0081-9.
- John P, Wei Y, Liu W, Du M, Guan F, Zang X. The B7x immune checkpoint pathway: from discovery to clinical trial. Trends Pharmacol Sci. 2019;40(11):883–896. doi:10.1016/j.tips.2019.09.008.
- Li J, Lee Y, Li Y, Jiang Y, Lu H, Zang W, Zhao X, Liu L, Chen Y, Tan H. et al. Co-inhibitory molecule B7 superfamily member 1 expressed by tumor-infiltrating myeloid cells induces dysfunction of anti-tumor CD8(+) T cells. Immunity. 2018;48(4):773–786.e5. doi:10.1016/j.immuni.2018.03.018.
- Peuker K, Strigli A, Tauriello DVF, Hendricks A, von Schönfels W, Burmeister G, Brosch M, Herrmann A, Krüger S, Nitsche J. et al. Microbiota-dependent activation of the myeloid calcineurin-NFAT pathway inhibits B7H3- and B7H4-dependent anti-tumor immunity in colorectal cancer. Immunity. 2022;55(4):701–717.e7. doi:10.1016/j.immuni.2022.03.008.
- Wang J, Sun J, Liu LN, Flies DB, Nie X, Toki M, Zhang J, Song C, Zarr M, Zhou X. et al. Siglec-15 as an immune suppressor and potential target for normalization cancer immunotherapy. Nat Med. 2019;25(4):656–666. doi:10.1038/s41591-019-0374-x.
- van de Wall S, Santegoets KCM, van Houtum EJH, Büll C, Adema GJ. Sialoglycans and siglecs can shape the tumor immune microenvironment. Trends Immunol. 2020;41(4):274–285. doi:10.1016/j.it.2020.02.001.
- Sun J, Lu Q, Sanmamed MF, Wang J. Siglec-15 as an emerging target for next-generation cancer immunotherapy. Clin Cancer Res. 2021;27(3):680–688. doi:10.1158/1078-0432.CCR-19-2925.
- Bordon Y. Inflammation: live long and prosper with Siglecs. Nat Rev Immunol. 2015;15(5):266–267. doi:10.1038/nri3851.
- Zheng M, Tian ZG. Liver-mediated adaptive immune tolerance. Front Immunol. 2019;10:2525. doi:10.3389/fimmu.2019.02525.
- Lu LC, Hsu C, Shao YY, Chao Y, Yen CJ, Shih IL, Hung YP, Chang C-J, Shen Y-C, Guo J-C. et al. Differential organ-specific tumor response to immune checkpoint inhibitors in hepatocellular carcinoma. J Liver Cancer. 2019;8(6):480–490. doi:10.1159/000501275.
- Nault JC, Villanueva A. Biomarkers for hepatobiliary cancers. Hepatology. 2021;73(Suppl 1):115–127. doi:10.1002/hep.31175.
- Ally A, Balasundaram M, Carlsen R, Chuah E, Clarke A, Dhalla N, Holt RA, Jones SJM, Lee D, Ma Y, et al. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169(7):1327–1341.e23. doi:10.1016/j.cell.2017.05.046.
- Racanelli V, Rehermann B. The liver as an immunological organ. Hepatology. 2006;43(Supplement 1):S54–62. doi:10.1002/hep.21060.
- Haubruck P, Colbath AC, Liu Y, Stoner S, Shu C, Little CB. Flow cytometry analysis of immune cell subsets within the murine spleen, bone marrow, lymph nodes and synovial tissue in an osteoarthritis model. J Vis Exp. 2020;(158). doi:10.3791/61008-v.
- Shi W, Wang Y, Zhang C, Jin H, Zeng Z, Wei L, Tian Y, Zhang D, Sun G. Isolation and purification of immune cells from the liver. Int Immunopharmacol. 2020;85:106632. doi:10.1016/j.intimp.2020.106632.
- Sun Y, Wu L, Zhong Y, Zhou K, Hou Y, Wang Z, Zhang Z, Xie J, Wang C, Chen D, et al. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell. 2021;184(2):404–421.e16. doi:10.1016/j.cell.2020.11.041.
- Hudson TJ, Anderson W, Aretz A, Barker AD, Bell C, Bernabé RR, Bhan MK, Calvo F, Eerola I, Gerhard, DS, Guttmacher A, et al. International network of cancer genome projects. Nature. 2010;464:993–998.
- Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn). 2015;19:A68–77. doi:10.5114/wo.2014.47136.
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi:10.1093/bioinformatics/btp616.
- McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288–4297. doi:10.1093/nar/gks042.
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902.e21. doi:10.1016/j.cell.2019.05.031.
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–386. doi:10.1038/nbt.2859.
- Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat Methods. 2017;14(3):309–315. doi:10.1038/nmeth.4150.
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14(10):979–982. doi:10.1038/nmeth.4402.
- Hou X, Chen C, He X, Lan X. Siglec-15 silencing inhibits cell proliferation and promotes cell apoptosis by inhibiting STAT1/STAT3 signaling in anaplastic thyroid carcinoma. Dis Markers. 2022;2022:1606404. doi:10.1155/2022/1606404.
- Angata T, Tabuchi Y, Nakamura K, Nakamura M. Siglec-15: an immune system Siglec conserved throughout vertebrate evolution. Glycobiology. 2007;17(8):838–846. doi:10.1093/glycob/cwm049.
- Ishida-Kitagawa N, Tanaka K, Bao X, Kimura T, Miura T, Kitaoka Y, Hayashi K, Sato M, Maruoka M, Ogawa T, et al. Siglec-15 protein regulates formation of functional osteoclasts in concert with DNAX-activating protein of 12 kDa (DAP12). J Biol Chem. 2012;287(21):17493–17502. doi:10.1074/jbc.M111.324194.
- Li TJ, Jin KZ, Li H, Ye LY, Li PC, Jiang B, Lin X, Liao Z-Y, Zhang H-R, Shi S-M, et al. SIGLEC15 amplifies immunosuppressive properties of tumor-associated macrophages in pancreatic cancer. Cancer Lett. 2022;530:142–155. doi:10.1016/j.canlet.2022.01.026.
- Doroshow DB, Bhalla S, Beasley MB, Sholl LM, Kerr KM, Gnjatic S, Wistuba II, Rimm DL, Tsao MS, Hirsch FR. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat Rev Clin Oncol. 2021;18(6):345–362. doi:10.1038/s41571-021-00473-5.
- Yu M, Chen Z, Zhou Q, Zhang B, Huang J, Jin L, Zhou B, Liu S, Yan J, Li X, et al. PARG inhibition limits HCC progression and potentiates the efficacy of immune checkpoint therapy. J Hepatol. 2022;77(1):140–151. doi:10.1016/j.jhep.2022.01.026.
- Hao JQ, Nong JY, Zhao D, Li HY, Su D, Zhou LJ, Dong YJ, Zhang C, Che NY, Zhang SC, et al. The significance of Siglec-15 expression in resectable non-small cell lung cancer. Neoplasma. 2020;67(6):1204–1222. doi:10.4149/neo_2020_200220N161.
- Quirino MWL, Pereira MC, Deodato de Souza MF, Pitta IDR, Da Silva Filho AF, Albuquerque MSS, Albuquerque APB, Martins MR, Pitta MGDR, Rêgo MJBDM, et al. Immunopositivity for Siglec-15 in gastric cancer and its association with clinical and pathological parameters. Eur J Histochem. 2021;65(1):65. doi:10.4081/ejh.2021.3174.